Abstract
1. Scope
The intestinal microbiota transforms a wide range of available substrates, including polyphenols. Microbial catabolites of polyphenols can contribute in significant ways to the health promoting properties of their parent polyphenols. This work aimed to identify intestinal metabolites of xanthohumol (XN), a prenylated flavonoid found in hops (Humulus lupulus) and beer, as well as to identify pathways of metabolism of XN in the gut.
2. Methods and results
To investigate intestinal metabolism, XN and related prenylated flavonoids, isoxanthohumol (IX) and 8-prenylnaringenin (8PN) were added to growing cultures of intestinal bacteria, Eubacterium ramulus and E. limosum. We used liquid chromatography coupled with mass spectrometry to identify metabolites of the flavonoids from the cultures. The metabolic capacity of E. limosum appears to be limited to O-demethylation. Evidence from our study indicates that E. ramulus hydrogenates XN to form α,β-dihydroxanthohumol (DXN) and metabolizes the potent phytoestrogen 8PN into the chalcones, O-desmethylxanthohumol (DMX) and O-desmethyl-α,β-dihydroxanthohumol (DDXN).
3. Conclusion
Microbial metabolism is likely to affect both activity and toxicity of XN and derivatives. This study along with others highlights that attention should be focused on metabolites, in particular, products of intestinal microbial metabolism.
Keywords: Xanthohumol; 8-prenylnaringenin; α,β-dihydroxanthohumol; intestinal metabolism; Eubacterium ramulus
Graphical abstract
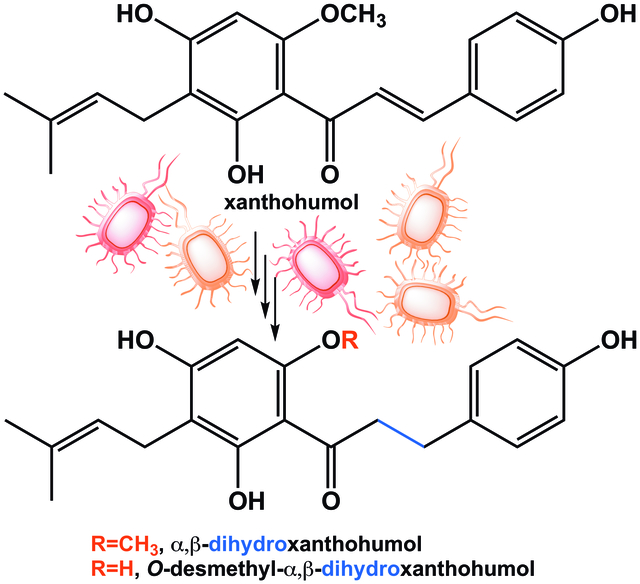
Xanthohumol, a bioactive flavonoid found in hops and beer, exerts anti-inflammatory, antiglycemic, antihyperlipidemic effects in animal models of metabolic syndrome. Gut microbial metabolism of xanthohumol has long been recognized to produce estrogenic metabolites, notably 8-prenylnaringenin. This in vitro study demonstrates that 8-prenylnaringenin is not the end-product of metabolism by gut microbes. We show that Eubacterium ramulus is capable of hydrogenating xanthohumol and 8-prenylnaringenin to form α,β-dihydro-xanthohumol and O-desmethyl-α,β-dihydroxanthohumol. These findings shed new light on the perceived pro-estrogenicity of xanthohumol.
1. Introduction
Hops (Humulus lupulus) are traditionally used as an essential ingredient in beer brewing providing bitterness and flavor to beer. They contain a fair amount of secondary plant metabolites including prenylated chalcones, which have various health promoting properties. Xanthohumol is the main prenylflavonoid found in hops (0.1–1% on dry weight); other chalcones can also be detected, but they occur at 10 to 100-fold lower concentrations [1]. XN receives much attention as it exhibits anti-obesity effects [2], anticancer properties [3–5], and, improves cognitive function [6]. In vitro and animal studies have established that XN is non-enzymatically converted into its flavanone isomer, isoxanthohumol (IX), and enzymatically into 8-prenylnaringenin (8PN), 6-prenylnaringenin (6PN) and desmethylxanthohumol (DMX) [7,8]. Therefore, the metabolism of XN might play a sizeable role in the bioactivity of the prenylated chalcone. Concern has been raised about the amount of 8PN produced from intake of XN, 8PN being a well-known phytoestrogen, which demonstrated in vivo estrogenic activity [9,10]. 8PN also prevents bone loss in rats [11], inhibits angiogenesis [12] and metastasis [13], and exhibits anti-androgenic activity [14].
Recent human studies indicate that plasma concentrations and urinary excretions of microbial metabolites can exceed those of the original compound, especially for polyphenols that reach the colon as they are not easily absorbed in the upper gastrointestinal tract [15,16]. For instance, naringenin, the non-prenylated analogue of 8PN, shows intensive microbial biotransformation in the intestine, including ring cleavage and dehydroxylation, thereby forming simple phenolics absorbed from the colon [17]. The emerging consensus is that the gut microbiota may play a crucial role in the potential health benefits of flavonoids [18,19]. There are only a few studies, however, investigating the influence of the intestinal microbial community in the metabolism of XN and derivatives [8,20–22].
The human gut houses 1014 microorganisms [23] regrouped in an estimate of 500 to 1000 species with an enormous metabolic potential. Far beyond the fermentation of non-digestible food compounds, the gut microbiota is implicated in developmental, immunological, and nutritional host functions and have a profound impact on human health [24]. Each portion of the gastrointestinal tract has a distinctive microbial community, but the majority of the organisms are in the distal tract, where the density of some species may account for 35 – 50% of the contents [25,26]. Data from several studies suggest the Bacteroidetes and Firmicutes represent the two most dominant bacterial phyla in the human gut [27,28], although Firmicutes are believed to be the most diverse group. In line with its enormous diversity, the Firmicutes in the gastrointestinal tract perform several different functions ranging from health promoting effects exerted by some probiotics such as Lactobacillus spp. to pathogenic effects caused by Clostridium difficile [29]. Many species of the genera Clostridium, Blautia, Dorea, Eubacterium, Ruminococcus, Anaerostipes, Roseburia, and Coprococcus belong to the Firmicutes phylum and have been reported to affect their hosts’ gut health in several ways. For instance, butyrate-producing Firmicutes, such as E. rectale, E. hallii and E. ramulus, can utilize a wide range of polysaccharides, suggesting that these bacteria make an important contribution to colonic fermentation of dietary components [30].
The genus Eubacterium is one of the most abundant genera of the human gastrointestinal microbiota, with densities of up to 1010 colony forming units (CFU)/g of intestinal content [29]. By cleaving off the methyl group of IX, E. limosum is able to activate the flavanone in vitro and in vivo to 8PN [8,20]. Similar to E. limosum, E. ramulus is a strictly anaerobic bacterium found to be present in the gastrointestinal tract of most individuals. The organism is a gram-positive, non-sporulating and chain-forming rod first described by Moore and Holdeman in 1974 [31]. E. ramulus occurs at approximately 108 CFU/g dry feces in humans [32] and may be considered a key organism for flavonoid degradation in the human intestine as it is involved in the conversion of several flavonols/flavanonols, flavones/flavanones, as well as their corresponding chalcones and dihydrochalcones. E. ramulus catalyzes the isomerization, the reduction but also the cleavage of these flavonoids ring system [33,34], giving rise to the corresponding hydroxyphenylacetic and hydroxyphenylpropionic acids, as well as acetate and butyrate.
To gain more information on the impact of the gut microbiota on the bioavailability and activity of XN and metabolites upon ingestion, in this report we investigated the metabolic potential of E. ramulus in monocultures or co-cultured with E. limosum under in vitro conditions. Our findings demonstrate that 8PN is not the end-product of XN metabolism by gut microbes and shed new light on the perceived pro-estrogenicity of XN.
2. Materials and methods
2.1. Chemicals
XN (99+% purity) was a gift from Hopsteiner, Inc. (New York, NY, USA). IX, 8PN and DMX were isolated and purified from hops as described by Stevens et al., [35,36]. DXN was synthesized from XN as described by Miranda et al., [37]. 13C-labeled O-desmethyl-α,β-dihydroxanthohumol (13C-DDXN) was synthesized following a protocol described in supporting information (Figure S1). HPLC grade acetonitrile, ethyl acetate, methanol and water were purchased from EMD Millipore (Gibbstown, NJ, USA). Dimethyl sulfoxide (DMSO) ACS reagent grade was obtained from Sigma-Aldrich Chemical Company (St Louis, MO, USA).
2.2. Fermentation conditions and dosage information
All fermentation experiments were performed in 10 mL Cooked Meat Medium, K19 from Hardy Diagnostics (Santa Maria, CA, USA). E. ramulus ATCC®29099 and E. limosum ATCC®8486 were purchased from ATCC (Manassas, VA, USA). Stock solutions (5 mM) of the different prenylflavonoids (X, IX, and 8PN) were prepared in DMSO. Time-dependent conversion experiments were carried out in fresh chopped meat medium by adding 10 μL stock solutions/mL of medium and 0.5 mL of an overnight culture of E. ramulus diluted to an optical density of 0.25. The final flavonoid concentration in the incubation mixtures was 50 μM, which is similar to the concentrations used by others in microbial metabolism studies of flavonoids incubated in vitro with E. limosum [8] or E. ramulus [34]. For mono- and co-culture experiments, 0.5 mL of an overnight culture of growing E. ramulus and/or E. limosum was added to chopped meat medium and incubated anaerobically for 3 days. Control experiments in absence of bacteria were performed by adding 10 μL stock solutions/mL of fresh chopped meat medium. Control experiments in absence of the flavonoids were performed by adding 0.5 mL of an overnight culture of bacteria to 10 mL of fresh medium. Day 0 concentrations were measured as a ‘100%’ basis for the calculation of recovery rates by incubating substrate flavonoids in sterile chopped meat medium for less than 2 hours prior to extraction and LC-MS/MS analysis.
Tubes were filled with incubation mixtures and placed in an anaerobic jar (GasPak 100 anaerobic systems from BD, Franklin Lakes, NJ, USA). After sealing the jar, the anaerobic environment was generated, maintained, and monitored using a BD GasPak Anaerobe container system with CO2 indicator tablet (BD). If O2 enters the jar via a leak, the indicator tablet changes color from white to blue, thereby ensuring that an anaerobic environment is achieved. All cultures were incubated at 37°C for the duration of the experiments. All experiments were performed in triplicate.
2.3. Chemical analysis
2.3.1. Extraction protocol
At the end of the incubation period, a liquid/liquid extraction with ethyl acetate was performed. 1 mL of each bacterial culture was sampled, vortexed, then centrifuged at 5,000 g for 15min, and the supernatant extracted with 2 mL of ethyl acetate. The ethyl acetate phase was collected and dried under reduced pressure using a desiccator. The extracts were dissolved in 300 μL of methanol and transferred into HPLC vials stored at −80°C until LC-MS/MS analysis.
2.3.2. LC-MS/MS identification of metabolites
Separation and identification of the metabolites and residual substrates were achieved using HPLC performed on a Shimadzu LC system (Shimadzu; Columbia, MD, USA) coupled to a hybrid triple quadrupole-linear ion trap mass spectrometer (3200 QTRAP; AB Sciex; Concord, Canada) equipped with an electrospray ionization source. The source temperature was set at 600°C with a needle voltage of - 4.5 kV. MS/MS acquisition was performed in a negative mode, with collision energy set at - 40 V. Nitrogen was used as the source gas, curtain gas, and collision gas.
Characterization of 13C-DDXN was performed on a hybrid triple quadrupole-linear ion trap mass spectrometer (4000 QTRAP; AB Sciex; Concord, Canada) with an electrospray ionization source. The collision energy was set at −30 V.
Chromatographic separations were carried out on a C18 reversed phase column (Phenomenex Inc., Torrance, CA, USA). The column size was 150×2 mm with a particle size of 5μm. The flow rate was 0.2 mL/min and the injection volume was 10 μL. The HPLC method used a gradient composed of solvent A – water and solvent B – acetonitrile, both acidified with 0.1% (v/v) formic acid. The elution gradient was as follows: 0 – 20 min, from 42% to 52% B; increase from 52% to 90% B over the next 5 min; held at 90% B from 25 to 30 min; then, instant shift to 42% B and run for another 6 min to equilibrate the column. Peaks were identified by comparison of the retention times and product ions with those of reference compounds. Data processing was performed using PeakView Software v1.2 (AB Sciex).
2.3.3. Quantitative analysis
For quantitative analyses, the samples were analyzed on AB Sciex 4000 QTRAP using an Agilent C8 reversed phase column (2.1 × 50 mm, 3.5 μm), an injection volume of 1 μL and a mobile phase flow of 0.4 mL/min. The solvent gradient started at 30% B and increased to 60% B until 1.5 min, was held at 60% from 1.5–2.5 min, increased to 100% until 3 min, was held at 100% from 3–3.8 min, decreased to 30% at 3.9 min, and the column was equilibrated until 6.0 min. The solvents and other acquisition parameters were kept the same. Selected reaction monitoring (SRM) transitions for quantification were: m/z 353→119, m/z 353→233 for XN and IX; m/z 339→119, m/z 339→219 for 8PN; m/z 355→191, m/z 355→249 for DXN; m/z 341→191, m/z 341→235 for DDXN; m/z 342→192, m/z 342→236 for 13C-DDXN. Quantification was performed using AB Sciex MultiQuant Software v3.0.2. External 10-point calibration curves were prepared for all the flavonoids (R values > 0.995).
Because the synthesized amount of 13C-DDXN was insufficient to create an accurate standard curve based on mass, we used 1H NMR for absolute concentration determination. A known amount of DXN was added to 13C-DDXN in deuterated chloroform (Cambridge Isotopes, MA, USA) and analyzed on a 700 MHz NMR spectrometer (Bruker, MA, USA). The peak areas of the proton resonances arising from the single hydrogen atom on ring A of the flavonoids were measured for both analytes, 13C-DDXN and DXN. The ratio of the area of individual resonances of 13C-DDXN to that of the calibrant, DXN, is equal to the ratio of their concentrations. This ratio was used to generate a calibration curve for DDXN by LC-MS/MS.
3. Results
3.1. Incubation of XN, IX, and 8PN with E. ramulus
Considering the role of E. ramulus in intestinal microbial degradation of flavonoids, the current study sought to investigate the metabolism of XN and related flavonoids by the bacterium. Therefore, XN, IX and 8PN were added to growing cultures of E. ramulus and incubated under anaerobic conditions.
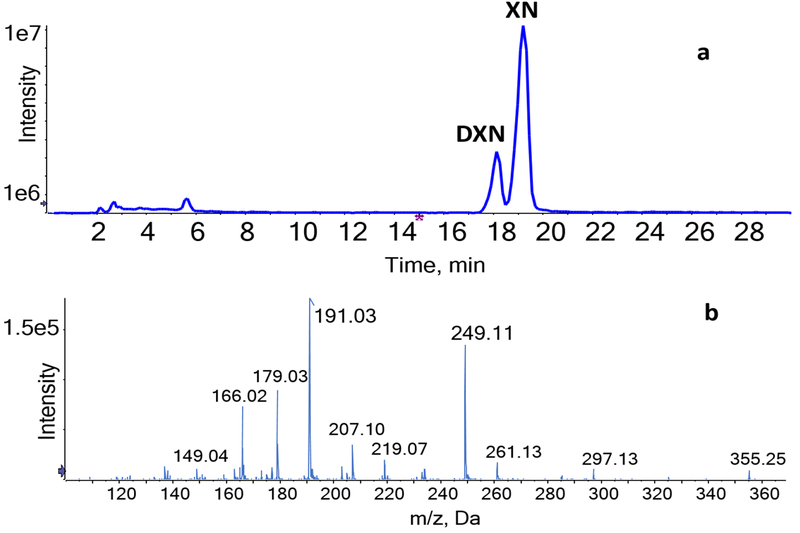
After incubation with XN, LC-MS/MS analysis detected a metabolite peak eluting at 17.9 min in the samples (Fig. 1a). This metabolite was absent in sterile chopped meat medium incubated with XN and in bacterial cultures grown without XN for the same duration. MS analysis of XN metabolite showed a molecular ion peak of m/z 355 [M-H]–. The product ion spectrum of this molecular ion shows high intensities of m/z 191 and characteristic product ion of m/z 249 (Fig. 1b). Based on the retention time and product ion spectrum, the novel microbial metabolite of XN was identified as α,β-dihydroxanthohumol (DXN), a prenylated chalcone previously reported as a minor constituent of hops [38] and which we reported to be devoid of intrinsic estrogenicity [37].
Figure 1.
(a) Total ion chromatogram (TIC) of XN and DXN after incubation of XN with E. ramulus. (b) HPLC-ESI-MS/MS product ion spectrum of DXN (m/z 355) in negative ion mode.
To study the kinetics of E. ramulus conversion of XN into DXN, the strain was incubated with XN for various periods of time over 5 days. The concentrations of metabolites of XN, IX and DXN were measured and their recovery rates calculated as molar percentages of the day 0 concentrations of the substrates (Table 1). Concentrations of XN on day 0 were averaging 20 μM. We attributed the decreased recovery to binding of the flavonoids to proteins from the cooked meat medium. The liquid-liquid extraction did not provide a representative sample of the medium as it could not account for the solid portion of the medium (i.e., meat pieces). The recovery rates of IX, formed by spontaneous isomerization of XN, were lower than 4% in all the incubations containing XN. The concentrations of IX remained consistent regardless of the presence of a bacterium. At day 1, DXN was not quantifiable but days 3 and 5 showed an increase in the average recovery rate from 4.6% to 6.2%.
Table 1:
Conversion of XN by E. ramulus.
| Time (days) | E. ramulus + XN | ||
|---|---|---|---|
| XN | DXN | IX | |
| 1 | 93.8% (1.44%) | ND* | 3.3% (0.03%) |
| 3 | 50.5% (2.57%) | 4.6% (0.27%) | 3.5% (0.15%) |
| 5 | 30.7% (4.11%) | 6.2% (0.89%) | 3.1% (0.08%) |
XN (50 μM) was incubated for 5 days with E. ramulus. Disappearance of XN, production of DXN and IX were monitored at days 1, 3 and 5. On day 0, the average recovery of XN was 20 μM. Results are presented as mean (+SEM) percentage recovery of XN, IX and DXN calculated as molar percentages of the day 0 concentrations of XN.
ND: not detectable.
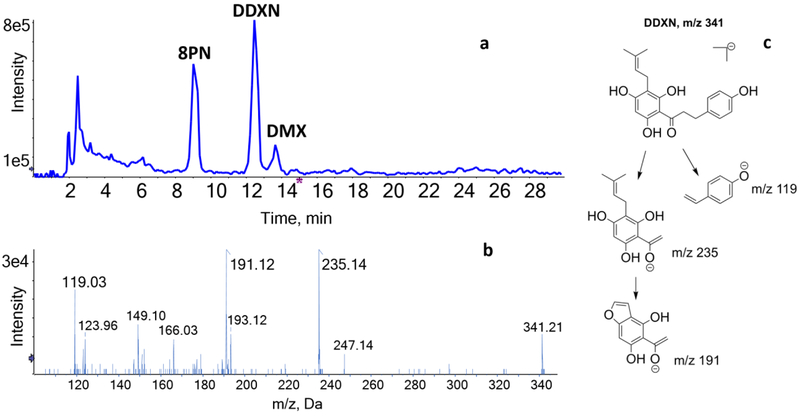
In samples derived from E. ramulus cultures incubated with 8PN, LC-MS/MS analysis yielded two metabolites whose peaks eluted at 12.2 and 13.6 min (Fig. 2a), although the peak at 13.6 min was visible in only two of the replicates. These metabolites were absent in the control samples comprising of sterile chopped meat medium incubated with 8PN and in bacterial cultures incubated without 8PN for the same duration. Based on the retention time of m/z 339 [M-H]– at 13.6 min and the product ion pattern corresponding to those of authentic DMX, we identified one of the metabolites as DMX.
Figure 2.
(a) Total ion chromatogram (TIC) of 8PN, DMX and DDXN after incubation of 8PN with E. ramulus. (b) HPLC-ESI-MS/MS product ion spectrum of DDXN (m/z 341) in negative ion mode. (c) Proposed structures for the major MS/MS product ions of DDXN in negative ion mode.
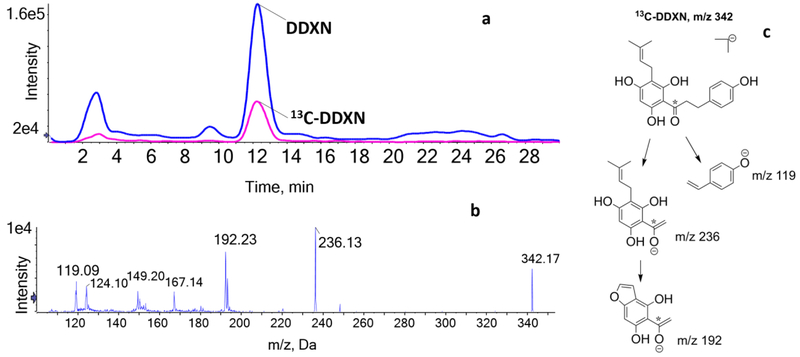
The peak with m/z 341 [M-H]– eluting at 12.2 min corresponded to the molecular formula C20H22O5. Upon MS-MS fragmentation of the MH– ion, the following fragments were most prominent: m/z 235 [A-ring C13H15O4–], m/z 191 [A-ring C10H7O4–], and m/z 119 [B-ring C8H7O–] (Fig. 2b). We propose a fragmentation pattern and structures for the major product ions observed in negative mode (Fig. 2c). To confirm this metabolite identity, we spiked the product of 8PN conversion with synthetic 13C-DDXN. The retention time of 13C-DDXN (Fig. 3a) and its fragmentation pattern (Fig. 3b) matched those of the metabolite with m/z 341 [M-H]–. We identified this metabolite with m/z 341 [M-H]– as DDXN, an O-demethylated and hydrogenated metabolite of XN. The proposed fragmentation pattern of 13C-DDXN (Fig. 3c) is in concordance with that of DDXN.
Figure 3.
(a) Extracted ion chromatogram (XIC) of DDXN (m/z 341) and 13C-DDXN (m/z 342). (b) HPLC-ESI-MS/MS of 13C-DDXN in negative ion mode (m/z 342). (c) Proposed structures for the major MS/MS product ions of 13C-DDXN in negative ion mode (* = 13C).
To establish the time course of 8PN conversion, a separate experiment was performed by incubating DMX with E. ramulus. This resulted in the metabolic conversion of DMX into DDXN by the bacterium (Fig. S2). DMX was consequently identified as an intermediate product in the conversion of 8PN to DDXN. The isomerization of 8PN to the chalcone DMX, the first step in the metabolism of the flavanone is followed by the reduction of DMX’s α,β-double bond to form DDXN, which is in good agreement with described transformation of tricyclic flavanones by E. ramulus [33]. Recovery rates of 8PN and DDXN from their respective substrates are reported in Table 2. 8PN recovery was consistently low when incubated with E. ramulus, probably due to its conversion to DMX and DDXN, whose concentrations were over 3-fold higher than that of 8PN.
Table 2:
Conversion of 8PN by E. ramulus and IX by E. ramulus and E. limosum co-cultures.
| Starting metabolite | |||
| IX | 8PN | DDXN | |
| E. ramulus | |||
| 8PN | ND* | 15.5% (1.64%) | 55.7% (24.59%) |
| E. ramulus + E. limosum | |||
| IX | 7.6% (2.17%) | 34.5% (1.99%) | 63.3% (8.08%) |
8PN (50 μM) was added to a culture of E. ramulus and IX (50 μM), to a co-culture of E. ramulus and E. limosum. After an incubation period of 3 days, the concentrations of IX, 8PN and DDXN were measured. On day 0, the average recovery of 8PN and IX was 17 μM and 20 μM, respectively. The results are presented as mean (+SEM) percentage recovery of IX, 8PN and DDXN calculated as molar percentages of the day 0 concentrations of the flavonoids.
ND: not detectable.
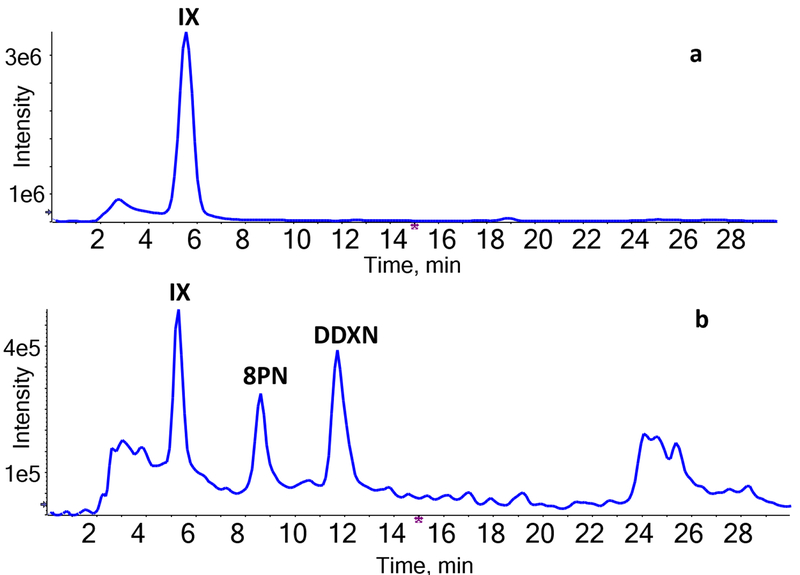
In contrast to XN and 8PN, IX showed no metabolic conversion when incubated with E. ramulus (Fig. 4a). Together, our studies showed that E. ramulus selectively reduces XN, yielding the dihydrochalcone DXN, and it metabolizes 8PN into DDXN.
Figure 4.
Total ion chromatogram (TIC) of IX, 8PN, and DDXN after incubation of IX with (a) E. ramulus, (b) E. limosum and E. ramulus.
3.2. Incubation of XN, IX, and 8PN with E. ramulus and E. limosum
In co-culture experiments involving E. ramulus and E. limosum, the strains were incubated with XN, IX or 8PN under the same anaerobic conditions. The cultures containing XN showed production of DXN, similar to the incubation of XN with E. ramulus. From these observations we conclude that XN and DXN are not further metabolized by E. limosum, which is consistent with the absence of metabolite formation from XN and DXN upon exposure to monocultures of E. limosum. The co-cultures produced DMX and DDXN from 8PN, consistent with the metabolic profile obtained from the incubation of 8PN with E. ramulus, indicating that E. limosum cannot metabolize 8PN. Monocultures of IX with E. limosum showed conversion to 8PN as previously reported [8]. In the co-cultures containing IX, LC-MS/MS analysis showed metabolites peaks corresponding to 8PN and DDXN (Fig. 4b), providing evidence that E. ramulus was able to metabolize 8PN produced upstream by E. limosum. In the co-cultures of E. ramulus and E. limosum, more than 90% of IX was converted by the bacteria. IX final concentrations were low, while 8PN recovery was around 35% and DDXN recovery rates around 63% of the day 0 concentrations of IX (Table 2).
In regard to the previously reported ability of E. ramulus to cleave the ring system of flavonoids, we analyzed for in vitro production of the corresponding hydroxyphenylacetic acid, 4-hydroxyphenylacetic acid, and hydroxyphenylpropionic acid, 3- (4-hydroxyphenyl)-propionic acid. However, by using HPLC-ESI-MS and authentic standards, we were unable to confirm cleavage of the flavonoids by E. ramulus (data not shown).
4. Discussion
While various effects of polyphenols have been demonstrated in animal models of diseases and in humans, the discrepancy between micromolar concentrations used in cell culture models to achieve a certain biological effect and the circulating concentrations in animals and humans in the nanomolar range resulting from efficacious doses of polyphenols remains largely unexplained [39]. During the past few years, the field of polyphenol research has shifted towards examining their metabolism and the biological activity of their metabolites. Discovery of the influence of the gut microbiota on human health [40,41] raises the question whether the health properties attributed to polyphenols are due to prebiotic-like effects or to their metabolites generated by the gut microbiome.
Although beer is the most important dietary source of XN and related polyphenols, XN generally occurs at minor concentrations. While IX is usually the most abundant prenylflavonoid in beer due to thermal isomerization during the brewing process [8]. The free 2’-hydroxy group in hop chalcones sets these compounds up for intramolecular Michael-type addition, thereby converting them into their corresponding flavanones [35]. The possibility that XN and its isomer, IX, could act as pro-estrogens was demonstrated by several authors [42,43]. IX can be metabolized in vitro by human liver microsomes to form the most potent phytoestrogen known to date, 8PN [44,45], and the pharmacokinetics of XN and metabolites in rats suggests rapid isomerization of XN to IX followed by hepatic demethylation of IX to produce 8PN [7]. Possemiers and co-workers estimated that 35% of ingested IX is converted to 8PN in the gut through O-demethylation of IX by E. limosum [8]. However, reduction of the enone C-C double bond of XN by E. ramulus results in DXN, which, in contrast to XN cannot undergo the subsequent isomerization to flavanone IX. This hydrogenation renders the conversion of XN to IX, 8PN or any other flavanone impossible. DXN has been proven to have an extremely low affinity for the estrogen receptors and to not induce cell proliferation in MCF-7 breast cancer cells [37]. Although most of the beneficial health effects of flavonoids are attributed to their antioxidant properties, and hydrogenation such as occurring in DXN results in a decrease of the antioxidant activity [46], DXN has greater steady-state concentrations, and retains the beneficial effects of XN on dysfunctional glucose metabolism and impaired cognitive function [37]. On the other hand, by metabolizing 8PN, E. ramulus possibly reduces its estrogenicity. The reaction starts with isomerization of 8PN leading to formation of the corresponding chalcone, DMX, which is subsequently reduced into DDXN.
The diversity of the gut microbiome and the high complexity of its metabolic potential render single bacterial cultures only partially informative regarding the transformation of a given molecule. To broaden our understanding of the flavonoids gut metabolism and to verify whether E. ramulus utilizes prenylated flavonoids substrates made available by another bacterium, we performed co-cultures experiments with E. limosum. This gut microbe is capable of O-demethylation of methoxy derivatives of the isoflavonoids, biochanin A, formononetin and glycitein [47], and is involved in the production of active estrogen-like compound, enterodiol [48]. Most importantly, E. limosum catalyzes the formation of phytoestrogen 8PN by O-demethylation of IX [8]. Our study demonstrated that E. ramulus metabolizes 8PN made available by E. limosum into DDXN, thereby decreasing the amount of 8PN available for absorption. These reactions could explain the low levels of 8PN found in physiological samples of mice exposed to XN [49]. The antioxidant properties of DMX have been investigated [50] but little to none is currently known about the biological properties or lack thereof of DDXN. So far, E. limosum O-demethylase has been reported to specifically metabolize isoflavones [47] and flavanones [8], explaining its lack of ability to accept chalcone XN and dihydrochalcone DXN as substrates.
The ability of E. ramulus to catalyze the selective hydrogenation of XN and DMX, thereby preventing further cyclization, interferes with the formation of estrogenic flavanone 8PN in vivo. Enzymes responsible for the isomerization and reduction of flavonoids by E. ramulus, chalcone isomerase and enoate reductase, have been reported. Herles et al. [51] showed a chalcone isomerase from E. ramulus catalyzes the conversion of naringenin chalcone to naringenin and demonstrated the ability of the enzyme to metabolize other chalcones such as isoliquiritigenin and butein, which have different hydroxylation patterns. In addition to this enzymatic reaction, E. ramulus chalcone isomerase is responsible for the reverse reaction, transforming flavanones including naringenin, eriodictyol and homoeriodictyol into their corresponding chalcones [52]. While an enoate reductase reduces the α,β-double bond in chalcones catalyzing their conversion into corresponding dihydrochalcones [52]. In our study, the conversion of 8PN into DMX is likely to be mediated by bacterial chalcone isomerase and the conversion of XN into DXN as well as DMX into DDXN by an enoate reductase from E. ramulus.
In addition to both of these enzymes, Schoefer and co-workers identified a hydrolase encoded by E. ramulus involved in the metabolism of flavonoids [53]. Enzymatic cleavage of C–C bonds in flavonoids appears to be characteristic of gut microbiota and can take place in the heterocyclic C-ring of flavonols, flavones, flavanones, and anthocyanidins, as well as in the aromatic ring A of flavanols [39]. This enzyme called phloretin hydrolase can cleave the C-C ring of several dihydrochalcones including phloretin, thereby producing phenolic acids. However, our results show no detectable formation of hydroxyphenylacetic and hydroxyphenylpropionic acids from C-ring cleavage of the prenylated flavonoids by E. ramulus.
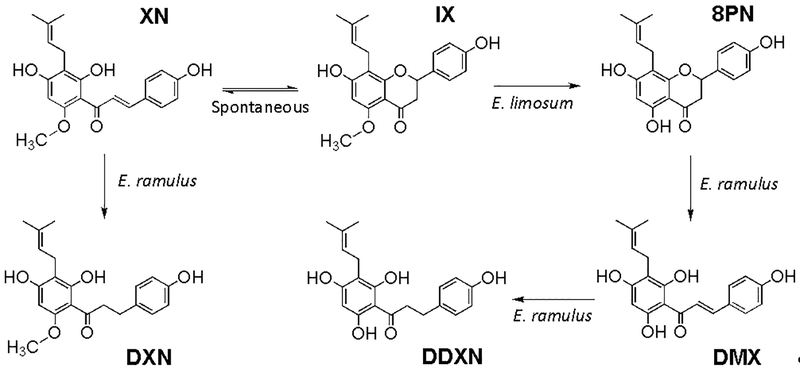
Based on our current findings and previous knowledge of XN metabolism, we propose a mechanism of degradation of XN by intestinal bacteria E. ramulus and E. limosum as shown in Fig. 5.
Figure 5.
Metabolism of XN and related prenylated flavonoids by gut bacteria E. ramulus and E. limosum. XN is spontaneously converted into IX, which is converted to 8PN by E. limosum [8]. E. ramulus is responsible for the formation of DXN from XN and DDXN from 8PN.
5. Conclusion
Complete metabolism of polyphenols in the gut involves a vast diversity of microbes. Although it has become clear that understanding the complex metabolic fate of dietary polyphenols is crucial to maximize the overall health effects from their intake, the diversity and abundance of the human microbiota, as well as cultivation-related problems present an experimental challenge. Furthermore, in vivo metabolism could largely fluctuate from one individual to another due to interindividual differences in the gut microbiota composition. Our investigation showed that XN, 8PN and DMX are readily metabolized by E. ramulus in vitro. This highlights that focus should remain on investigating intestinal microbial metabolism, which is likely to affect both activity and toxicity of ingested molecules.
Supplementary Material
Acknowledgement
This work was supported by the Linus Pauling Institute, the OSU College of Pharmacy, Hopsteiner, Inc., New York, the OSU Foundation Buhler-Wang Research Fund, and the National Institutes of Health (Grants R01AT009168 and R01AT010271).
Abbreviations
- 8PN
8-prenylnaringenin
- DDXN
Desmethyl-α,β-dihydroxanthohumol
- DMX
Desmethylxanthohumol
- DXN
α,β-dihydroxanthohumol
- IX
Isoxanthohumol
- XN
Xanthohumol
Footnotes
Conflict of interests: The authors declare no conflict of interest.
Supporting information accompanies this paper
References
- [1].Stevens JF, Page JE, Phytochemistry 2004, 65, 1317. [DOI] [PubMed] [Google Scholar]
- [2].Legette LL, Luna AYM, Reed RL, Miranda CL, Bobe G, Proteau RR, Stevens JF, Phytochemistry 2013, 91, 236. [DOI] [PubMed] [Google Scholar]
- [3].Gerhauser C, Alt A, Heiss E, Gamal-Eldeen A, Klimo K, Knauft J, Neumann I, Scherf HR, Frank N, Bartsch H, Becker H, Mol. Cancer Ther 2002, 1, 959. [PubMed] [Google Scholar]
- [4].Dietz BM, Kang YH, Liu G, Eggler AL, Yao P, Chadwick LR, Pauli GF, Farnsworth NR, Mesecar AD, Van Breemen RB, Bolton JL, Chem. Res. Toxicol 2005, 18, 1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Goto K, Asai T, Hara S, Namatame I, Tomoda H, Ikemoto M, Oku N, Cancer Lett. 2005, 219, 215. [DOI] [PubMed] [Google Scholar]
- [6].Zamzow DR, Elias V, Legette LL, Choi J, Stevens JF, Magnusson KR, Behav. Brain Res. 2014, 275, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Legette L, Ma L, Reed RL, Miranda CL, Christensen JM, Rodriguez‐Proteau R, Stevens JF, Mol. Nutr. Food Res 2012, 56, 466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Possemiers S, Heyerick A, Robbens V, De Keukeleire D, Verstraete W, Agric J. Food Chem. 2005, 53, 6281. [DOI] [PubMed] [Google Scholar]
- [9].Milligan S, Kalita J, Pocock V, Heyerick A, De Cooman L, Rong H, De Keukeleire D, Reproduction 2002, 123, 235. [PubMed] [Google Scholar]
- [10].Diel P, Thomae RB, Caldarelli A, Zierau O, Kolba S, Schmidt S, Schwab P, Metz P, Vollmer G, Planta Med. 2004, 70, 39. [DOI] [PubMed] [Google Scholar]
- [11].Miyamoto M, Matsushita Y, Kiyokawa A, Fukuda C, Sugano M, Akiyama T, Planta Med. 1998, 64, 516. [DOI] [PubMed] [Google Scholar]
- [12].Pepper MS, Hazel SJ, Hümpel M, Schleuning WD, J. Cell. Physiol 2004, 199, 98. [DOI] [PubMed] [Google Scholar]
- [13].Rong H, Boterberg T, Maubach J, Stove C, Depypere H, Van Slambrouck S, Serreyn R, De Keukeleire D, Mareel M, Bracke M, Eur. J. Cell Biol 2001, 80, 580. [DOI] [PubMed] [Google Scholar]
- [14].Zierau O, Morrissey C, Watson RWG, Schwab P, Kolba S, Metz P, Vollmer G, Planta Med. 2003, 69, 856. [DOI] [PubMed] [Google Scholar]
- [15].Manach C, Hubert J, Llorach R, Scalbert A, Mol. Nutr. Food Res 2009, 53, 1303. [DOI] [PubMed] [Google Scholar]
- [16].van Dorsten FA, Grün CH, van Velzen EJ, Jacobs DM, Draijer R, van Duynhoven JP, Mol. Nutr. Food Res 2010, 54, 897. [DOI] [PubMed] [Google Scholar]
- [17].Rechner AR, Smith MA, Kuhnle G, Gibson GR, Debnam ES, Srai SKS, Moore KP, Rice-Evans CA, Free Radical Biol. Med 2004, 36, 212. [DOI] [PubMed] [Google Scholar]
- [18].Kemperman RA, Bolca S, Roger LC, Vaughan EE, Microbiology 2010, 156, 3224. [DOI] [PubMed] [Google Scholar]
- [19].Crozier A, Jaganath IB, Clifford MN, Nat. Prod. Rep 2009, 26, 1001. [DOI] [PubMed] [Google Scholar]
- [20].Possemiers S, Bolca S, Grootaert C, Heyerick A, Decroos K, Dhooge W, De Keukeleire D, Rabot S, Verstraete W, Van de Wiele T, Nutr J 2006, 136, 1862. [DOI] [PubMed] [Google Scholar]
- [21].Possemiers S, Rabot S, Espìn JC, Bruneau A, Philippe C, González-Sarrías A, Heyerick A, Tomás-Barberán FA, De Keukeleire D, Verstraete W, J. Nutr 2008, 138, 1310. [DOI] [PubMed] [Google Scholar]
- [22].Hanske L, Loh G, Sczesny S, Blaut M, Braune A, Mol. Nutr. Food Res 2010, 54, 1405. [DOI] [PubMed] [Google Scholar]
- [23].Fujimura KE, Slusher NA, Cabana MD, Lynch SV, Expert Rev. Anti infe 2010, 8, 435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Bolca S, Van de Wiele T, Possemiers S, Curr. Opin. Biotechnol 2013, 24, 220. [DOI] [PubMed] [Google Scholar]
- [25].Salminen S, Isolauri E, Salminen E, Antonie Van Leeuwenhoek 1996, 70, 347. [DOI] [PubMed] [Google Scholar]
- [26].Williamson G, Clifford MN, Br. J. Nutr 2010, 104, S48. [DOI] [PubMed] [Google Scholar]
- [27].Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Nature 2010, 464, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA, Science 2005, 308, 1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Rajilić-Stojanović M, de Vos WM, FEMS Microbiol. Rev 2014, 38, 996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Louis P, Flint HJ, FEMS Microbiol. Lett 2009, 294, 1. [DOI] [PubMed] [Google Scholar]
- [31].Moore WEC, Holdeman LV, J. Appl. Microbiol 1974, 27, 961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Simmering R, Pforte H, Jacobasch G, Blaut M, FEMS Microbiol. Ecol 2002, 40, 243. [DOI] [PubMed] [Google Scholar]
- [33].Schneider H, Blaut M, Arch. Microbiol 2000, 173, 71. [DOI] [PubMed] [Google Scholar]
- [34].Braune A, Gütschow M, Engst W, Blaut M, Appl. Environ. Microbiol 2001, 67, 5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Stevens JF, Ivancic M, Hsu VL, Deinzer ML, Phytochemistry 1997, 44, 1575. [Google Scholar]
- [36].Stevens JF, Taylor AW, Nickerson GB, Ivancic M, Henning J, Haunold A, Deinzer ML, Phytochemistry 2000, 53, 759. [DOI] [PubMed] [Google Scholar]
- [37].Miranda CL, Johnson LA, Montgolfier O, Elias VD, Ullrich LS, Hay JJ, Paraiso IL, Choi J, Reed RL, Revel JS, Kioussi C, Sci. Rep 2018, 8, 613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Etteldorf N, Etteldorf N, Becker H, Zeitschrift für Naturforschung C 1999, 54, 610. [Google Scholar]
- [39].Stevens JF, Maier CS, Phytochem. Rev 2016, 15, 425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Nature 2009, 457, 480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Guinane CM, Cotter PD, Therap. Adv. Gastroenterol 2013, 6, 295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Coldham NG, Horton R, Byford MF, Sauer MJ, Food Addit. Contam 2002, 19, 1138. [DOI] [PubMed] [Google Scholar]
- [43].Schaefer O, Bohlmann R, Schleuning WD, Schulze-Forster K, Hümpel M, Agric J. Food Chem. 2005, 53, 2881. [DOI] [PubMed] [Google Scholar]
- [44].Milligan SR, Kalita JC, Pocock V, Van De Kauter V, Stevens JF, Deinzer ML, Rong H, De Keukeleire D, J. Clin. Endocrinol. Metab 2000, 85, 4912. [DOI] [PubMed] [Google Scholar]
- [45].Nikolic D, Li Y, Chadwick LR, Pauli GF, Van Breemen RB, J. Mass Spectrom. 2005, 40, 289. [DOI] [PubMed] [Google Scholar]
- [46].Tronina T, Bartmańska A, Filip-Psurska B, Wietrzyk J, Popłoński J, Huszcza E, Bioorg. Med. Chem 2013, 21, 2001. [DOI] [PubMed] [Google Scholar]
- [47].Hur HG, Rafii F, FEMS Microbiol. Lett 2000, 192, 21. [DOI] [PubMed] [Google Scholar]
- [48].Chen JX, Deng CY, Zhang YT, Liu ZM, Wang PZ, Liu SL, Qian W, Yang DH, Appl. Microbiol. Biotechnol 2016, 100, 9111. [DOI] [PubMed] [Google Scholar]
- [49].Miranda CL, Elias VD, Hay JJ, Choi J, Reed RL, Stevens JF, Arch. Biochem. Biophys 2016, 599, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Miranda CL, Stevens JF, Ivanov V, McCall M, Frei B, Deinzer ML, Buhler DR, Agric J. Food Chem. 2000, 48, 3876. [DOI] [PubMed] [Google Scholar]
- [51].Herles C, Braune A, Blaut M, Arch. Microbiol 2004, 181, 428. [DOI] [PubMed] [Google Scholar]
- [52].Gall M, Thomsen M, Peters C, Pavlidis IV, Jonczyk P, Grünert PP, Beutel S, Scheper T, Gross E, Backes M, Geißler T, Angew. Chem., Int. Ed 2014, 53, 1439. [DOI] [PubMed] [Google Scholar]
- [53].Schoefer L, Braune A, Blaut M, Appl. Environ. Microbiol 2004, 70, 6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.