Significance
Polo-like kinase 4 (PLK4), a known cell cycle regulatory protein, localizes to the centrosome during S-phase and facilitates centrosome duplication. We show that phospho-serine305-PLK4 is subsequently redistributed to kinetochores (metaphase and anaphase), midzone during cleavage furrow formation (anaphase and telophase), and midbody (cytokinesis) during the cell cycle, and is required for cell division. Inhibition of PLK4 kinase activity prevents cytokinesis, leading to polyploidy and vulnerability to anaphase or mitotic catastrophe. The role of PLK4 in cytokinesis is consistent with its role as a potential target for cancer therapy.
Keywords: polo-like kinase 4, cell cycle regulation, cytokinesis, centrosome, midbody
Abstract
The mitotic protein polo-like kinase 4 (PLK4) plays a critical role in centrosome duplication for cell division. By using immunofluorescence, we confirm that PLK4 is localized to centrosomes. In addition, we find that phospho-PLK4 (pPLK4) is cleaved and distributed to kinetochores (metaphase and anaphase), spindle midzone/cleavage furrow (anaphase and telophase), and midbody (cytokinesis) during cell division in immortalized epithelial cells as well as breast, ovarian, and colorectal cancer cells. The distribution of pPLK4 midzone/cleavage furrow and midbody positions pPLK4 to play a functional role in cytokinesis. Indeed, we found that inhibition of PLK4 kinase activity with a small-molecule inhibitor, CFI-400945, prevents translocation to the spindle midzone/cleavage furrow and prevents cellular abscission, leading to the generation of cells with polyploidy, increased numbers of duplicated centrosomes, and vulnerability to anaphase or mitotic catastrophe. The regulatory role of PLK4 in cytokinesis makes it a potential target for therapeutic intervention in appropriately selected cancers.
The use of targeted cancer therapeutics that inhibit specific molecular alterations to which some cancers are “addicted” (1) has become a reality (2–4). Development of such therapies has led to substantial progress in cancer treatments for specific patient subgroups whose cancers contain these molecular alterations. However, the current spectrum of adult malignancies with novel therapeutic “targets” remains limited while the mainstay for cancer treatment continues to be cytotoxic chemotherapy. Most nonspecific chemotherapeutic agents are predominantly antimitotic drugs frequently prescribed in a “one-size-fits-all” fashion based on the histology and extent of disease dissemination. A great deal is now known about the network of mitotic serine/threonine kinases that regulate the process of cell division, and there are growing opportunities to bring more focused approaches to the disruption of cell division, particularly in aneuploid cancer cells. The requirements of segregating complex genomes of aneuploid malignant cells into viable daughter cells produces vulnerability that could be exploited to therapeutic advantage.
A number of mitotic kinases are critically important in regulating specific portions of the cell cycle. These include members of the polo-like kinase (PLK) family, the cyclin-dependent kinase (CDK) family, the aurora family, and the “never in mitosis A” (NIMA) family, as well as those kinases implicated in mitotic checkpoints (Bub1, BubR1, and TTK/Esk/Mps1), mitotic exit, and cytokinesis. Accumulating evidence suggests that PLK4 is not only an important cell-cycle regulatory protein but is also a potential therapeutic target. Expression-array analyses identified PLK4 as one of 16 serine-threonine kinases associated with poor prognosis in breast cancer patients (5). PLK4 was also identified as a therapeutic target for cancer by using high-throughput screens that involve genome-wide small inhibitory RNA (siRNA) viability analyses of multiple human cancer cell lines in which PLK4 depletion significantly decreased viability in 8 of 21 cancer cell lines tested and had only a limited effect on normal immortalized cell lines (6, 7). In addition, the Broad Institute DepMap Web site shows 497 of 517 cancer cell lines to be dependent on PLK4 by using CRISPR-Cas9 methods (https://depmap.org/portal/gene/PLK4?tab=overview) (8). Furthermore, PLK4 mRNA levels are increased in human tumor specimens relative to paired normal tissue, and these elevated levels correlate with poor survival (9). Finally, an inhibitor of PLK4 kinase activity, CFI-400945, has been shown to inhibit lung cancer growth in mice (9).
PLK4 functions at the intersection of mitotic and DNA damage pathways (10), and aberrant expression is associated with centriole increases and centrosome dysfunction (11, 12). This may affect control of cell division and play a role in genomic instability. An increase in centrosome number is frequently observed in aneuploid cancers, in which it is proposed to play a causal role in genome instability and tumor development as well as in tumor progression and aggressiveness (13). PLK4 is found on human chromosome 4q28, a region that frequently undergoes loss in hepatocellular carcinoma (14). Although full deletion of PLK4 leads to embryonic lethality (15), PLK4+/− heterozygous mice are viable but have ∼15 times greater probability of developing spontaneous liver and lung cancers than PLK4+/+ homozygous littermates (16).
Members of the PLK family (PLK1–5) have an amino-terminal serine/threonine kinase domain and two to three carboxyl-terminal polo-boxes (PBs) that interact with other proteins. PLK4 is unique among the PLK family and shares only limited homology with other family members (17, 18). Other family members have two PBs, whereas PLK4 has one C-terminal PB and two cryptic PBs (CPBs) that function as PBs without a linker region to facilitate PLK4 homodimerization and autophosphorylation (19). The PLK4 cryptic PBs bind the centrosome proteins (CEPs) CEP152 and CEP192 that direct intact PLK4 to this organelle. Other critical domains of PLK4 are composed of proline (P), aspartate (D), glutamate (E), serine (S), and threonine (T) amino acid residues with a uniquely abundant distribution (i.e., PEST domains) (20). These conserved PEST domains have been demonstrated to serve as constitutive or conditional proteolytic signaling sequences and function to regulate enzymatic protein cleavage activities (21). Currently, PLK4 is recognized as having an essential role in centriole duplication but not in regulating cytokinesis (7, 11, 17, 18, 20, 22–26).
Because expression array analyses identified PLK4 as a serine-threonine kinase associated with a poor prognosis in breast (5) and lung (9) cancer patients and because PLK4 was identified as a therapeutic target for cancer by using high-throughput screens (6, 7) coupled with the established role of PLK4 in centrosome duplication and, therefore, regulation of cell division, we investigated the role of PLK4 in cell cycle regulation and as a potential new target for therapy with an anti-PLK4 kinase inhibitor, CFI-400945 (27–29).
Results
Localization of Phospho-PLK4 to Centrosomes.
It is generally accepted that turnover of PLK4 kinase is strictly controlled to prevent aberrant centriole increase or “amplification” (30). This turnover is achieved by an autoregulatory mechanism in which PLK4 homodimers are autophosphorylated at residues in a downstream regulatory element located carboxyl-terminal to its catalytic kinase domain (amino acids 285 and 289). Phosphorylated PLK4 is reported to be recognized by the SCF (S phase kinase-associated protein 1–cullin–F-box) complex, ubiquitinated, and targeted to the proteasome for degradation (18, 22, 30). Although PLK4 is reported to be restricted to the centrosome (7, 17, 18, 22), the subcellular distribution of autophosphorylated PLK4 has not been characterized. We therefore developed a phosphospecific antibody to the serine 305 residue of PLK4, located within the first of three PLK4 PEST domains, to track the localization of this form of PLK4. Similar to published reports, antibodies recognizing the C-terminal PB domain were localized to centrosomes of human ovarian (Fig. 1), breast, and colorectal cancer cell lines throughout the cell cycle (7, 17, 18, 22, 30); however, the phospho-serine305-PLK4–specific antibody also localized to kinetochores (metaphase and anaphase), the central spindle midzone or the cleavage furrow (anaphase and telophase), and midbody (cytokinesis) depending on cell-cycle phase (Fig. 2). Similar results were obtained by using a second phospho-S305-PLK4 antibody (14299).
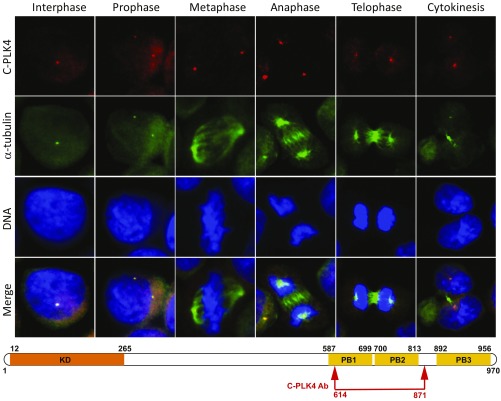
Fig. 1.
PLK4 is localized to centrosomes throughout S-phase, G2, and M-phase during the cell cycle when an antibody against C-terminal PB domains is used for immunofluorescence (IF). IF microscopy using anti-PLK4 rabbit polyclonal antibody (C-PLK4, ab137398; Abcam; to aa 614–871; red IF) and an anti–α-tubulin antibody (green IF) in synchronized cell lines permitted identification of PLK4 in centrosomes throughout the cell cycle (interphase, prophase, metaphase, anaphase, telophase, and cytokinesis). Results are illustrated for OVCAR3 ovarian cancer cells photographed at 400× original magnification. (Lower) Approximate location of the commercially available (C-PLK4) antibody used for IF above. Please note differences in PLK4 distribution compared with Fig. 2 based on phosphorylation status and epitopes recognized.
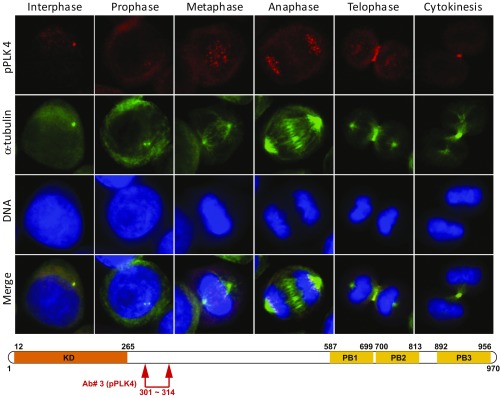
Fig. 2.
Localization of phospho-PLK4 to centrosomes, kinetochores, and midbody varies with the phase of the cell cycle when an antibody to the N-terminal PEST domain is used. Phospho-serine305-PLK4 is identified first in centrosomes of interphase and prophase cells followed by kinetochores (metaphase and anaphase) and, subsequently, the cleavage furrow (telophase) and midbody (cytokinesis) of synchronized cells by immunofluorescence (IF) microscopy using an anti–phospho-serine305-PLK4 rabbit polyclonal antibody (our Ab#3 to a PLK4-serine305 peptide; red IF). Anti–α-tubulin antibody (green IF) identifies centrosomes, spindle apparatus, and midbody. DAPI stain (blue) identifies DNA. Results are illustrated for OVCAR3 ovarian cancer cells photographed at 400× original magnification. (Lower) Schematic diagram illustrates approximate location (arrows) of our rabbit polyclonal antibody (Ab#3, anti-phospho-PLK4) for phospho-PLK4. Similar results were obtained with a second, independently produced anti–phospho-serine305-PLK4 antibody (14299) from the laboratory of G.P.N.
Cleavage of Phospho-PLK4 and Localization to Kinetochores, Midzone/Cleavage Furrow, and Midbody During M-Phase.
Western immunoblot analyses using antibodies to an epitope centered on cysteine 458 (as detailed later) showed that PLK4 displayed the expected electrophoretic mobility of a 97-kDa protein (Fig. 3 and SI Appendix, Fig. S1). In contrast, the major protein recognized by anti–phospho-PLK4 (serine305) displayed a mobility of a 50–60-kDa protein in breast, ovarian, and colorectal cancer cell lines, presumably because of protein cleavage rather than degradation of the entire PLK4 molecule (Fig. 3 and SI Appendix, Fig. S1).
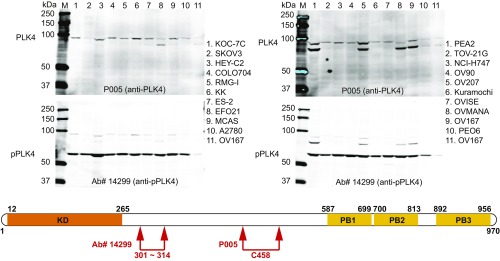
Fig. 3.
Western immunoblot of 19 different ovarian cell lines and one colorectal (NCI-H747) cancer cell line demonstrates full-length PLK4 (Upper Left and Upper Right) and phospho-PLK4 (Lower Left and Lower Right). (Upper) Western blots demonstrate full-length (97-kDa) PLK4 with a commercially available anti-PLK4 antibody (P005; Cell Signaling 3258, recognizing an epitope centered on the cysteine458 amino acid). (Lower) Western blots were performed by using the same filter after washing with buffer and reprobing with an anti–phospho-serine305-PLK4 antibody (Ab#14299). Please note that the phospho-PLK4 in the lower filter is of substantially smaller size, ∼60 kDa. The difference is appreciated as the full-length signals were not completely removed by stripping in the lower filters. Cultured cells were not synchronized with regard to cell cycle. We confirmed that the 60-kDa band corresponded to pPLK4 by removing this band from the gel and performing amino acid sequence analyses. We also confirmed that a second anti–phospho-PLK4 antibody (Ab#3) from our laboratory recognized phospho-PLK4 by using immunoprecipitation assays followed by PAGE with silver staining to identify the band recognized on Western blot and, finally, MS to assess the amino-acid sequence of protein band (amino-acid sequence provided in Results). SI Appendix, Fig. S1 shows results in 23 additional colorectal and 9 breast cancer cell lines. (Lower) Schematic illustration of PLK4 amino-acid sequence shows the relative locations of epitopes recognized by the two antibodies used for Western immunoblot analyses (Upper), one from the laboratory of G.P.N. recognizing phospho-serine305-PLK4, produced in collaboration with Cell Signaling Technology, and the second commercially available recognizing cysteine458-PLK4.
To confirm the specificity of our antibodies for two anti-PLK4 antibodies and our phospho-serine305-PLK4 antibody, we used various approaches. The first approach used competition studies with different PLK4 peptides or recombinant protein, as described in Materials and Methods, and these peptides succeeded in quenching the immunofluorescence staining observed with each antibody.
The second approach, used to confirm the specificity of a commercially available anti-PLK4 antibody recognizing an epitope centered on cysteine 458 and our anti–phospho-PLK4 Ab#3, involved siRNA directed to PLK4 in HCT-116 human colorectal cancer cells, OVCAR3 human ovarian cancer cells, and CAL51 human breast cancer cells to inhibit expression of PLK4 and demonstrate that localization with our anti–phospho-PLK4 antibody was specifically dependent on expression of PLK4. In each of these cell lines, we found a greater than 90% suppression of PLK4 mRNA, a greater than 90% suppression of full-length PLK4 (detected by a PLK4 antibody detecting a central PLK4 epitope surrounding cysteine 458), and a greater than 90% suppression of phospho-PLK4 detected with our anti–phospho-PLK4 antibody in the centrosomes or in the midbodies (SI Appendix, Fig. S2).
The third approach, also to confirm specificity of our anti–phospho-PLK4 antibody Ab#3, used lysates from OV90 ovarian cancer cells and NCI-H716 colorectal cancer cells for affinity chromatography with cross-linked anti–phospho-serine305-PLK4 antibody to separate antibody-bound and associated proteins from other proteins in the lysate. The proteins bound by the column were released and separately collected. The collected proteins were separated by polyacrylamide gel electrophoresis (PAGE) and identified by silver staining of the gel. One silver-stained band of ∼50–60 kDa, corresponding to a similarly sized band recognized in a parallel Western immunoblot, was detected. This band was excised for mass spectrometry (MS) amino acid sequence analyses. The sequence analyses yielded a single peptide sequence (LKMPHEKHYTLCGTPN) that corresponded uniquely to the primary amino acid sequence of PLK4 from residue 161 through 176, thus demonstrating specificity of our anti–phospho-serine305-PLK4 antibody for PLK4 protein and providing supportive evidence that the proteins detected by this antibody in centrosomes, kinetochores, cleavage furrow, and midbodies are phospho-PLK4.
The observed spatial-temporal differences in subcellular distribution of phospho-PLK4 have not been reported previously (10, 11, 22). To date, one other group has described PLK4 localization outside of centrosomes (at the central spindle) and provided evidence for a cytokinesis defect in PLK4−/− cells (14, 15, 31). This lack of detection of the noncentrosome localizations of PLK4 is likely caused at least in part by inherent difficulties in characterizing a low-abundance protein like PLK4, and by the frequent use of antibodies targeting the carboxyl terminus of PLK4 as well as exogenous PLK4 to overexpress the protein and overcome these challenges (11). To address these problems, we developed immunofluorescence techniques for localization of PLK4 by modifying a previously described method involving the use of sodium dodecyl sulfate (SDS) as an antigen retrieval technique (32). This permitted localization of endogenous PLK4 in frozen tissues, probably because of the denaturing effect of SDS on the secondary structure of PLK4 protein, or through heat with high pH (9.0) for localization in fixed cell lines and tissues. Our use of synchronized cells also proved to be a distinct advantage for characterization of a protein with apparently multiple functions in different subcellular locations during different phases of the cell cycle.
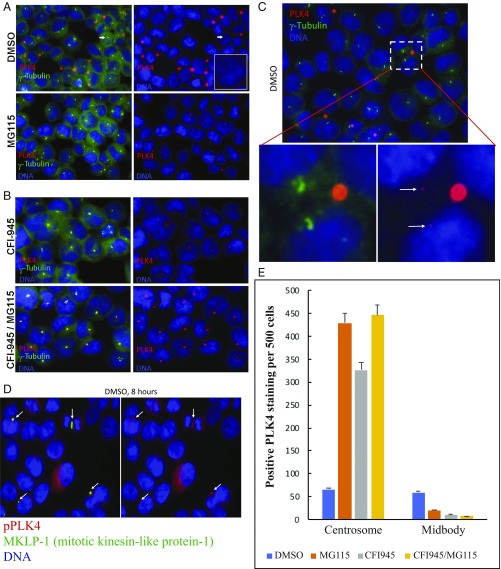
To address whether PLK4 cleavage is required for the noncentrosome localization of PLK4, we used the inhibitor MG115, a potent, reversible inhibitor of chymotrypsin-like and caspase-like activities, as well as calpains and various lysosomal cathepsins (33). Short-term treatment with MG115 caused accumulation of PLK4 in centrosomes and prevented localization of PLK4 to the midbody (Fig. 4 A and C). After a 4-h treatment with MG115, the number of cells with PLK4-positive centrosomes increased from 66 to 430 of 500 cells, a greater than sixfold increase (Fig. 4E). In addition, the intensity of each individual centrosome-related signal was substantially increased, facilitating identification (e.g., compare “DMSO” with “MG115” in Fig. 4 A and C). In contrast, the number of cells with PLK4-positive midbodies decreased from 59 to 20 of 500 cells, a >50% reduction after only a 4-h MG115 treatment, with substantially less PLK4 per labeled midbody (Fig. 4 A and E). These results strongly support the concept that activated/phosphorylated PLK4 is cleaved into smaller fragments, perhaps removing the PB domains that are known to bind the STIL, CEP152, and CEP192 proteins in centrosomes. This would facilitate redistribution of an amino-terminal fragment of PLK4 to the spindle midzone/cleavage furrow and midbody, a distribution we have demonstrated by colocalization of PLK4 with MKLP-1 (mitotic kinesin-like protein-1), a known central spindle/midbody protein (Fig. 4D).
Fig. 4.
Immunofluorescence localization of PLK4 to centrosomes, cleavage furrow, and midbody is altered compared with control (A, DMSO) by treatment with MG-115 protease inhibitor (A, MG115), CFI-400945 PLK4 kinase inhibitor (B, CFI-945), or both (B, CFI-945/MG115). Identification of PLK4 localization to centrosomes (A, DMSO, arrow) is improved by higher magnification (C) to illustrate size difference between centrosomes (arrows) and (substantially larger) midbodies. (D) PLK4 localization to midbodies is confirmed by colocalization with mitotic kinesin-like protein-1 (MKLP-1), a midbody protein. (E) Relative distribution of PLK4 to centrosomes and midbodies with DMSO, MG115, CFI-945, or MG115 and CFI945 treatment. Additional descriptive details are provided in the legend to SI Appendix, Fig. S4.
Transient Transfection with PLK4 siRNA and “Kinase-Dead” PLK4.
To confirm that PLK4 is required for duplication of centrosomes in our models (11, 30, 34), we performed experiments using siRNAs targeting PLK4 to down-regulate PLK4 proteins and a PLK4-specific small molecule inhibitor to inhibit its kinase activity. There was a >90% suppression of PLK4 mRNA following 24 h treatment with PLK4 siRNA, but no similar reduction following treatment with a scrambled control siRNA (SI Appendix, Figs. S2 and S3). In contrast, as expected, there was no reduction in PLK4 mRNA after 24 h of treatment with the PLK4 inhibitor CFI-400945 (SI Appendix, Fig. S3A). Although the majority of unsynchronized HCT-116 colorectal cancer cells treated with scrambled control siRNA contained two centrosomes, indicating that centrosome duplication occurred in the majority of cells during this time interval, those treated with PLK4 siRNA contained only one centrosome, confirming the dependency on PLK4 for centrosome duplication in these cells (SI Appendix, Fig. S3 B–D). Similar results were obtained with OVCAR3 human ovarian cancer cells and CAL51 human breast cancer cells (SI Appendix, Fig. S2). These experiments also confirmed the specificity of our anti–phospho-PLK4 antibody for protein product encoded by the PLK4 mRNA, as described earlier (SI Appendix, Fig. S2 C vs. D and E vs. F).
Transient transfection of HCT-116 cells with a kinase-dead (KD) PLK4 cDNA (PLK4-K41M) sequence or a green fluorescence protein (GFP)-tagged wild-type PLK4 cDNA resulted in production of tumor cells that showed centrosome amplification with both transfectants (SI Appendix, Fig. S4). However, transfectants overexpressing the PLK4-KD had predominantly multiple nuclei or multilobed nuclei, whereas wild-type PLK4 transfectants contained only single nuclei in tumor cells (SI Appendix, Fig. S4). The presence of multiple nuclei and multilobed nuclei in tumor cells transfected with KD PLK4 is consistent with a role for PLK4 kinase activity in completing cytokinesis.
Inhibition of PLK4 Kinase Activity with CFI-400945.
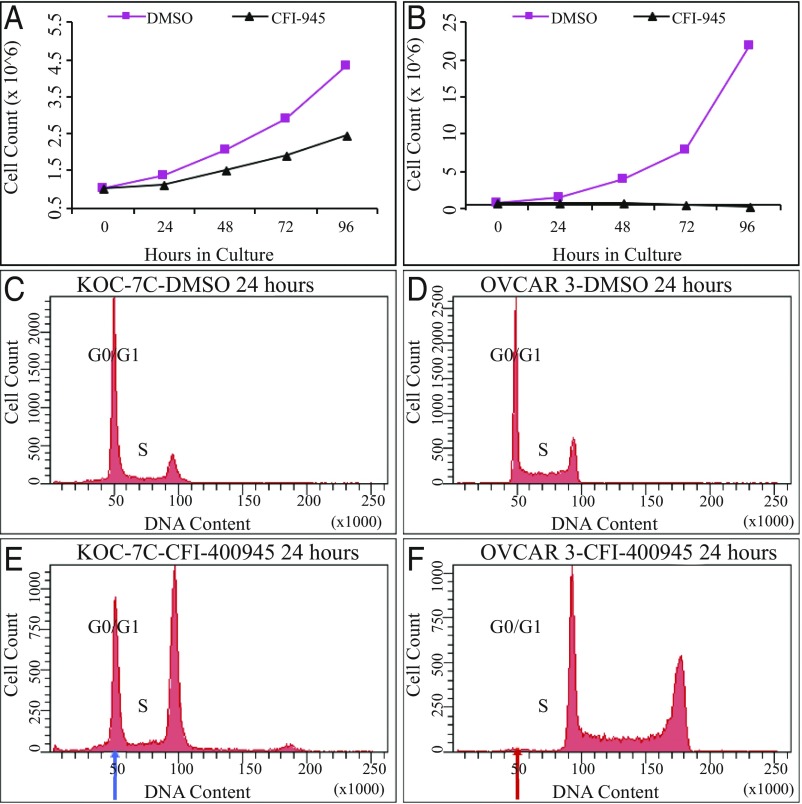
To address whether PLK4 kinase activity is required for completion of cytokinesis, we used CFI-400945, a small-molecule inhibitor of PLK4 kinase activity. A series of small-molecule PLK4 kinase inhibitors have been developed and characterized during validation studies as clinical therapeutic candidates. One of these, CFI-400945 (27–29), is a potent and selective inhibitor of PLK4 (Ki = 0.26 nM, IC50 = 2.8 nM) relative to other PLK family members (PLK1, PLK2, and PLK3, IC50 > 50 μM). Treatment of breast, ovarian, and colorectal cancer cell lines with CFI-400945 caused a reduction in the number of PLK4 midbody-positive tumor cells (Fig. 4 and SI Appendix, Fig. S5), inhibition of proliferative activity (Fig. 5), and acquisition of polyploid nuclei (Fig. 5 E and F and SI Appendix, Fig. S5 B, D, and F). The degree of proliferative suppression in cell culture (Fig. 5 A and B) was proportional to acquisition of polyploid nuclei by flow cytometric analysis (Fig. 5 E and F). For example, 21 different ovarian cancer cell lines with substantial suppression of cell proliferation (Fig. 5B vs. Fig. 5A), as well as 24 human colorectal cancer cell lines and 32 human breast cancer cell lines, also had the initial G0/G1 modal DNA peak shifted almost completely from the baseline G0/G1 DNA content (Fig. 5D) to the position of the original G2/M DNA peak after 24 h treatment (Fig. 5F), demonstrating a cause-and-effect relationship. The extent to which CFI-400945 treatment inhibited cell proliferation was variable within the cell line panel. Cell lines with only partial suppression of proliferative activity (Fig. 5A) showed an incomplete shift in the modal DNA index from the G0/G1 DNA content to the G2/M (double) DNA content (Fig. 5 C and E). These data are consistent with an inhibitory effect of CFI-400945 on cytokinesis following DNA synthesis that permits a doubling of the DNA content of individual tumor cells without cellular abscission into two daughter cells.
Fig. 5.
Ovarian cancer cell growth in culture with and without CFI-400945 PLK4 inhibitor: KOC-7C compared with OVCAR3. (A) Cell proliferation for KOC-7C ovarian cancer cells without (DMSO) and with CFI-400945 PLK4 inhibitor (CFI-945) showing modest reduction in cell proliferation. N at hour 0 = 1 × 106 cells. (B) Cell proliferation of OVCAR3 ovarian cancer cells without (DMSO) and with CFI-400945 PLK4 inhibitor (CFI-945) showing substantial inhibition of cell proliferation. N at hour 0 = 1 × 106 cells. (C) Flow cytometry of control KOC-7C ovarian cancer cells after 24 h in culture (DMSO). (D) Flow cytometry of control OVCAR3 ovarian cancer cells after 24 h in culture (DMSO). (E) Flow cytometry of CFI-400945 inhibitor (50 nM)-treated OVCAR3 ovarian carcinoma cells after 24 h, with large residual population of cells retaining baseline modal DNA index (blue arrow), with only partial response to inhibitor. (F) Flow cytometry of CFI-400945 inhibitor (50 nM)-treated OVCAR3 ovarian cancer cells demonstrating no residual population of cells retaining the baseline modal DNA index (red arrow), indicating a complete response to inhibitor. Of note, the differential response observed here is not the result of variations in the population doubling time of the cell lines (KOC-7C = 21.00 h, OVCAR-3 = 19.49 h). Movies S1 and S2 show videomicroscopy of OVCAR3 cells with and without PLK4 CFI-400945 inhibitor.
To confirm this possibility through direct observation, we used time-lapse videomicroscopy of CFI-400945 inhibitor-treated compared with control cells. We demonstrated that PLK4 inhibitor-treated cells were able to round up from the culture dish surface, initiate, and complete M-phase with acquisition of two nuclei or one much enlarged nucleus; however, PLK4 inhibitor-treated cells were not able to undergo cellular abscission and therefore could not separate into daughter cells (Movies S1–S6). Among PLK4 inhibitor-treated cells, this led to the accumulation of large cells containing multiple nuclei or exceptionally large nuclei (Movies S2, S4, and S6). These morphological changes contrasted with the appearance of untreated control cells, which rapidly rounded up from the culture surface and dissolved the nuclear envelope, with chromosomal condensation followed by separation of chromosomes with formation of two separate nuclei and abscission of the cell into two daughter cells (Movies S1, S3, and S5). In untreated control cells, this resulted in an overall increased number of cells in the flask, whereas PLK4 inhibitor-treated cells did not significantly increase in number and individual cells appeared substantially larger, with each cell having two individual nuclei or significantly enlarged nuclei (Movies S1–S6).
Discussion
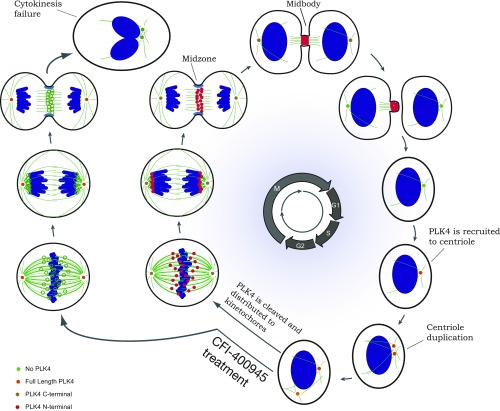
Our results suggest that PLK4, like PLK1, is involved in the precise spatial-temporal control of cell division with sequential localization of the protein to centrosomes, kinetochore, and cleavage furrow/midbody facilitating microtubule organization at centrosomes and cellular abscission at the midbody (Fig. 6). This differs from the predominant current model that has the role of PLK4 restricted to centrosome duplication (7, 11, 17–19, 22, 24–26) and phosphorylation of selected proteins, such as Cdc25C (35). Interestingly, many of the genes related to the chromosome passenger complex (CPC) are not only localized in some of the same subcellular organelles (kinetochore, cleavage furrow, and midbody) during M-phase (36), but are also coexpressed with PLK4, as are multiple members of the anaphase promoting complex/cyclosome (APC/C). Although the ubiquitin–proteasome pathway is the major proteolytic system in mammalian cells, MG115 also inhibits protease activity in other cellular organelles in addition to proteasomes (33). Therefore, we can only speculate about the cellular site of proteolytic cleavage for PLK4, although we believe PLK4 cleavage associated with redistribution of phospho-PLK4 during M-phase to kinetochores, midzone/cleavage furrow, and midbody is not likely to be associated with the proteasome.
Fig. 6.
Schematic diagram illustrates PLK4 distribution and functions throughout the cell cycle. The cellular images to the right illustrate changes associated with progression through the cell cycle beginning with interphase/G1 at the extreme right. During the transition from G1 to S-phase, PLK4 is localized to the centrosome. Subsequently, PLK4 expression, but not kinase activity, is required for centrosome duplication. PLK4 is localized to both centrosomes. Throughout M-phase, antibodies to C-terminal PLK4 sites localize the protein continuously to both centrosomes. However, antibodies to phospho-serine305-PLK4 localize N-terminal PLK4 first to the kinetochore (metaphase and anaphase), then to the central spindle of the cleavage furrow (telophase), and finally to the midbody (cytokinesis), where PLK4 kinase activity is required for cellular abscission at the conclusion of M-phase. CFI-400945 treatment: the cellular images on the left side of the schematic drawing show the changes related to treatment with the PLK4 kinase inhibitor CFI-400945, which prevents phosphorylation of PLK4. As a result, movement of PLK4 to the kinetochores, cleavage furrow, and midbody is inhibited; cellular abscission is prevented; and daughter cells are not able to separate from one another, forming larger cells with polyploid nuclei or double nuclei. Adapted by permission of ref. 18, Springer Nature: Nature Reviews Molecular Cell Biology, copyright 2014; and by permission of ref. 49, Springer Nature: Nature Cell Biology, copyright 2012.
A role in regulation of cytokinesis positions phospho-PLK4 as a potential therapeutic target for cancer therapy. Treatment with the CFI-400945 PLK4 inhibitor was associated with a near-complete loss of PLK4-positive midbodies, suggesting the requirement for PLK4 phosphorylation and PLK4 cleavage to facilitate migration to the cleavage furrow and midbody. Indeed, midbodies were markedly reduced or absent in CFI-400945–treated cells, suggesting that PLK4 plays an important role in midbody formation and function (Fig. 6). Although centrosome duplication is an important PLK4 function, aberrant centrosome duplication caused by PLK4 alterations are unusual. Based on our findings, we consider PLK4’s role in cytokinesis to be the primary functional activity that makes PLK4 a potential target for cancer therapeutics.
Off-target effects of the CFI-400945 PLK4 inhibitor, including those on aurora kinase B (37), are not likely to account for the cellular changes we observe. The biochemical 50% inhibitory concentration (i.e., IC50) of CFI-400945 for PLK4 is 2.8 nM, whereas, for AURKB, it is 98 nM, a 35-fold differential (38). In addition, use of known AURKB inhibitors, such as AZD1152, in our laboratory demonstrates divergent cellular changes when tested in human colorectal cancer cell lines, including differential effects on cell proliferation, divergent effects in washout assays (39), and divergent effects in outgrowth assays (39).
Interestingly, the CFI-400945 kinase inhibitor interfered with cytokinesis in a fashion that varied among the cell lines evaluated. Those cell lines that sustained a variable amount of proliferative activity despite CFI-400945 treatment had a subpopulation of cells that maintained the original modal (G0/G1) DNA index to a variable degree. These “resistant” cell lines may represent a subpopulation of cancer cells that function as a cancer stem cell subpopulation or a subpopulation of cancer cells with a functional alternative pathway to facilitate cytokinesis. These studies in multiple cell lines will be described in a subsequent report.
Cells can undergo diverse fates according to their status at anaphase (40). Proper segregation of chromosomes in mitosis leads to generation of two genetically identical daughter cells. Alternatively, gradual degradation of cyclin-B in the presence of prolonged spindle checkpoint activation causes cells to exit mitosis without dividing chromosomes in anaphase, a phenomenon termed cell “slippage” (40). Cells that exit mitosis via slippage enter G1 as tetraploid cells and may continue to cycle, senesce, or undergo apoptosis. Finally, anaphase catastrophe occurs when a cell with multiple centrosomes fails to coalesce the centrosomes into two spindle poles and enters anaphase with a multipolar spindle. Segregation of chromosomes to more than two daughter cells characteristically leads to cell death. Alternatively, mitotic catastrophe relates to cell death following defective cell-cycle checkpoints and the generation of aneuploid (or polyploid) cells that are not able to successfully sort the complex genome into daughter cells and undergo cell death.
“Cell fate decisions” are typically made during the process of cell division. It is hypothesized that a tumor cell reaches a critical point at which it must “decide” between continued cell division, apoptosis, or the capacity for stem-cell renewal. Our findings suggest that PLK4 inhibition prevents cell proliferation and/or induces cell death in bulk tumor cells and cancer stem cells. A rational therapeutic approach can be based on the idea that inhibition of PLK4 diminishes the potential for mitotic coordination and regulation in the context of aneuploidy. We hypothesize that, when centrosome regulation and/or the ability to complete cytokinesis is inhibited, the balance will be tilted toward anaphase (or mitotic) collapse and away from the potential for continued division or the selection of the cancer stem cell renewal fate.
In addition, the PLK4 inhibitor CFI-400945 may synergize with other drugs such as standard chemotherapeutic agents that act on other members of a PLK4-coexpression network, regulating the cell cycle. For example, an agent that promotes anaphase catastrophe, such as CFI-400945, with a microtubule-targeting drug may prove an attractive combination regimen. Taxanes target microtubules and disrupt normal timing of mitosis by delaying the spindle assembly checkpoint (SAC) activity. Moreover, the PLK4-coexpression network includes topoisomerase II-alpha (TOP2A) and Mps1/TTK, both of which have drugs targeting their activity and are available for combination testing (irinotecan and CFI-401870). Inhibitors of other anaphase catastrophe pathways, such as CDK2 (41) (Seliciclib/CYC202/R-roscovitine) or Mps1/TTK (6), might be effective in combination with CFI-400945. Preliminary results already suggest synergy between CFI-400945 and irinotecan (TOP2A) and between CFI-400945 and CFI-401870 (TTK/MPS1).
Based on our observations, PLK4 has a regulatory role in centrosome duplication and may function as an integrative protein that localizes to different subcellular organelles including the centrosome, kinetochore, cleavage furrow, and midbody during different portions of the cell cycle (interphase, prophase, metaphase, anaphase, telophase, and cytokinesis) with catalytic activity that can be modulated by interactions with different intracellular inhibitors and activators in each of these locations that include a role in cell abscission during cytokinesis.
Materials and Methods
Tissue Culture.
We used breast cancer (HCC1569, HCC1954, BT549, MDA-MB-415, HCC202, MCF7, T47D, CAL51), ovarian cancer (KOC-7C, SKOV3, HEYC2, COLO704, RMG-1, KK, ES-2, EFO21, MCAS, A2780, OV167, PEA2, TOV-21G, OV90, OV207, Kuramochi, OVISE, OVMANA, PEO6, CaOv-3, OV177, OVCAR3, OVCAR5, OVSAHO, OVTOKO, PEO14), colorectal cancer (HCT-116, HCT-15, DLD-1, COLO 205, COLO320DM, COLO320HSR, NCI-H747, NCI-H716, COLO201, LS1034, SNU-C2B, SNU-C1, LS513, LS411N, ATR-FLOX, LS174t, WiDr, RKO, SK-CO1, HCT-8, LS-180, HT-29, C2Bbe1, SW480), and immortalized breast epithelial (MCF-10–2A, MCF-12A) cell lines from the American Type Culture Collection (ATCC) or the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) for our experiments grown as described elsewhere (42–44). Cultured cells were synchronized as needed by using single or double thymidine blockade. For experiments with CFI-400945, the PLK4 inhibitor was added to medium at 10 nM, 50 nM, 100 nM, or 500 nM concentrations and compared with results with DMSO controls.
Anti–Phospho-Serine305-PLK4 Antibody.
The rabbit anti–phospho-serine305-PLK4 antibody (Ab#3) detecting phosphorylated S305-PLK4 was produced by a commercial vendor (Agro-Bio) with double-affinity purification of rabbit anti-serum after immunization with synthetic peptide (SISG{pS}LFDKRRLLC) coupled to keyhole limpet hemocyanin, as described by others (20).
Competition Studies with PLK4 Peptides.
Competition studies were performed for each rabbit anti-PLK4 antibody (ab137398; Abcam; catalog no. 3258; Cell Signaling Technology) or anti–phospho-PLK4 antibody [our Ab#3, produced commercially by Agro-Bio with SISG{pS}LFDKRRLLC as peptide immunogen (20)] using different PLK4 or phospho-PLK4 peptides. The peptide used for competition with antibody 3258 (P005) included the antigenic site of our antibody (PLK4 Blocking Peptide, 11842S; Cell Signaling Technology) at a concentration of 2 μg/mL (a 10-fold excess compared with the antibody concentration). The peptide used for competition with antibody ab137398 (C-PLK4) was full-length PLK4 (P4771, recombinant PLK4 protein from Abnova) at a concentration of 10 μg/mL (a 10-fold excess compared with the antibody concentration). The peptide used for competition with our Ab#3, anti–phospho-PLK4, was the peptide SISG{pS}LFDKRRLLC (courtesy of Jackie Mason, University of Toronto, Toronto, ON, Canada) at a concentration of 10 μg/mL (a 10-fold excess compared with the antibody concentration). These peptides were added to the diluent of the anti-PLK4 antibodies before performing the immunofluorescence procedure on the cells.
Immunoprecipitation.
Cell lysates were prepared by treating 10 million cells with 1 mL Tris-buffered saline (TBS) solution, 350 mM NaCl, and 0.3% Nonidet P-40 (FNN0021; Life Technologies) plus Protease Inhibitor Mixture (P8340; Sigma) on ice for 30 min. The lysate was cleared by centrifuging at 16,000 × g for 20 min at 4 °C. Immunoprecipitation of the lysates was performed with column-based technology by using cross-linking and spin column sample-handling (Pierce Crosslink IP Kit, no. 26147; Thermo Scientific). Rabbit polyclonal anti–phospho-serine305-PLK4 antibody was cross-linked to protein A/G-coated agarose beads with disuccinimidyl suberate (DSS). The prepared lysates were precleared by using control beads. The antibody resin was incubated with the cell lysate overnight at 4 °C, allowing the antibody:antigen complex to form. The beads were washed (0.025 M Tris, 0.15 M NaCl, 0.001 M EDTA, 1% Nonidet P-40, 5% glycerol, pH 7.4) to remove nonbound components of the sample. The bound antigen was recovered by dissociation from the antibody with acidic elution buffer (pH 2.8) twice. The procedure was performed in a microcentrifuge spin cup, allowing eluted solutions to be fully separated from the agarose resin by centrifugation. Only antigen is eluted by the procedure, facilitating protein identification with subsequent MS without interference from antibody fragments.
SDS/PAGE and Silver Staining of Gel.
Eluted protein from immunoprecipitation and column chromatography was separated by electrophoresis on polyacrylamide gels at 100 V for 90 min or until the samples were sufficiently separated. The gel was rinsed in water, fixed for 30 min (methanol and acetic acid solution), washed twice for 20 min each (400 mL deionized distilled water), and stained for 20 min by using a commercially available silver staining solution (NH4NO3 and AgNO3; silver stain plus, 161–0449; Bio-Rad). Staining was stopped with 5% acetic acid (15 min) followed by washing with deionized distilled water (5 min). The silver-stained bands of interest were identified by a Western immunoblot processed in parallel to identify bands recognized by rabbit polyclonal anti–phospho-serine305-PLK4. The identified bands were excised from the gel and processed as described as follows.
Digestion of Immunoprecipitates Separated by SDS/PAGE and MS.
After reduction and alkylation using dithiothreitol (DTT) and iodoacetamide (IAA), excised gel bands were digested into peptides by using trypsin. The resulting peptides were acidified, desalted using a Sep-Pak C18 column, and then subjected to liquid chromatography (LC)–MS/MS sequencing and data analysis. The identification of released PLK4 interaction was carried out by an LC/MS system consisting of an Eksigent NanoLC Ultra 2D and Thermo Fisher Scientific LTQ Orbitrap XL. Proteome Discoverer 1.3 (Thermo Fisher Scientific) was used for protein identification by using Sequest algorithms. The following criteria were followed. The variable modifications used were carbamidomethylation (C) and oxidation (M). Searches were conducted against UniProt or an in-house customer database. We also validated the identifications by manual inspection of the mass spectra (45).
Western Immunoblot.
Western immunoblot analyses were performed as described previously (46) using antibodies recognizing PLK4 (3258; Cell Signaling Technology and Abcam), phospho-PLK4 (Ab#3 and 14299), γ-tubulin (clone GTU-88; Sigma), and α-tubulin (DM1A mouse monoclonal antibody T6199; Sigma).
Immunofluorescence.
The subcellular localization of various proteins was performed with immunofluorescence using antibodies recognizing PLK4 (3258; Cell Signaling Technology and Abcam), phospho-PLK4 (Ab#3 and 14299; Agro-Bio and Cell Signaling), γ-tubulin, α-tubulin, kinetochores, and mitotic kinesin-like protein-1 (MKLP1; mouse monoclonal sc-390113; Santa Cruz Biotechnology). An SDS antigen retrieval technique (32) was used with frozen or fresh cells, whereas heat-induced antigen retrieval with high pH (9.0) was used with fixed, paraffin-embedded cell lines.
Proliferation Assays.
Proliferation assays were performed by using 2 × 105 cells per T75 flask over an 8-d period according to standard protocols (47).
Flow Cytometry.
Analyses of DNA content and cell cycle were performed with flow cytometry (48).
PLK4 siRNA Knockdown.
Depletion of PLK4 was performed by using Trilencer-27 siRNA duplexes against PLK4 (SR307314; OriGene) at a final concentration of 20 nM with Universal scrambled negative control siRNA duplex (SR30004; OriGene) as control. Transfections were performed by using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s forward transfection protocol. Briefly, 240 pmol of siRNA and 54 μL of Lipofectamine RNAiMAX were used to prepare RNAi duplex-Lipofectamine RNAiMAX complexes in 1,800 μL of Opti-MEM I medium (Invitrogen). Cells were cultured in T-75 flasks until 50% confluence was achieved before treatment. Cells were collected 24–96 h after transfection for real-time PCR analysis and immunofluorescent staining to monitor PLK4 knockdown.
Transient Transfection Assays.
Transient transfections were performed with FuGENE HD Transfection Reagent according to the manufacturer’s recommendations (Promega). Briefly, one part of PLK4-WT or PLK4-KD plasmid DNA was combined with three parts of FuGENE HD Transfection Reagent and incubated for 15 min at room temperature. Ten microliters of the FuGENE HD Transfection Reagent/DNA mixture were added to each well of 12-well plates containing 10,000 cells per well in 600 μL growth medium. The cells were cultured for 24–48 h and assayed 24–48 h after transfection.
Time-Lapse Videomicroscopy.
A Zeiss inverted, spinning confocal microscope containing a stage-top incubator was used to perform time-lapse video microscopy. HCT116 or OVCAR3 cells were seeded in growth medium in Glass Bottom Microwell Dishes (MatTek). After attaching to the glass surface for 24 h, the cells were synchronized with a thymidine block for 12–16 h. The cells were then released into fresh medium containing 50 nM CFI-400945 or the same quantity of DMSO before being positioned on the stage. The same protocol was adhered to when preparing CAL51 cells for acquisition but by using MatTekII four-well chamber slides.
Time-lapse video microscopy of the HCT116 and OVCAR3 cells was performed with a cell observer (Zeiss) at constant conditions of 37 °C and 5% CO2. Phase-contrast images were acquired every minute for 18 h by using a 10× phase-contrast objective (Zeiss) and an AxioCam HRm camera with Zeiss AxioVision 4.7 software. Time-lapse video microscopy of the CAL51 cells was performed with an AxioObserver (Zeiss) at constant conditions of 37 °C and 5% CO2. Differential interference contrast (DIC) microscopic images were acquired every minute for 22.15 h by using a 20× objective and an Axiocam 506 camera with Zeiss Zen Pro software.
Supplementary Material
Acknowledgments
We thank Ivonne Villalobos for administrative support; Caihong Xia for technical support; Joseph Rodgers, PhD [University of Southern California (USC)], for permission to use his videomicroscope and for his assistance with the CAL51 videomicroscopy experiment; Wendy Fantl, PhD (Stanford University) for characterizing and providing the 14299 anti–phospho-PLK4 antibody; and Cheng-Ming Chuong, MD, PhD (USC), for critically reviewing the manuscript. Flow cytometry was performed in the USC Flow Cytometry Core Facility, proteomic studies were performed in the USC Proteomics Core, and videomicroscopy was performed in the USC Imaging Core, which are supported in part by National Cancer Institute Cancer Center Shared Grant Award P30CA014089 and the USC Office of the Provost, Dean’s Development Funds, Keck School of Medicine of USC. This study was supported by California Institute for Regenerative Medicine Grants DR1-01477 (Subaward Agreement 1559-S-NA228) and DR3-07067 (Subaward Agreement 1559-S-SA669); the Dr. Miriam & Sheldon G. Adelson Medical Research Foundation (M.F.P., J.S.B., and D.J.S.); Breast Cancer Research Foundation Grant BCRF-18-132 (to M.F.P.); Tower Cancer Research Foundation Jessica M. Berman Senior Investigator Award 006886-0001; Avon Foundation for Women Grant 02-2015-062 (to M.F.P.); and the endowed Harold E. Lee Chair for Cancer Research (to M.F.P.).
Footnotes
Conflict of interest statement: The intellectual properties of the PLK4 inhibitor CFI400945 is owned by the University Health Network (Toronto) and is in the process of being transferred to a company in which T.W.M. has equity. Observations for a role of polo-like kinase 4 on cytokinesis supported by CFI-400945 inhibition were disclosed to the University of Southern California Stevens Institute in March 2016. D.J.S. is an employee of BioMarin and has stock or other ownership in Pfizer. Research grants from the following companies have been provided to authors’ institutions: Cepheid (M.F.P.), Eli Lilly & Company (M.F.P.), Novartis Pharmaceuticals (M.F.P. and D.J.S.), F. Hoffmann La-Roche Ltd. (M.F.P. and J.S.B.), Pfizer (D.J.S.). M.F.P. holds a consulting or advisory role with honoraria with Karyopharm Therapeutics, Puma Biotechnology, Biocartis, Eli Lilly & Company, Novartis Pharmaceuticals, and F. Hoffmann–La Roche. D.J.S. holds a consulting or advisory role with honoraria with Eli Lilly & Company, and Novartis Pharmaceuticals. Pfizer, BioMarin, and Novartis also provided travel, accommodations, or expenses to D.J.S. S.D., Y.Z., R.G., G.P.N., N.O., and M.P. declare no potential conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1818820116/-/DCSupplemental.
References
- 1.Weinstein IB. (2002) Cancer. Addiction to oncogenes–The Achilles heal of cancer. Science 297:63–64. [DOI] [PubMed] [Google Scholar]
- 2.Slamon D, et al. ; Breast Cancer International Research Group (2011) Adjuvant trastuzumab in HER2-positive breast cancer. N Engl J Med 365:1273–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shaw AT, et al. (2014) Ceritinib in ALK-rearranged non-small-cell lung cancer. N Engl J Med 370:1189–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldman JM. (2012) Ponatinib for chronic myeloid leukemia. N Engl J Med 367:2148–2149. [DOI] [PubMed] [Google Scholar]
- 5.Finetti P, et al. (2008) Sixteen-kinase gene expression identifies luminal breast cancers with poor prognosis. Cancer Res 68:767–776. [DOI] [PubMed] [Google Scholar]
- 6.Brough R, et al. (2011) Functional viability profiles of breast cancer. Cancer Discov 1:260–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mason JM, et al. (2014) Functional characterization of CFI-400945, a polo-like kinase 4 inhibitor, as a potential anticancer agent. Cancer Cell 26:163–176. [DOI] [PubMed] [Google Scholar]
- 8.Tsherniak A, et al. (2017) Defining a cancer dependency map. Cell 170:564–576.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawakami M, et al. (2018) Polo-like kinase 4 inhibition produces polyploidy and apoptotic death of lung cancers. Proc Natl Acad Sci USA 115:1913–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ha GH, Breuer EK (2012) Mitotic kinases and p53 signaling. Biochem Res Int 2012:195903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA (2005) The polo kinase Plk4 functions in centriole duplication. Nat Cell Biol 7:1140–1146. [DOI] [PubMed] [Google Scholar]
- 12.Kuriyama R, Bettencourt-Dias M, Hoffmann I, Arnold M, Sandvig L (2009) Gamma-tubulin-containing abnormal centrioles are induced by insufficient Plk4 in human HCT116 colorectal cancer cells. J Cell Sci 122:2014–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Godinho SA, et al. (2014) Oncogene-like induction of cellular invasion from centrosome amplification. Nature 510:167–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosario CO, et al. (2010) Plk4 is required for cytokinesis and maintenance of chromosomal stability. Proc Natl Acad Sci USA 107:6888–6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hudson JW, et al. (2001) Late mitotic failure in mice lacking Sak, a polo-like kinase. Curr Biol 11:441–446. [DOI] [PubMed] [Google Scholar]
- 16.Ko MA, et al. (2005) Plk4 haploinsufficiency causes mitotic infidelity and carcinogenesis. Nat Genet 37:883–888. [DOI] [PubMed] [Google Scholar]
- 17.de Cárcer G, Manning G, Malumbres M (2011) From Plk1 to Plk5: Functional evolution of polo-like kinases. Cell Cycle 10:2255–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zitouni S, Nabais C, Jana SC, Guerrero A, Bettencourt-Dias M (2014) Polo-like kinases: Structural variations lead to multiple functions. Nat Rev Mol Cell Biol 15:433–452. [DOI] [PubMed] [Google Scholar]
- 19.Slevin LK, et al. (2012) The structure of the plk4 cryptic polo box reveals two tandem polo boxes required for centriole duplication. Structure 20:1905–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sillibourne JE, et al. (2010) Autophosphorylation of polo-like kinase 4 and its role in centriole duplication. Mol Biol Cell 21:547–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rechsteiner M, Rogers SW (1996) PEST sequences and regulation by proteolysis. Trends Biochem Sci 21:267–271. [PubMed] [Google Scholar]
- 22.Holland AJ, Cleveland DW (2014) Polo-like kinase 4 inhibition: A strategy for cancer therapy? Cancer Cell 26:151–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wong YL, et al. (2015) Cell biology. Reversible centriole depletion with an inhibitor of polo-like kinase 4. Science 348:1155–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McLamarrah TA, et al. (2018) An ordered pattern of Ana2 phosphorylation by Plk4 is required for centriole assembly. J Cell Biol 217:1217–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aydogan MG, et al. (2018) A homeostatic clock sets daughter centriole size in flies. J Cell Biol 217:1233–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gemble S, Basto R (2018) Fast and furious . . . or not, Plk4 dictates the pace. J Cell Biol 217:1169–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laufer R, et al. (2013) The discovery of PLK4 inhibitors: (E)-3-((1H-Indazol-6-yl)methylene)indolin-2-ones as novel antiproliferative agents. J Med Chem 56:6069–6087. [DOI] [PubMed] [Google Scholar]
- 28.Sampson PB, et al. (2014) The discovery of polo-like kinase 4 inhibitors: Identification of (1R,2S)-2-(3-((E)-4-(((cis)-2,6-dimethylmorpholino)methyl)styryl)-1H-indazol-6-yl)-5′-methoxyspiro[cyclopropane-1,3′-indolin]-2′-one (CFI-400945) as a potent, orally active antitumor agent. J Med Chem 58:147–169. [PubMed] [Google Scholar]
- 29.Sampson PB, et al. (2014) The discovery of polo-like kinase 4 inhibitors: Design and optimization of spiro[cyclopropane-1,3′[3H]indol]-2′(1′H)-ones as orally bioavailable antitumor agents. J Med Chem 58:130–146. [DOI] [PubMed] [Google Scholar]
- 30.Sillibourne JE, Bornens M (2010) Polo-like kinase 4: The odd one out of the family. Cell Div 5:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leung GC, et al. (2002) The Sak polo-box comprises a structural domain sufficient for mitotic subcellular localization. Nat Struct Biol 9:719–724. [DOI] [PubMed] [Google Scholar]
- 32.Brown D, et al. (1996) Antigen retrieval in cryostat tissue sections and cultured cells by treatment with sodium dodecyl sulfate (SDS). Histochem Cell Biol 105:261–267. [DOI] [PubMed] [Google Scholar]
- 33.Kisselev AF, Goldberg AL (2001) Proteasome inhibitors: From research tools to drug candidates. Chem Biol 8:739–758. [DOI] [PubMed] [Google Scholar]
- 34.Bettencourt-Dias M, et al. (2005) SAK/PLK4 is required for centriole duplication and flagella development. Curr Biol 15:2199–2207. [DOI] [PubMed] [Google Scholar]
- 35.Bonni S, Ganuelas ML, Petrinac S, Hudson JW (2008) Human Plk4 phosphorylates Cdc25C. Cell Cycle 7:545–547. [DOI] [PubMed] [Google Scholar]
- 36.Ruchaud S, Carmena M, Earnshaw WC (2007) Chromosomal passengers: Conducting cell division. Nat Rev Mol Cell Biol 8:798–812. [DOI] [PubMed] [Google Scholar]
- 37.Oegema K, Davis RL, Lara-Gonzalez P, Desai A, Shiau AK (2018) CFI-400945 is not a selective cellular PLK4 inhibitor. Proc Natl Acad Sci USA 115:E10808–E10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sampson PB, et al. (2015) The discovery of polo-like kinase 4 inhibitors: Identification of (1R,2S).2-(3-((E).4-(((cis).2,6-dimethylmorpholino)methyl)styryl). 1H.indazol-6-yl)-5′-methoxyspiro[cyclopropane-1,3′-indolin]-2′-one (CFI-400945) as a potent, orally active antitumor agent. J Med Chem 58:147–169. [PubMed] [Google Scholar]
- 39.Degenhardt Y, et al. (2010) Sensitivity of cancer cells to Plk1 inhibitor GSK461364A is associated with loss of p53 function and chromosome instability. Mol Cancer Ther 9:2079–2089. [DOI] [PubMed] [Google Scholar]
- 40.Galimberti F, Thompson SL, Ravi S, Compton DA, Dmitrovsky E (2011) Anaphase catastrophe is a target for cancer therapy. Clin Cancer Res 17:1218–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Galimberti F, et al. (2010) Targeting the cyclin E-Cdk-2 complex represses lung cancer growth by triggering anaphase catastrophe. Clin Cancer Res 16:109–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Finn RS, et al. (2009) PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res 11:R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Konecny GE, et al. (2011) Expression of p16 and retinoblastoma determines response to CDK4/6 inhibition in ovarian cancer. Clin Cancer Res 17:1591–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mouradov D, et al. (2014) Colorectal cancer cell lines are representative models of the main molecular subtypes of primary cancer. Cancer Res 74:3238–3247. [DOI] [PubMed] [Google Scholar]
- 45.Zhou Y, et al. (2011) Rapid and enhanced proteolytic digestion using electric-field-oriented enzyme reactor. J Proteomics 74:1030–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Slamon DJ, et al. (1989) Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science 244:707–712. [DOI] [PubMed] [Google Scholar]
- 47.Wang C, et al. (2007) Abelson interactor protein-1 positively regulates breast cancer cell proliferation, migration, and invasion. Mol Cancer Res 5:1031–1039. [DOI] [PubMed] [Google Scholar]
- 48.Ormerod MG. (2000) Flow Cytometry. A Practical Approach (Oxford Univ Press, Oxford: ). [Google Scholar]
- 49.Fededa JP, Gerlich DW (2012) Molecular control of animal cell cytokinesis. Nat Cell Biol 14:440–447. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.