Significance
Protein flexibility is essential for the robustness of biological systems, yet the dynamics underlying this flexibility are difficult to observe, because they are small, fast, and stochastic. Photoprotection in plants is critical for robust growth under highly variable sunlight, but the complexity of photosynthetic proteins means that identifying conformational states and dynamics responsible is challenging. Here, we develop a method using the correlation function of the fluorescence lifetime to characterize multiple dynamical processes in single proteins. By applying this method to the protein Light-Harvesting Complex Stress Related 1 (LHCSR1), we identify two local protein motions that control quenching of excess sunlight, which is a photoprotective effect. Our analytical approach enables a structure-based understanding of the photoprotective mechanisms in green plants.
Keywords: single-molecule fluorescence spectroscopy, photosynthetic light harvesting, nonphotochemical quenching, protein dynamics
Abstract
Biological systems are subjected to continuous environmental fluctuations, and therefore, flexibility in the structure and function of their protein building blocks is essential for survival. Protein dynamics are often local conformational changes, which allows multiple dynamical processes to occur simultaneously and rapidly in individual proteins. Experiments often average over these dynamics and their multiplicity, preventing identification of the molecular origin and impact on biological function. Green plants survive under high light by quenching excess energy, and Light-Harvesting Complex Stress Related 1 (LHCSR1) is the protein responsible for quenching in moss. Here, we expand an analysis of the correlation function of the fluorescence lifetime by improving the estimation of the lifetime states and by developing a multicomponent model correlation function, and we apply this analysis at the single-molecule level. Through these advances, we resolve previously hidden rapid dynamics, including multiple parallel processes. By applying this technique to LHCSR1, we identify and quantitate parallel dynamics on hundreds of microseconds and tens of milliseconds timescales, likely at two quenching sites within the protein. These sites are individually controlled in response to fluctuations in sunlight, which provides robust regulation of the light-harvesting machinery. Considering our results in combination with previous structural, spectroscopic, and computational data, we propose specific pigments that serve as the quenching sites. These findings, therefore, provide a mechanistic basis for quenching, illustrating the ability of this method to uncover protein function.
Biological systems have evolved sophisticated protein structures to carry out a wide range of functions. The relationship between structure and function is often studied based on well-resolved, static structural models, such as those from X-ray crystallography and electron microscopy. However, protein conformational dynamics lead to time-varying structures, which emerge in response to environmental and thermal perturbations and therefore, are critical to survival under natural conditions (1, 2). These dynamics occur across spatial, temporal, and energetic scales (3). Most commonly, the dynamics are fast and thus, local. Local dynamics can take place simultaneously and asynchronously across several discrete sites in an individual protein (2, 3), providing multifunctionality to the system if individually controlled. These dynamics are obscured in conventional structural measurements due to ensemble averaging. Single-molecule spectroscopy has emerged as a powerful approach to access conformational dynamics and their impact on function in various biological systems (4). However, fast (i.e., microsecond), local, and multiple dynamical processes have remained challenging to resolve.
The current state-of-the-art is characterization of dynamics via changes in fluorescence using either change point finding (CPF) (5–9) or intensity correlation function analysis (10). CPF analysis bins the photon data, obscuring fast dynamics. In contrast, intensity correlation function analysis characterizes fluctuations in the photon arrival rate, accessing dynamics down to microseconds. However, the fluorescence lifetime is a powerful indicator of conformation for chromoproteins and for lifetime-based FRET measurements, yet it is ignored in intensity-based analyses.
2D fluorescence lifetime correlation (2D-FLC) analysis was recently introduced as a method to both resolve fast dynamics and use fluorescence lifetime information (11, 12). In this analysis, substates are identified based on the correlation function of the lifetime directly from the photon stream data without binning. This approach accesses dynamics down to the microsecond timescale and multiple dynamical processes occurring in parallel on different timescales (13). However, previous implementations had three major limitations. (i) States were required to be separated in lifetime by an order of magnitude for robust estimation. (ii) Only simple dynamic processes associated with one fluorescence emitter could be characterized. Because the analysis was applied to diffusing ensembles, the photons from different molecules were interspersed. This interspersal prevents separation of multiple independent dynamics within individual molecules, even if they occur. (iii) A model function to describe multiple dynamic components was lacking. As a result, the 2D-FLC analysis has been unable to separate small changes in fluorescence lifetime (i.e., local conformational dynamics) and characterize multiple conformational dynamics occurring in parallel at discrete sites within the protein, limiting the applicability of the method.
One class of chromoproteins that exhibits multiple and rapid dynamics is photosynthetic Light-Harvesting Complexes (LHCs). LHCs absorb light and transfer the energy to a reaction center, where photoelectric conversion occurs. Under high light (sunny), LHCs also dissipate excess energy as heat through a process known as nonphotochemical quenching (NPQ), which is activated by conformational changes (14, 15). In unicellular algae and mosses, Light-Harvesting Complex Stress Related 1 (LHCSR1) is the LHC responsible for dissipation (16–27). Single-molecule spectroscopy of LHCSR1 revealed frequent transitions between bright (photoactive) and dim (quenched) states (9). These transitions emerge from conformational dynamics that vary the configuration between pigments, likely chlorophylls a (Chls a), which are the fluorescence emitters, and carotenoids, which can quench Chl a (28). A pH drop and conversion of carotenoids from violaxanthin (Vio) into zeaxanthin (Zea) are both induced in the organism under high light (20, 29). Single-molecule experiments showed that the conformational dynamics changed with both pH and carotenoid conversion, resulting in enhanced quenching efficiency (9). In contrast, an LHC primarily responsible for light harvesting (LHCB1) lacked these dynamics (9). While these results demonstrated that the conformational dynamics of LHCSR1 control quenching, the underlying dynamics were unresolved due to limited time resolution that averaged over multiple dynamical processes. Furthermore, the switching mechanism and quenching site remained unclear, as LHCSR1 contains eight Chls a and four carotenoids (24), where each Chl a–carotenoid pair can be a quenching site.
In this study, we generalize the 2D-FLC analysis for complex states and dynamics by introducing a preestimation of lifetime state, which enables robust and reliable separation of states with closely spaced lifetimes, and developing a model correlation function to understand multiple independent dynamics. Application of the generalized analysis to single-molecule data from immobilized LHCSR1 and LHCB1 resolves previously hidden conformations of these proteins, including their fluorescence brightness and lifetime, and it identifies and characterizes the associated multiple dynamical processes. Notably, we discovered millisecond and microsecond photoprotective dynamics occurring at two different sites in parallel within LHCSR1. Within the context of structural models (30), we identified the likely molecular origin of the dynamics, uncovering a molecular basis for the mechanism of NPQ in photosynthetic LHCs. This advance enables the study of chromoproteins as described here as well as FRET-based studies of complex systems.
Results
Determination of the Correlation Function of the Fluorescence Lifetime.
2D-FLC analysis uncovers protein conformations that exhibit different fluorescence lifetimes (i.e., amounts of quenching) and the transitions between them. To study conformations with closely spaced lifetimes, the analysis required two steps, which are shown in Fig. 1. First, the number of states and their lifetimes were estimated from the lifetime distribution, and second, these states were used for the 2D-FLC analysis. The introduction of step 1, reported here, enhanced the reproducibility, stability, and applicability to complex systems compared with previous implementations of the 2D-FLC analysis.
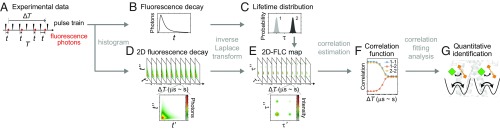
Fig. 1.
2D-FLC analysis of a two-state model system. (A) Schematic of the photon stream, where each photon (red circles) is detected at a measurement time T and an emission time t after excitation (black triangles). (B) The fluorescence decay profile is a histogram of t for all photons and is inverse Laplace transformed to generate the fluorescence lifetime distribution in C. (D) The 2D histogram of (, ), which are the emission times for two photons separated by as in A, at from to s. (E) The 2D-FLC maps from inverse Laplace transform of the 2D histograms using the lifetime distribution as a global variable. (F) The correlation function is extracted from the 2D-FLC maps, where autocorrelations (1–1, 2–2) are diagonal peaks and cross-correlations (1–2) are cross-peaks. (G) Analysis of the correlation function within the context of structural models enables assignment to sites in the protein. Details are described in SI Appendix, sections S1–S3 and Fig. S2.
In step 1, the fluorescence decay profile (Fig. 1B) is a histogram of the emission times t, which are the photon arrival times relative to excitation (Fig. 1A). The decay profile is fitted with the maximum entropy method to obtain a fluorescence lifetime distribution. The fluorescence lifetime distribution exhibits peaks that reflect the number and associated lifetimes of the states, which are shown for a two-state model system in Fig. 1C.
In step 2 of the 2D-FLC analysis, each photon pair separated by an interval of , as shown in Fig. 1A, provides a set of emission times (, ) that are histogrammed for all pairs in the photon stream to generate a 2D fluorescence decay (2D-FD) profile (e.g., Fig. 1D for s). The 2D-FDs are inverse Laplace transformed by fitting with the lifetime states identified (two states in the case of LHCSR1 and LHCB1) (a discussion of the analysis with three lifetime states is in SI Appendix, Fig. S8) to generate the 2D-FLC map (e.g., Fig. 1E for s). In the 2D-FLC map, the diagonal peaks are the autocorrelation of individual lifetime states, whereas the off-diagonal peaks are the cross-correlation from processes, such as interconversion between states.
The global fitting of 2D-FDs at various values gives the temporal dependence of the correlation intensity (Fig. 1F), i.e., the correlation function , which contains the fluorescence intensity of each state and the transition rates between states. In a simple two-state system, the peaks all exhibit a sigmoidal profile as illustrated in Fig. 1F. The intensity of diagonal peaks decreases at a time corresponding to the on time of the states, and the intensity of the cross-peaks increases at a time corresponding to interconversions between the states. In more complex systems, the peaks exhibit additional features as discussed in more detail below.
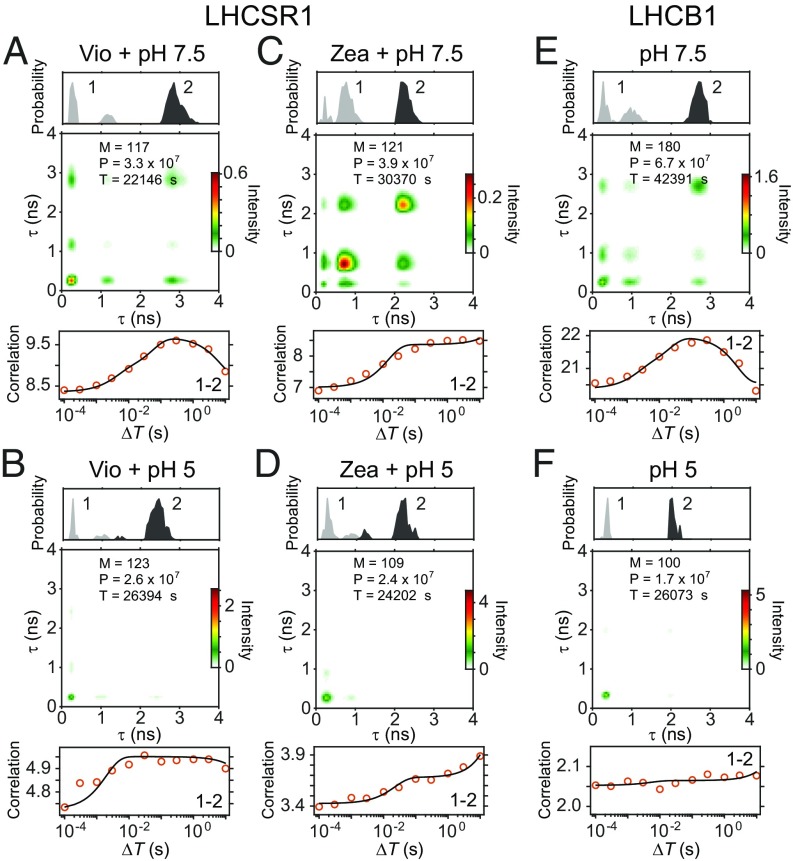
2D-FLC analysis with two lifetime states was performed for LHCSR1s and LHCB1s. As shown in Fig. 2 A–F, Top, the fluorescence lifetime distribution of all samples exhibited two peaks at 1 and 2–3 ns, corresponding to states 1 and 2, respectively. While the photophysics of these states includes a contribution from triplet states, for the relatively low excitation fluence used here, triplets do not affect the states identified through the fluorescence lifetime distribution (details are described in SI Appendix, section S10). However, even at low fluences, quenching from singlet–triplet annihilation decreases the rate of fluorescence emission (31–33) and therefore, likely plays a role in the relative intensities observed. Conversion from Vio to Zea shortened the lifetime of state 2 (Fig. 2C, Top). 2D-FLC maps in Fig. 2 A–F, Middle exhibit diagonal peaks for state 1 in all samples. Diagonal peaks for state 2 and off-diagonal peaks were also observed at pH 7.5 (Fig. 2 A, C, and E, Middle). However, on pH drop, only the diagonal peak of state 1 was observed (Fig. 2 B, D, and F, Middle). Moreover, conversion from Vio to Zea and pH drop changed the correlation temporal profile as shown in Fig. 2 B and C, Bottom, indicating changes in dynamics.
Fig. 2.
2D-FLC of single LHCSR1 and LHCB1 under different conditions. Lifetime distribution (Top), 2D-FLC map at s (Middle), and cross-correlation curve (Bottom) in LHCSR1-V-7.5 (A), LHCSR1-V-5 (B), LHCSR1-Z-7.5(C), LHCSR1-Z-5 (D), LHCB1-V-7.5 (E), and LHCB1-V-5 (F). The total numbers of molecules (M) and photons (P) and the sum of measurement times (T) used in the analysis are shown in Middle. The cross-correlation curve (1–2) corresponds to the off-diagonal element of . The fitting curve (black) was determined using the model function . All results for auto- and cross-correlations are shown in SI Appendix, Fig. S9.
Identification of Multiple Independent Dynamics Associated with NPQ in LHCs.
The complex profile of the correlation curves (Fig. 2 A–F, Bottom and SI Appendix, Fig. S9) is different from a sigmoidal correlation curve found for a simple two-state system with one dynamic component (examples are in SI Appendix, Fig. S3), suggesting that the dynamics in LHCSR1 and LHCB1 are generated from multiple components. Furthermore, the base cross-correlation at the shortest s can report on the free energy landscape as illustrated in the simulations shown in SI Appendix, Fig. S4 and discussed in SI Appendix, section S5. The base cross-correlation is zero for multiple dynamics that occur on a simple free energy landscape (i.e., a single emissive site within a protein). The base cross-correlation is nonzero for multiple dynamics that occur separately on multiple free energy wells (i.e., each corresponds to dynamics for one of multiple emissive sites within a protein). As shown in Fig. 2 A–F, Bottom, the base cross-correlation is nonzero for LHCSR1 and LHCB1. Given the number of Chl a in the protein, we assign this to be from multiple emitters, each giving rise to a dynamic component. The correlation fitting analysis can estimate these dynamic properties as illustrated in Fig. 1G.
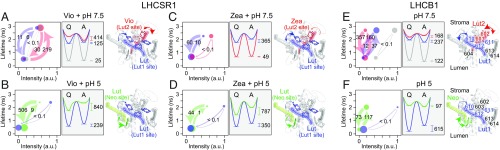
Fig. 3 shows diagrams of the intensities, lifetimes, and transition rates of states 1 and 2 for LHCSR1 and LHCB1 (all parameters are summarized in SI Appendix, Table S1, and SDs for LHCSR1-V-7.5 are in SI Appendix, Fig. S13 and Table S2). These states and dynamics were extracted from fitting to the correlation model function as shown as solid lines in Fig. 2 A–F, Bottom and SI Appendix, Fig. S9. was developed for a system composed of multiple dynamic components (SI Appendix, section S3 has details). To reproduce the profile of the correlation curve, three dynamic components on different timescales were needed for LHCSR1-V-7.5 and LHCB1-7.5 (SI Appendix, Fig. S10), and two were sufficient for the other samples. This fit, therefore, separated multiple dynamical processes, which were averaged out in previous analyses, and identified fast dynamics, which could not be resolved in the previous studies (9, 34). Resolution of these processes uncovered pigment dependence of the conformational dynamics. To demonstrate the reliability of the extracted dynamics, SDs in the dynamic components of LHCSR1-V-7.5 data were calculated. The SDs in the transition rates were 15.5–48.9% (25.3% on average), leading to deviations of 16.1% in the free energy difference and 8.0% in the state population (SI Appendix, section S8, Fig. S13, and Table S2). These deviations are significantly smaller than the differences between the components, confirming the validity of our separation of multiple processes. As shown in Fig. 3 and discussed in more detail below, Zea enrichment suppresses the active-biased component, and the pH drop stabilizes the quenched state. Both of these changes enhance the quenching efficiency.
Fig. 3.
Dynamics of LHCSR1 and LHCB1. Schematic of protein dynamics (Left), free energy surface (Center), and structural model (Right) in LHCSR1-V-7.5 (A), LHCSR1-V-5 (B), LHCSR1-Z-7.5(C), LHCSR1-Z-5 (D), LHCB1-V-7.5 (E), and LHCB1-V-5 (F). Transitions occur between short-lifetime [quenched (Q)] and long-lifetime [active (A)] states. In the schematics, circle size and arrow thickness reflect the relative population and transition rate between the states, respectively. The transition rate (in 1/s) is labeled on each arrow. The free energy surface (in wavenumbers) was calculated as described in SI Appendix, section S7. All dynamics and free energy parameters are listed in SI Appendix, Table S1. The components in blue, red, and green are likely conformational fluctuations of the pigments in blue, red, and green, respectively, as illustrated in the structural model. As shown in E and F, Chls a cluster into Chl a610–611–612 (blue), Chl a602–603 (red), Chl a613–614 (gray), and Chl a604 (green). These clusters neighbor carotenoids, Lut1 (blue), Lut2 (red), Neo (green), and Vio (gray).
Discussion
Structural Assignment of Dynamic Sites in LHCB1.
By fitting the correlation curves of LHCSR1 and LHCB1, we quantitatively estimated the number of dynamic components, fluorescence lifetime and intensity in the active and quenched states, and transition rates between the states. By considering our results in combination with information from X-ray crystallography, previous spectroscopic measurements, and computational data, it is possible to speculate as to the structural origin of the dynamic components. The conformational states, dynamics, and their likely origin are illustrated in Fig. 3. The quenching sites proposed are consistent with our data and previous data (24, 35–40). However, given the complexity of the system, other sites may alternatively be partially or fully responsible for the observed dynamics. We initially discuss potential quenching sites in LHCB1 in the context of its crystal structure (30), which provide clues as to those in LHCSR1.
As shown in SI Appendix, Fig. S11, LHCB1 contains eight Chls a, six Chls b, four carotenoids, two luteins (Luts; Lut1 and Lut2), neoxanthin (Neo), and Vio (30, 41). The Chls a are organized into three clusters, Chl a610–611–612, Chl a602–603, and Chl a613–614, as well as monomeric Chl a604 (nomenclature from ref. 30), although our preparation may lack Chl a611, Chl b607, and Vio. The Chls a contain the lowest energy levels, which are the emissive sites that appear as dynamic components in the 2D-FLC analysis (42–44).
The 2D-FLC analysis identified three dynamic components in LHCB1 (Fig. 3E): active-biased (blue), quenched-biased (red), and almost static (gray) dynamics. The active-biased component (Fig. 3E, blue) exhibits high fluorescence intensity in the active state and large energy dissipation in the quenched state. This component likely corresponds to the Lut1/Chl a610–(611)–612 site (37, 45). This cluster is the main energy sink (42–44) and would, if quenched, produce the significant change in fluorescence intensity observed. Additionally, Lut1 and Chl a612 interact strongly (38, 39), which would further enhance the change in intensity. Furthermore, computational results found that the Lut1/Chl a612 interaction has the largest and thus, slowest fluctuations in accordance with the slower dynamics observed for this component (35). The fast quenched-biased component (Fig. 3E, red) likely corresponds to the Lut2/Chl a602–603 site (37, 45), which shows higher flexibility and thus, faster dynamics in molecular dynamics (MD) simulations (36). Furthermore, the weaker interaction of Lut2 and Chl a603 compared with that of Lut1 and Chl a612 (35) would be expected to produce a smaller change in fluorescence intensity between the active and quenched states as observed in the fast quenched-biased component. In this way, dynamic rates can be used in parallel to predicted quenching yields to assign the components to likely sites. However, as additional structural and spectroscopic information emerges, these assignments can be either further validated or refined. The static component (Fig. 3E, gray) likely includes unquenched emitters as the active state, i.e., Chl a613–614 cluster and Chl a604 (37), and photobleached/degraded complexes as the quenched state.
In LHCB1 at low pH, the 2D-FLC analysis identified two dynamic components, both of which have an enhanced quenching efficiency (Fig. 3F). The static component is stabilized into the quenched state (Fig. 3F, blue) and likely corresponds to the Lut1/Chl a610–(611)–612 site because of the ability of Lut1 to quench the Chl a as discussed above. The equilibrium free energy difference shifted toward the quenched state by more than 800 cm−1, possibly through displacement of Lut1 toward the Chl a (38, 46). A contribution from Lut2, which is also a major candidate for quenching (39), might be included in or even dominate the static quenched component if the Lut2 is shifted toward the Chl a602–603 cluster. At low pH, the change in the polarizability of the detergent shell around the hydrophobic core of the molecule likely causes a small decrease in the diameter of LHCB1 (47, 48). The decrease in diameter decreases the distance between the pigments, which would increase quenching throughout the protein, making assignment of quenching sites particularly challenging. The active-biased dynamic component (Fig. 3F, green) likely corresponds to the Chl a604 quenched by neighboring Neo, because the free energy landscape of this component exhibits a different pH dependence in LHCB1 and LHCSR1 (i.e., the quenched state is stabilized in LHCSR1). As discussed below, it is the region around Neo that is affected by protonation and therefore, would differ between the two proteins. The conformational change around Neo has been proposed to red shift the neighboring Chl a604 (42). In this case, the Chl a604 becomes an energy sink instead of Chl a602–603 and Chl a613–614. Calculations showed that Neo can quench Chl a604 (39), although it is likely to only partially quench as observed, because of the higher energy level and longer lifetime of its S1 state compared with the other carotenoids (49).
Multiple Independent Regulatory Dynamics in LHCSR1.
The 2D-FLC analysis identified three dynamic components in LHCSR1-V-7.5 (Fig. 3A): active-biased fast dynamics (red), quenched-biased slow dynamics (blue), and an almost static component (gray). LHCSR1 contains eight Chl binding sites, six of which are homologous to those in LHCB1 (17), that each embed Chl a and four carotenoids but with two Lut in the Lut1 and Neo sites and two Vio in the Lut2 and Vio sites (25). Similar to LHCB1-7.5, in LHCSR1-V-7.5, the two dynamic components (Fig. 3A, red and blue) are likely to be associated with conformational fluctuations of the carotenoids in the Lut1 and Lut2 sites, while the static component (Fig. 3A, gray) would be due to unquenched emitters as well as photobleached/degraded complexes. In Zea-enriched LHCSR1 (Fig. 3C), one of two components is almost static (Fig. 3C, red), and the other is more quenched biased (Fig. 3C, blue), suggesting that Zea binding suppresses the active-biased dynamics and shifts the equilibrium slightly toward the quenched side. The active-biased component (Fig. 3A, red) likely corresponds to Vio in the Lut2 site interacting with neighboring Chl a, because on conversion from Vio to Zea, this component was suppressed, shifting the overall system toward the quenching side. Accordingly, the quenched-biased component (Fig. 3A, blue) likely corresponds to Lut1. The higher hydrophobicity of Zea can tighten the binding in the Lut2 site and compact the transmembrane helixes, which could stabilize the quenched state (50).
Under high light, the thylakoid lumen acidifies (29), protonating residues on the luminal loop of LHCSR1, which causes conformational changes and quenching. Thus, at low pH, the specific effect of protonatable residues is present along with the effect of the change in the detergent shell (51). In particular, protonation is thought to twist Neo (40, 46), turning on quenching at Lut1 (38). The pH drop increases the overall quenching efficiency, primarily by stabilizing the quenched state in the component associated with the Lut1 site (Fig. 3B, blue) as seen in LHCB1, consistent with the observation of electron transfer quenching between Lut and Chl a in LHCSR1 (24). Ensemble LHCSR1-V-5 results showed that the main contribution to quenching originates from Lut (24), and therefore, the other quenched-biased component (Fig. 3B, green) likely corresponds to the other Lut in the Neo site. The pH-induced conformational change (25, 51, 52) seems to cause the energy sink in the system to change, likely allowing the use of Lut in the Neo site instead of Vio in the Lut2 site as the quencher. The dynamics of Lut in the Neo site are strongly biased toward the quenched side in contrast to those of Neo in LHCB1 (Fig. 3F, green). The hydrophobicity of Lut compared with Neo (50) can lead to tighter binding closer to the Chl a energy sink and thus, more efficient quenching of excitation energy. Conversion from Vio to Zea caused little change in the dynamics (Fig. 3D), confirming that the Lut1 and Lut in the Neo site rather than Vio are the main energy quenchers at low pH (24).
The photophysical mechanism of NPQ remains debated, with four primary proposals: (i) energy transfer from Chl to the carotenoid S1 state (24, 38), (ii) charge transfer between Chl and carotenoid (24, 53–55), (iii) a mixed Chl–carotenoid excitonic state (56–58), and (iv) charge transfer between Chls (59). Furthermore, these mechanisms may occur cooperatively, such as has been observed in LHCSR1 for the first two mechanisms (24). Because both of these mechanisms involve interaction between Chl and carotenoids, they can be controlled by the quenching sites proposed. As additional structural and spectroscopic information about the mechanism emerges, the proposed sites may be validated or refined.
Enhancement of NPQ on Photobleaching.
If the quenching required exceeds the capacity of NPQ, the LHC can photobleach. The 2D-FLC analysis as a function of illumination time (SI Appendix, section S9 and Figs. S14–S19) revealed that initial photobleaching stabilizes the quenched state, which may provide another mechanism for photoprotection. Additionally, in LHCSR1-V-7.5, the active-biased component associated with the Vio in the Lut2 site preferentially photobleaches, whereas in LHCSR1-Z-7.5, initial photobleaching is delayed, indicating that Zea contributes resistance to photobleaching.
Conclusions
As demonstrated here, the developed 2D-FLC analysis applied to single-molecule data separated and resolved the dynamics that underlie the quenching of LHCs, characterizing the energetics and dynamics for each component. This enables speculation as to the Chls a and carotenoids within the LHC responsible for quenching, suggesting a molecular origin of photoprotection in LHCSR1. The structural models proposed in this study are the most probable ones based on existing spectroscopic, biochemical, and computational data. In each case, results from one or more of these tools strongly favor the assigned quenching site as discussed above. However, as additional data become available, the proposed sites may be validated or refined. In particular, this knowledge is a useful benchmark for future work, such as point mutation analyses, MD simulations, and comparative studies of carotenoid composition, which have been a powerful tool to disentangle the contributions of individual pigments (34). Because the in vitro system does not fully mimic or capture the in vivo complexity and behaviors, the results reported here should complement, help interpret, and be interpreted in the context of information from the intact organism. Applying the generalized 2D-FLC analysis to single-molecule data is a powerful tool to investigate multifunctionality in proteins. This approach enables simultaneous visualization of dynamics on timescales ranging from microseconds to seconds, accessing previously hidden aspects of biological systems.
Materials and Methods
Preparation of and single-molecule spectroscopy on LHCSR and LHCB1 were performed as described previously (9) and in SI Appendix, section S10. Detailed procedures for the 2D-FLC analysis are described in SI Appendix, sections S1–S3.
Supplementary Material
Acknowledgments
We thank Dr. Justin Caram for helpful comments. The work was supported by NIH Grant NIH9P41EB015871 for 2D-FLC development and Center for Quantum Molecular Design Phase I Center for Chemical Innovation Grant CHE-1740645 from the NSF for application to LHCs. Work was also supported by the Marie Curie Actions Initial Training Networks Solar Energy to Biomass (SE2B) (Grant 675006-SE2B to R.B and A.P.) and by the Research Projects of National Relevance (PRIN) “HARVEST” (Grant 201795SBA3-004 to L.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1821207116/-/DCSupplemental.
References
- 1.Krüger TPJ, Ilioaia C, Horton P, Alexandre MTA, van Grondelle R (2014) How protein disorder controls non-photochemical fluorescence quenching. Non-Photochemical Quenching and Energy Dissipation in Plants, Algae and Cyanobacteria, Advances in Photosynthesis and Respiration, eds Demmig-Adams B, Garab G, Adams W III, Govindjee (Springer, Dordrecht, The Netherlands), Vol 40, pp 157–185.
- 2.Kondo T, Chen WJ, Schlau-Cohen GS (2017) Single-molecule fluorescence spectroscopy of photosynthetic systems. Chem Rev 117:860–898. [DOI] [PubMed] [Google Scholar]
- 3.Hofmann C, Aartsma TJ, Michel H, Köhler J (2003) Direct observation of tiers in the energy landscape of a chromoprotein: A single-molecule study. Proc Natl Acad Sci USA 100:15534–15538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brucale M, Schuler B, Samorì B (2014) Single-molecule studies of intrinsically disordered proteins. Chem Rev 114:3281–3317. [DOI] [PubMed] [Google Scholar]
- 5.Watkins LP, Yang H (2005) Detection of intensity change points in time-resolved single-molecule measurements. J Phys Chem B 109:617–628. [DOI] [PubMed] [Google Scholar]
- 6.Goldsmith RH, Moerner WE (2010) Watching conformational- and photodynamics of single fluorescent proteins in solution. Nat Chem 2:179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schlau-Cohen GS, Wang Q, Southall J, Cogdell RJ, Moerner WE (2013) Single-molecule spectroscopy reveals photosynthetic LH2 complexes switch between emissive states. Proc Natl Acad Sci USA 110:10899–10903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schlau-Cohen GS, et al. (2015) Single-molecule identification of quenched and unquenched states of LHCII. J Phys Chem Lett 6:860–867. [DOI] [PubMed] [Google Scholar]
- 9.Kondo T, et al. (2017) Single-molecule spectroscopy of LHCSR1 protein dynamics identifies two distinct states responsible for multi-timescale photosynthetic photoprotection. Nat Chem 9:772–778. [DOI] [PubMed] [Google Scholar]
- 10.Digman MA, Gratton E (2011) Lessons in fluctuation correlation spectroscopy. Annu Rev Phys Chem 62:645–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishii K, Tahara T (2013) Two-dimensional fluorescence lifetime correlation spectroscopy. 1. Principle. J Phys Chem B 117:11414–11422. [DOI] [PubMed] [Google Scholar]
- 12.Ishii K, Tahara T (2013) Two-dimensional fluorescence lifetime correlation spectroscopy. 2. Application. J Phys Chem B 117:11423–11432. [DOI] [PubMed] [Google Scholar]
- 13.Otosu T, Ishii K, Tahara T (2015) Microsecond protein dynamics observed at the single-molecule level. Nat Commun 6:7685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lutz W, Bassi R, Kruse O (2016) Multi-level light capture control in plants and green algae. Trends Plant Sci 21:55–68. [DOI] [PubMed] [Google Scholar]
- 15.Ruban AV. (2016) Nonphotochemical chlorophyll fluorescence quenching: Mechanism and effectiveness in protecting plants from photodamage. Plant Physiol 170:1903–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graham P, et al. (2009) An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature 462:518–521. [DOI] [PubMed] [Google Scholar]
- 17.Bonente G, et al. (2010) Analysis of LhcSR3, a protein essential for feedback de-excitation in the green alga Chlamydomonas reinhardtii. PLoS Biol 9:e1000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alboresi A, Gerotto C, Giacometti GM, Bassi R, Morosinotto T (2010) Physcomitrella patens mutants affected on heat dissipation clarify the evolution of photoprotection mechanisms upon land colonization. Proc Natl Acad Sci USA 107:11128–11133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tokutsu R, Minagawa J (2013) Energy-dissipative supercomplex of photosystem II associated with LHCSR3 in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 110:10016–10021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pinnola A, et al. (2013) Zeaxanthin binds to light-harvesting complex stress-related protein to enhance nonphotochemical quenching in Physcomitrella patens. Plant Cell 25:3519–3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pinnola A, et al. (2015) Heterologous expression of moss light-harvesting complex stress-related 1 (LHCSR1), the chlorophyll a-xanthophyll pigment-protein complex catalyzing non-photochemical quenching, in Nicotiana sp. J Biol Chem 290:24340–24354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pinnola A, et al. (2015) Light-harvesting complex stress-related proteins catalyze excess energy dissipation in both photosystems of Physcomitrella patens. Plant Cell 27:3213–3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maruyama S, Tokutsu R, Minagawa J (2014) Transcriptional regulation of the stress-responsive light harvesting complex genes in Chlamydomonas reinhardtii. Plant Cell Physiol 55:1304–1310. [DOI] [PubMed] [Google Scholar]
- 24.Pinnola A, et al. (2016) Electron transfer between carotenoid and chlorophyll contributes to quenching in the LHCSR1 protein from Physcomitrella patens. Biochim Biophys Acta 1857:1870–1878. [DOI] [PubMed] [Google Scholar]
- 25.Pinnola A, et al. (2017) Functional modulation of LHCSR1 protein from Physcomitrella patens by zeaxanthin binding and low pH. Sci Rep 7:11158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kosuge K, et al. (2018) LHCSR1-dependent fluorescence quenching is mediated by excitation energy transfer from LHCII to photosystem I in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 115:3722–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pinnola A, Bassi R (2018) Molecular mechanisms involved in plant photoprotection. Biochem Soc Trans 46:467–482. [DOI] [PubMed] [Google Scholar]
- 28.Holleboom C-P, Walla PJ (2014) The back and forth of energy transfer between carotenoids and chlorophylls and its role in the regulation of light harvesting. Photosynth Res 119:215–221. [DOI] [PubMed] [Google Scholar]
- 29.Johnson MP, Zia A, Ruban AV (2012) Elevated pH restores rapidly reversible photoprotective energy dissipation in Arabidopsis chloroplasts deficient in lutein and xanthophyll cycle activity. Planta 235:193–204. [DOI] [PubMed] [Google Scholar]
- 30.Liu Z, et al. (2004) Crystal structure of spinach major light-harvesting complex at 2.72 Å resolution. Nature 428:287–292. [DOI] [PubMed] [Google Scholar]
- 31.Rutkauskas D, Novoderezkhin V, Cogdell RJ, van Grondelle R (2004) Fluorescence spectral fluctuations of single LH2 complexes from Rhodopseudomonas acidophila strain 10050. Biochemistry 43:4431–4438. [DOI] [PubMed] [Google Scholar]
- 32.Gruber JM, Chmeliov J, Krüger TPJ, Valkunas L, Van Grondelle R (2015) Singlet–triplet annihilation in single LHCII complexes. Phys Chem Chem Phys 17:19844–19853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gruber JM, et al. (2016) Photophysics in single light-harvesting complexes II: From micelle to native nanodisks. Proc SPIE 9714:97140A. [Google Scholar]
- 34.Tutkus M, Chmeliov J, Rutkauskas D, Ruban AV, Valkunas L (2017) Influence of the carotenoid composition on the conformational dynamics of photosynthetic light-harvesting complexes. J Phys Chem Lett 8:5898–5906. [DOI] [PubMed] [Google Scholar]
- 35.Liguori N, Periole X, Marrink SJ, Croce R (2015) From light-harvesting to photoprotection: Structural basis of the dynamic switch of the major antenna complex of plants (LHCII). Sci Rep 5:15661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liguori N, et al. (2017) Different carotenoid conformations have distinct functions in light-harvesting regulation in plants. Nat Commun 8:1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Di Valentin M, Biasibetti F, Ceola S, Carbonera D (2009) Identification of the sites of chlorophyll triplet quenching in relation to the structure of LHC-II from higher plants. Evidence from EPR spectroscopy. J Phys Chem B 113:13071–13078. [DOI] [PubMed] [Google Scholar]
- 38.Ruban AV, et al. (2007) Identification of a mechanism of photoprotective energy dissipation in higher plants. Nature 450:575–578. [DOI] [PubMed] [Google Scholar]
- 39.Fox KF, et al. (2017) The carotenoid pathway: What is important for excitation quenching in plant antenna complexes?. Phys Chem Chem Phys 19:22957–22968. [DOI] [PubMed] [Google Scholar]
- 40.Belgio E, Duffy CDP, Ruban AV (2013) Switching light harvesting complex II into photoprotective state involves the lumen-facing apoprotein loop. Phys Chem Chem Phys 15:12253–12261. [DOI] [PubMed] [Google Scholar]
- 41.Barros T, Kühlbrandt W (2009) Crystallisation, structure and function of plant light-harvesting complex II. Biochim Biophys Acta 1787:753–772. [DOI] [PubMed] [Google Scholar]
- 42.Müh F, El-Amine Madjet M, Renger T (2010) Structure-based identification of energy sinks in plant light-harvesting complex II. J Phys Chem B 114:13517–13535. [DOI] [PubMed] [Google Scholar]
- 43.Schlau-Cohen GS, et al. (2009) Pathways of energy flow in LHCII from two-dimensional electronic spectroscopy. J Phys Chem B 113:15352–15363. [DOI] [PubMed] [Google Scholar]
- 44.Novoderezhkin V, Marin A, van Grondelle R (2011) Intra- and inter-monomeric transfers in the light harvesting LHCII complex: The Redfield–Förster picture. Phys Chem Chem Phys 13:17093–17103. [DOI] [PubMed] [Google Scholar]
- 45.Lampoura SS, Barzda V, Owen GM, Hoff AJ, van Amerongen H (2002) Aggregation of LHCII leads to a redistribution of the triplets over the central xanthophylls in LHCII. Biochemistry 41:9139–9144. [DOI] [PubMed] [Google Scholar]
- 46.Pandit A, et al. (2013) An NMR comparison of the light-harvesting complex II (LHCII) in active and photoprotective states reveals subtle changes in the chlorophyll a ground-state electronic structures. Biochim Biophys Acta 1827:738–744. [DOI] [PubMed] [Google Scholar]
- 47.Bassi R, Silvestri M, Dainese P, Moya I, Giacometti GM (1991) Effects of a non-ionic detergent on the spectral properties and aggregation state of the light-harvesting chlorophyll a/b protein complex (LHCII). J Photochem Photobiol B, Biol 9:335–353. [Google Scholar]
- 48.Moya I, Silvestri M, Vallon O, Cinque G, Bassi R (2001) Time-resolved fluorescence analysis of the photosystem II antenna proteins in detergent micelles and liposomes. Biochemistry 40:12552–12561. [DOI] [PubMed] [Google Scholar]
- 49.Polívka T, Sundström V (2004) Ultrafast dynamics of carotenoid excited states- from solution to natural and artificial systems. Chem Rev 104:2021–2072. [DOI] [PubMed] [Google Scholar]
- 50.Ruban AV, Johnson MP (2010) Xanthophylls as modulators of membrane protein function. Arch Biochem Biophys 504:78–85. [DOI] [PubMed] [Google Scholar]
- 51.Ballottari M, et al. (2016) Identification of pH-sensing sites in the Light Harvesting Complex Stress-Related 3 protein essential for triggering non-photochemical quenching in Chlamydomonas reinhardtii. J Biol Chem 291:7334–7346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liguori N, Roy LM, Opacic M, Durand G, Croce R (2013) Regulation of light harvesting in the green alga Chlamydomonas reinhardtii: The C-terminus of LHCSR is the knob of a dimmer switch. J Am Chem Soc 135:18339–18342. [DOI] [PubMed] [Google Scholar]
- 53.Holt NE, et al. (2005) Carotenoid cation formation and the regulation of photosynthetic light harvesting. Science 307:433–436. [DOI] [PubMed] [Google Scholar]
- 54.Ahn TK, et al. (2008) Architecture of a charge-transfer state regulating light harvesting in a plant antenna protein. Science 320:794–797. [DOI] [PubMed] [Google Scholar]
- 55.Dall’Osto L, et al. (2017) Two mechanisms for dissipation of excess light in monomeric and trimeric light-harvesting complexes. Nat Plants 3:17033. [DOI] [PubMed] [Google Scholar]
- 56.van Amerongen H, van Grondelle R (2001) Understanding the energy transfer function of LHCII, the major light-harvesting complex of green plants. J Phys Chem B 105:604–617. [Google Scholar]
- 57.Bode S, et al. (2009) On the regulation of photosynthesis by excitonic interactions between carotenoids and chlorophylls. Proc Natl Acad Sci USA 106:12311–12316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liao P-N, et al. (2011) Two-photon study on the electronic interactions between the first excited singlet states in carotenoid- tetrapyrrole dyads. J Phys Chem A 115:4082–4091. [DOI] [PubMed] [Google Scholar]
- 59.Müller MG, et al. (2010) Singlet energy dissipation in the photosystem II light-harvesting complex does not involve energy transfer to carotenoids. Chemphyschem 11:1289–1296. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.