Significance
Phosphate (Pi) is essential for life; thus, serum Pi level is kept constant under tight regulation by fibroblast growth factor (FGF) 23. Conversely, serum FGF23 levels are also controlled by sensing Pi alterations in serum, but this Pi-sensing mechanism remains elusive. In this study, we found that unliganded FGFR is activated by high Pi, leading to an increase in serum FGF23 level by skeletal induction of an FGF23 O-glycosylation enzyme that results in FGF23 proteolytic protection. Thus, the present study elucidates a Pi-sensing mechanism in the control of serum FGF23 levels and provides a molecular basis for a better understanding of hypo- or hyperphosphatemic diseases.
Keywords: phosphate sensor, fibroblast growth factor receptor 1, fibroblast growth factor 23
Abstract
Fibroblast growth factor (FGF) 23 produced by bone is a hormone that decreases serum phosphate (Pi). Reflecting its central role in Pi control, serum FGF23 is tightly regulated by serum Pi alterations. FGF23 levels are regulated by the transcriptional event and posttranslational cleavage into inactive fragments before its secretion. For the latter, O-glycosylation of FGF23 by GALNT3 gene product prevents the cleavage, leading to an increase in serum FGF23. However, the molecular basis of Pi sensing in the regulation of serum FGF23 remains elusive. In this study, we showed that high Pi diet enhanced the skeletal expression of Galnt3, but not Fgf23, with expected increases in serum FGF23 and Pi in mice. Galnt3 induction by high Pi was further observed in osteoblastic UMR 106 cells, and this was mediated by activation of the extracellular signal-regulated kinase (ERK) pathway. Through proteomic searches for the upstream sensor for high Pi, we identified one subtype of the FGF receptor (FGFR1c), which was phosphorylated by high Pi in the absence of FGFs. The mode of unliganded FGFR activation by high Pi appeared different from that of FGFR bound to a canonical FGFR ligand (FGF2) when phosphorylation of the FGFR substrate 2α and ERK was monitored. Finally, we showed that an FGFR inhibitor and conditional deletion of Fgfr1 in osteoblasts/osteocytes abrogated high Pi diet-induced increases in serum FGF23 and femoral Galnt3 expression in mice. Thus, these findings uncover an unrecognized facet of unliganded FGFR function and illustrate a Pi-sensing pathway involved in regulation of FGF23 production.
Phosphate (Pi) is a key mineral component in numerous cellular events and hard tissues, and the serum Pi level and Pi balance in the body are well controlled, regardless of excess or deficient dietary intake. There are several physiological Pi regulators that maintain a proper balance of Pi in the body, and dysregulation of this system has been reported to result in diseases associated with deranged mineral and bone metabolism (1). Among these regulators, fibroblast growth factor (FGF) 23 is known to play a central role in regulating serum Pi level (2). FGF23 is one of the FGF family members that mediates a wide range of biological processes in both developing and adult vertebrates and is considered an endocrine FGF from its mode of physiological actions (3). As FGF23 has low affinity for heparan sulfate, it needs α-Klotho for FGF receptor (FGFR) binding to elicit its biological actions (4). FGF23 is mainly produced by osteoblasts/osteocytes and reduces serum Pi by inhibiting proximal tubular Pi reabsorption and suppressing intestinal Pi absorption by decreasing the 1,25-dihydroxyvitamin D [1,25(OH)2D] level (5–7). Excessive and deficient FGF23 results in hypo- and hyperphosphatemic diseases, respectively, indicating its significance in Pi control (1). Thus, FGF23 levels are finely managed by changes in serum Pi levels. FGF23 levels are regulated by both the expression in bone and posttranslational modification of the FGF23 protein (8, 9). A portion of the FGF23 protein is proteolytically cleaved into inactive fragments before secretion. We showed that mucin-type O-linked glycosylation of the FGF23 protein prevents this cleavage and increases the level of biologically active full-length FGF23 (10). The GALNT3 gene encodes an enzyme that initiates the attachment of an O-linked glycan to the FGF23 protein (11). Although this posttranslational modification of FGF23 appears pivotal in the regulation of serum FGF23 level, its molecular mechanisms in response to serum Pi alternations are unclear.
Here, we found that skeletal Galnt3, but not Fgf23, was induced with an increase in the serum level of FGF23 in mice fed a high Pi diet. This Galnt3 induction by high Pi was also observed in an osteoblastic cell line, coupled with activation of unliganded FGFR (FGFR1c). The mode of activation of unliganded FGFR by high Pi was different from that of FGF2-liganded FGFR, and activated FGFR led to the extracellular signal-regulated kinase (ERK) pathway-mediated induction of Galnt3. These findings illustrate the unrecognized function of FGFR and uncover the presence of a Pi-sensing system that posttranslationally regulates FGF23 production.
Results
A High Pi Diet and High Extracellular Pi Increase FGF23 Levels by Preventing the Processing of FGF23 Protein.
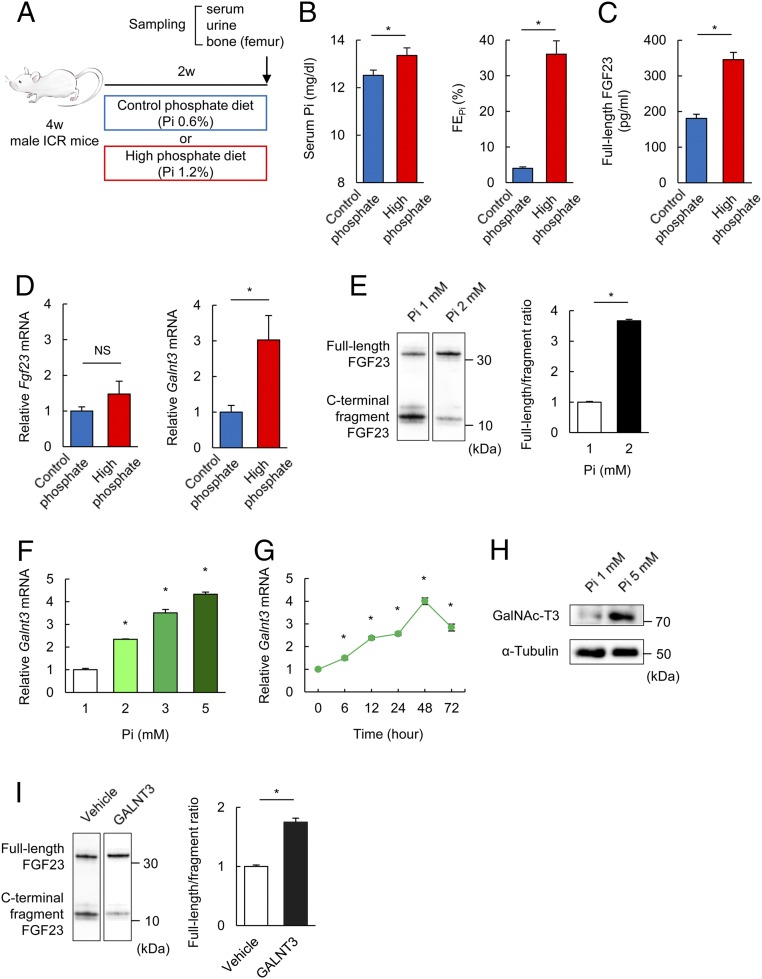
Four-week-old male ICR mice were fed a control (0.6%: CP) and high (1.2%: HP) Pi diet for 2 wk and were then killed (Fig. 1A). There were no differences in serum urea nitrogen, creatinine, calcium (Ca) levels and fractional excretion of Ca (FECa) after 2 wk (SI Appendix, Fig. S1 A–D). The serum Pi level and fractional excretion of Pi (FEPi) were significantly higher in the HP group than the CP group (Fig. 1B). The serum concentration of full-length biologically active FGF23 in the HP group was significantly higher than that in the CP group (Fig. 1C). In contrast to our predictions, HP did not increase Fgf23 mRNA but increased Galnt3 mRNA in the femur (Fig. 1D). These results suggest that FGF23 production is regulated through Galnt3 induction in response to dietary HP intake in intact animals.
Fig. 1.
Pi increases FGF23 levels by preventing the processing of the FGF23 protein via the gene product of Galnt3. (A) Schema of the experimental procedure. (B and C) Serum Pi, FEPi (B) and serum full-length FGF23 (C) in mice fed a CP or HP diet for 2 wk. (D) Fgf23 and Galnt3 mRNA expression in femurs of mice fed a CP or HP diet for 2 wk. (E) Immunoblotting with an antibody to FGF23 secreted into the media from UMR106 cells overexpressing His-tagged human FGF23 after immunoprecipitation with anti-His-tag antibody under 1 or 2 mM Pi for 48 h (Left). The Right shows the quantified ratio of full-length protein/the C-terminal fragment of FGF23 protein. (F) Galnt3 mRNA expression under various extracellular Pi concentrations for 48 h in UMR106 cells. (G) Galnt3 mRNA expression with 5 mM Pi in UMR106 cells was measured at the indicated times. (H) Immunoblotting with an antibody to GalNAc-T3 encoded by Galnt3 from the lysates of UMR106 cells under 1 or 5 mM Pi for 48 h. (I) Immunoblotting with an antibody to FGF23 secreted into the media from UMR106 cells overexpressing human GALNT3 together with His-tagged FGF23 under 1 mM Pi for 48 h (Left). The Right shows the quantified ratio of full-length/C-terminal fragment of FGF23 protein. Data represent the mean ± SEM. (B–D) n = 19 mice per group; *P < 0.05 by Student’s t test; NS, not significant. (E and I) n = 3 per group; *P < 0.05 by Student’s t test. (F and G) n = 3 per group; *P < 0.05 by ANOVA with a post hoc Dunnett’s test compared with 1 mM of extracellular Pi (F) or 0 h (G). (E, H, and I) Data are presented as a representative image.
To further assess these in vivo observations, we tested them in vitro using the UMR106 osteoblastic cell line. Because there are no cell lines available that produce FGF23 protein to examine the effects of high extracellular Pi on the processing of the FGF23 protein, His-tagged human FGF23 was overexpressed in UMR106 cells for further in vitro assays. Full-length FGF23 was detected at higher ratios under high extracellular Pi conditions (2 mM) than under low Pi conditions (1 mM) when the ratio of the full-length protein vs. the cleaved C-terminal fragment was estimated in the culture media of UMR106 cells (Fig. 1E). Galnt3 mRNA expression was also enhanced under high extracellular Pi in a dose-dependent manner in the UMR106 cells, and Galnt3 induction peaked at 48 h (Fig. 1 F and G). High extracellular Pi also enhanced the expression of the GalNAc-T3 protein encoded by Galnt3 (Fig. 1H). Furthermore, when human GALNT3 and FGF23 were overexpressed, the ratio of the full-length protein vs. the C-terminal fragment of the FGF23 protein was increased (Fig. 1I). These results indicate that high extracellular Pi increases Galnt3 expression and thereby prevents the processing of FGF23 protein.
It was previously reported that the processing of FGF23 protein is mediated by protease like Furin and the processing is further enhanced by phosphorylation of FGF23 protein by a kinase [family with sequence similarity 20, member C (FAM20C)] (8). Therefore, the roles of these two factors in the Pi response in FGF23 processing were assessed. High phosphate diet and high extracellular phosphate did not change expression of Furin in the femur and UMR106 cells (SI Appendix, Fig. S2 A and B), while Fam20c expression was enhanced by high phosphate diet and high extracellular phosphate (SI Appendix, Fig. S2 C and D). From the observation that the serum full-length FGF23 was increased by high phosphate diet, the roles of FAM20C and Furin in the Pi response appeared marginal.
Collecting these findings together, we assumed that that Galnt3, but not Fgf23, is a Pi-responsive gene and that the induction of Galnt3 expression by high extracellular Pi must be mediated by a Pi-sensing mechanism. Therefore, we used Galnt3 expression as a marker to explore the Pi-sensing mechanism in bone.
High Extracellular Pi Induces Galnt3 Expression Through Activation of the ERK Pathway.
We next addressed the mechanism by which extracellular Pi induces Galnt3 expression. As Pi level alterations did not affect the half-life of Galnt3 mRNA (SI Appendix, Fig. S3), we reasoned that Galnt3 is induced by Pi at the transcriptional level. However, as Galnt3 induction by Pi peaked at 48 h (Fig. 1G), we presumed the involvement of intermediary molecules. To address this point, we performed a DNA microarray analysis using UMR106 cells with or without high Pi treatment for 6 h. We focused on 17 genes that appeared inducible at the transcriptional level by high Pi (Fig. 2A), and the ERK pathway was selected (Fig. 2B). Among the members of this pathway, three genes encoding transcription factors related to the ERK pathway, early growth response 1 (Egr1), ETS variant 4 (Etv4), and ETS variant 5 (Etv5), were further identified because the ERK pathway was previously reported to be activated by high Pi (12, 13). Indeed, all three genes were induced by high Pi (Fig. 2C), while the addition of sulfate, which increased the osmotic pressure to a similar extent as Pi, did not induce these genes. Most notably, silencing of Egr1 and Etv5 by small interfering RNA (siRNA) aborted the induction of Galnt3 by high Pi (Fig. 2D), confirming the significance of these transcriptional activators in Galnt3 induction by high Pi.
Fig. 2.
The up-regulation of Galnt3 by Pi requires activation of the ERK pathway and induction of Egr1 and Etv5. (A and B) The results of DNA microarray analysis in a volcano plot using UMR106 cells with or without 5 mM Pi treatment for 6 h (A, Left). The table shows 17 genes significantly induced by Pi (A, Right). These included genes for several transcription factors, ERK pathway components, and bone-related proteins (B). (C) Egr1, Etv4, and Etv5 mRNA expression by 5 mM Pi or 5 mM sulfate for 6 h evaluated by qPCR. (D) Galnt3 mRNA expression by 5 mM Pi for 12 h compared with 1 mM Pi under siRNA-mediated silencing of Egr1, Etv4, and Etv5. (E and F) Immunoblotting with an antibody to phosphorylated ERK1/2 from the lysates of UMR106 cells treated with 1 or 5 mM Pi, 100 ng/mL FGF2, or 5 mM sulfate for 15 min (E) or treated with 1, 3, or 5 mM Pi (F). (G) ERK activation under various extracellular Pi concentrations for 24 h evaluated by SRE luciferase assays. (H) Galnt3 mRNA expression by 5 mM Pi for 48 h with or without the MEK1/2 inhibitor U0126. Data represent the mean ± SEM. (C, D, and G) n = 3 per group; *P < 0.05 by ANOVA with a post hoc Tukey’s test (C and D) or with a post hoc Dunnett’s test compared with 1 mM of extracellular Pi (G). (E and F) Data are presented as a representative image. (H) n = 3 per group; *P < 0.05 compared with 1 mM Pi without U0126, #P < 0.05 compared with 1 mM Pi with U0126, †P < 0.05 compared with 5 mM Pi with U0126 cells by ANOVA with a post hoc Tukey’s test.
We then asked if high Pi activates the ERK pathway by immunoblotting activated ERK1/2 in UMR106 cells. Activation of ERK1/2 (the phosphorylated form) was induced by high Pi as well as a known activator (FGF2) in the UMR106 cells (Fig. 2E). High extracellular Pi induced the phosphorylation of ERK1/2 in a dose-dependent manner, as shown by an immunoblotting assay (Fig. 2F) or a serum response element (SRE) luciferase reporter assay designed to monitor the activity of the ERK signaling pathway (Fig. 2G). Furthermore, the ERK kinase (MEK) 1/2 inhibitor U0126 attenuated the effect of high Pi on Galnt3 induction (Fig. 2H), further supporting our findings that high Pi induces Galnt3 through activation of the ERK pathway. However, high Pi did not induce ERK phosphorylation in other nonbone rat cell lines (the L6 myoblastic cell line and the JCT19 fibroblastic cell line) (SI Appendix, Fig. S4 A and B). Since it is still unclear how the high Pi signal results in ERK activation, we further searched for an upstream factor that activates the ERK pathway by high Pi.
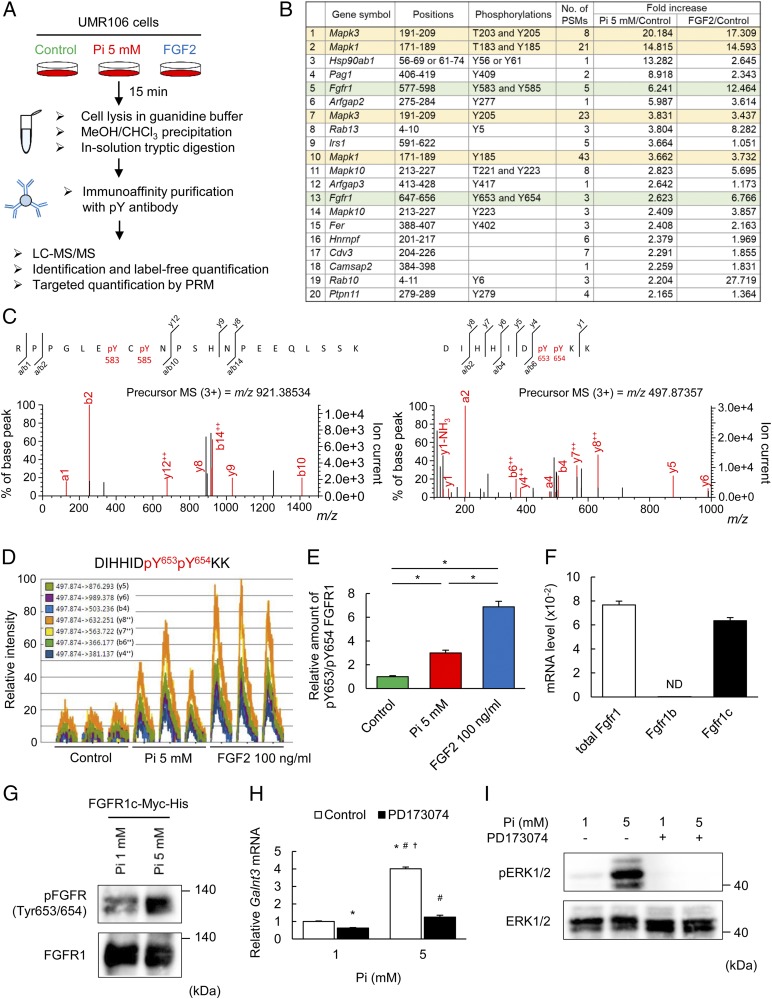
Activation of the FGF Receptor by High Pi Induces Galnt3.
Since the ERK pathway is activated as a downstream signal inducer of activated transmembrane receptors in general, we focused on receptor tyrosine kinases (RTKs) (14). As RTKs are phosphorylated at tyrosine residues upon activation, phosphorylated tyrosine residues were searched by a proteomic approach. From lysates of UMR106 cells treated with high Pi, tryptic peptides were immunopurified by an antiphosphotyrosine antibody and subjected to LC-MS/MS analysis (Fig. 3A). Fig. 3B shows the results of the identification and label-free quantification of the top 20 peptides with phosphotyrosine in the order of fold increase by high Pi (the whole data set is shown in SI Appendix, Table S1). ERK1 and ERK2, encoded by Mapk3 and Mapk1, respectively, were the top two proteins. This analysis led us to identify FGFR1 as the only RTK and a phosphorylation target of high Pi. The identification of FGFR1 is supported by previous reports suggesting the involvement of FGFR in cellular responses to extracellular Pi (12, 15). Six tyrosine residues, tyrosines 653, 583, 463, 766, 585, and 654, are known to be sequentially phosphorylated in this order upon the FGFR1 activation (16, 17). In addition, phosphorylation of two tyrosine residues, 653 and 654, is important for generating an activation loop and dramatically increasing tyrosine kinase activity of FGFR1 (3). In our study, there were two types of FGFR1 peptides, one with phosphorylated tyrosines 583 and 585 and the other with phosphorylated tyrosines 653 and 654, suggesting that FGFR1 was activated by Pi (Fig. 3C). We further performed targeted quantification using parallel reaction monitoring (PRM), which enabled highly specific and accurate quantification of multiple peptides simultaneously (whole dataset is shown in SI Appendix, Table S2). The phosphorylated form of FGFR1 at tyrosines 653 and 654 was significantly increased approximately threefold by high Pi (Fig. 3 D and E). However, the protein level of the phosphorylated FGFR1 at tyrosines 583 and 585 was increased, but its increase was statistically not significant (SI Appendix, Fig. S5 A and B). FGF2, a canonical FGFR ligand that induces FGFR phosphorylation, induced phosphorylation of tyrosines 653 and 654, as expected, but this change was more robust than that of high Pi (Fig. 3E). The results of quantification of other peptides by PRM analysis are shown in SI Appendix, Fig. S5 C and D. The protein level of ERK2, which is encoded by Mapk1, with phosphorylated threonine 183 and tyrosine 185 was increased by both high Pi and FGF2 (SI Appendix, Fig. S5C). FGFR is known to activate several intracellular signaling molecules, such as phosphoinositide 3-kinase (PI3K)-protein kinase B (Akt) and phospholipase Cγ (PLCγ)-calcineurin, in addition to ERK (18). However, unlike FGF2, high Pi was not effective in increasing the protein level of phosphorylated PLCγ (SI Appendix, Fig. S5D). Expectedly, the calcineurin inhibitor FK506 showed no effect on Galnt3 induction by high Pi (SI Appendix, Fig. S6A). Likewise, high Pi did not induce phosphorylation of Akt (SI Appendix, Fig. S6B), and PI3K inhibitor wortmannin had no effect on Galnt3 induction (SI Appendix, Fig. S6C). These results suggest that high Pi specifically activates the ERK pathway downstream of FGFR1.
Fig. 3.
FGFR1c functions as a Pi-sensing mechanism. (A) Schematic diagram of the proteomics workflow. UMR106 cells were treated with 5 mM Pi or 100 ng/mL FGF2 for 15 min. After immunoaffinity purification with an anti-phosphotyrosine antibody, each sample was analyzed by LC-MS/MS. Three biological replicates for each sample were prepared, and an accurate amount of the proteins was measured by targeted MS. (B) The results of identification and label-free quantification of the top 20 peptides with phosphotyrosine in the order of fold increase induced by high Pi. (C) Phosphorylation of tyrosines 583 and 585 and tyrosines 653 and 654 of FGFR1 was demonstrated by the MS/MS spectrum of the m/z 921.38534 and 497.87357 ions in tryptic phophopeptides, respectively. Experiments were independently repeated two times with similar results. (D) Extracted ion chromatograms of transitions of the FGFR1 peptide with phosphorylated tyrosines 653 and 654. Each peptide in three independently prepared samples was analyzed by targeted MS using the PRM method. (E) The amount of FGFR1 peptide with phosphorylated tyrosines 653 and 654 normalized to the amount of cyclin-dependent kinase 2 (CDK2) peptide with phosphorylated tyrosine 15 in the same sample. (F) The expression level of FGFR1 subtypes evaluated by qPCR. (G) Immunoblotting with phosphorylated FGFR (tyrosine 653/654) from the lysates of UMR106 cells overexpressing Fgfr1c under 1 or 5 mM Pi for 15 min. (H) Galnt3 mRNA expression with 1 or 5 mM Pi for 48 h with or without PD173074. (I) Immunoblotting with an antibody to phosphorylated ERK1/2 from the lysates of UMR106 cells under 1 or 5 mM Pi, with or without PD173074 for 15 min. Data represent the mean ± SEM. (E) n = 3 per group; *P < 0.05 by ANOVA with a post hoc Tukey’s test. (F) n = 3 per group. ND, not detected. (G and I) Data are presented as a representative image. (H) n = 3 per group; *P < 0.05 compared with 1 mM Pi without PD173074, #P < 0.05 compared with 1 mM Pi with PD173074, †P < 0.05 compared with 5 mM Pi with PD173074 by ANOVA with a post hoc Tukey’s test.
UMR106 cells endogenously express only FGFR1c (Fig. 3F), one of the two FGFR1 subtypes (FGFR1b and FGFR1c) (3), at a low protein level. When Fgfr1c was overexpressed in UMR106 cells, increased phosphorylation of FGFR1c was observed by immunoblotting with an antibody to phosphorylated FGFR under high extracellular Pi (Fig. 3G). The FGFR inhibitor PD173074 aborted both Galnt3 induction and ERK phosphorylation by high Pi (Fig. 3 H and I). Similarly, silencing of Fgfr1 also blunted the enhanced expression of Galnt3 and the enhanced phosphorylation of ERK by high Pi (SI Appendix, Fig. S7 A and B).
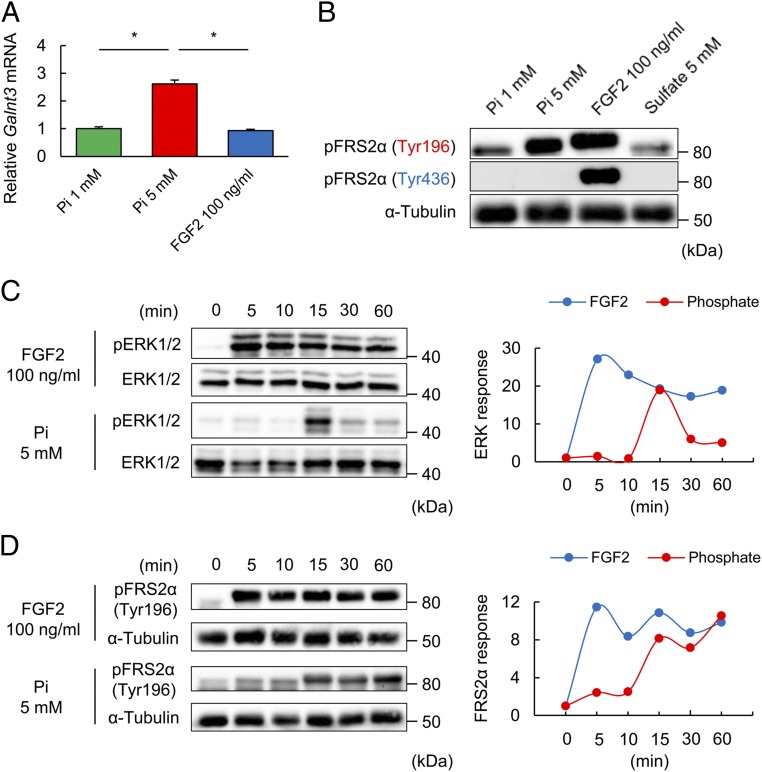
Galnt3 Induction Is Mediated by Activated FGFR with High Pi but Not a Canonical FGF Receptor Ligand.
As high Pi potently induced Galnt3 via the activated FGF receptor, we tested whether a canonical FGFR ligand was also effective in inducing Galnt3. While one of the canonical FGFR ligands, FGF2, activated the ERK pathway (Fig. 2E), as expected from previous reports, FGF2 tended to be inactive in Galnt3 induction (Fig. 4A). To explore the molecular basis of the difference in FGFR activation between high Pi and FGF2, we further assessed the phosphorylation sites of FGFR substrate 2α (FRS2α), which is known to be a phosphorylation substrate of activated FGFR and is necessary for the activation of the downstream ERK pathway (18, 19). Consistent with previous findings, tyrosines 196 and 436 of FRS2α were phosphorylated by FGF2 stimulation. However, phosphorylation by high Pi stimulation was evident only at tyrosine 196 (Fig. 4B). When ERK activation was monitored in a time course, activation of ERK by high Pi was transient, unlike that by FGF2 (Fig. 4C). In addition, the time course of the activation of FRS2α differed between high Pi and FGF2 (Fig. 4D). Thus, these findings elucidate the FGFR activation by Pi in which the Pi signal is transmitted into ERK pathway stimulation, leading to Galnt3 induction.
Fig. 4.
Pi and FGF2 differentially activate the FGFR1-FRS2α-ERK pathway. (A) Galnt3 mRNA expression under 1 or 5 mM Pi or 100 ng/mL FGF2 for 48 h. (B) Immunoblotting with phosphorylated FRS2α at tyrosine 196 or tyrosine 436 from the lysates of UMR106 cells under 1 or 5 mM Pi, 100 ng/mL FGF2, or 5 mM sulfate for 15 min. (C and D) The time course of ERK1/2 phosphorylation (C, Left) or FRS2α phosphorylation at Tyr196 (D, Left) by 100 ng/mL FGF2 or 5 mM Pi. The Right shows the phosphorylated ERK1/2 (C, Right) or phosphorylated FRS2α levels (D, Right). Data represent the mean ± SEM. (A) n = 3 per group; *P < 0.05 by ANOVA with a post hoc Tukey’s test. (B–D) Data are presented as a representative image.
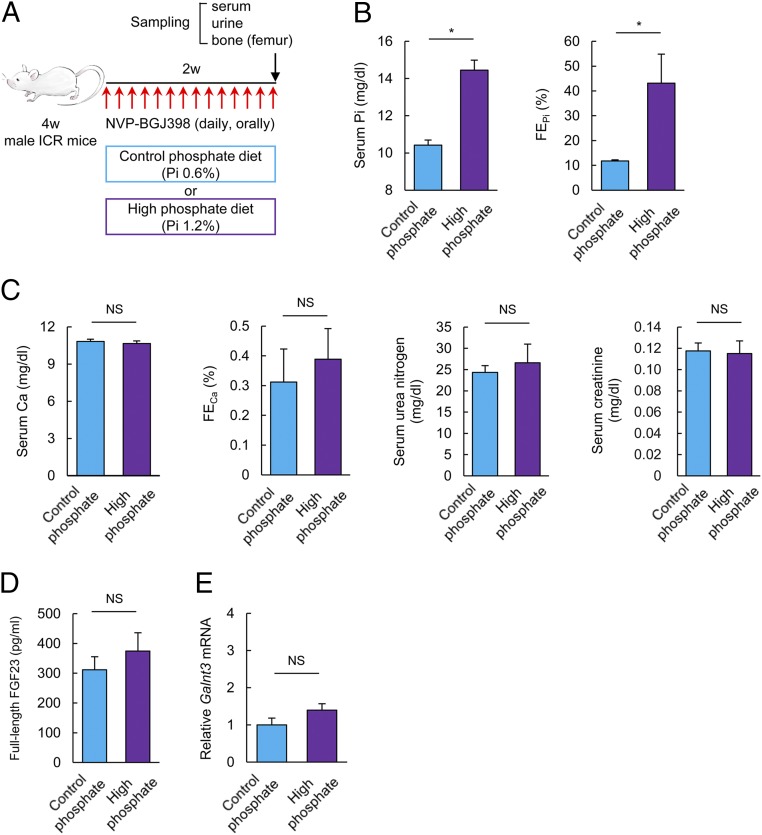
An FGFR Inhibitor Attenuates Galnt3 Up-Regulation by a High Pi Diet in Intact Animals.
We addressed whether activated the FGFR-ERK pathway indeed transmits extracellular Pi signals into Galnt3 induction in intact animals. Enhanced phosphorylation of ERK in the femur by a high phosphate diet was visible in the whole tissue extracts by immunoblotting (SI Appendix, Fig. S8), but neither phosphorylation of FRS2α nor GalNAc-T3 was detectable, presumably due to low protein expression levels. Then, an FGFR inhibitor, NVP-BGJ398, was given to mice fed a CP or HP diet for 2 wk (Fig. 5A). Except for serum Pi and FEPi (Fig. 5B), the levels of the other serum components (Fig. 5C) were unchanged by the Pi dietary contents. As a note, the baseline Pi level in mice in this experiment was lower than that in the previous experiment (compare Fig. 1B and Fig. 5B). Although the serum Pi level was significantly higher in mice fed an HP diet than in mice fed a CP diet under NVP-BGJ398 treatment, neither the serum level of full-length FGF23 nor Galnt3 mRNA in the femur showed a clear increase (compare Fig. 5D with Fig. 1C for serum FGF23 level, and Fig. 5E with Fig. 1D for Galnt3 in the femur), confirming the proposed function of FGFR in Galnt3 induction.
Fig. 5.
FGFR functions as a Pi sensor in vivo. (A) Schema of the experimental procedure. (B–D) Serum Pi, FEPi (B), serum Ca, FECa, serum urea nitrogen, serum creatinine (C), and serum full-length FGF23 (D) in mice fed a CP or HP diet for 2 wk under the administration of NVP-BGJ398. (E) Galnt3 mRNA expression in the femurs of mice fed a CP or HP diet for 2 wk under the administration of NVP-BGJ398 was evaluated by qPCR. Data represent the mean ± SEM. (B–E) n = 4 mice per group; *P < 0.05 by Student’s t test; NS, not significant.
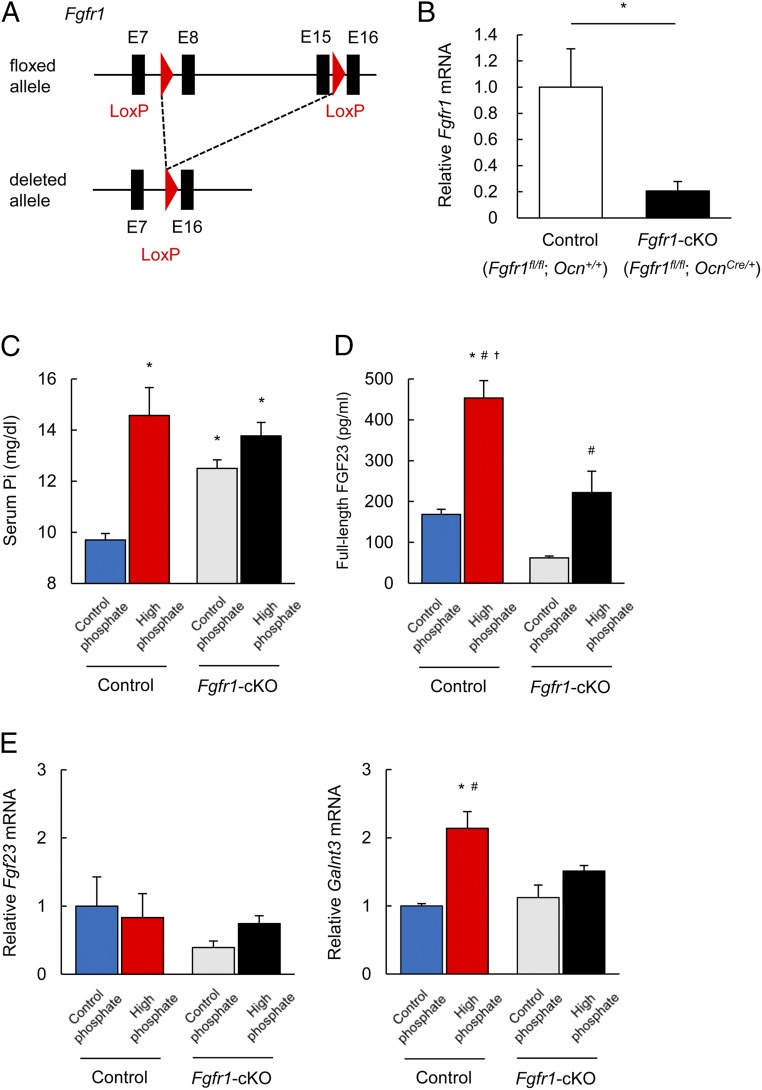
A Selective Ablation of Fgfr1 in Osteoblasts/Osteocytes Aborts Galnt3 Up-Regulation by a High Pi Diet.
We have selectively ablated Fgfr1 (Fgfr1-cKO) by crossing Osteocalcin (Ocn)-Cre mice (20) with floxed Fgfr1 mice (21) (Fig. 6A). The expression of Fgfr1 mRNA in the femur was much lower in Fgfr1-cKO mice (Fgfr1fl/fl; OcnCre/+) than that in control mice (Fgfr1fl/fl; Ocn+/+) (Fig. 6B). The serum Pi level was significantly higher not only in control and Fgfr1-cKO mice fed a HP diet, but also in Fgfr1-cKO mice fed a CP diet compared with that in control mice fed a CP diet (Fig. 6C). The increase of serum full-length FGF23 was blunted in Fgfr1-cKO mice fed a HP diet (Fig. 6D). Although there was no significant difference in Fgf23 mRNA expression, the enhancement of Galnt3 mRNA expression by a HP diet in Fgfr1-cKO mice was attenuated (Fig. 6E). These results again support the importance of FGFR1 in the regulation of Galnt3 expression by Pi.
Fig. 6.
Responses to high Pi diet in Fgfr1-cKO mice. (A) Genomic structure of the floxed and deleted Fgfr1 alleles. (B) Fgfr1 mRNA expression in the femurs of 8-wk-old male control (Fgfr1fl/fl; Ocn+/+) and Fgfr1-cKO (Fgfr1fl/fl; OcnCre/+) mice. (C and D) Serum Pi (C) and serum full-length FGF23 (D) in control and Fgfr1-cKO mice fed a CP or HP diet for 10 d. (E) Fgf23 and Galnt3 mRNA expression in the femurs of control and Fgfr1-cKO mice fed a CP or HP diet for 10 d were evaluated by qPCR. Data represent the mean ± SEM. (B) n = 4 mice per group; *P < 0.05 by Student’s t test. (C–E) n = 4–7 mice per group; *P < 0.05 compared with control mice fed a CP diet, #P < 0.05 compared with Fgfr1-cKO mice fed a CP diet, †P < 0.05 compared with Fgfr1-cKO mice fed a HP diet by ANOVA with a post hoc Tukey’s test.
Discussion
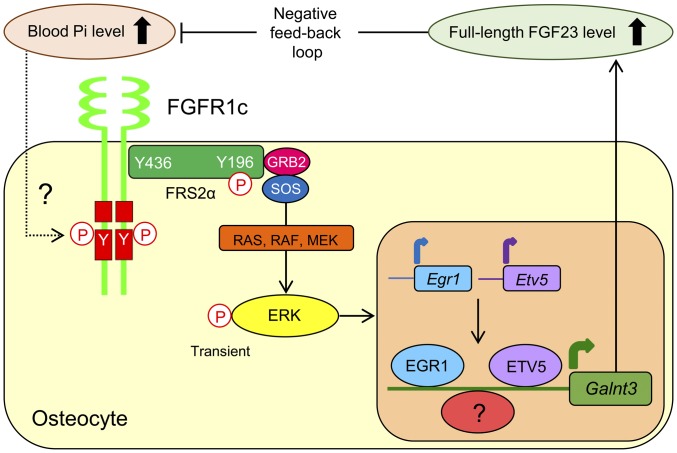
The production of active FGF23 is regulated in response to alterations in serum Pi level, although the molecular basis of Pi sensing in FGF23 production is unclear. In the present study, we found that a high Pi diet in mice enhanced the expression of Galnt3 but not Fgf23 in the femur with an increase in the serum level of full-length active FGF23 (Fig. 1 C and D). As the serum level of active full-length FGF23 was up-regulated, O-glycosylation of FGF23 by the Galnt3 product appeared to be enhanced. To further address the mechanism of Galnt3 induction by high Pi, we used an in vitro osteoblastic UMR106 cell culture model. By proteomic analysis and gene expression array analysis, high extracellular Pi was shown to activate unliganded FGFR1c, leading to Galnt3 induction via the ERK pathway, at least in part through two transcriptional activators (EGR1 and ETV5) (Fig. 7). However, when phosphorylation of downstream FRS2α and ERK was monitored, the mode of FGFR1c activation by high Pi was different from that by FGF2 (Fig. 4 B–D). Thus, the present findings illustrate a Pi-sensing pathway through FGFR1c in the regulation of FGF23 production. Finally, we showed that an FGFR inhibitor or selective deletion of Fgfr1 in osteoblasts/osteocytes abrogated high Pi diet-induced increases in serum concentration of active full-length FGF23 and femoral Galnt3 expression in mice (Figs. 5 D and E and 6 D and E). We, therefore, could suggest the significance of FGFR1 in a Pi-sensing mechanism in vivo. This idea is further supported by previous findings that some activating mutations in FGFR1 cause high FGF23 levels and hypophosphatemia in patients with osteoglophonic dysplasia (22). Pi has been reported to be involved in several other biological responses in addition to regulating FGF23 levels, such as PTH secretion, chondrocyte apoptosis, and the development of vascular diseases (23–25). In this respect, it is important to investigate whether FGFR1c also senses high Pi in these responses.
Fig. 7.
Proposed model of the Pi-sensing mechanism in bone cells. The detailed activation mechanism of FGFR1c is still unclear. EGR1 and ETV5, as transcription factors, are necessary for the induction of Galnt3 by Pi; however, other unknown factors may also be required to induce Galnt3.
Several humoral factors such as parathyroid hormone (PTH) and 1,25(OH)2D have been shown to control FGF23 expression and FGF23 levels (26–28). PTH was already reported to suppress Galnt3 expression (25), but the high phosphate diet rather increased PTH (SI Appendix, Fig. S9A). Furthermore, 1,25(OH)2D3 enhanced Fgf23 expression in UMR106 cells, as already reported (SI Appendix, Fig. S9B) (26), but 1,25(OH)2D3 had no effect on Galnt3 expression in UMR106 cells (SI Appendix, Fig. S9C). Therefore, we presume that PTH and 1,25(OH)2D are unlikely to facilitate increases in Galnt3 expression and serum level of full-length FGF23 by high phosphate.
Pi sensing in terms of Galnt3 induction was observed in the intact femur, and ERK activation for Galnt3 induction was evident only in the osteoblastic cell line but not in the other cell lines derived from nonbone tissues (SI Appendix, Fig. S4 A and B). It appears unlikely that the lack of Pi sensing in ERK activation in the other cell line is simply due to the absence of FGFR1c since we observed that overexpression of FGFR1c in that nonosteoblastic cell line was still not capable of activating the ERK pathway (SI Appendix, Fig. S4C). It is more likely that an unknown factor(s) in osteoblastic cells facilitates Pi sensing to stimulate the ERK pathway for Galnt3 induction through FGFR1c. This unknown factor(s) might account for the osteoblastic cell type-specific sensing of high Pi for FGFR1c activation. As FGFR1c isoforms preferentially interact with FGFR ligands that are secreted from epithelial tissues (18), the activity of FGFR ligands secreted from epithelial cells might be modulated when FGFR1c expression in mesenchymal lineage cells is activated by local or serum Pi levels. As chondrocytes, muscle cells and adipocytes are also differentiated from mesenchymal stem cells, such as osteoblasts, FGFR1c isoforms in these cell types might be activated by high Pi. This possibility could be tested by other assay systems to evaluate FGF actions, such as those in developing embryos of animal models.
In this study, EGR1 and ETV5 were identified as transcriptional activators that induce Galnt3 expression when FGFR1c was activated by high Pi (Fig. 2D). However, it is still unclear at this stage if other transcription factors are also required to activate the Galnt3 promoter. In a previous report concerning the promoter function of GALNT3 in a human breast cancer cell line (MCF-7) (29), only several binding sites for transcriptional activators such as SP1 and AP2 were identified in the promoter region. Thus, to date, no information on the GALNT3 promoter function in Pi sensing is available. In this regard, it is interesting to delineate and assess the binding sites for EGR1 and ETV5 in the GALNT3 promoter as an enhancer to address the impact of EGR1 and ETV5 on GALNT3 induction by high Pi. It is also possible that another signaling pathway(s) might facilitate the promoter function of Galnt3, such as the ERK pathway, as shown in this study. In terms of a screening system to sense extracellular Pi as well as other extracellular stimuli, the promoters of Galnt3 may be useful in further studies.
FGFR is known to form either homodimers to facilitate binding of paracrine FGFs or dimers with coreceptors, such as Klothos, for binding of endocrine FGFs such as FGF23 (18). Phosphorylation of FGFRs is known to be coupled with FGFR activation by FGFR ligand binding, while the strength of FGFR activation is different depending on FGFR ligand types (30). Although we found that unliganded FGFR1c was activated by high Pi, the activation process of FGFR1c still remains elusive. On the other hand, the dimerization of the unliganded FGFR model has been proposed by several reports (30–32). For dimerization of unliganded FGFR at the cell surface, the transmembrane (TM) domain, rather than the extracellular and the intracellular tyrosine kinase domains, is known to be pivotal (30, 31). High-resolution crystal structures of FGFR TM domains have already provided a view of the whole TM domain as composed of complement associations of α-helix loops on each domain (33). The significance of the TM domain for receptor activation is further implicated by the clinical observations by detecting genetic mutations in the TM and its adjacent domains of FGFR1 in patients with osteoglophonic dysplasia, who suffer from high FGF23 levels and hypophosphatemia by hyperactivation of the FGFR-mediated signaling. In addition, by an approach using the Förster resonance energy transfer (FRET)-based technique (31), FGFR1 with these mutations was shown to be prone to form FGFR1 dimerization compared with intact FGFR1 in the absence of canonical FGFR1 ligands. In the present study, high Pi was found to induce phosphorylation at the specific residues in the intracellular domain of FGFR1c, presumably resulting in an alteration of FGFR1c protein structure such as stabilizing interaction between the TM domains. Thus we assume at this stage, based on our present findings and these past reports, that FGFR1c activation by high Pi is mediated by potentiation of dimerization of unliganded FGFR1c by receptor phosphorylation.
We observed activation of FGFR1c by high Pi and characterized a cascade to transmit high Pi signals into Galnt3 induction. Furthermore, we identified the ERK pathway as an FGFR1c downstream signaling pathway among several known downstream pathways activated by FGFR (18), although we cannot exclude the possibility that the downstream signaling pathways other than AKT and PLCγ may be stimulated by activated FGFR1c by high Pi. Further biochemical assays revealed that the mode of FGFR1c activation by high Pi is not identical to that by the FGFR canonical ligand (FGF2). These findings suggest the possibility that the structural form of activated FGFR1c induced by high Pi is different from that by canonical ligands (18) and hence interacts with unknown factors located on the cell membrane and/or in the cytosol, as FGF2 failed to induce Galnt3 (Fig. 4A), regardless of activation of the same downstream ERK pathway. Further dissection of the molecular basis of FGFR1c activation by high Pi is clearly required for a better understanding of Pi sensing for FGF23 production. Recent studies indicated the involvement of sodium-dependent Pi transporter PiT2 encoded by Slc20a2 in the regulation of FGF23 by phosphate (34, 35). However, a high phosphate diet did not induce the expression of Slc20a2 in the femur (SI Appendix, Fig. S10A). In addition, silencing Slc20a2 in UMR106 cells did not suppress the induction of Galnt3 by high extracellular phosphate (SI Appendix, Fig. S10 B and C).
In conclusion, our results demonstrate the importance of posttranslational modification of the FGF23 protein in the regulation of FGF23 production. These observations are corroborated by the fact that inactivating mutations in GALNT3 cause hyperphosphatemic familial tumoral calcinosis owing to impaired actions of FGF23 by enhanced proteolytic processing of its protein (36).
Materials and Methods
Materials.
Stimulation by Pi was performed using 0.1 M Pi buffer, pH 7.4, containing Na2HPO4 and NaH2PO4. FGF2 was purchased from R&D Systems. U0126, wortmannin, and PD1703074 were purchased from Wako. FK506 was purchased from Sigma-Aldrich. Calcitriol [1,25(OH)2D3] was purchased from Carbosynth.
Mice and Treatment.
All animal procedures were approved by the Animal Research Committee at Tokushima University. ICR mice were purchased from Japan SLC. Four-week-old male ICR mice were fed a CP diet containing 0.6% Pi (16012703, Research Diets) or a HP diet containing 1.2% Pi (11101203, Research Diets) for 2 wk. Each diet included 0.5% Ca, and there were no differences in any other component. NVP-BGJ398 (Selleckchem) was formulated as a suspension in PEG300/D5W (2:1, vol/vol) and orally administered daily for 2 wk at 15 mg/kg (37). We generated conditional knockout mice for Fgfr1 (Fgfr1-cKO) using Ocn-Cre mice. Ocn-Cre mice were originally purchased from The Jackson Laboratory. The floxed Fgfr1 mice were obtained from Juha Partanen, University of Helsinki, Helsinki, Finland (21). The floxed Fgfr1 mice and Ocn-Cre mice were maintained in C57BL/6N background. The floxed Fgfr1 allele was detected by PCR with specific primers 5′-AATAGGTCCCTCGACGGTATC-3′ and 5′-CTGGGTCAGTGTGGACAGTGT-3′. The wild-type Fgfr1 allele was detected with primers 5′-CCCCATCCCATTTCCTTACCT-3′ and 5′-TTCTGGTGTGTCTGAAAACAGCT-3′. Ocn-Cre transgenes were detected with specific primers 5′-CAAATAGCCCTGGCAGATTC-3′ and 5′-TGATACAAGGGACATCTTCC-3′. Eight-week-old male Fgfr1-CKO mice were fed a CP diet containing 0.6% Pi or a HP diet containing 1.2% Pi for 10 d.
Animals were killed and blood was collected by cardiac puncture. Before killing, urine samples were collected using metabolic cages. The femur was removed, and the epiphysis and adherent soft tissues were cut away. Bone marrow was flushed with saline. The cleaned femur was rapidly frozen in liquid nitrogen.
Cell Culture and in Vitro Experiments.
The UMR106 rat osteosarcoma cell line was obtained from ATCC. UMR106 cells were cultured in DMEM (Wako) supplemented with 10% FBS (Mediatech) and penicillin/streptomycin/amphotericin B (Wako). Cell cultures were maintained at 37 °C with 5% CO2. For experiments involving stimulation by several reagents, UMR106 cells were plated at 200,000 per well in 12-well plates in DMEM with 10% FBS and antibiotics. After reaching 80% confluence, cells were cultured in medium with 1% FBS and stimulated with each reagent. At 50% confluence, UMR106 cells were transfected with expression vectors using FuGENE HD Transfection Reagent (Promega).
Full-Length FGF23 and Intact PTH Measurements.
Full-length FGF23 concentrations were measured by the FGF23 ELISA kit (Kainos) (2, 38). Serum intact PTH concentrations were measured by the mouse PTH 1-84 ELISA kit (Immutopics).
RNA Analysis.
Collected femur samples were soaked in RNAiso Plus (TaKaRa) and homogenized with TissueLyser II (Qiagen). Total RNA from the homogenates and UMR106 cells were extracted with a NucleoSpin RNA system (Machrey-Nagel). First-strand cDNA was synthesized from total RNA with PrimeScript RT Master Mix (TaKaRa). RT-qPCR was performed using FastStart Essential DNA Green Master (Roche) and LightCycler 96 System (Roche). The gene expression level was normalized to that of glyceraldehyde-3-phosphate dehydrogenase (Gapdh).
Immunoblot Analysis.
UMR106 cells were lysed in RIPA buffer (50 mM Hepes, 150 mM NaCl, 1% Nonidet P-40, 0.5% Na-deoxycholate, 0.1% SDS, 1 mM EDTA) with Complete Mini Protease Inhibitor Mixture (Roche). For immunoblot analysis of phosphorylated proteins, RIPA buffer was also supplemented with PhosSTOP (Roche) to inhibit phosphatase activity. His-tagged human FGF23 protein secreted into culture media of UMR106 cells was purified using the MagneHis Protein Purification System (Promega). Frozen femur samples were subjected to protein extraction using drill homogenizer. SDS/PAGE was performed using 10% polyacrylamide gels. After electrophoresis, proteins were transferred to polyvinylidene difluoride (PVDF) membranes. Membranes were incubated with anti-FGF23 C-terminal antibody (a gift from Kyowa Hakko Kirin Co., 1:2,000), anti-GalNAc-T3 antibody (Aviva Systems Biology, 1:1,000), anti-phospho-ERK1/2 (Thr202/Tyr204) antibody (Cell Signaling Technology, 1:1,666), anti-ERK1/2 antibody (Cell Signaling Technology, 1:4,000), anti-phospho-Akt (Ser473) antibody (Cell Signaling Technology, 1:2,000), anti-Akt antibody (Cell Signaling Technology, 1:1,000), anti-phospho-FGFR (Tyr653/654) antibody (Cell Signaling Technology, 1:1,000), anti-FGFR1 antibody (Cell Signaling Technology, 1:1,000), anti-phospho-FRS2α (Tyr196) antibody (Cell Signaling Technology, 1:2,000), anti-phospho-FRS2α (Tyr436) antibody (Cell Signaling Technology, 1:2,000), and anti-α-Tubulin antibody (Sigma-Aldrich, 1:4,000) followed by horseradish peroxidase-conjugated secondary antibodies. Protein bands were visualized with Clarity Western ECL Substrate (Bio-Rad) and the ChemiDoc Touch Imaging System (Bio-Rad). Image Lab software (Bio-Rad) was used to quantify the intensity of bands.
DNA Microarray Analysis.
DNA microarray analysis was performed using UMR106 cells with or without 5 mM Pi treatment for 6 h. Gene expression was tested using SurePrint G3 Rat GE Ver2.0 8 × 60K (Agilent Technologies). Data were analyzed using GeneSpring software (Agilent Technologies) and normalized. The statistical analysis was performed using Mann–Whitney unpaired tests.
Gene Silencing.
Duplexed siRNA of each target gene (Egr1, Etv4, Etv5, Fgfr1, and Slc20a2) and its scrambled siRNA as a negative control were designed and purchased from Bioneer. The siRNA oligonucleotides were transfected using Lipofectamine RNAiMAX reagent (Invitrogen).
Luciferase Reporter Assays.
The activation of the ERK signaling pathway was analyzed using a pGL4.33 vector that contained a SRE (Promega). Luciferase activities were measured with a Dual-Luciferase Reporter Assay System (Promega) and a GloMax Discover Multimode Microplate Reader (Promega). As a reference to normalize the transfection efficiency, the pRL-TK vector (Promega) was cotransfected in all experiments.
Identification and Quantification of Tyrosine-Phosphorylated Peptides by LC-MS/MS Analysis.
A previously described method was used with some modifications (39–41). Unstimulated UMR106 cells or cells stimulated with 5 mM Pi or 100 ng/mL FGF2 for 15 min were lysed in buffer containing 6 M guanidine-HCl, 100 mM Tris⋅HCl, pH 8.0, 2 mM DTT, and PhosSTOP. The lysates were sonicated and cleared by centrifugation at 15,000 × g at 4 °C for 15 min. Proteins (each 1.0 mg) were reduced with 5 mM DTT for 30 min and then alkylated with 27.5 mM iodoacetamide for 30 min in the dark. After methanol/chloroform precipitation, proteins were dissolved in 100 µL of 0.1% RapiGest SF (Waters) in 50 mM triethylammonium bicarbonate buffer. The proteins were digested with 10 μg of trypsin (MS-Grade, Thermo Fisher Scientific). After 16 h at 37 °C, tyrosine-phosphorylated peptides were enriched using the PTMScan Phospho-Tyrosine Rabbit mAb (P-Tyr-1000) kit (Cell Signaling Technology) in the presence of 1 mg/mL Pefabloc SC (Sigma-Aldrich) in accordance with the manufacturer’s instructions. Peptides were eluted with 150 μL of 60% ACN/0.1% TFA and desalted using GL-Tip SDB (GL Sciences). The eluates were evaporated in a SpeedVac concentrator (Thermo Fisher Scientific).
LC-MS/MS analysis of the resultant peptides was performed on an EASY-nLC 1200 UHPLC connected to a Q Exactive Plus mass spectrometer through a nanoelectrospray ion source (Thermo Fisher Scientific). The peptides were separated on a 75-µm inner diameter × 150 mm C18 reversed-phase column (Nikkyo Technos) with a linear gradient from 4 to 28% acetonitrile (ACN) for 0–100 min followed by an increase to 80% ACN during 100–110 min. The mass spectrometer was operated in a data-dependent acquisition mode with a top 10 MS/MS method. MS1 spectra were measured with a resolution of 70,000, an AGC target of 1 × 106 and a mass range from 350 to 1,500 m/z. MS/MS spectra were triggered at a resolution of 17,500, an AGC target of 5 × 104, an isolation window of 2.0 m/z, a maximum injection time of 60 ms, and a normalized collision energy of 27. Dynamic exclusion was set to 10 s. Raw data were directly analyzed against the SwissProt database restricted to Rattus using Proteome Discoverer version 2.2 (Thermo Fisher Scientific) for identification and label-free precursor ion quantification. The search parameters were as follows: (i) trypsin as an enzyme with up to one missed cleavage; (ii) precursor mass tolerance of 10 ppm; (iii) fragment mass tolerance of 0.02 Da; (iv) carbamidomethylation of cysteine as a fixed modification; and (v) oxidation of methionine and phosphorylation of serine, threonine, and tyrosine as variable modifications. Peptides were filtered at a false-discovery rate of 1% using the percolator node. Normalization was performed such that the total sum of abundance values for each sample over all peptides was the same. The MS proteomics data have been deposited in the ProteomeXchange Consortium via the jPOST partner repository (https://repository.jpostdb.org) with the dataset identifier PXD010790 (42).
Several selected peptides, including FGFR1 with phosphorylated tyrosine residues, were measured by PRM, an MS/MS-based targeted quantification method using high-resolution MS. Targeted MS/MS scans were acquired by a time-scheduled inclusion list at a resolution of 70,000, an AGC target of 2 × 105, an isolation window of 4.0 m/z, a maximum injection time of 500 ms, and a normalized collision energy of 27. Time alignment and relative quantification of the transitions were performed with PinPoint version 1.4 (Thermo Fisher Scientific). The MS proteomics data for PRM analysis have been deposited in the ProteomeXchange Consortium via the jPOST partner repository with the dataset identifier PXD010791.
Analysis of mRNA Stability.
The analytical method was previously described (43). Galnt3 mRNA levels in UMR106 cells were analyzed by RT-qPCR at 0, 1, 2, and 4 h after treatment with actinomycin D. The half-life of the mRNA was calculated by a method previously described.
Statistics.
All data are presented as the mean ± SEM. The results were analyzed for significant differences using Student’s t test or ANOVA followed by a Dunnett’s or Tukey’s multiple comparison post hoc test. Significance was set at P < 0.05.
Supplementary Material
Acknowledgments
We thank Dr. Juha Partanen (University of Helsinki) for providing the floxed Fgfr1 mice. We also thank M. Kawano, N. Miura, R. Sakai, and T. Ozaki for technical assistance; and H. Takeichi, K. Hata, and M. Iwata for secretarial assistance. This work was supported by KAKENHI Grant-in-Aid for Young Scientists 18K15980 from the Japan Society for the Promotion of Sciences (JSPS) (to Y.T.), by KAKENHI Grant-in-Aid for Scientific Research 16H05327 from JSPS (to T.M.), and by the Japan Agency for Medical Research and Development under Grant 17ek0109150h0003 (to S.F.). This work was also supported by the Support Center for Advanced Medical Sciences, Institute of Biomedical Sciences, Tokushima University Graduate School.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. O.W.M. is a guest editor invited by the Editorial Board.
Data deposition: The MS proteomics data have been deposited to the ProteomeXchange Consortium via the jPOST partner repository with dataset identifiers PXD010790 and PXD010791.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1815166116/-/DCSupplemental.
References
- 1.Fukumoto S., FGF23-FGF receptor/Klotho pathway as a new drug target for disorders of bone and mineral metabolism. Calcif. Tissue Int. 98, 334–340 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Shimada T., et al. , Targeted ablation of Fgf23 demonstrates an essential physiological role of FGF23 in phosphate and vitamin D metabolism. J. Clin. Invest. 113, 561–568 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ornitz D. M., Itoh N., The fibroblast growth factor signaling pathway. Wiley Interdiscip. Rev. Dev. Biol. 4, 215–266 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Urakawa I., et al. , Klotho converts canonical FGF receptor into a specific receptor for FGF23. Nature 444, 770–774 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Shimada T., et al. , FGF-23 is a potent regulator of vitamin D metabolism and phosphate homeostasis. J. Bone Miner. Res. 19, 429–435 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Feng J. Q., et al. , Loss of DMP1 causes rickets and osteomalacia and identifies a role for osteocytes in mineral metabolism. Nat. Genet. 38, 1310–1315 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu S., et al. , Pathogenic role of Fgf23 in Hyp mice. Am. J. Physiol. Endocrinol. Metab. 291, E38–E49 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Tagliabracci V. S., et al. , Dynamic regulation of FGF23 by Fam20C phosphorylation, GalNAc-T3 glycosylation, and furin proteolysis. Proc. Natl. Acad. Sci. U.S.A. 111, 5520–5525 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song L., Linstedt A. D., Inhibitor of ppGalNAc-T3-mediated O-glycosylation blocks cancer cell invasiveness and lowers FGF23 levels. eLife 6, e24051 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frishberg Y., et al. , Hyperostosis-hyperphosphatemia syndrome: A congenital disorder of O-glycosylation associated with augmented processing of fibroblast growth factor 23. J. Bone Miner. Res. 22, 235–242 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Bennett E. P., et al. , Control of mucin-type O-glycosylation: A classification of the polypeptide GalNAc-transferase gene family. Glycobiology 22, 736–756 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamazaki M., et al. , Both FGF23 and extracellular phosphate activate Raf/MEK/ERK pathway via FGF receptors in HEK293 cells. J. Cell. Biochem. 111, 1210–1221 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Kimata M., et al. , Signaling of extracellular inorganic phosphate up-regulates cyclin D1 expression in proliferating chondrocytes via the Na+/Pi cotransporter Pit-1 and Raf/MEK/ERK pathway. Bone 47, 938–947 (2010). [DOI] [PubMed] [Google Scholar]
- 14.McKay M. M., Morrison D. K., Integrating signals from RTKs to ERK/MAPK. Oncogene 26, 3113–3121 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Nishino J., et al. , Extracellular phosphate induces the expression of dentin matrix protein 1 through the FGF receptor in osteoblasts. J. Cell. Biochem. 118, 1151–1163 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Furdui C. M., Lew E. D., Schlessinger J., Anderson K. S., Autophosphorylation of FGFR1 kinase is mediated by a sequential and precisely ordered reaction. Mol. Cell 21, 711–717 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Lew E. D., Furdui C. M., Anderson K. S., Schlessinger J., The precise sequence of FGF receptor autophosphorylation is kinetically driven and is disrupted by oncogenic mutations. Sci. Signal. 2, ra6 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goetz R., Mohammadi M., Exploring mechanisms of FGF signalling through the lens of structural biology. Nat. Rev. Mol. Cell Biol. 14, 166–180 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gotoh N., Regulation of growth factor signaling by FRS2 family docking/scaffold adaptor proteins. Cancer Sci. 99, 1319–1325 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang M., et al. , Osteoblast-specific knockout of the insulin-like growth factor (IGF) receptor gene reveals an essential role of IGF signaling in bone matrix mineralization. J. Biol. Chem. 277, 44005–44012 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Trokovic R., et al. , FGFR1 is independently required in both developing mid- and hindbrain for sustained response to isthmic signals. EMBO J. 22, 1811–1823 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.White K. E., et al. , Mutations that cause osteoglophonic dysplasia define novel roles for FGFR1 in bone elongation. Am. J. Hum. Genet. 76, 361–367 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silver J., Naveh-Many T., Phosphate and the parathyroid. Kidney Int. 75, 898–905 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Papaioannou G., et al. , Raf kinases are essential for phosphate induction of ERK1/2 phosphorylation in hypertrophic chondrocytes and normal endochondral bone development. J. Biol. Chem. 292, 3164–3171 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jono S., et al. , Phosphate regulation of vascular smooth muscle cell calcification. Circ. Res. 87, E10–E17 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Saito H., et al. , Circulating FGF-23 is regulated by 1α,25-dihydroxyvitamin D3 and phosphorus in vivo. J. Biol. Chem. 280, 2543–2549 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Fan Y., et al. , Parathyroid hormone 1 receptor is essential to induce FGF23 production and maintain systemic mineral ion homeostasis. FASEB J. 30, 428–440 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hori M., Kinoshita Y., Taguchi M., Fukumoto S., Phosphate enhances Fgf23 expression through reactive oxygen species in UMR-106 cells. J. Bone Miner. Metab. 34, 132–139 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Nomoto M., et al. , Structural basis for the regulation of UDP-N-acetyl-alpha-D-galactosamine: Polypeptide N-acetylgalactosaminyl transferase-3 gene expression in adenocarcinoma cells. Cancer Res. 59, 6214–6222 (1999). [PubMed] [Google Scholar]
- 30.Sarabipour S., Hristova K., Mechanism of FGF receptor dimerization and activation. Nat. Commun. 7, 10262 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Comps-Agrar L., Dunshee D. R., Eaton D. L., Sonoda J., Unliganded fibroblast growth factor receptor 1 forms density-independent dimers. J. Biol. Chem. 290, 24166–24177 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin C. C., et al. , Inhibition of basal FGF receptor signaling by dimeric Grb2. Cell 149, 1514–1524 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Bocharov E. V., et al. , Structure of FGFR3 transmembrane domain dimer: Implications for signaling and human pathologies. Structure 21, 2087–2093 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bon N., et al. , Phosphate-dependent FGF23 secretion is modulated by PiT2/Slc20a2. Mol. Metab. 11, 197–204 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bon N., et al. , Phosphate (Pi)-regulated heterodimerization of the high-affinity sodium-dependent Pi transporters PiT1/Slc20a1 and PiT2/Slc20a2 underlies extracellular Pi sensing independently of Pi uptake. J. Biol. Chem. 293, 2102–2114 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Topaz O., et al. , Mutations in GALNT3, encoding a protein involved in O-linked glycosylation, cause familial tumoral calcinosis. Nat. Genet. 36, 579–581 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Guagnano V., et al. , Discovery of 3-(2,6-dichloro-3,5-dimethoxy-phenyl)-1-{6-[4-(4-ethyl-piperazin-1-yl)-phenylamino]-pyrimidin-4-yl}-1-methyl-urea (NVP-BGJ398), a potent and selective inhibitor of the fibroblast growth factor receptor family of receptor tyrosine kinase. J. Med. Chem. 54, 7066–7083 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Yamazaki Y., et al. , Increased circulatory level of biologically active full-length FGF-23 in patients with hypophosphatemic rickets/osteomalacia. J. Clin. Endocrinol. Metab. 87, 4957–4960 (2002). [DOI] [PubMed] [Google Scholar]
- 39.Ishikawa E., et al. , Protein kinase D regulates positive selection of CD4+ thymocytes through phosphorylation of SHP-1. Nat. Commun. 7, 12756 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abe Y., Nagano M., Tada A., Adachi J., Tomonaga T., Deep phosphotyrosine proteomics by optimization of phosphotyrosine enrichment and MS/MS parameters. J. Proteome Res. 16, 1077–1086 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Motani K., Kosako H., Activation of stimulator of interferon genes (STING) induces ADAM17-mediated shedding of the immune semaphorin SEMA4D. J. Biol. Chem. 293, 7717–7726 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okuda S., et al. , jPOSTrepo: An international standard data repository for proteomes. Nucleic Acids Res. 45, D1107–D1111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Graham J. R., Hendershott M. C., Terragni J., Cooper G. M., mRNA degradation plays a significant role in the program of gene expression regulated by phosphatidylinositol 3-kinase signaling. Mol. Cell. Biol. 30, 5295–5305 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.