Significance
World food supply depends on improving grain yield and quality, which are determined by accumulation of starch and proteins in maize endosperm, respectively. Because initiation of synthesis of the two compounds occurs 8 to 10 d after pollination in starchy endosperm cells, regulatory factors must control their coordinated accumulation. This work shows that two maize endosperm-specific transcription factors can coordinate the accumulation of starch and proteins by regulating the expression of key starch biosynthetic enzymes and the major seed proteins.
Keywords: maize endosperm, gene regulation, starch synthesis, protein
Abstract
Grain starch and protein are synthesized during endosperm development, prompting the question of what regulatory mechanism underlies the synchronization of the accumulation of secondary and primary gene products. We found that two endosperm-specific NAC transcription factors, ZmNAC128 and ZmNAC130, have such a regulatory function. Knockdown of expression of ZmNAC128 and ZmNAC130 with RNA interference (RNAi) caused a shrunken kernel phenotype with significant reduction of starch and protein. We could show that ZmNAC128 and ZmNAC130 regulate the transcription of Bt2 and then reduce its protein level, a rate-limiting step in starch synthesis of maize endosperm. Lack of ZmNAC128 and ZmNAC130 also reduced accumulation of zeins and nonzeins by 18% and 24% compared with nontransgenic siblings, respectively. Although ZmNAC128 and ZmNAC130 affected expression of zein genes in general, they specifically activated transcription of the 16-kDa γ-zein gene. The two transcription factors did not dimerize with each other but exemplified redundancy, whereas individual discovery of their function was not amenable to conventional genetics but illustrated the power of RNAi. Given that both the Bt2 and the 16-kDa γ-zein genes were activated by ZmNAC128 or ZmNAC130, we could identify a core binding site ACGCAA contained within their target promoter regions by combining Dual-Luciferase Reporter and Electrophoretic Mobility Shift assays. Consistent with these properties, transcriptomic profiling uncovered that lack of ZmNAC128 and ZmNAC130 had a pleiotropic effect on the utilization of carbohydrates and amino acids.
Endosperms, up to 90% of cereal grains, not only support embryo and seedling development but also provide the primary resource of carbohydrate and protein for humans and livestock. Carbohydrate is mainly stored as starch granules (SGs) in amyloplasts of endosperm cells. Prolamins, the most abundant storage proteins in most grains, are deposited in protein bodies (PBs) or protein storage vacuoles (PSVs). Therefore, SGs and PBs/PSVs are the most abundant organelles in grain endosperm cells. Due to the importance of human food, they have been comprehensively studied (1, 2).
Maize (Zea mays) prolamins, also called zeins, account for ∼60 to 70% of endosperm proteins (1). Maize endosperm contains four types of prolamins, α-, β-, γ-, and δ-zeins, which are encoded by a large set of genes (3, 4). Because their expression is strictly regulated in a temporal and spatial pattern, the transcriptional regulation has been widely investigated by identifying conserved cis-elements in their promoters and corresponding transcription factors (TFs). Several conserved cis-elements have been found in promoters of most zein genes, like the prolamin-box (P-box) (TGTAAAG), Opaque2-box (O2-box) (TCCACGT), or O2-like box (TTTACGT) (5). The corresponding TFs have also been successively identified. O2, an endosperm-specific bZIP TF, binds to the O2-box to transactivate the expression of 22-kDa α-zein and 15-kDa β-zein genes (6–8). Recently, a genome-wide strategy of ChIP-Seq combined with differential expression analysis also identified other O2 binding motifs, like TGACGTGG (9, 10). O2 can regulate the expression of most zeins except for 16-kDa γ-zein, illustrating a central role for O2-regulating zein expression. Another well-known endosperm-specific DNA-binding with one finger (Dof) TF is the Prolamin-Box Binding Factor (PBF). PBF specifically binds the P-box to regulate the expression of 22-kDa α-zein and 27-kDa γ-zein genes (5, 11). Besides O2 and PBF, an additional four TFs were recently identified to regulate the expression of zeins. Three of them are bZIP TFs: i.e., O2 heterodimerizing proteins (OHP1 and OHP2) and ZmbZIP22 (12, 13). OHP1 and OHP2 can recognize an O2-like box in the promoters of the 27-kDa γ-zein and 22-kDa α-zein whereas ZmbZIP22 binds to the ACAGCTCA box in the 27-kDa γ-zein promoter, forming a complex with O2, PBF, OHP1/OHP2 (13). ZmMADS47, a MADS box-containing TF, binds the CATGT motif in the promoters of α-zein and 50-kDa γ-zein, but its interaction with O2 is required for the transcriptional activation of these genes (14). However, there are still examples of missing regulatory factors: for instance, Dzr1, regulating δ-zein accumulation (15) or the transcriptional activators of 16-kDa γ-zein.
Moreover, how are synthesis and accumulation of both zeins and starch coordinated at the filling stage from 10 to 35 d after pollination (DAP) (16–18)? Unlike the synthesis of zeins, the starch biosynthetic pathway is more complicated and involves carbohydrate metabolism, such as the oxidative pentose phosphate pathway (2). Indeed, there is less knowledge about the transcriptional regulation of starch biosynthetic genes in maize or other crops although O2 not only regulates the expression of zeins but also affects the regulation of other storage compounds (9, 10). Direct evidence for O2 modulating starch synthesis is that O2 binds the O2-box in the promoter of SSIII to regulate its expression (19). PBF binds the core elements of P-box near O2-box in the promoter of SSIII to coordinate its expression with O2. The synergetic regulatory role of PBF and O2 also applies to zein genes (19). The previous studies also tried to identify TFs regulating synthetic starch genes by coexpression analysis and biochemistry experiments, like the Electrophoretic Mobility Shift Assay (EMSA) and the Dual-Luciferase Reporter (DLR) assay. There have been candidate TFs, including one barley WRKY transcription factor of SUSIBA2, and three maize transcription factors of ZmNAC36, ZmbZIP91, and ZmEREB156, although their regulatory function on starch synthesis remains to be validated (20–23).
In the search for TFs that regulate both starch biosynthetic and zein genes, we took an approach different to the above studies by accounting for gene amplification. We investigated RNA expression at early stages of endosperm development and found that the two TFs, ZmNAC128 and ZmNAC130, are specifically and strongly expressed at the filling stage. Because of the high identities of ZmNAC128 and ZmNAC130 in amino acid sequence, we used RNA interference (RNAi) to reduce their expression because it overcomes gene amplification and is dominant (24). Indeed, nacRNAi leads to a shrunken kernel phenotype with significant reduction of starch and protein accumulation during endosperm development, which would not have been detected by a single mutant screen. Molecular and biochemistry evidence uncovered that ZmNAC128 and ZmNAC130 coordinate the accumulation of starch and protein through the transcriptional regulation of at least the Bt2 and the 16-kDa γ-zein genes.
Results
Shrunken Kernel Phenotype by Knockdown of ZmNAC128 and ZmNAC130.
A total of 112 putative proteins were annotated as the NAC superfamily in the maize B73 V4 genome although the spatiotemporal transcriptome atlas showed that only two NAC TFs, ZmNAC128 and ZmNAC130, were strongly and specifically expressed at the filling stage of endosperm cells (16, 25). The two genes appear to have arisen from the allotetraploidization of maize and are located on chromosome 3 (ZmNac128) and chromosome 8 (ZmNAC130). The latter generated a tandemly duplicated copy, ZmNAC118 (SI Appendix, Fig. S1). However, only ZmNAC128 and ZmNAC130, but not ZmNAC118, are strongly expressed in the developing endosperm, suggesting that they possibly possess the same molecular function (SI Appendix, Fig. S1). We also verified their expression pattern by real-time qPCR (SI Appendix, Fig. S2). Phylogenetic analysis placed ZmNAC128 and ZmNAC130 within a sister branch, and alignment of their protein sequences showed that there were high sequence similarities in the NAC domain and C-terminal regions (SI Appendix, Figs. S3 and S4). A yeast transactivation assay showed that their C-terminal regions possess transactivation activity (SI Appendix, Fig. S5). These results and their common ancestry suggested that the two NAC TFs might have redundant functions at the endosperm filling stage.
To investigate such a redundancy, a five-amino acids deletion within the conserved NAC domain of ZmNAC130 (nac130) was generated with the CRISPR-Cas9 technology (SI Appendix, Figs. S4 and S6 A and B). No change of zein and nonzein protein accumulation or a kernel phenotype was apparent in this nac130 mutant (SI Appendix, Fig. S6 D and E). Therefore, an RNAi transgenic construct was used to knock down the expression of both ZmNAC128 and ZmNAC130 in a dominant fashion (SI Appendix, Fig. S6). Two independent nacRNAi events (#2 and #4) were obtained and backcrossed to three inbred lines (B73, Mo17, and W64A) for two generations. Real-time qPCR results showed that expression of ZmNAC128 and ZmNAC130 are significantly reduced in the two nacRNAi lines, #2 and #4 (SI Appendix, Fig. S7). Mature nacRNAi transgenic cobs of B73 backcrosses exhibited a shrunken phenotype of kernels compared with the plump kernel phenotype of nontransgenic seeds (NT) (Fig. 1A and SI Appendix, Fig. S8). The same shrunken kernel phenotype was observed with nacRNAi transgenic cobs from backcrosses with Mo17 and W64A (SI Appendix, Fig. S8). Because the transgenic kernels exhibited the same shrunken phenotype for each transgenic event and in different inbred backcrosses, nacRNAi#4 in B73 background was used as the genetic material for all subsequent experiments.
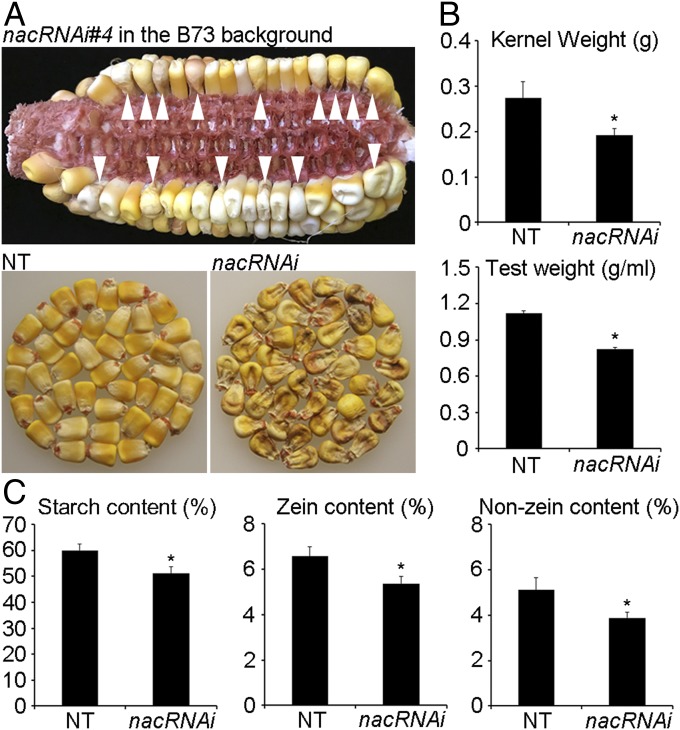
Fig. 1.
The phenotypes of nacRNAi. (A) Kernel phenotypes of nacRNAi#4 in the B73 background. In the Upper, the white arrowheads point out the transgenic nacRNAi kernels in the cob. The Lower Left and Right represent the nontransgenic (NT) and nacRNAi kernels from the same cob, respectively. (B) Kernel weight (Upper) and test weight (Lower) of NT and nacRNAi seeds. The data are measured from the three mature cobs of nacRNAi#4 in the B73 background with ±SD. g, gram; g/mL, gram per milliliter. (C) The content of starch, zeins, and nonzeins in the mature kernels of NT and nacRNAi siblings. Percent represents mg per 100 mg mature dry kernels. In B and C, the asterisk represents a significant difference from NT (Student’s t test, P < 0.05).
Reduction of Starch and Protein in nacRNAi.
Kernel weight (KW) and test weight (TW) were considered as indicators of endosperm filling for grain yield and quality. Compared with NT with 0.27 gram of KW and 1.12 g/mL TW, KW and TW of nacRNAi siblings were reduced by ∼30% and 26%, respectively (Fig. 1B). Therefore, knockdown of ZmNAC128 and ZmNAC130 affected the filling of endosperm, causing a shrunken kernel phenotype with a significant reduction of KW and TW. To investigate the mechanism underlying the shrunken kernel phenotype, starch and protein compartmentalization in nacRNAi kernels was analyzed. Transmission electron microscopy (TEM) was employed to observe the development of SGs and PBs at the filling-stage endosperm cells. The TEM observation showed that SGs were apparently smaller in the 20-DAP nacRNAi endosperm cells than those in NT (SI Appendix, Fig. S9A). PBs in 20-DAP endosperm cells did not show a difference in size and shape between NT and nacRNAi siblings, but their number was reduced by 40% in nacRNAi (SI Appendix, Fig. S9 B and C). Furthermore, the content of starch and total seed proteins (including zeins and nonzeins) in mature dry seeds was measured. In 100 mg of mature seed flour of NT, there was 60.0 ± 2.4 mg of starch, 6.6 ± 0.4 mg of zein proteins, and 5.1 ± 0.5 mg of nonzein proteins. Compared with NT, the content of starch, zeins, and nonzeins was reduced by ∼15%, 18%, and 24% in mature kernels of nacRNAi siblings, respectively (Fig. 1C). Clearly, the two NAC TFs reduced starch and protein accumulation.
ZmNAC128 and ZmNAC130 as TFs for the Bt2 Gene.
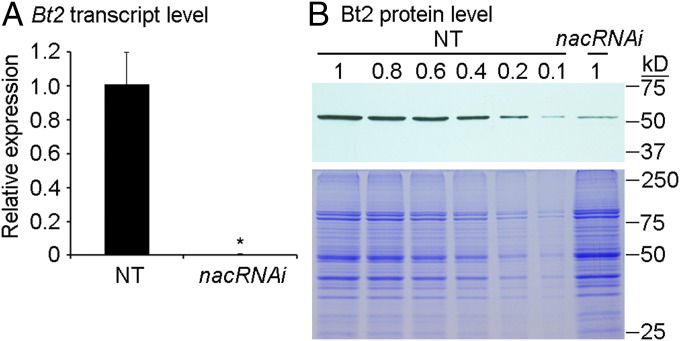
Because nacRNAi affected the size of SGs and starch content, we determined the expression of genes contributing to starch biosynthesis in 20-DAP endosperms by real-time qPCR (SI Appendix, Fig. S10A). Transcript levels of seven starch synthetic genes were significantly down-regulated by an at least twofold threshold. These down-regulated genes included Bt2 (brittle2) encoding adenosine diphosphate glucose pyrophosphorylase (AGPase) small subunit, SS1 encoding starch synthase 1, Zpu1 encoding pullulanase-type starch-debranching enzyme 1, Sus1 encoding sucrose synthase 1, Sbe2b encoding starch branching enzyme 2b, Sbe1 encoding starch branching enzyme 1, and GBSS1 (also called Waxy) encoding granule-bound starch synthase 1. Among them, the transcript level of Bt2 was the most reduced by about 200-fold in nacRNAi compared with NT (Fig. 2A). Consistent with the real-time qPCR result, immunoblotting further showed that the protein accumulation of Bt2 dropped by 80 to 90% in the 20-DAP nacRNAi endosperm compared with NT (Fig. 2B). Because AGPase, which is composed of Bt2 and Sh2 (AGPase large subunit), is the limiting enzyme for starch synthesis in maize endosperm, loss of Bt2 could explain the shrunken phenotype of nacRNAi kernels. To determine whether this occurred through direct interaction of the two NAC TFs with the Bt2 promoter, we performed the DLR assay in Arabidopsis protoplast cells. The constructs of both reporter and effector were transiently introduced into the same protoplast cells to measure the ratio of fluorescent signals of the two types of luciferases. The Renilla luciferase (REN) gene was driven by the 35S promoter as an internal control, and firefly luciferase (LUC) was driven by the 1.5- kilo base pair (kbp) fragment upstream from the start codon of Bt2 (SI Appendix, Fig. S11A). Compared with the control, LUC activity was increased by 38-fold in the presence of NAC128 and by 100-fold in presence of NAC130. Although both together increased LUC activity by only 72-fold (SI Appendix, Fig. S11B), these assays suggested that the two NAC TFs were functionally independent for the regulation of Bt2, albeit with different binding or transactivation activity.
Fig. 2.
Transcript and protein levels of Bt2 in 20-DAP endosperms of NT and nacRNAi. (A) Real-time qPCR analysis of the Bt2 expression. The data from three replicates are illustrated with ±SD. The asterisk represents a significant difference from NT (Student’s t test, P < 0.05). (B) Immunoblotting analysis of the Bt2 protein. The Upper displays the immunoblotting result. The protein loading amount in NT is diluted by 0.8-, 0.6-, 0.4-, 0.2-, and 0.1-fold. The Lower is a piece of CBB-staining replicated gel as the loading control.
Transcriptional Activation of the 16-kDa γ-Zein Gene by ZmNAC128 and ZmNAC130.
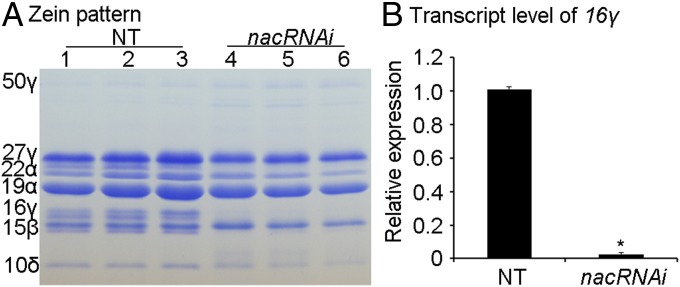
The Coomassie Brilliant Blue (CBB)-staining protein gel showed that zein proteins were apparently reduced in nacRNAi whereas 16-kDa γ-zein was almost not visible in the CBB-staining protein gel of nacRNAi kernel (Fig. 3A). Consistent with the reduction of zeins, real-time qPCR detection showed that the transcript levels of most zeins, except for z1B, were significantly down-regulated by more than twofold in 20-DAP nacRNAi endosperm compared with NT (SI Appendix, Fig. S10B). The transcript level of the 16-kDa γ-zein gene was reduced by even 40-fold (Fig. 3B), indicating that ZmNAC128 and ZmNAC130 could directly regulate the 16-kDa γ-zein gene. Therefore, we also conducted the DLR assay with the 16-kDa γ-zein promoter. Compared with the control, LUC activity driven by 16-kDa γ-zein promoter is increased by 735-fold with ZmNAC128 and by 1,568-fold with ZmNAC130. Similar to the Bt2 promoter, the LUC activity driven by the 16-kDa γ-zein promoter is not stronger in the presence of both NAC128 and NAC130 (SI Appendix, Fig. S11C).
Fig. 3.
The expression of 16-kDa γ-zein in nacRNAi. (A) The accumulation of zein proteins in NT and nacRNAi. Samples 1 and 4, 2 and 5, and 3 and 6 are, respectively, from the three cobs of B73 background. 50γ, 50-kDa γ-zein; 27γ, 27-kDa γ-zein; 22α, 22-kDa α-zein; 19α, 19-kDa α-zein; 16γ, 16-kDa γ-zein; 15β, 15-kDa β-zein; 10δ, 10-kDa δ-zein. (B) Transcript levels of 16-kDa γ-zein in 20-DAP endosperms of NT and nacRNAi. The data from three replicates are illustrated with ±SD. The asterisk represents a significant difference from NT (Student’s t test, P < 0.05).
ACGCAA as a Primary Cis-Element Binding Site of ZmNAC128 and ZmNAC130.
Given their specific binding to different promoters, the question arose whether there was a common cis-element binding site for ZmNAC128 and ZmNAC130, which would be consistent with the high identity of amino acid sequences of the two TFs (SI Appendix, Fig. S4). Therefore, a DLR assay with EMSA was performed to narrow down the cis-element binding site (Fig. 4 and SI Appendix, Fig. S12).
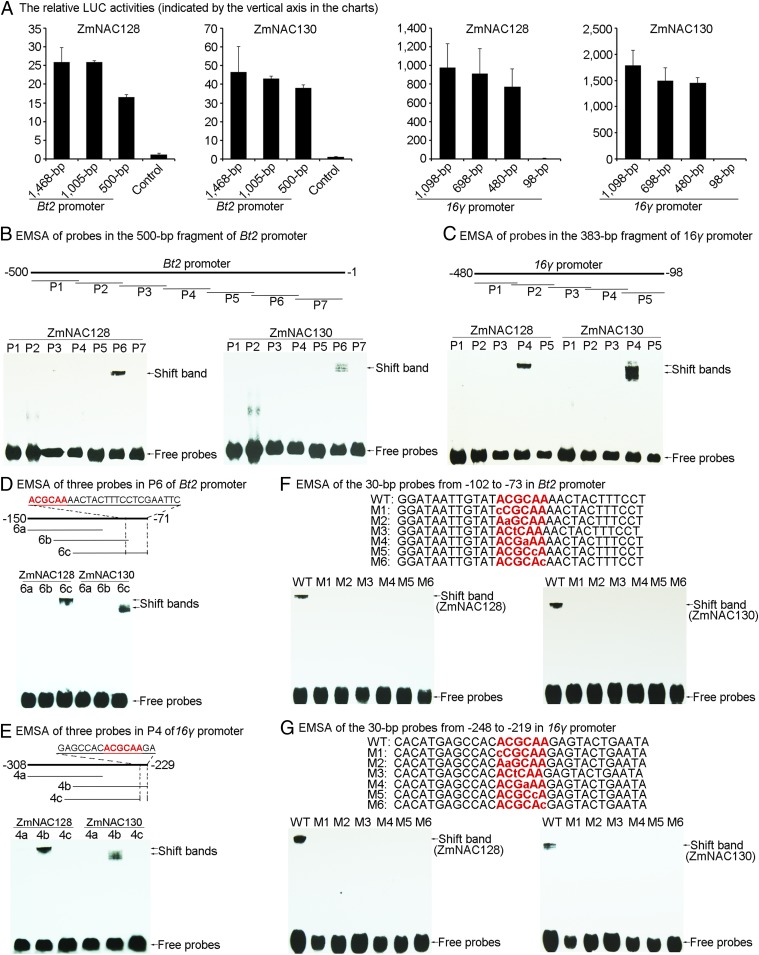
Fig. 4.
Identification of the cis-element binding site of ZmNAC128 and ZmNAC130. (A) Transactivation of different truncated promoters of Bt2 and 16-kDa γ-zein (16γ) by ZmNAC128 and ZmNAC130. The length of all promoters is calculated from the first nucleotide of the start codon (ATG). In the transactivation assay of Bt2 promoter, Control represents a 496-bp sequence downstream from the start codon. The relative LUC activity is the LUC-to-REM ratio, which is measured by using the same assay described in SI Appendix, Fig. S11. The promoter sequences of Bt2 and 16-kDa γ-zein are in SI Appendix, Fig. S12, and all of the probe sequences are in Dataset S2. (B) EMSA of five probes in the 500-bp promoter region of Bt2 from −500 to −1 with ZmNAC128 and ZmNAC130. (C) EMSA of five probes in the 383-bp promoter region of 16γ from −480 to −98 with ZmNAC128 and ZmNAC130. (D) EMSA of three probes truncated from P6 in the Bt2 promoter with ZmNAC128 and ZmNAC130. (E) EMSA of three probes truncated from P4 in the 16γ promoter with ZmNAC128 and ZmNAC130. (F) EMSA of ZmNAC128 and ZmNAC130 with a series of 30-bp probes of the Bt2 promoter from −102 to −73. (G) EMSA of ZmNAC128 and ZmNAC130 with a series of 30-bp probes of the 16γ promoter from −248 to −219. In F and G, The 6-bp box of ACGCAA center in the 30-bp probe (WT) and a series of mutant probes are produced by 1-bp mutations in the 6-bp box. The 6-bp consensus sequence (ACGCAA) in the promoters of Bt2 and 16-kDa γ-zein is highlighted in the red and bold format.
For the Bt2 promoter, four fragments upstream from the start codon different in length were used. The result showed that, when the Bt2 promoter was shortened to a 500-bp promoter fragment, neither ZmNAC128 or ZmNAC130 could transactivate LUC activity, compared with strong transactivation activities of 1,468-bp-, 1,005-bp-, and 500-bp-length promoter fragments, placing the cis-element in the Bt2 promoter in the 500-bp region from −500 to −1 upstream from the start codon (Fig. 4A). The DLR assay was also performed to shorten the target region of the 16-kDa γ-zein promoter. The cis-element in the 16-kDa γ-zein promoter was placed within the 383-bp region from −480 to −98 upstream from the start codon (Fig. 4A).
Then, the 500-bp promoter of Bt2 was divided into seven 80-bp fragments (Fig. 4B). After labeling the 3′ end of the seven fragments using biotin, we performed EMSA with purified recombinant His-ZmNAC128 and His-ZmNAC130. The shifted bands in the lane of the sixth probe (P6) indicated that ZmNAC128 and ZmNAC130 bound P6 to form retarded bands in the gel (Fig. 4B). The 383-bp promoter fragment of the 16-kDa γ-zein gene was divided into five 80-bp probes. ZmNAC128 and ZmNAC130 retarded the fourth probe (P4) as shown by band shifts (Fig. 4C), locating the binding sites within the 80-bp fragments of the Bt2 and 16-kDa γ-zein promoter, respectively.
Furthermore, the 80-bp fragments of Bt2 and 16-kDa γ-zein promoter were further divided into the three probes, respectively. The EMSA results of the three probes in the P6 of Bt2 promoter showed that only P6c was retarded by ZmNAC128 and ZmNAC130 (Fig. 4D). The cis-element of ZmNAC128 and ZmNAC130 was finally located within a 15-bp sequence in the Bt2 promoter (Fig. 4D). The P4b of 16-kDa γ-zein promoter was retarded by ZmNAC128 and ZmNAC130, narrowing down the cis-element into a 25-bp sequence in the 16-kDa γ-zein promoter (Fig. 4E). Common to both is a 6-bp consensus sequence of ACGCAA.
To validate the consensus sequence ACGCAA in the two promoters as the cis-element of ZmNAC128 and ZmNAC130, we subjected ACGCAA to point mutation analysis (Fig. 4 F and G). Every point mutation of ACGCAA abolished the binding interaction with ZmNAC128 and ZmNAC130, confirming the 6-bp sequence of ACGCAA as the cis-element binding site. Having the same cis-element binding site also explained the redundancy of ZmNAC128 and ZmNAC130.
Metabolism of Carbohydrates and Amino Acids in the Presence of nacRNAi.
To investigate which other genes might be affected in the absence of the two NAC TFs, we performed transcriptomic analysis of 20-DAP nacRNAi endosperm as described in Materials and Methods (26). The principal component analysis showed that the three biological replicates of nacRNAi and NT were clustered into one group (SI Appendix, Fig. S13A). Furthermore, 2,138 up-regulated and 2,218 down-regulated genes were identified by a threshold of P value < 0.05 and twofold change (SI Appendix, Fig. S13B and Dataset S1). Furthermore, the 2,218 down-regulated genes were subjected to the enrichment analysis of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. The result showed that the enriched pathways center on the metabolisms of carbohydrates and amino acids (SI Appendix, Fig. S13C). Three or four carbohydrate metabolic pathways were also significantly down-regulated in the presence of nacRNAi, including starch and sucrose metabolism, glycolysis, gluconeogenesis, fructose, and mannose metabolism, which correlated with the reduction of SG size and starch content. Biosynthesis of alanine, tryptophan, arginine, proline, histidine, phenylalanine, tyrosine, valine, leucine, isoleucine, lysine, and asparagine was significantly down-regulated in developing nacRNAi endosperm, which interferes with protein synthesis in developing endosperm. Such a broad selection of amino acids explains also the reduction of both zein and nonzein proteins in mature nacRNAi kernels compared with NT.
Discussion
Coordinated Regulation of Zein and Starch Biosynthetic Genes.
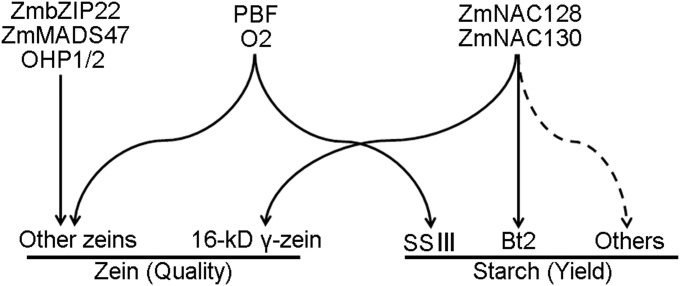
Maize endosperm accumulates starch and proteins in a spatiotemporal pattern, requiring coordinated regulation of gene expression. Previously, it has been shown that mutations in the starch biosynthetic pathway could also affect transcription levels of zein genes (18). A mutation of bt2 increased transcripts of α-zein and starch biosynthetic genes (Sh1, Sh2, etc.) in the developing endosperm. Here, we could show a direct regulatory effect through transcriptional activation by two specific TFs (Fig. 5). Moreover, reduction of Bt2 transcription in the nacRNAi lines did not increase transcripts of zein genes (SI Appendix, Fig. S10). On the contrary, the expression of most zein genes and multiple starch biosynthetic genes was significantly down-regulated in nacRNAi compared with NT. Failure of expressing the two endosperm-specific NAC TFs had no compensatory effect, as shown with reduction of zeins by RNAi in Illinois-High-Protein maize (27). Given that ZmNAC128 and ZmNAC130 transactivate the Bt2 and 16-kDa γ-zein genes directly via a common DNA binding site (ACGCAA), we now could also ask whether such a binding site existed in promoters of other target genes, whose transcript levels were reduced in nacRNAi. Indeed, of 15 starch biosynthetic genes besides Bt2 (SI Appendix, Fig. S10), six other genes contained such a conserved binding site within their 1-kbp-length promoter regions. The six potential target genes include Zpu1, GBSS1, Sh2, SS5 encoding starch synthesis 5, ISA2 encoding isoamylase-type starch-debranching enzyme 2, and SS2a encoding starch synthase 2a. Interestingly, the cis-element ACGCAA also exists within 500 bp of the promoters of the 50-kDa γ-zein and 15-kDa β-zein genes. Therefore, the lower reduction of transcript levels of other genes than the Bt2 and 16-kDa γ-zein genes in nacRNAi could be due to the mosaic structure of promoters containing multiple cis-acting enhancer sequences, requiring other TFs for full transcriptional activation.
Fig. 5.
A proposed model for ZmNAC128 and ZmNAC130 regulating the synthesis of zeins and starch.
Syntenic alignments between cereal genomes has shown that TFs like O2 have been conserved throughout cereal evolution (28). Indeed, rice OsbZIP58 like maize O2 controls rice storage accumulation by regulating the expression of starch biosynthetic and storage-protein genes (29). Analysis of protein sequences of ZmNAC128 and ZmNAC130 could also identify two related NAC TFs in rice, Os01g01470 and Os01g29840, which have 85% similarity. Syntenic analysis of chromosomal regions of rice and the homeologous regions in maize showed that the four NAC TFs are collinear (SI Appendix, Fig. S1). Moreover, the rice spatiotemporal transcriptome atlas showed that Os01g01470 and Os01g29840 are specifically and strongly expressed in rice developing seeds (30). Due to the conservation of regulatory mechanisms through the evolution of the grasses, each species can serve as a reference for the others (31).
The 16-kDa γ-Zein as a Regulator of the PB Initiation.
In developing endosperm cells, the four types of α-, β-, γ-, and δ-zeins are deposited within the endoplasmic reticulum lumen to form the orderly spherical accretions called PBs. RNAi against different zeins indicate that 27-kDa γ-zein RNAi has 60% reduction of PB number and is not involved in protein body filling (32). Combinations of different zein RNAi lines (without 27-kDa γ-zein RNAi) have a weak effect on PB initiation except that the combination of 27-kDa γ-zein RNAi with α-zeinRNAi;50-kDa γ-zeinRNAi;15-kDa δ-zeinRNAi has a 30% reduction in PB number (32). Therefore, 27-kDa γ-zein plays an important role in PB initiation. There are the three (50-, 27-, 16-kDa) γ-zein genes in maize. Deletion of 27- and 50-kDa γ-zein genes still allowed 10% of PBs to be formed compared with wild type (33), suggesting that additional proteins could be involved in the initiation of PBs. Here, we found that nacRNAi dramatically reduced the accumulation of 16-kDa γ-zein and PB number in the developing endosperm cells of nacRNAi, but the PB size and shape apparently remained unchanged. Because 16-kDa γ-zein gene was likely the result from unequal crossing-over with loss of the 50- and 27-kDa γ-zein genes after allotetraploidization (3), its function was probably similar in PB formation. For instance, maize Mucronate mutation forms a misfolded 16-kDa γ-zein that leads to irregular PB formation, but it could be rescued to restore normal PB by silencing the expression of the mutant 16-kDa γ-zein gene (34, 35). Although 16-kDa γ-zein lost a large part of its N-terminal domain compared with 27-kDa γ-zein that was able to promote PB formation (36, 37), a recent study showed that 16-kDa γ-zein acquired a new function in PB assembly (38). Furthermore, the 16-kDa γ-zein promoter lost all known conserved cis-elements present in other zein promoters (5) but acquired the cis-element ACGCAA, recognized by the corresponding NAC TFs, thereby restoring its endosperm-specific expression and synchronization with starch storage.
Functional Redundancy of ZmNAC128 and ZmNAC130.
The in vivo DLR assay has been used to determine whether two TFs have a cooperative effect, requiring physical interaction between them (13, 19, 39). For example, interaction between O2 and PBF on the expression of zein, PPDK, and SSIII genes can be illustrated with DLR assays (19, 39). However, ZmNAC128 or ZmNAC130 alone can already transactivate the Bt2 and 16-kDa γ-zein promoters to full strength (SI Appendix, Fig. S11). We further performed yeast-two-hybrid (Y2H) and bimolecular fluorescence complementation (BiFC) assays to determine whether the two NAC TFs can interact with each other. Because their C-terminals have transactivation activity, the Y2H assay used their NAC domains linked in the vector of BD to test the interactions with their full-length, N- or C-terminal regions linked in the vector of AD (SI Appendix, Fig. S14). The Y2H results showed that there was no interaction. Similar to the Y2H results, the BiFC assay only detected weak interaction between NLUC-NAC128 and CLUC-NAC128 (SI Appendix, Fig. S15). Therefore, ZmNAC128 and ZmNAC130 are functionally redundant. This finding is also consistent with the gene balance hypothesis. In contrast to a previously described cochaperone, there was no selection against the duplication after allotetraploidization because of the absence of protein–protein interaction (40). Furthermore, the two homeologous regions in the maize genome exhibited a large expansion on chromosome 3 or contraction on chromosome 8, including a segmental inversion between the NAC118/NAC130 and its flanking genes, a pattern previously described for the diploidization of the maize genome (41). This process might have also prevented the loss of the duplicated copy. Interestingly, in rice, a segmental duplication achieved the same purpose of having two rice NAC TFs (SI Appendix, Fig. S1), indicating some selective pressure on redundancy.
Materials and Methods
Genetic materials and molecular procedures are described in SI Appendix, SI Materials and Methods. The correct splicing information for Bt2 is based on cDNA analysis (42).
Supplementary Material
Acknowledgments
We thank Janine R. Shaw (University of Florida) for drawing our attention to the cDNA data of the Bt2 gene. This research was supported by the Selman A. Waksman Chair in Molecular Genetics (to J.M.).
Footnotes
The authors declare no conflict of interest.
Data deposition: RNA-Seq data reported in this paper have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE127525).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1904995116/-/DCSupplemental.
References
- 1.Larkins B. A., Wu Y., Song R., Messing J., “Maize seed storage proteins” in Maize Kernel Development, B. A. Larkins, Ed. (CABI, Boston, MA, 2017), pp. 175–189.
- 2.Hannah L. C., Boehlein S., “Starch biosynthesis in maize endosperm” in Maize Kernel Development, B. A. Larkins, Ed. (CABI, Boston, MA, 2017), pp. 149–159.
- 3.Xu J. H., Messing J., Organization of the prolamin gene family provides insight into the evolution of the maize genome and gene duplications in grass species. Proc. Natl. Acad. Sci. U.S.A. 105, 14330–14335 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong J., et al. , Analysis of tandem gene copies in maize chromosomal regions reconstructed from long sequence reads. Proc. Natl. Acad. Sci. U.S.A. 113, 7949–7956 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu Y., Messing J., Rapid divergence of prolamin gene promoters of maize after gene amplification and dispersal. Genetics 192, 507–519 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmidt R. J., Burr F. A., Burr B., Transposon tagging and molecular analysis of the maize regulatory locus Opaque-2. Science 238, 960–963 (1987). [DOI] [PubMed] [Google Scholar]
- 7.Schmidt R. J., Ketudat M., Aukerman M. J., Hoschek G., Opaque-2 is a transcriptional activator that recognizes a specific target site in 22-kD zein genes. Plant Cell 4, 689–700 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ueda T., et al. , Mutations of the 22- and 27-kD zein promoters affect transactivation by the Opaque-2 protein. Plant Cell 4, 701–709 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li C., et al. , Genome-wide characterization of cis-acting DNA targets reveals the transcriptional regulatory framework of Opaque2 in maize. Plant Cell 27, 532–545 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhan J., et al. , Opaque-2 regulates a complex gene network associated with cell differentiation and storage functions of maize endosperm. Plant Cell 30, 2425–2446 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vicente-Carbajosa J., Moose S. P., Parsons R. L., Schmidt R. J., A maize zinc-finger protein binds the prolamin box in zein gene promoters and interacts with the basic leucine zipper transcriptional activator Opaque2. Proc. Natl. Acad. Sci. U.S.A. 94, 7685–7690 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Z., Yang J., Wu Y., Transcriptional regulation of zein gene expression in maize through the additive and synergistic action of Opaque2, prolamine-box binding factor, and O2 heterodimerizing proteins. Plant Cell 27, 1162–1172 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li C., Yue Y., Chen H., Qi W., Song R., The ZmbZIP22 transcription factor regulates 27-kD γ-zein gene transcription during maize endosperm development. Plant Cell 30, 2402–2424 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiao Z., et al. , ZmMADS47 regulates zein gene transcription through interaction with Opaque2. PLoS Genet. 12, e1005991 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai J., Messing J., Increasing maize seed methionine by mRNA stability. Plant J. 30, 395–402 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Chen J., et al. , Dynamic transcriptome landscape of maize embryo and endosperm development. Plant Physiol. 166, 252–264 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keeling P. L., Myers A. M., Biochemistry and genetics of starch synthesis. Annu. Rev. Food Sci. Technol. 1, 271–303 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Giroux M. J., Boyer C., Feix G., Hannah L. C., Coordinated transcriptional regulation of storage product genes in the maize endosperm. Plant Physiol. 106, 713–722 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Z., Zheng X., Yang J., Messing J., Wu Y., Maize endosperm-specific transcription factors O2 and PBF network the regulation of protein and starch synthesis. Proc. Natl. Acad. Sci. U.S.A. 113, 10842–10847 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang H., et al. , Sucrose and ABA regulate starch biosynthesis in maize through a novel transcription factor, ZmEREB156. Sci. Rep. 6, 27590 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J., et al. , ZmbZIP91 regulates expression of starch synthesis-related genes by binding to ACTCAT elements in their promoters. J. Exp. Bot. 67, 1327–1338 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Zhang J., et al. , Novel role of ZmaNAC36 in co-expression of starch synthetic genes in maize endosperm. Plant Mol. Biol. 84, 359–369 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Sun C., et al. , A novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. Plant Cell 15, 2076–2092 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Segal G., Song R., Messing J., A new opaque variant of maize by a single dominant RNA-interference-inducing transgene. Genetics 165, 387–397 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li G., et al. , Temporal patterns of gene expression in developing maize endosperm identified through transcriptome sequencing. Proc. Natl. Acad. Sci. U.S.A. 111, 7582–7587 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Z., Messing J., Transcriptome and differential gene expression analysis for nacRNAi and non-transgenic sibling. The National Center for Biotechnology Information Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/submission/update/?acc=GSE127525. Deposited 28 February 2019.
- 27.Wu Y., Messing J., RNA interference can rebalance the nitrogen sink of maize seeds without losing hard endosperm. PLoS One 7, e32850 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu J. H., Messing J., Diverged copies of the seed regulatory Opaque-2 gene by a segmental duplication in the progenitor genome of rice, sorghum, and maize. Mol. Plant 1, 760–769 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Wang J. C., Xu H., Zhu Y., Liu Q. Q., Cai X. L., OsbZIP58, a basic leucine zipper transcription factor, regulates starch biosynthesis in rice endosperm. J. Exp. Bot. 64, 3453–3466 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawahara Y., et al. , Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6, 4 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garcia N., Zhang W., Wu Y., Messing J., Evolution of gene expression after gene amplification. Genome Biol. Evol. 7, 1303–1312 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo X., et al. , Nonredundant function of zeins and their correct stoichiometric ratio drive protein body formation in maize endosperm. Plant Physiol. 162, 1359–1369 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuan L., Dou Y., Kianian S. F., Zhang C., Holding D. R., Deletion mutagenesis identifies a haploinsufficient role for γ-zein in Opaque2 endosperm modification. Plant Physiol. 164, 119–130 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu Y., Messing J., Rescue of a dominant mutant with RNA interference. Genetics 186, 1493–1496 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim C. S., et al. , The maize Mucronate mutation is a deletion in the 16-kDa gamma-zein gene that induces the unfolded protein response. Plant J. 48, 440–451 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Prat S., Pérez-Grau L., Puigdomènech P., Multiple variability in the sequence of a family of maize endosperm proteins. Gene 52, 41–49 (1987). [DOI] [PubMed] [Google Scholar]
- 37.Mainieri D., et al. , Protein body formation in the endoplasmic reticulum as an evolution of storage protein sorting to vacuoles: Insights from maize γ-zein. Front. Plant Sci. 5, 331 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mainieri D., et al. , Maize 16-kD γ-zein forms very unusual disulfide-bonded polymers in the endoplasmic reticulum: Implications for prolamin evolution. J. Exp. Bot. 69, 5013–5027 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang J., Ji C., Wu Y., Divergent transactivation of maize storage protein zein genes by the transcription factors Opaque2 and OHPs. Genetics 204, 581–591 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garcia N., Messing J., TTT and PIKK complex genes reverted to single copy following polyploidization and retain function despite massive retrotransposition in maize. Front. Plant Sci. 8, 1723 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bruggmann R., et al. , Uneven chromosome contraction and expansion in the maize genome. Genome Res. 16, 1241–1251 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hannah L. C., et al. , Maize genes encoding the small subunit of ADP-glucose pyrophosphorylase. Plant Physiol. 127, 173–183 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.