Abstract
Colinearity of a large region from barley (Hordeum vulgare) chromosome 5H and rice (Oryza sativa) chromosome 3 has been demonstrated by mapping of several common restriction fragment-length polymorphism clones on both regions. One of these clones, WG644, was hybridized to rice and barley bacterial artificial chromosome (BAC) libraries to select homologous clones. One BAC from each species with the largest overlapping segment was selected by fingerprinting and blot hybridization with three additional restriction fragment-length polymorphism clones. The complete barley BAC 635P2 and a 50-kb segment of the rice BAC 36I5 were completely sequenced. A comparison of the rice and barley DNA sequences revealed the presence of four conserved regions, containing four predicted genes. The four genes are in the same orientation in rice, but the second gene is in inverted orientation in barley. The fourth gene is duplicated in tandem in barley but not in rice. Comparison of the homeologous barley and rice sequences assisted the gene identification process and helped determine individual gene structures. General gene structure (exon number, size, and location) was largely conserved between rice and barley and to a lesser extent with homologous genes in Arabidopsis. Colinearity of these four genes is not conserved in Arabidopsis compared with the two grass species. Extensive similarity was not found between the rice and barley sequences other than within the exons of the structural genes, and short stretches of homology in the promoters and 3′ untranslated regions. The larger distances between the first three genes in barley compared with rice are explained by the insertion of different transposable retroelements.
The grass family is the fourth largest family of flowering plants (approximately 10,000 species) and includes wheat (Triticum spp.), maize (Zea mays), rice (Oryza sativa), barley (Hordeum vulgare), and oats (Avena sativa). This diverse family exhibits a large variation in DNA content. Rice is a diploid species with a relatively small genome (430 Mb), whereas barley and many other diploid Triticeae spp. contain 4,000 to 8,000 Mb of DNA in their nuclei (Bennett and Leitch, 1995). These differences in DNA content indicate that a single chromosome from barley contains more DNA than one complete haploid rice genome.
Despite the enormous differences in genome size, comparative RFLP maps have revealed a remarkable conservation of gene content and gene order among the cereal genomes (Gale and Devos, 1998). Early DNA renaturation studies had indicated that the larger plant genomes contain more repetitive DNA and that some of this repetitive DNA was intermixed with genes (Flavell and Smith, 1976). The sequencing of a 280-kb segment around the adh1-F gene in maize, a large genome grass (2,400 Mb), showed that intergenic regions were mainly composed of retrotransposons inserted within each other. These retro-elements accounted for more than 60% of the DNA in the adh1-F interval and more than 70% of the total maize genome (SanMiguel et al., 1996; SanMiguel and Bennetzen, 1998). Comparison of the orthologous region in a close relative with a smaller genome, sorghum (Sorghum bicolor; 750 Mb), showed a complete absence of detected retro-elements. Only genes identified within this interval were evolutionarily conserved, indicating an extensive differentiation in the intergenic regions of these species (Chen et al., 1997).
The diploid Triticeae genomes are even larger than the maize genome (barley = 5,000 Mb; rye [Secale cereale] = 8,000 Mb) and repetitive sequences represent more than 75% of the genome (Flavell and Smith, 1976; Suoniemi et al., 1996; Gribbon et al., 1999; Vicient et al., 1999). Panstruga et al. (1998) sequenced a 60-kb region in barley around the mlo locus and found three genes plus retrotransposons and complex repeat structures that accounted for one-quarter of the DNA in this interval. Similar repeat structures accounted for more than one-half of the DNA sequence in another 66-kb region in barley around the rar1 locus (Shirasu et al., 2000). Even though these and other preliminary sequencing efforts in Triticeae species (Wei et al., 1999; Stein et al., 2000; Lagudah et al., 2001) are providing a first picture of the organization of these large genomes, no large segment of Triticeae DNA sequence has been compared yet with its orthologous region in rice. Preliminary comparisons of this sort will help indicate to what degree the large amount of rice sequencing generated by the Rice Genome Project will be useful for positional cloning and genetic characterization in the Triticeae.
This paper presents the DNA sequence and sequence comparison of an orthologous region of the rice and barley genomes. Our data show that comparative sequence analyses of this type can greatly assist sequence annotation, including gene identification and characterization of gene structure. More importantly, these studies will help indicate the nature, frequency, and timing of different types of sequence evolution in these two important grass lineages.
RESULTS
Clone Identification
Thirty-seven thousand and 300,000 clones from rice and barley bacterial artificial chromosome (BAC) libraries, respectively, were screened for individual clones that contained homology to WG644. Four clones with homology were identified in each library and clones 635P2 (barley) and 36I5 (rice) were selected for sequencing because they appeared to have the greatest degree of overlap. Both BACs hybridized also with rice RFLP clones R2311 (two copies in 36I5) and R2628 and barley subclone UCW12 from the plasmid rescue of BAC 635P2. Barley subclone UCW12 was positioned on a Triticum monococcum genetic map (Dubcovsky et al., 1996) and found to be completely linked to WG644. RFLP markers R2311 and R2628 were also found to be closely linked to WG644 in different rice genetic maps (Maroof et al., 1996; Harushima et al., 1998; Sarma et al., 1998; Kato et al., 1999).
BAC Sequencing
Rice BAC 36I5 was sequenced at different levels of coverage (5×, 10×, 15×, and 20×; PHRED > 20) and assembled after each round of sequence to evaluate the best balance between initial coverage and final finishing efforts. The number of gaps and “problems,” defined as areas covered by a single subclone or areas with a base quality below 1 in 10,000 error rate, decreased with the increasing coverage (5×, six gaps and 216 problems; 10×, three gaps and 80 problems; 15×, two gaps and 43 problems; and 20×, two gaps and 39 problems). Although the gap sizes were reduced, other problems were not efficiently solved by increasing coverage from 15× to 20×. One problem region on this BAC, containing two duplicated regions in inverted orientation from the end of gene 4, adjacent to a tandem of five large repeats, was not fully resolved, resulting in two contigs of 66,700 and 5,812 bp. We report here the sequence of the first 50,000 bp with a predicted error rate lower than 1 in 100,000 bp (GenBank accession no. AY013245).
Barley BAC 635P2 was completely assembled, starting from a 13× redundant set of shotgun sequences (PHRED scores > 20). Difficult regions such as G- and C-rich areas and gaps in the region of two nested BARE-1 retrotransposons in barley BAC 635P2 were closed using a combination of primer walking and drhodamine and dGTP big dye terminator chemistries. The complete sequence was 102,433 bp and has a predicted error rate lower than 1 in 100,000 bp (GenBank accession no. AY013246).
Colinear Gene Region
The first step in the sequence analysis of these two colinear BACs was the delimitation of regions that are conserved and unique between the rice and barley sequences. Unique regions were annotated individually for each BAC, but conserved regions were annotated simultaneously in both BACs and compared with the information provided by the rice-barley sequence alignment and six-frame translation of the homologous regions.
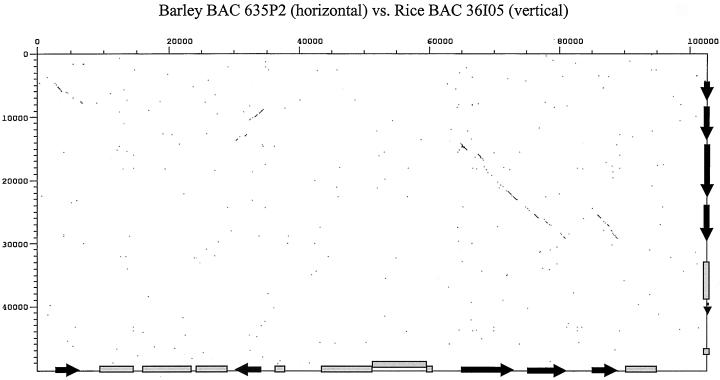
Comparison of the complete sequence of 36I5 with the complete sequence of 635P2 using the program Dotter revealed the presence of four conserved regions (Fig. 1). The same regions were detected by the gene prediction programs Genescan and GeneMark.hmm and by BLAST and GeneSeqer searches of the expressed sequence tag (EST) databases, suggesting that they correspond to gene regions. No similarity was found between the rice and barley sequence outside the genes with the exception of short stretches of homology in the promoters and 3′-untranslated regions and non-colinear Stowaway transposable elements.
Figure 1.
Sequence comparison of barley BAC 635P2 with the first 50 kb of rice BAC 36I5 using the program Dotter (word 25, similarity 80). The location of genes and large transposable elements in rice (vertical) and barley (horizontal) are indicated by arrows and boxes, respectively. A more detailed annotation is presented in Figure 2.
The conserved regions span 30-kb at the 5′ end of the rice sequence and the complete 102 kb of the barley sequence. The greater physical length of the four-gene region on the barley BAC was due to larger intergenic spaces than in rice. Distances between genes 1 and 2 (rice = 1.4 kb; barley = 24.0 kb), and between genes 2 and 3 (rice = 0.6 kb; barley = 30.5 kb) were greatly expanded in barley by the insertion of different transposable elements (see below). Gene 4 was duplicated in barley (gene 4a and 4b) but not in rice (Figs. 1 and 2).
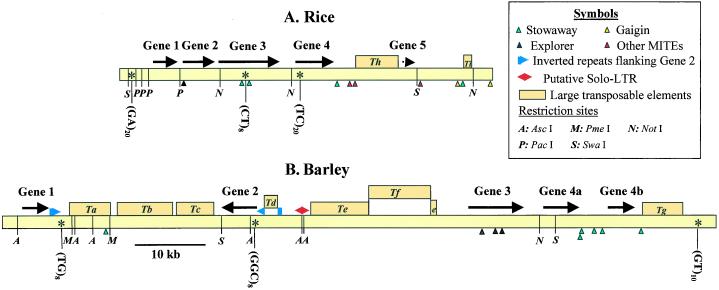
Figure 2.
Organization of repetitive and conserved sequences in barley BAC 635P2 and the first 50 kb of rice BAC36I5. Genes are represented by arrows, transposable elements by boxes, and MITEs by triangles. Ta and Tb, Copia-like retrotransposon; Tc, non-LTR retroelement; Td, terminal-inverted repeat element; Te and Tf, nested BARE-1 retrotransposons; Tg, MuDR-like element; Th, Copia-like retrotransposon; Ti, nonautonomous Mutator element. SSRs with eight or more repeats are indicated by an asterisk.
The four genes are in the same predicted transcriptional orientation in rice, but the second gene is in inverted orientation in barley (Figs. 1 and 2). The relative inversion of gene 2 was confirmed experimentally. All fragments observed on a pulse field agarose gel with digestions of both BACs with eight-cutter restriction enzymes and all possible double digestions agreed with the restriction maps predicted by the current sequence assembly (Fig. 2). An additional gel replica was prepared with DNA from both BACs digested with eight six-cutter restriction enzymes, several of them with restriction sites between exons 3 and 9 from gene 2. Filter replicas from the six- and eight-cutter restriction enzymes were hybridized with two probes from genes 2 and 1 probe from gene 1 to analyze putative short inversions. The first probe for gene 2 included exons 1 to 3 and the second one exons 9 to 13. Restriction fragments detected by hybridization with the three probes were all predicted by the current sequence assembly, suggesting that the relative inversion of gene 2 is not the product of an incorrect sequence assembly.
Two areas of inverted homology were identified flanking barley gene 2 (6,893–7,457 and 38,348–35,605 bp; Fig. 2). These inverted repeats might have been associated with a recombination event between nonorthologous repeated sequences located in opposite orientation at both sides of gene 2, resulting in the inversion of this barley gene.
The four Arabidopsis genes with the highest similarity with the four genes identified within the rice and barley BACs (Table I) seemed to be not colinear in Arabidopsis. The closest Arabidopsis homologues for rice and barley genes 1, 2, and 4 (Table I) are all located on Arabidopsis chromosome 5, but on three different BACs positioned approximately 10,000 kb from each other on the physical map (BAC F14F8 = 5,400–5,500 kb; MSL3 = 23,700–23,800; MXF12 = 14,400–14,500). The closest homolog for barley and rice gene 3 maps to Arabidopsis chromosome 1 (BAC F4N2 = 25,100–25,200 kb).
Table I.
BLASTP comparisons between the barley predicted protein, the rice predicted protein, and the two most similar Arabidopsis proteins
| Predicted Barley Gene | Predicted Rice
|
Arabidopsis Gene | BLASTP
|
cDNA sAcc. No. | BLASTN
|
Exons Aligned | |||
|---|---|---|---|---|---|---|---|---|---|
| Score | Expect | Score | Expect | Score | Expect | ||||
| Gene 1 | 562 | e-159 | F14F8.20 | 421 | e-118 | BE429317 Wheat | 315 | 2e-83 | Start to 2 |
| 322 aa | F3N23.2 | 281 | 6e-76 | BE704834 Rye | 961 | 0 | 2 to stop | ||
| Gene 2 | 758 | 0 | MSL3.6 | 503 | e-142 | BE494453 Rye | 555 | e-155 | Start to 5 |
| 423 aa | T16K5.10 | 120 | 2e-27 | BE413026 Barley | 1,439 | 0 | 2 to 11 | ||
| AI941761 Maize | 123 | 2e-25 | 10 to stop | ||||||
| Gene 3 | 1,403 | 0 | S1K1 | 797 | 0 | AW672216 Sorghum | 190a | 7e-48 | 1 to 6 |
| 825 aa | K7L4.2 | 269 | 7e-72 | BE186368 Maize | 182 | 5e-43 | 10 to 13 | ||
| AU031105 Rice | 168 | 8e-39 | 14 to 16 | ||||||
| BE475909 Maize | 248 | e-62 | 16 to stop | ||||||
| Gene 4 | 1,150 | 0 | MXF12.6 | 909 | 0 | BE493856 Rye | 856 | 0 | 1 to 7 |
| 644 aa | F14M4.17 | 368 | e-102 | AW428933 Tomato | 70 | 4e-9 | 6 to 9 | ||
| AW758716 Pinus | 76 | 7e-11 | 8 to 10 | ||||||
| BE357922 Sorghum | 726 | 0 | 10 to 16 | ||||||
| BE357994 Sorghum | 426 | e-116 | 15 to 17 | ||||||
| BE040236 Rice | 222 | 5e-55 | 16 to stop | ||||||
| Gene 4b | 1,101 | 0 | MXF12.6 | 891 | 0 | ||||
| 644 aa | |||||||||
| Rice gene 5 | — | — | — | C27091 Rice | 432 | e-118 | 1 | ||
| 296 aa | C74904 Rice | 292 | 3e-76 | 1 | |||||
| C27330 Rice | 312 | 2e-82 | 2 | ||||||
BLASTN comparisons between the barley predicted cDNA and the dbEST database. The last column includes the predicted exons confirmed by alignment with cDNA clones. Gene 5 has been found only in rice.
TBLASTX score.
An additional search of the Arabidopsis database was performed using TBLASTN and the four Arabidopsis predicted proteins concatenated in a single file. The objective of this search was to explore the possibility of genes, which are colinear between the grasses and Arabidopsis but which have diverged considerably in sequence. This search showed that the Arabidopsis gene MXF12.5 adjacent to the closest Arabidopsis homologue for barley gene 4 (MXF12.6, Table I) was similar to barley gene 3, although with BLAST scores (score = 70) one order of magnitude lower than the S1K1 closest homolog (score = 797). Significant BLAST scores were also obtained between two grass genes and the Arabidopsis BAC MSL3. However, in this case the closest homolog of gene 2 (MSL3.6) was more than 20 kb apart from the Arabidopsis gene showing similarity with barley gene 3 and that similarity was very low (score 67.6 Expect 5e-10). These weak similarities to gene 3 in the proximity of the closest Arabidopsis homologues to genes 2 and 4 are more likely related to the abundance of kinase genes similar to S1K1 than to true conserved pairs of colinear genes.
The Nonoverlapping Region from Rice BAC 36I5
Gene predictions in the 20-kb of the BAC 36I5 contiguous sequence that does not overlap with BAC 635P2 were more difficult because of the absence of comparable barley sequence. However, both Genescan and GeneSeqer predicted an initiation codon at 39,753 bp and a terminal exon at 40,748 bp tentatively designated as gene 5. The presence of a gene in this region was further supported by the significant similarity between three nonoverlapping segments from this gene with three rice cDNA clones (C27091 from 11–280 bp, 95% identical; C74904 from 320–519 bp, 92% identical; and C27330 from 707–883 bp, 95% identical) (Table I). Alignment with C74904 suggested different exon-intron boundaries to those predicted by Genescan. Assembly of a protein from the putative coding regions was performed and analysis of the putative protein by BLASTP and TBLASTN produced no matches to known proteins.
Gene Structure
The comparison of the colinear barley and rice sequence not only delimited the genes within the genomic sequence but also helped to establish their structure.
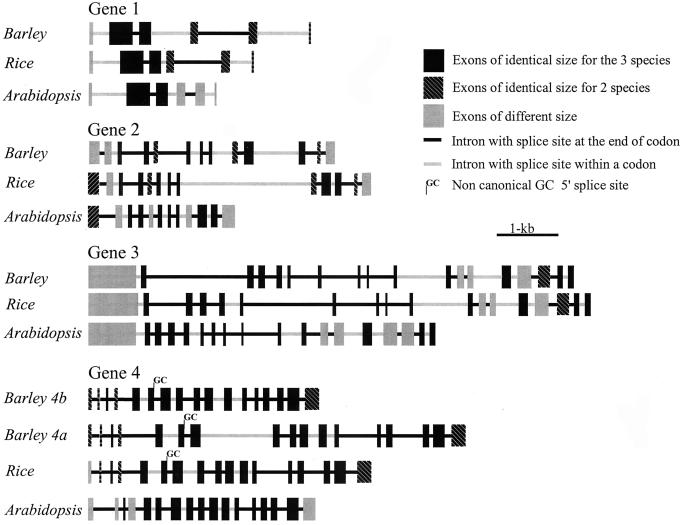
The predicted protein from barley gene 1 has a high level of similarity with the predicted rice protein (93% similarity, Table I) and with the closest Arabidopsis homologue (gene F14F8.20, 353 aa, 73% similarity). All three genes are divided in six exons (Fig. 3). Exon 2 has identical size in all three species and exons 3 to 6 have identical length in barley and rice. However, analysis of the Arabidopsis genomic sequence revealed the existence of alternative GT (19 bp downstream from the current annotation) and AG (71 bp upstream from the current annotation) splice sites between exons 3 and 4. If these alternative splice sites are used, the third exon of the predicted Arabidopsis protein would have the same length as the rice and barley exon 3 and the overall similarity between Arabidopsis and barley predicted proteins would improve from 73% to 81%. This alternative annotation is presented in Figure 3. Gene structure in barley was confirmed by the absence of gaps in the alignment of the DNA sequence from the predicted barley cDNA (concatenated predicted exons) with a cDNA from Triticum durum (BE429317, 97% identical) and a cDNA from cold stressed winter rye (BE704834, 96% identical, Table I). The closest Arabidopsis homologue has been annotated as a putative mitochondrial carrier protein by the presence of a mitochondrial energy transfer protein signature (CAC01763).
Figure 3.
Structure of the genes found in the regions conserved between rice BAC 36I5 and barley BAC 635P2 and the closest Arabidopsis homologues (Table I). Exons (boxes) and introns (lines) are proportional to their length. Exons of identical length in the three species are indicated in black and those of identical length in rice and barley in black with white diagonal lines.
The predicted protein from barley gene 2 also has a high level of similarity with the predicted rice protein (88% similarity, Table I) and with the closest Arabidopsis homologue (gene MSL3.6, 425 aa, 72% similarity, Table I). The rice and barley genes are divided in 13 exons, 10 of which are of identical length (Fig. 3). The Arabidopsis gene has seven exons of identical length to the grass species but has one exon less. The last exon of the Arabidopsis gene showed 79% similarity to the last two exons from rice and its length (210 bp) was identical to the sum of the lengths of the last two exons in rice (57 and 153 bp). Analysis of the Arabidopsis genomic sequence showed the presence of alternative splice sites that would improve the similarity between the Arabidopsis predicted protein and the predicted proteins from grasses. An alternative GT splice site 87 bp downstream of the annotated end of exon 1 would determine an exon of identical length to rice (Fig. 3). The use of alternative splice sites for exon 2 (166 bp from annotated AG and 76 bp downstream from annotated GT) would greatly improve similarity with predicted exon 2 from grasses. The predicted Arabidopsis exon 5 for gene 2 has six amino acids less than the predicted rice or barley protein at the start of exon 5. The use of an alternative splice site 18 bp downstream from the one that has been annotated in Arabidopsis would result in the addition of six amino acids (VPKVKQ) that are very similar to those found in rice and barley (VAKIKQ and VSKIKQ, respectively). These three modifications of the Arabidopsis annotation would improve the overall similarity between the Arabidopsis and barley predicted proteins from 72% to 75%. This alternative annotation is presented in Figure 3. The predicted gene structure in barley has been confirmed by the alignment of the predicted nucleotide sequence of the barley cDNA based on the genomic sequence with cDNAs from rye (BE494453, 96% identical), barley (BE413026, 99% identical), and maize (AI941761, 81% identical; Table I). The presence of this sequence in different cDNA libraries related to flowering may provide an indication of its function. The closest Arabidopsis homologue has been annotated as a putative cleavage stimulation factor subunit 1 (BAB10643).
Gene 3 is the largest of this interval with 790 amino acids in barley and 804 in rice. The translated proteins show 88% similarity (Table I) and are both divided in 18 exons, 14 of them of identical length (Fig. 3). The closest Arabidopsis homologue (gene S1K1, 63% similarity with barley predicted protein, Table I) has 13 exons of identical length to rice and barley but has five additional predicted exons after exon 18. However, 30-bp downstream from the annotated donor splice site of Arabidopsis exon 18, a stop codon is present in the same position as in barley and rice. The 10 amino acids predicted for these additional 30 bp (CDTLRTILRL) are identical to the last 10 amino acids in rice and barley gene 3. The predicted amino acids from the additional five exons from the Arabidopsis protein produced no significant hits with the non-redundant and SwissProt databases, and therefore require further experimental confirmation. These last five exons are not included in Figure 3. The 18 exons predicted for barley gene 3 were confirmed by alignment of the predicted cDNA with different cDNA clones from sorghum (AW672216, 76% identical), maize (BE186368, 89% identical and BE475909, 88% identical), and rice (AU031105, 88% identical with barley and 98% identical with rice predicted cDNA) (Table I). The stop codon and the polyadenylation signal (409 bp downstream from the stop codon) were confirmed by alignment of the genomic sequence with maize EST BE475909 and rice EST AU031106 (97% identical with rice), respectively. The closest Arabidopsis homologue has been annotated as a Ser/Thr kinase with homology to yeast and mammalian stress activated kinases (AC008262).
The predicted barley protein from gene 4 showed a high similarity with the predicted rice protein (95% similarity) and with the closest Arabidopsis homologue (gene MXF12.6, 655 aa, 86% similarity, Table I). The rice, barley, and Arabidopsis genes are organized in 17 exons, 16 of which are of identical length in rice and barley, and 13 of which are identical in the three species (Fig. 3). The predicted gene structure was supported by the perfect alignment of the predicted rice and barley cDNA with six different overlapping cDNAs from rye, sorghum, rice, tomato (Lycopersicon esculentum), and pine (Table I). Gene 4 includes RFLP markers WG644 and R2311 (Harushima et al., 1998; Sarma et al., 1998; Kato et al., 1999). The closest Arabidopsis homologue has been annotated as an ABC transporter-like protein (BAB10828).
Barley genes 4a and 4b are duplications of an ancestral barley gene 4. The predicted 4a and 4b proteins have 17 exons of identical length (Fig. 3). The percentage of conserved amino acids between barley genes 4a and 4b (95%) is identical to that observed between barley gene 4a and rice gene 4 (95%) suggesting that this duplication is almost as old as the rice-barley divergence or that duplication has accelerated divergence between the two copies. Rice gene 4 showed lower similarity with barley gene 4b (92%) than with barley gene 4a (95%). This suggests that barley gene 4a may be either the true orthologue of rice gene 4 (if gene 4b from rice was deleted) or the barley copy that conserved a similar function to rice gene 4 (if the duplication never occurred in the rice lineage and the two barley copies diverged in their function).
The ends of exon 6 in barley genes 4a and 4b and rice gene 4 are not absolutely clear because the closest canonical GT 5′-splice sites force three different deletions/insertions that disagree with the boundary of homology between the three genes. The use of an alternative conserved GC 5′-splice site simultaneously eliminates the insertion/deletions in all three genes, determines exons of similar length with Arabidopsis and provides a perfect adjustment to the boundary of the barley-rice homology. This alternative GC 5′-splice site, predicted by comparative sequence analysis, was confirmed by the perfect alignment of the border between exons 6 and 7 with a rye cDNA (BE493856, BLAST score 829 E = 0). A similar non-canonical GC 5′-splice site has been reported in a comparison between the mouse and human GALT genes (Batzoglou et al., 2000).
BLASTN comparisons between rice and barley genomic sequences from gene regions successfully identified 123 of the 125 exons annotated in the colinear region. Almost no similarity was found in the intron regions, except a few base pairs flanking the exon-intron boundaries. The number of base pairs from each end of the significantly similar segments detected by BLASTN and the closest exon-intron boundary (including AG-GT splice sites) was calculated for the 208 exon-intron boundaries annotated in the colinear region. The average number of base pairs between these two points varied between 0 and 68 bp with an average of 9.8 bp and an sd of 11.8 bp. Once a larger number of comparisons between rice and barley genes become available, the distribution of these distances can be used to assign a probability to gene prediction programs based on rice-Triticeae comparisons.
Only exon 5 from gene 2 and exon 1 from gene 4 were not detected by BLASTN between the rice and barley genomic sequences. Exons 14 and 15 from gene 3 were detected but merged in a single segment by BLASTN. These four exons were detected by the gene prediction program GeneMark.hmm in both rice and barley. GeneMark.hmm (using the rice model) and Genescan (using the maize model) detected 56% and 22% of the 54 exons present in the rice genes. The number of exons predicted by these two programs in the barley genes (total 71 exons) was slightly higher than the number predicted in rice (GeneMark.hmm 62%, Genescan 32%), but significantly smaller than the number of exons detected by the rice-barley sequence comparison (98.4%).
Short stretches of homology between rice and barley were found upstream of the start codons of genes 1 and 3 using BLASTN. A region of 37 nucleotides (86% identical) was found 285 and 217 bp upstream of the start codon in rice and barley genes 1, respectively. Two conserved regions of 24 (94% identical) and 38 nucleotides (95% identical) were identified upstream of the start codons of gene 3 in rice (50 and 120 bp) and barley (41 and 91 bp). No significant homology was found between the promoter regions of barley genes 4a and 4b, or between any of the promoter regions in the grass genes and the closest Arabidopsis homologues. The conserved regions in genes 1 and 3 provide good starting points to delimit sequences with relevant biological function within the promoter regions.
Intergenic Regions
The organization of the different retroelements in these two BACs is shown in Figure 2. Coordinates for the different transposable elements are available in the annotated sequences in GenBank (AY013245 and AY013246). The termini of retrotransposons and some terminal inverted repeat (TIR) elements such as Mutator (Bennetzen, 1996) are large enough and contain enough features to robustly ascertain exact insertion sites using only same sequence comparisons. However, other transposable elements are more variable in this aspect and coordinates for the boundaries required additional identification methods.
The larger distance between genes 1 and 2 in barley compared with rice is due in part to the insertion of two Copia-like retrotransposons and a non-long terminal repeat (LTR) retroelement (Ta, Tb, Tc; Fig. 2). These three transposable elements account for 18 of the 22-kb difference in size between the corresponding barley and rice intergenic region. The larger distance between genes 2 and 3 in barley compared with rice, similarly, is due to the insertion of a TIR element (Td, Fig. 2) and two BARE-1 retroelements (Te and Tf, Fig. 2). BARE-1-Tf is inserted into the 3′-LTR of the BARE-1-Te in opposite orientation (Fig. 2). A putative solo LTR of a BAGY-2 retrotransposon (Shirasu et al., 2000) was found upstream of the two nested BARE-1 elements. These elements account for 21 kb of the 29-kb difference between barley and rice in the region between genes 2 and 3. An additional 5-kb transposable element with homology to MuDR was found downstream of barley gene 4b (Tg, Fig. 2). Eight Stowaway miniature inverted transposable elements (MITEs) were found in the introns and intergenic regions of barley genes 3, 4a, and 4b.
One large Copia-like retrotransposon (6 kb) was found between rice genes 4 and 5 (Th, Fig. 2). A nonautonomous Mutator transposable element and 11 MITEs were also found in this 50,000 bp of rice BAC 36I5.
Simple Sequence Repeats
BAC sequencing provides the opportunity to find SSR markers for specific regions of the genome. Simple sequence repeats (SSR), each with eight or more repeats, were identified in the rice and barley BACs (Fig. 2).
In rice BAC 36I5, one dinucleotide SSR was found in an intergenic region and two other dinucleotide SSRs were located within introns of genes 3 and 4. The long (TC)20 SSR located between exons 1 and 2 in gene 4 seems to be a good candidate to develop a PCR marker for this region of the rice genome by designing specific primers in the conserved flanking exons.
In barley BAC 635P2, two short dinucleotide SSRs were found in intergenic regions at the beginning and end of the BAC. The trinuclotide SSR located within the first exon of gene 2 is not a perfect repeat (GGC)3(GAC)(GGC)4 and no differences in size were found in this region between the barley cDNA and closely related cDNAs from rye (BE494453) and wheat (BE400296), suggesting that these repeats will not yield a polymorphic SSR.
DISCUSSION
Gene Discovery and Gene Structure
Sequence comparison between colinear rice and barley BACs 36I5 and 635P2 exemplifies the potential of comparative genomics as an independent method for gene identification to validate predictions made by gene finding programs (Avramova et al., 1996). Because no similarity was found between the intergenic regions, a visual comparison of the complete rice and barley BAC sequences using a dotplot immediately delimited the gene regions (Fig. 1). With the exception of rapidly evolving gene families (for instance, disease resistance genes; Leister et al., 1998), this method may provide a fast way to identify Triticeae genes once the complete rice sequence becomes available.
Sequence comparison was useful not only to identify the genes present in the region but also to provide valuable information for the determination of gene structure. Predicted gene structures were validated in most cases by comparisons with EST databases. A similar approach has been used in human-mouse comparisons of orthologous regions to accurately identify coding exons (Batzoglou et al., 2000). The percentage of exons identified by the BLASTN comparison of rice and barley gene regions (98.4%) was significantly higher than the percentage identified by GeneMark.hmm (59.2%) and Genescan (28%). However, four exons were correctly predicted by GeneMark.hmm and missed or merged by the rice-barley comparison, suggesting that a combination of gene finding programs, search of the EST databases, and comparative sequence analyses will provide the best strategy to determine gene content and structure.
None of the gene prediction programs were able to detect the non-canonical 5′-“GC” splice site at the end of exon 6 in gene 4. The alternative GC splice was first predicted by the comparative sequence analysis and then confirmed by the alignment of the predicted sequence with two different cDNAs. It would be useful to establish the frequency of these non-canonical sites in plant genes to assign them a correct probability in gene-finding programs.
If exon-intron boundaries were randomly distributed among the 3 bp of a codon, one-third of them would be expected to be between codons and two-thirds within codons. However, 66% of the observed exon-intron boundaries were located between codons and only 34% within codons (Fig. 3), indicating a non-random distribution. It is interesting to point out that the 16 exon-intron boundaries found within a codon were all conserved in Arabidopsis and both grass species, suggesting the possibility of some biological constraints in their modification.
Even though the comparisons between barley and rice genomic sequence were the most useful to determine exon-intron boundaries, comparisons with Arabidopsis homologues were useful to polish the gene annotation. Arabidopsis annotation can also benefit from the comparison with grasses. For instance, we found five regions (gene 1, exons 3 and 4; gene 2, exons 1, 2, and 5; and gene 3, exon 18) where data from the grass species suggested alternative annotations for the Arabidopsis genes that would increase similarity with the predicted grass proteins.
Gene Islands
The expected average distance between genes in barley is approximately 240 kb, based on the size of the barley genome (5,000 Mb; Arumuganathan and Earle, 1991) and assuming that barley has about as many genes (21,000) as predicted for Arabidopsis (Bevan et al., 1998). However, the average gene density in barley BAC 635P2 is approximately one gene every 20 kb, approximately 12 times higher than the expected genome average. A very similar gene density was observed in the sequence of two other chromosome regions in barley (Panstruga et al., 1998; Shirasu et al., 2000), suggesting that many barley genes are clustered in islands of higher-than-average gene densities. Gene clustering was also observed in several wheat regions (Feuillet and Keller, 1999; Lagudah et al., 2001), suggesting that this is a common feature of the large Triticeae genomes.
The existence of gene clusters was also suggested by studies based on preparative centrifugation in Cs2SO4 density gradients (Barakat et al., 1997). Barakat and coworkers (1997) designated these regions as “gene space” and suggested that they have a particular GC composition. However, the GC content of barley BAC 635P2 (44.2%) is outside the interval determined for barley “gene space” (45.8%–46.7%) according to Barakat et al. (1997). The GC content of BAC 635P2 is very similar to the average base pair composition of the barley (42.8%–45.6%) and wheat (45.7%) genomes (Melzer and Kleinhofs, 1987; Lagudah et al., 2001), suggesting that the base pair composition of the gene-rich regions is not significantly different from the average genome base pair composition in the Triticeae.
The base-composition of the 635P2 BAC (A = 27.6%, T = 28.2%, C = 21.8%, and G = 22.5%) is also very similar to the composition of the Copia-like retro-element BARE-1 (A = 25.8%, T = 28.1%, C = 21.4%, and G = 24.8%), which represents approximately 3% of the barley genome (Vicient et al., 1999). In wheat, 13% of the random sequences from 2 Mb of sheared T. monococcum genomic DNA showed significant similarity with sequences of a set of known Copia-like retro-elements (Lagudah et al., 2001). The large number of copies of these transposable elements suggests that their own base composition can have a large effect on the average base-composition of the complete genome.
Genome Size and Intergenic Regions
It is possible to speculate that the massive increase in genome size in the Triticeae may have also resulted in a tendency to have larger introns. However, the number of corresponding introns that were larger in barley than in rice was very similar to the number of introns that were larger in rice than in barley. Twenty-five introns were larger in barley and 23 were larger in rice. If the total length of the introns is considered, two genes were larger in rice and two in barley (Fig. 3). These two results suggest that most of the contributions to the differences in genome size between barley and rice occurred outside the genes. There was also a similar number of corresponding introns that were larger in rice than in Arabidopsis (27 introns) as vice versa (26 introns were larger in Arabidopsis), although the total length of the introns was larger in rice for the four genes analyzed here (Fig. 3). These results further demonstrate that expansion and contraction of the introns was not affected by the differential expansion-contraction of the genomes outside of the genes but suggest that large insertions within the introns are more frequent or are retained longer in these grass species than in Arabidopsis. A larger number of genes should be compared to test this hypothesis.
The large difference in size between the rice and barley colinear gene regions was mainly due to the insertion of different layers of retroelements in the intergenic regions. Similar results were reported before in comparisons between colinear BACs from the small genomes of rice and sorghum with the colinear BAC from the larger genome of maize (Chen et al., 1997). The sequence of a 240-kb interval of the maize genome around the Adh1 gene (SanMiguel et al., 1996) showed 21 retrotransposons and two solo-LTRs, but only three small non-LTR retroelements (retroposons) and no large TIR elements (Tikhonov et al., 1999). The presence of a MuDR-related element in this 100-kb of barley DNA sequence may suggest a higher frequency of this element family in barley compared with maize (Bennetzen, 1996). However, larger regions should be compared to confirm this hypothesis.
As found in other studies (Bureau and Wessler, 1994; Tikhonov et al., 1999), most MITEs were associated with genes, inserting into introns or just upstream or downstream of them (Fig. 2). However, one Stowaway MITE was inserted into the 3′-LTR of retrotransposon Ta (Fig. 2). Such events appear to be relatively rare, and this is the first of which we are aware.
Cursory examination of the LTR sequences of the rice and barley retrotransposons discovered here suggests that deletions may be more common in rice than in barley. In the eight LTRs of the four intact retrotransposon of 635P2 only one insertion/deletion (indel) larger than 30 bp was found (corresponding to the insertion of the above mentioned Stowaway element into the Ta LTR). Yet, a large deletion of 521 bp and a 5-bp insertion/deletion were the only differences found between the sequences of the 36I5 retrotransposon's LTRs.
Rearrangement of the Rice, Barley, and Arabidopsis Genomes
Rice and barley have diverged for approximately 50 million years since they shared a common grass ancestor. During that time, a relatively small number of large chromosomal rearrangements have differentiated their nuclear genomes (Devos and Gale, 1997). However, these comparative mapping analyses have a 5- to 10-cM sensitivity that would miss most small chromosomal rearrangements. Our comparative sequence analysis of a four-gene segment indicates a single gene inversion and a gene duplication that differentiates these two genomes in this region. As this region represents less than 0.01% of either of these genomes, this suggests that thousands of such small re-arrangements may be present. From analysis of these two lineages alone, we cannot predict where or when these rearrangements occurred, but a broader characterization of other grass genomes should provide this information.
A comparison to the Arabidopsis genome suggests that genes from this region have been retained in the approximate 240 million years since these three species diverged from a common Angiosperm ancestor. Although none of the Arabidopsis gene pairs appear to be tightly linked, three of them are on the same chromosome, suggesting the possibility that most of the re-arrangements involved cis events like inversions, duplications, or deletions. A similar lack of colinearity between rice and Arabidopsis was previously reported by Devos et al. (1999). Despite differing only 3- to 5-fold in divergence time, Arabidopsis appears to have many fold more rearrangements relative to the grasses. This observation suggests that either most of these rearrangements occurred between the divergence times of Arabidopsis/grasses and barley/rice or that most of the rearrangements occurred specifically in the lineage that gave rise to Arabidopsis. Comparative genetic mapping studies have suggested that Arabidopsis may have had an unusually high frequency of chromosomal re-arrangement during evolution (Paterson et al., 1996, 2000), possibly a secondary outcome of its unusually small genome size.
MATERIALS AND METHODS
BAC Selection
Colinearity between the long arm of rice chromosome 3 and the long arm of homeologous group 5 in the Triticeae has been studied extensively because of the presence of vernalization and frost tolerance genes Vrn-1 and Fr-1 in wheat and barley (Hordeum vulgare) and heading date gene Hd6 in rice (Dubcovsky et al., 1998; Sarma et al., 1998; Kato et al., 1999; Sutka et al., 1999; Yamamoto et al., 2000). RFLP marker WG644 was mapped in this colinear region in rice, barley, and wheat (Kleinhofs et al., 1993; Dubcovsky et al., 1998; Sarma et al., 1998) and was used to screen the Morex barley BAC library (Yu et al., 2000) and the Nipponbare rice HindIII BAC library (http://www.genome.clemson.edu/orders/lib_desc/nippon.html). Positive BACs found in both libraries were fingerprinted with restriction enzyme HindIII to determine the approximate position of WG644 within the selected BACs. DNA was transferred to nylon membranes and hybridized with clone WG644 to confirm that all the BACs were from the same locus.
Sequencing
DNA from BACs 36I5 (rice) and 635P2 (barley) was extracted using the Large Construct Kit (Qiagen USA, Valencia, CA) and sheared with a HydroShear device (Genemachines, Inc., San Carlos, CA ) to two different average sizes of 2 and 9 kb. The sheared fragments were blunt-ended, dephosphorylated, and “A” tails added by incubation with Taq polymerase. Inserts were then ligated into pCR4TOPO using the TA cloning system. Clones were sequenced from both directions using big dye terminator chemistry and run on an ABI3700 capillary sequencer. Base calling and quality assessment were done using PHRED (Ewing and Green, 1998), assembled by PHRAP and edited with CONSED (Gordon et al., 1998). Gaps were filled by a combination of primer walking and shotgun sequencing of subclones with extremes at both sides of the sequencing gaps. Final error rate was estimated using CONSED.
Sequence Analysis
Complete sequences from the rice and barley BACs were compared using the computer program Dotter (Sonnhammer and Durbin, 1995) to identify the conserved regions. Open reading frames were predicted using the rice training set of the gene-finding program GeneMark.hmm (Lukashin and Borodovsky, 1998; http://genemark.biology.gatech.edu/GeneMark/) and the maize (Zea mays) training set for Genescan (Burge and Karlin, 1997; http://genes.mit.edu/GENSCAN.html). Sequences from the BACs were also compared with NCBI dbEST and non-redundant databases using BLASTN, BLASTX, and TBLASTX algorithms (Altschul et al., 1997). Gene structure was determined by a combination of the two gene prediction programs mentioned above with GeneSeqer (Usuka et al., 2000; http://gremlin3.zool.iastate.edu/cgi-bin/prg/gs.cgi, maize splice site model) that generates splicing alignments of significant ESTs with the BAC genomic sequence.
Information from the gene-prediction and gene-splicing programs was combined with the identification of significantly similar segments between the barley and rice genomic sequence using BLASTN. These conserved regions, generally limited to the exons, were translated into proteins to adjust AG/GT exon-intron boundaries to the reading frames. The complete predicted cDNA and predicted protein were determined from the annotated structure of the gene. The predicted cDNA was aligned with the significant ESTs to confirm the annotated exon-intron boundaries. The predicted protein was used to search for the closest homologous (and, hence, possibly orthologous) Arabidopsis gene. The predicted Arabidopsis gene structure was then used to further polish the gene structure determined by the previous methods.
The positions of transposable elements were determined by a combination of FASTA, BLAST, and NETBLAST searches to the GenBank/EMBL nonredundant database and TIGR (http://www.tigr.org/tdb/rice/blastsearch.shtml), homology searches to known transposable elements, same sequence comparisons, and orthologous sequence comparisons. The sequences of both BACs were compared with themselves (same sequence comparison) and each other (orthologous sequence comparison) using COMPARE, DOTPLOT, STEMLOOP, REPEATS, and GAPs (Wisconsin Package Version 10.1, Genetics Computer Group, Madison, WI).
Restriction Map
Restriction maps of 36I5 and 635P2 were constructed to experimentally validate computer sequence assembly. This experimental confirmation was important to determine the effect of large retroelements with large inverted repeats on the assembly algorithms. BACs were individually digested with 8-bp specificity restriction enzymes AscI, NotI, PacI, Pme I, and Swa I. All possible single and double digestions were analyzed for the restriction enzymes with one or more sites within this BAC. Restriction fragments were separated by pulse field electrophoresis in 1% (w/v) agarose gels (12°C, 14 h, 200 V, pulse 0.2–13 s). Gel filter replicas were hybridized with [α-32P]-labeled clones R2311, R2628, UCW12, Exons 1 to 3 from rice gene 2, exons 9 through 13 from rice gene 2, and BAC vector pBELOBAC11 (CVU51113). The two different segments from gene 2 were used to test the inversion of this gene.
ACKNOWLEDGMENTS
The authors thank J. Emberton and D. Murphy for excellent technical assistance and G. Tranquilli and M. Helguera for preliminary work in the identification of the colinear BACs.
Footnotes
This work was supported in part by the National Science Foundation Plant Genome Program (grant no. 9975793) and by the U.S. Department of Agriculture National Research Initiative (grant no. 2000–1678 to J.D.).
LITERATURE CITED
- Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumuganathan K, Earle ED. Nuclear DNA content of some important plant species. Plant Mol Biol Rep. 1991;9:208–218. [Google Scholar]
- Avramova Z, Tikhonov A, SanMiguel P, Jin YK, Liu CN, Woo SS, Wing RA, Bennetzen JL. Gene identification in a complex chromosomal continuum by local genomic cross-referencing. Plant J. 1996;10:1163–1168. doi: 10.1046/j.1365-313x.1996.10061163.x. [DOI] [PubMed] [Google Scholar]
- Barakat A, Carels N, Bernardi G. The distribution of genes in the genomes of Gramineae. Proc Natl Acad Sci USA. 1997;94:6857–6861. doi: 10.1073/pnas.94.13.6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzoglou S, Pachter L, Mesirov JP, Berger B, Lander ES. Human and mouse gene structure: comparative analysis and application to exon prediction. Genome Res. 2000;10:950–958. doi: 10.1101/gr.10.7.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MD, Leitch IJ. Nuclear DNA amounts in Angiosperms. Ann Bot. 1995;76:113–176. [Google Scholar]
- Bennetzen JL. The Mutator transposable element system of maize. Curr Topics Microbiol Immunol. 1996;204:195–229. doi: 10.1007/978-3-642-79795-8_9. [DOI] [PubMed] [Google Scholar]
- Bevan M, Bancroft I, Bent E, Love K, Goodman H, Dean C, Bergkamp R, Dirkse W, Van Staveren M, Stiekema W. Analysis of 1.9 Mb of contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature. 1998;391:485–488. doi: 10.1038/35140. [DOI] [PubMed] [Google Scholar]
- Bureau TE, Wessler SR. Stowaway: a new family of inverted repeat elements associated with the genes of both monocotyledonous and dicotyledonous plants. Plant Cell. 1994;6:907–916. doi: 10.1105/tpc.6.6.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Chen M, SanMiguel P, Oliveira ACd, Woo S-S, Zhang H, Wing RA, Bennetzen JL. Microcolinearity in sh2-homologous regions of the maize, rice, and sorghum genomes. Proc Natl Acad Sci USA. 1997;94:3431–3435. doi: 10.1073/pnas.94.7.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos KM, Beales J, Nagamura Y, Sasaki T. Arabidopsis-rice: will colinearity allow gene prediction across the eudicot-monocot divide? Genome Res. 1999;9:825–829. doi: 10.1101/gr.9.9.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos KM, Gale MD. Comparative genetics in the grasses. Pl Mol Biol. 1997;35:3–15. [PubMed] [Google Scholar]
- Dubcovsky J, Lijavetzky D, Appendino L, Tranquilli G. Comparative RFLP mapping of Triticum monococcum genes controlling vernalization requirement. Theor Appl Genet. 1998;97:968–975. [Google Scholar]
- Dubcovsky J, Luo M-C, Zhong G-Y, Bransteiter R, Desai A, Kilian A, Kleinhofs A, Dvorak J. Genetic map of diploid wheat, Triticum monococcum L., and its comparison with maps of Hordeum vulgare L. Genetics. 1996;143:983–999. doi: 10.1093/genetics/143.2.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using PHRED: II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Feuillet C, Keller B. High gene density is conserved at syntenic loci of small and large grass genomes. Proc Natl Acad Sci USA. 1999;96:8265–8270. doi: 10.1073/pnas.96.14.8265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell RB, Smith DB. Nucleotide sequence organization in the wheat genome. Heredity. 1976;37:231–252. [Google Scholar]
- Gale MD, Devos KM. Comparative genetics in the grasses. Proc Natl Acad Sci USA. 1998;95:1971–1974. doi: 10.1073/pnas.95.5.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P. CONSED: a graphical tool for sequencing finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Gribbon BM, Pearce SR, Kalendar R, Schulman AH, Paulin L, Jack P, Kumar A, Flavell AJ. Phylogeny and transpositional activity of Ty1-copia group retrotransposons in cereal genomes. Mol Gen Genet. 1999;261:883–891. doi: 10.1007/pl00008635. [DOI] [PubMed] [Google Scholar]
- Harushima Y, Yano M, Shomura P, Sato M, Shimano T, Kuboki Y, Yamamoto T, Lin SY, Antonio BA, Parco A. A high-density rice genetic linkage map with 2275 markers using a single F-2 population. Genetics. 1998;148:479–494. doi: 10.1093/genetics/148.1.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Miura H, Sawada S. Comparative mapping of the wheat Vrn-1 region with the rice Hd-6 region. Genome. 1999;42:204–209. [Google Scholar]
- Kleinhofs A, Kilian A, Saghai MA, Biyashev RM, Hayes P, Chen FQ, Lapitan N, Fenwick A, Blake TK, Kanazin V. A molecular, isozyme and morphological map of the barley (Hordeum vulgare) genome. Theor Appl Genet. 1993;86:705–712. doi: 10.1007/BF00222660. [DOI] [PubMed] [Google Scholar]
- Lagudah E, Dubcovsky J, Powell W (2001) Wheat genomics. Pl Physiol Biochem (in press)
- Leister D, Kurth J, Laurie DA, Yano M, Sasaki T, Devos K, Graner A, SchulzeLefert P. Rapid reorganization of resistance gene homologues in cereal genomes. Proc Natl Acad Sci USA. 1998;95:370–375. doi: 10.1073/pnas.95.1.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukashin AV, Borodovsky M. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 1998;26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroof MAS, Yang GP, Biyashev RM, Maughan PJ, Zhang Q. Analysis of the barley and rice genomes by comparative RFLP linkage mapping. Theor Appl Genet. 1996;92:541–551. doi: 10.1007/BF00224556. [DOI] [PubMed] [Google Scholar]
- Melzer JM, Kleinhofs A. Molecular genetics of barley. In: Yasuda S, Konishi T, editors. Proceedings of the Fifth International Barley Genetics Symposium. Okayama, Japan: Sanyo Press Co., Ltd.; 1987. pp. 481–491. [Google Scholar]
- Panstruga R, Buschges R, Piffanelli P, SchulzeLefert P. A contiguous 60 kb genomic stretch from barley reveals molecular evidence for gene islands in a monocot genome. Nucleic Acids Res. 1998;26:1056–1062. doi: 10.1093/nar/26.4.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Burow MD, Draye X, Elsik CG, Jiang CX, Katsar CS, Lan TH, Lin YR, Ming RG. Comparative genomics of plant chromosomes. Plant Cell. 2000;12:1523–1539. doi: 10.1105/tpc.12.9.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Lan TH, Reischmann KP, Chang C, Lin YR, Liu SC, Burow MD, Kowalski SP, Katsar CS, Delmonte TA. Toward a unified genetic map of higher plants, transcending the monocot-dicot divergence. Nat Genet. 1996;14:380–382. doi: 10.1038/ng1296-380. [DOI] [PubMed] [Google Scholar]
- SanMiguel P, Bennetzen JL. Evidence that a recent increase in maize genome size was caused by the massive amplification of intergene retrotransposons. Ann Bot. 1998;82:37–44. [Google Scholar]
- SanMiguel P, Tikhonov A, Jin Y-K, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer PS, Edwards KJ, Lee M, Avramova Z. Nested retrotransposons in the intergenic regions of the maize genome. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- Sarma RN, Gill BS, Sasaki T, Galiba G, Sutka J, Laurie DA, Snape JW. Comparative mapping of the wheat chromosome 5A Vrn-A1 region with rice and its relationship to QTL for flowering time. Theor Appl Genet. 1998;97:103–109. [Google Scholar]
- Shirasu K, Schulman AH, Lahaye T, Schulze-Lefert P. A contiguous 66-kb barley DNA sequence provides evidence for reversible genome expansion. Genome Res. 2000;10:908–915. doi: 10.1101/gr.10.7.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer ELL, Durbin R. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene. 1995;167:1–10. doi: 10.1016/0378-1119(95)00714-8. [DOI] [PubMed] [Google Scholar]
- Stein N, Feuillet C, Wicker T, Schlagenhauf E, Keller B. Subgenome chromosome walking in wheat: a 450-kb physical contig in Triticum monococcum L. spans the Lr10 resistance locus in hexaploid wheat (Triticum aestivum L.) Proc Natl Acad Sci USA. 2000;97:13436–13441. doi: 10.1073/pnas.230361597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suoniemi A, Anamthawat-Jonsson K, Arna T, Schulman AH. Retrotranposon BARE-1 is a major dispersed component of the barley (Hordeum vulgare L.) genome. Pl Mol Biol. 1996;30:1321–1329. doi: 10.1007/BF00019563. [DOI] [PubMed] [Google Scholar]
- Sutka J, Galiba G, Vagujfalvi A, Gill BS, Snape JW. Physical mapping of the Vrn-A1 and Fr1 genes on chromosome 5A of wheat using deletion lines. Theor Appl Genet. 1999;99:199–202. [Google Scholar]
- Tikhonov AP, SanMiguel PJ, Nakajima Y, Gorenstein ND, Bennetzen JL, Avramova Z. Colinearity and its exceptions in orthologous adh regions of maize and sorghum. Proc Natl Acad Sci USA. 1999;96:7409–7414. doi: 10.1073/pnas.96.13.7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usuka J, Zhu W, Brendel V. Optimal spliced alignment of homologous cDNA to a genomic DNA template. Bioinformatics. 2000;16:203–211. doi: 10.1093/bioinformatics/16.3.203. [DOI] [PubMed] [Google Scholar]
- Vicient CM, Kalendar R, Anamthawat-Jonsson K, Suoniemi A, Schulman AH. Structure, functionality, and evolution of the BARE-1 retrotransposon of barley. Genetica. 1999;107:53–63. [PubMed] [Google Scholar]
- Wei F, Gobelman-Werner K, Morroll SM, Kurth J, Mao L, Wing R, Leister D, Schulze-Lefert P, Wise RP. The Mla (powdery mildew) resistance cluster is associated with three NBS-LRR gene families and suppressed recombination within a 240-kb DNA interval on Chromosome 5S (1HS) of barley. Genetics. 1999;153:1929–1948. doi: 10.1093/genetics/153.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Lin HX, Sasaki T, Yano M. Identification of heading date quantitative trait locus Hd6 and characterization of its epistatic interactions with Hd2 in rice using advanced backcross progeny. Genetics. 2000;154:885–891. doi: 10.1093/genetics/154.2.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Tomkins JP, Waugh R, Frisch DA, Kudrna D, Kleinhofs A, Brueggeman RS, Muehlbauer GJ, Wing RA. A bacterial artificial chromosome library for barley (Hordeum vulgare) Theor Appl Genet. 2000;101:1093–1099. [Google Scholar]