Abstract
Over 40 species of nonhuman primates host simian immunodeficiency viruses (SIVs). In natural hosts, infection is generally assumed to be nonpathogenic due to a long coevolutionary history between host and virus, although pathogenicity is difficult to study in wild nonhuman primates. We used whole-blood RNA-seq and SIV prevalence from 29 wild Ugandan red colobus (Piliocolobus tephrosceles) to assess the effects of SIV infection on host gene expression in wild, naturally SIV-infected primates. We found no evidence for chronic immune activation in infected individuals, suggesting that SIV is not immunocompromising in this species, in contrast to human immunodeficiency virus in humans. Notably, an immunosuppressive gene, CD101, was upregulated in infected individuals. This gene has not been previously described in the context of nonpathogenic SIV infection. This expands the known variation associated with SIV infection in natural hosts and may suggest a novel mechanism for tolerance of SIV infection in the Ugandan red colobus.
Keywords: red colobus, simian immunodeficiency virus, gene expression, CD101
Introduction
The effects of infectious pathogens on host physiology are often influenced by coevolution between host and pathogen. A prominent example is the human immunodeficiency viruses (HIVs), the causative agents of acquired immunodeficiency syndrome (AIDS), which have close relatives in nonhuman primates (Hirsch et al. 1989; Gao et al. 1999). In contrast to HIV, simian immunodeficiency virus (SIV) infection in natural hosts is generally considered nonpathogenic, reflecting a long history of coevolution favoring both benignness in the virus and adaption for tolerance by the host (Chahroudi et al. 2012; Jasinska et al. 2013). Despite there being over 40 species of nonhuman primates naturally infected with endemic strains of SIV, studies on the pathogenicity of SIV infection in natural hosts have been limited to chimpanzees and three species within the Cercopithecinae (African green monkey [AGM], sooty mangabey [SM], and mandrill [MND] ;Bosinger et al. 2009; Jacquelin et al. 2009; Keele et al. 2009; Apetrei et al. 2011; Ma et al. 2013; Greenwood et al. 2014). Recently, studies in SIVcpz-infected chimpanzees (Keele et al. 2009) and SIVmnd-1-infected MNDs (Greenwood et al. 2014) have demonstrated variation in the pathogenic effects of SIV infection in natural hosts, but to our knowledge no study has assessed the relationship between SIV infection and host gene expression patterns in a wild population of natural SIV hosts. It has become increasingly clear that the effects of SIV infection vary across natural hosts, and the assumption of nonpathogenic SIV infection in natural hosts warrants further testing in a wider breadth of taxa, particularly in understudied groups such as the Colobinae (Bosinger et al. 2013; Palesch et al. 2018).
Nonpathogenic SIV Infection
During acute SIV infection, both pathogenic and nonpathogenic infections are characterized by a rapid induction of Type I interferon response (signaling proteins produced in response to viral infections that initiate a signaling cascade of downstream antiviral effector proteins; Sadler and Williams 2008), upregulation of interferon-stimulated gene (ISG) expression, high viral loads, and a decrease in peripheral CD4+ T cell counts (reviewed by Chahroudi et al. [2012]). Nonpathogenic SIV infection is distinguished from pathogenic infection during the transition from acute to chronic phase, ∼21–40 days after infection (Mir et al. 2011). During this period, one of the hallmark features of nonpathogenic SIV infection is the rapid attenuation of ISG expression to baseline levels in the chronic phase (Bosinger et al. 2009; Jacquelin et al. 2009; Lederer et al. 2009). Additionally, natural hosts maintain high viral loads but recover CD4+ T cell homeostasis, though variation in the effects of SIV infection on CD4+ T cell homeostasis has been observed in natural hosts (Keele et al. 2009; Greenwood et al. 2014). Although the upregulation of ISGs persists in pathogenic SIV/HIV infection in macaques and humans (Paiardini and Müller-Trutwin 2013), rapid control of ISG expression occurs in natural hosts (AGMs and SMs) within 4–8 weeks postinfection (Bosinger et al. 2009; Jacquelin et al. 2009). The gene expression patterns underlying this downregulation of ISGs have only been investigated in experimentally infected captive populations of two species, AGMs and SMs. Research on AGMs and SMs suggests that natural hosts actively downregulate the innate and adaptive immune response following infection (Chahroudi et al. 2012), and studies have proposed negative regulatory mechanisms by which the acute immune response is attenuated through the upregulation of a small number of immunosuppressive genes, including IDO, LAG3, IL10, and LGALS3 in AGMs (Jacquelin et al. 2009) and ADAR in SMs (Bosinger et al. 2009).
SIVkrc Infection in the Ugandan Red Colobus
Natural populations of Ugandan red colobus (URC; Piliocolobus tephrosceles; Old World monkey subfamily Colobinae) have emerged as an important wild primate species for understanding the role of host biology in viral disease (Goldberg et al. 2008, 2009; Lauck et al. 2011, 2013; Bailey et al. 2014; Sibley et al. 2014; Bailey, Lauck, Ghai, et al. 2016; Bailey, Lauck, Sibley, et al. 2016; Ladner et al. 2016). SIV diversity in red colobus was first described in western red colobus (Piliocolobus badius) in Taï forest in Côte d’ Ivoire and established the red colobus as a potential reservoir for viral transmission to humans (Locatelli, Liegeois, et al. 2008). Full SIV genome characterization from a different western red colobus (Piliocolobustemminckii) from the Abuko Nature Reserve, the Gambia, showed that SIV lineages in red colobus were species specific (Locatelli, Lafay, et al. 2008). Goldberg et al. (2009) showed that URC in Kibale National Park, Uganda, are natural hosts for a novel SIV lineage designated SIVkrc.
Genomic studies of host responses to SIV infection in natural populations have been limited by the difficulty of obtaining and preserving high-quality biomaterials from animals that exist in remote locations and are often endangered. Transcriptomic approaches to understanding the effects of SIV infection on host gene expression patterns have therefore been limited to studies of captive populations of AGMs and SMs, in which the effects of experimental infection have been assessed with the use of microarrays (Bosinger et al. 2009; Jacquelin et al. 2009). Those studies provide valuable hypotheses, which can now be tested in wild populations of naturally infected hosts, to examine the effects of SIV infection in natural hosts within the ecological context of host–virus coevolution.
In this study, we used gene expression profiles from SIVkrc-infected and SIVkrc-uninfected URC from Kibale National Park, Uganda, to address three questions. 1) Do ISG expression patterns differ between naturally SIVkrc-infected and -uninfected URC? Both AGMs and SMs mount an acute ISG response that is attenuated in the chronic phase (Bosinger et al. 2009; Jacquelin et al. 2009). Given that signs of immunodeficiency are difficult to observe in URC, and our study subjects are presumed to be in the chronic phase of infection, we hypothesize that levels of ISG expression will be attenuated to baseline levels similar to uninfected URC. 2) Is the expression of immunosuppressive genes elevated in SIVkrc-infected URC relative to uninfected URC? Upregulation of immunosuppressive genes upon infection and sustained through the chronic phase has been observed in AGMs and SMs (Bosinger et al. 2009; Jacquelin et al. 2009). We hypothesize that SIVkrc-infected URC will show upregulation of immunosuppressive genes compared with uninfected URC. 3) Do SIVkrc-infected URC show CD4+ T cell depletion compared with uninfected URC? The effect of SIV infection on T cell subset populations varies across natural hosts, as AGMs and SMs maintain CD4+ T cell levels, whereas SIVmnd-1-infected MNDs and SIVcpz-infected chimpanzees show a loss of memory CD4+ T cells (Keele et al. 2009; Chahroudi et al. 2012; Greenwood et al. 2014), which provide immunological memory as part of the adaptive immune response (Harrington et al. 2008). Given the different SIV infection patterns in AGMs, SMs, MNDs, and chimpanzees, we test the null hypothesis that SIVkrc-infected URC will maintain CD4+ T cell levels comparable to uninfected URC.
This study represents the first high-resolution transcriptomic analysis of the relationship between SIV infection and host gene expression in a wild population of naturally infected SIV hosts. It is also the first study of host gene expression patterns in relation to SIV infection in a colobine and adds considerably to our current knowledge about variation in host patterns of gene expression across natural SIV hosts.
Materials and Methods
Ethics Statement
Animal use for this study followed the guidelines of the Weatherall Report (Weatherall 2006) on the use of NHPs in research. Our study was approved by the Uganda National Council for Science and Technology, Uganda Wildlife Authority, and the University of Wisconsin-Madison and University of Oregon Animal Care and Use Committees prior to the start of the study (University of Wisconsin-Madison IACUC #A3368-01; University of Oregon IACUC #15-06A). Biomaterials were shipped from Uganda to the United States under CITES permit #002290.
Study System and Sample Collection
This study is part of the Kibale EcoHealth Project, a long-term investigation of health and ecology across the human and wildlife community in and surrounding Kibale National Park, Uganda (0°13′-0°41′N, 30°19′-30°32′E; Goldberg et al. 2012). The URC, an arboreal Old World monkey in the subfamily Colobinae, is one of the 13 primate species found in Kibale (Struhsaker 2005). The Small Camp group is a well-habituated social group that has been a focus of the Kibale EcoHealth Project since 2005 (e.g., Chapman et al. 2005, 2012; Goldberg et al. 2008, 2009; Lauck et al. 2011, 2013; Simons et al. 2016). Samples for this study were collected from 29 adult URC (M = 21, F = 8) from the Small Camp group. Sample size considerations in differential expression analysis differ from classic ecological experiments as they are a trade-off between biological replicates and sequencing depth. Our sample size (infected n = 12; uninfected n = 17) exceeded both that of similar studies (Bosinger et al. 2009; Jacquelin et al. 2009), and recommendations from a benchmarking study on sample size for detecting differentially expressed genes at all fold changes (Schurch et al. 2016). Another benchmarking study by Liu et al. (2014) showed diminishing returns in the power to detect differentially expressed genes beyond a sequencing depth of 10M reads per replicate. Our study also exceeded that recommendation with a minimum of 19.6M reads, and average of 23.6M reads per replicate (see sequencing methods below).
Animals were immobilized in the field and blood plasma was collected as previously described (Lauck et al. 2011). Briefly, animals were anesthetized with a combination of xylazine (1.7 mg/kg) and ketamine (5.6 mg/kg) administered intramuscularly. The animals were then administered a reversal agent (atipamezole, 0.6 mg/kg) and returned to their social group following recovery. In addition, whole blood was collected using a modified PreAnalytiX PAXgene Blood RNA System protocol (Qiagen, Hilden, Germany). For each animal, ∼2.5 ml of blood was collected into a PAXgene Blood RNA tube, inverted ten times and stored at room temperature for 2–4 h. The blood and PAXgene buffer mixture was then aliquoted into 1-ml cryovials and stored at −20 °C overnight before being stored in liquid nitrogen. Samples were transported to the United States in an IATA-approved liquid nitrogen dry shipper and then transferred to −80 °C for storage until further processing.
The use of whole blood was motivated by several factors: 1) it allowed comparison to the only other studies of host gene regulatory responses to nonpathogenic SIV infection, as they also used whole blood (Bosinger et al. 2009; Jacquelin et al. 2009; Lederer et al. 2009), 2) whole blood is superior to PBMCs for transcriptomic analysis when storage is required as was the case for our study (Debey-Pascher et al. 2011), 3) global transcriptomic profiles from whole blood account for latent SIV reservoirs and may therefore be better for understanding immune responses to SIV infection (Kandathil et al. 2016), and 4) ISG expression profiles of SIV-infected SMs are largely concordant between whole blood and the lymph node (an important site of immune activity in response to SIV/HIV infection), enabling accurate inference of the broad transcriptomic responses to infection conserved between lymph nodes and whole blood (Bosinger et al. 2009).
Viral Prevalence and Load
We assessed SIVkrc infection prevalence and viral loads using both serology and quantitative reverse transcription PCR (RT-qPCR). SIVkrc prevalence was assessed from serum for each individual with an HIV-2 western blot assay (MP Diagnostics HIV-2 Western Blot version 1.2; MP Biomedicals, Singapore) capable of detecting divergent SIVs (Lauck et al. 2013).
Viral load was determined by RT-qPCR detection of both SIVkrc envelope (env) and long terminal repeat (LTR) sequences. Viral RNA was extracted from plasma using the QIAamp Viral RNA minikit (Qiagen, Valencia, CA). For the env qPCR assay, the primers SIVkrc env F2 5′ GCA AGG TAT GAG ATC CCY AGG CA 3′ and SIVkrc env R4, 5′ GYA AAG CCT GGG ACT GGA TCG T 3′ and probe SIVkrc PR2 5′ FAM-AGA TAC TAA GCC CAT TGC AGT TCC “T”GC AAA GCT CAG-SpC6 3′, where “T” is labeled with BHQ1, were used to detect a 127-bp sequence. For the LTR qPCR assay, the primers SIVkrc 5LTR F 5′ GAG GTC TGT GAA GAG TGC CG 3′ and SIVkrc 5LTR R 5′ CCT TGG GGA ACT AMA CCG TC 3′ and probe SIVkrc LTR PR 5′ FAM-CTG GCA CCT CCC TG “T” ACC AGC CGC CAG TCA-SpC6 3′, where “T” is labeled with BHQ1, were used to detect a 94-bp sequence. One microliter of RNA extract equivalent to 50-µl plasma was tested using the AgPath-ID One Step RT-PCR kit (Applied Biosystems, Foster City, CA) on a BioRad CFX96 iCycler (Hercules, CA) with a reverse transcriptase step at 45 °C for 15 min, denaturation at 95 °C for 10 min, followed by 55 cycles of 95 and 62 °C for 15 s each. Both env and LTR PCR products generated from one WB-positive URC (RC51) were cloned into the pCR 2.1 TOPO vector (Thermo Fisher, Waltham, MA) and RNA standards for the qPCR assays were synthesized in vitro using the RiboMAX large-scale RNA production system-T7 (Promega, Madison, WI) following the manufacturer’s instructions. Both assays could detect SIVkrc RNA in a linear range from 101 to 107 copies/reaction and could reliably detect five viral copies/reaction. All no-template and negative plasma controls always tested nonreactive.
RNA Extraction, Library Preparation, and Sequencing
Blood samples were thawed at room temperature for 2 h, and total RNA extraction followed the PreAnalytiX PAXgene Blood RNA Kit protocol with the exception that reagents were scaled for extraction of 1 ml rather than the full PAXgene Blood tube volume. Extracts were concentrated following the manufacturers protocol for the ZYMO Research RNA Clean and Concentrator kit (Irvine, CA). Alpha and beta globin mRNA was depleted from total RNA extracts following the manufacturer’s protocol for the GLOBINclear Kit (Waltham, MA). Prior to library preparation, integrity of globin-depleted total RNA was assessed on an AATI Fragment Analyzer (Ankeny, IA), and all 29 samples (RIN mean: 8.1, range: 6.6–9.2) were used for downstream analyses. Sequencing libraries were prepared from globin-depleted total RNA using the KAPA Biosystems Stranded mRNA-seq Kit (Wilmington, MA) and indexed with Illumina adapters for multiplexing all 29 individuals on a single lane. This library prep enriches for mRNA by capturing polyadenylated transcripts to the exclusion of rRNA. Libraries were sequenced on three replicate Illumina NextSeq 500 high output runs with single-end 150 base-pair reads at the Genomics & Cell Characterization Core Facility at the University of Oregon.
Mapping and Quantification
Demultiplexing, adapter trimming, and initial quality filtering were performed with default parameters within the Illumina NextSeq base calling software. Quality was assessed in FastQC v0.11.3 and reads for each individual were concatenated from three NextSeq runs. Additional quality filtering was performed with FASTX-toolkit, including trimming the first 15 base pairs from all reads, and removal of reads with quality score <Q20, and reads <25 base-pair length. Quality filtered RNA-seq reads were mapped to the URC genome (assembly accession: GCF_002776525.1) using STAR v.2.5 in two-pass mode with –outFilterMismatchNoverLmax 0.05. The number of reads mapping to each feature in the annotation was quantified using HTSeq v.0.9.1 (Anders et al. 2015) in intersection-nonempty mode with the –-stranded reverse option.
Data Visualization
We used multiple visualization methods to assess the degree to which SIVkrc infection separated our study subjects into distinct clusters. The feature counts matrix was preprocessed by removing low count genes for visualization. Genes with less than one count per million, calculated with CPM function in edgeR (Robinson et al. 2010), in at least three samples were removed. Missing values were treated as zeroes. The resulting data set was then normalized using the variance stabilizing transformation (VST) as implemented in DESeq2 v.1.16.1 (Anders and Huber 2010; Love et al. 2014) for visualization with hierarchical clustering, principal component analysis (PCA), and k-means clustering.
Hierarchical Clustering
Hierarchical clustering was performed using the 1,000 most variable genes (based on standard deviation ranks of all genes) with sample clustering based on the distance matrix (1 − r, where r is Pearson’s correlation coefficient). Hierarchical clustering was visualized as a dendrogram based on sample distances.
Principal Component Analysis
To identify whether infected and uninfected samples clustered together, we visualized our data in two-dimensional space using PCA. PCA graph was plotted using VST transformed data in R v.3.3.3 (R Development Core Team 2008). PCA loadings were attributed to genes (similar to expression values) and pathway analysis was conducted on the first four principal components using pgsea (Furge and Dykema 2006) in R v.3.3.3. The false discovery rates (FDRs; Benjamini and Hochberg 1995) of significant pathways are reported for each principal component.
K-Means Clustering
K-means clustering was used to cluster groups of genes based on their expression patterns in the blood transcriptome of infected and uninfected individuals. Genes were first ranked by standard deviation, and the top 2,000 most variable genes were used for clustering. The optimal number of clusters was determined based on the elbow method, wherein the addition of additional clusters does not substantially reduce the within groups sum of squares. The number of clusters was confirmed with t-SNE mapping (supplementary fig. S1, Supplementary Material online). Enriched Gene Ontology (GO) pathways were then identified for genes in each cluster using GSEA (Subramanian et al. 2005). For each cluster with significantly enriched pathways, a promoter analysis was conducted using a 300-bp region upstream of the transcription start site. For genes in significantly enriched pathways, the binding affinity of promoters to transcription factor binding sites was estimated using CIS-BP (Weirauch et al. 2014). Rather than using an arbitrary cutoff for a binary outcome of binding versus nonbinding, the best binding score for each transcription factor to every promoter sequence was used to test (Student’s t-test) for transcription factor binding motifs enriched in clusters. P values of enriched transcription factor binding motifs were corrected for multiple testing and FDR is reported.
Weighted Gene Coexpression Network Analysis
We identified coexpression networks and modules in SIVkrc-infected and -uninfected individuals using weighted gene coexpression network analysis (WGCNA; Langfelder and Horvath 2008). Coexpression networks were identified from the top 3,000 most variable genes. To reduce noise in the correlation matrix, we used a soft threshold based on the smallest power at which the scale-free topology index reached 0.9 and limited the minimum module size to 20 genes. Genes in each of the modules within each network were analyzed for enriched GO biological processes using gene set enrichment analysis. Gene set enrichment analysis was performed using GSEA as implemented in iDEP.53 (Ge et al. 2018) with a minimum and maximum geneset size filter of 10 and 2,000, respectively, and default FDR threshold of 0.2. GSEA was run in preranked mode with the GO biological process geneset background with the increased speed algorithm fsgea (Sergushichev 2016).
Differential Expression Analysis
We modeled the effects of SIVkrc infection on gene expression using a generalized linear model with a negative binomial distribution as implemented in DESeq2. Low count genes (e.g., those not typically expressed in blood) were not prefiltered for differential expression analysis as DESeq2 performs independent filtering to exclude genes with low power from FDR calculations, and by default optimizes the threshold to maximize the number of differentially expressed genes given a specified FDR (Love et al. 2014), thus allowing identification of potentially important differentially expressed genes that may be expressed at low levels. We modeled the effect of SIV infection on gene expression using a pairwise design (infected N = 12 vs. uninfected N = 17) with sex and RIN score as blocking factors. P values were corrected for multiple testing and the adjusted P values are reported as Q values (Storey and Tibshirani 2003). Because our sample size may have limited our power to detect differentially expressed genes, we followed Charruau et al. (2016) in setting a relatively liberal Q threshold of 0.2 for detecting candidate genes associated with infection. To determine possible functional enrichment in differentially expressed genes, we performed a GO term enrichment analysis in g:Profiler (Reimand et al. 2007) with minimum functional category and query size both set at 3, and with P values corrected using the default g:SCS threshold. We compared the differentially expressed genes to ISGs, chemokines, defensins, inhibitors, and antiviral restriction factors identified as being upregulated in acute infection and attenuated in the chronic phase in AGMs and SMs (Bosinger et al. 2009; Jacquelin et al. 2009) to assess if any of those genes were also upregulated in SIV-infected URC. We note that those two comparative studies were both performed with microarrays, and while the authors detail extensive efforts to ensure that AGM and SM hybridization to the arrays was comparable to rhesus macaque, the use of RNA-seq in our study provides an inherently unbiased view of transcriptome-wide patterns of gene expression. We also compared our differentially expressed genes to immunosuppressive genes that were upregulated in acute SIV infection and remained upregulated into the chronic phase of infection in AGMs and SMs. Finally, to identify immunosuppressive genes that might be upregulated in URC, but not AGMs or SMs, we compared our differentially expressed genes to a curated database of immunosuppressive genes in the Human Immunosuppression Gene Database (Liu et al. 2017).
Whole-Blood Deconvolution of Immune Cell Subsets
The effect of SIV infection on CD4+ T cell maintenance varies across AGMs, SMs, MNDs, and chimpanzees. Although AGMs and SMs maintain CD4+ T cell homeostasis, naturally infected MNDs and chimpanzees show a loss of memory CD4+ T cells (Keele et al. 2009; Greenwood et al. 2014). We obtained whole blood from a natural population in a remote field site and could therefore not perform immunohistochemistry and flow cytometry to isolate T cell subsets. To test the effect of SIVkrc infection on CD4+ T cell levels we estimated the relative abundance of immune cell subsets using the computational deconvolution method implemented in CIBERSORT (Newman et al. 2015). This is a robust method for determining the relative proportion of immune cell subsets (including T cells, memory B cells, natural killer cells, monocytes, macrophages, and neutrophils), and is superior to other deconvolution methods in accuracy, sensitivity to noise, and resolution of closely related cell types, particularly for whole-blood deconvolution (Newman et al. 2015; Wang et al. 2016). CIBERSORT was run with 1,000 permutations, and accurate deconvolution of cell types for each sample was assessed with goodness of fit. Differences in relative fractions of naïve, memory resting, memory activated, and total CD4+ T cells between SIVkrc-infected and -uninfected individuals were tested with a two-sample t-test.
Results
Sex Differences in SIVkrc Prevalence but Not Viral Load
To test the effects of SIVkrc infection on patterns of gene expression, we generated SIVkrc prevalence and viral load data from 29 individuals using two measures: 1) SIV antibody detection using HIV-2 western blot assays and 2) presence of SIV RNA envelope (env) and LTR sequences using two SIVkrc-specific RT-qPCR assays. Both measures of SIV infection (serology and RT-qPCR) were 100% concordant. SIVkrc infection prevalence in our study group was 41.4% (Wilson’s 95% confidence interval [CI] = 25.5–59.3%; Wilson 1927), with females having a substantially lower prevalence than males (F = 25% [CI = 7.1–59.1%], M = 52.6% [CI = 31.7–72.7%]; supplementary table S1, Supplementary Material online). Our observation of lower prevalence in females than males parallels that reported by Goldberg et al. (2009), and we observed an increase in overall prevalence of 18.8% since 2009. It should be noted that prevalence estimates are sensitive to sample size and the sex differences observed may be due to sample size constraints in our estimates. Differences observed in prevalence between males and females may also be the result of sex-bias in our dataset, as we have more than double the males than females in our sample. This type of sample bias could lead to a systematic underestimation in females relative to males. Finally, and we think most likely, this pattern could be due to the mode of transmission in URC. Aggressive behaviors are strongly skewed toward males in URC, therefore it is plausible that SIV transmission is higher in males due to the higher incidence of agonism-related injury (e.g., bites).
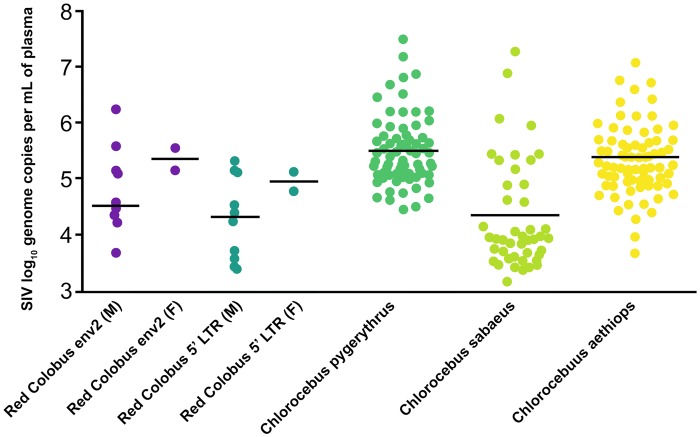
Viral load (reported as log10 genome copies per ml of plasma) estimates were similar between the env and LTR RT-qPCR assays (Mann–Whitney U = 94, Z = −1.24, P = 0.22; fig. 1) . There were no significant differences between males and females for either env (t[10]= 1.06, P = 0.31) or LTR (t[10] = 0.66, P = 0.26), and viral load for both measures were within the range of those reported using RT-qPCR of the viral int protein for three species of naturally SIV-infected AGMs (Ma et al. 2014; Bailey, Lauck, Ghai, et al. 2016; fig. 1).
Fig. 1.
—Viral load (reported as log10 genome copies per ml of plasma) generated with RT-qPCR of env and LTR for red colobus. Viral load data for Chlorocebus pygerythrus from Bailey, Lauck, Ghai, et al. (2016) and Chlorocebus sabaeus and Chlorocebus aethiops from Ma et al. (2014).
Sequence Data, Mapping, and Quantification
To identify gene expression differences between SIVkrc-infected and -uninfected URC, we quantified genome-wide gene expression from the same 29 URC individuals using RNA-seq. We generated 984.7 million 150 base-pair reads (average ∼33.95M reads per individual). We retained 871.2 million reads (average ∼30.04M reads per individual) after filtering. A high proportion of reads were successfully mapped to the most recent version of the annotated URC reference genome (GCF_002776525.1), with an average of 23.7M (78.3%, SD = 7.6) reads mapping uniquely, and average of 2.1M (6.6%, SD = 1.6) reads mapping to multiple locations. Of uniquely mapped reads, an average of 16.2M (67.8%, SD = 0.08) reads mapped to gene features in the red colobus annotation. Total mapped (combined unique and multimapped) reads averaged 25.7M (85.8%, SD = 8.5).
Individual Gene Expression Patterns Do Not Cluster According to Infection Status
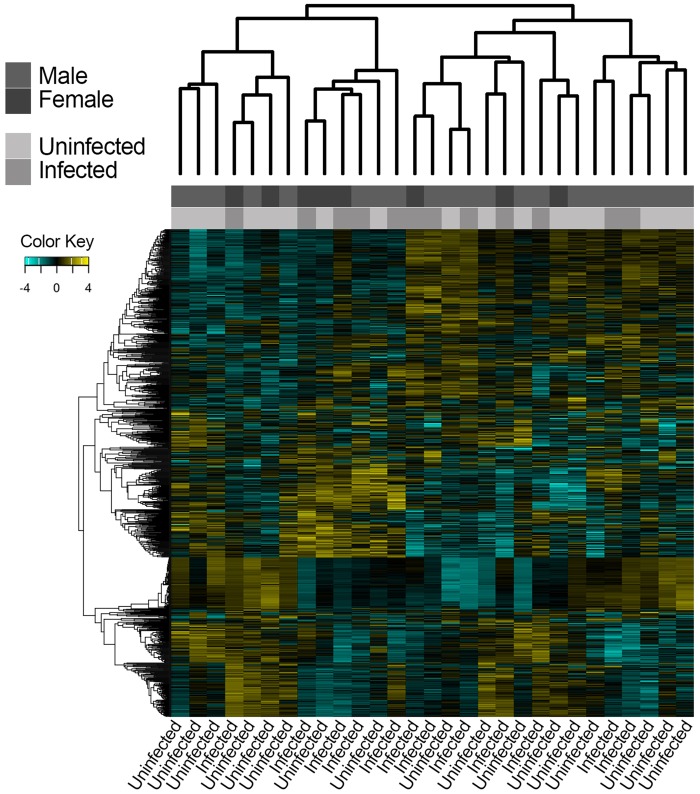
We used two-sample-based clustering methods to determine if individual gene expression patterns clustered according to infection status. After VST, a total of 17,583 genes were retained (filtered for genes with at least 1 CPM in at least 3 individuals) for clustering and visualization. Both hierarchical clustering based on sample distances (fig. 2) and PCA (fig. 3) indicated that individuals did not cluster according to infection status (infected vs. uninfected) or sex (M vs. F). Pathway analysis of loadings from PC1 found strong enrichment of pathways related to RNA processing (Q = 1 × 10e-292), cellular protein (Q = 1 × 10e-284), macromolecule localization (Q = 4 × 10e-287), and organelle organization (Q = 1 × 10e-293; supplementary table S2, Supplementary Material online). Pathway analysis of loadings from PC2 found that pathways related to ribosome biogenesis (Q = 1 × 10e-56) and ncRNA processing (Q = 1 × 10e-64) were enriched.
Fig. 2.
—Hierarchical clustering showing that individuals do not cluster according to infection status or sex.
Fig. 3.

—PCA of gene expression patterns showing that neither PC1 nor PC2 separate samples according to infection status or sex.
K-Means Clustering of Genes Identifies Fewer Enriched Immune Pathways in SIVkrc-Infected Individuals than Uninfected Individuals
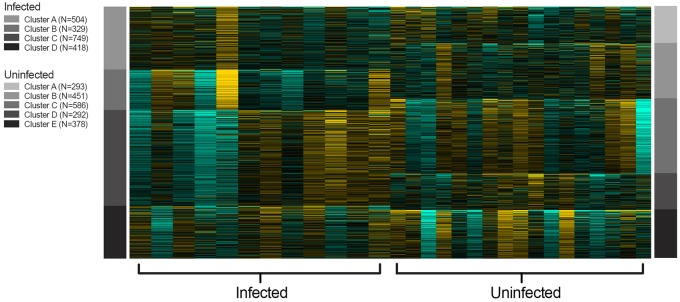
We used gene clustering to assess if infected individuals had more gene clusters enriched for immune-related pathways than uninfected individuals. K-means clustering identified five clusters of correlated genes in uninfected individuals (fig. 4), and four of the five (A, C, D, and E) were enriched primarily with immune-related pathways (supplementary table S3A, Supplementary Material online). Four clusters were identified (A–D) in infected individuals, of which two clusters (C and D) were enriched with immune-related pathways (supplementary table S3B, Supplementary Material online). Promoter analysis of clusters in uninfected individuals found 17 enriched transcription factor binding motifs in three clusters (B–D). Enriched binding motifs consisted of four transcription factor families: C2H2 ZF, bHLH, Rel, and SMAD (supplementary table S4A, Supplementary Material online). Promoter analysis in infected individuals found 38 enriched transcription factor binding motifs, consisting of seven transcription factor families: C2H2 ZF, E2F, Homeodomain, MBD, Myb/SANT, bHLH, Rel (supplementary table S4B, Supplementary Material online). Three transcription factor families (C2H2 ZF, bHLH, Rel) found in uninfected individuals were also found in infected individuals, suggesting global enrichment of those three families in the blood transcriptome of URC, regardless of infection status. Four transcription factor families were unique to infected individuals (MBD, Myb/SANT, Homeodomain, and E2F) and may play a role in the downregulation of the immune response during infection, as the specific transcription factors of three of the four (MBD, Myb/SANT, and E2F) families are transcriptional repressors.
Fig. 4.
—K-means clustering of top 2,000 most highly variable genes. K-means identified four and five gene clusters in infected and uninfected individuals, respectively.
Global Signatures of Immune Activity in the Blood Transcriptome
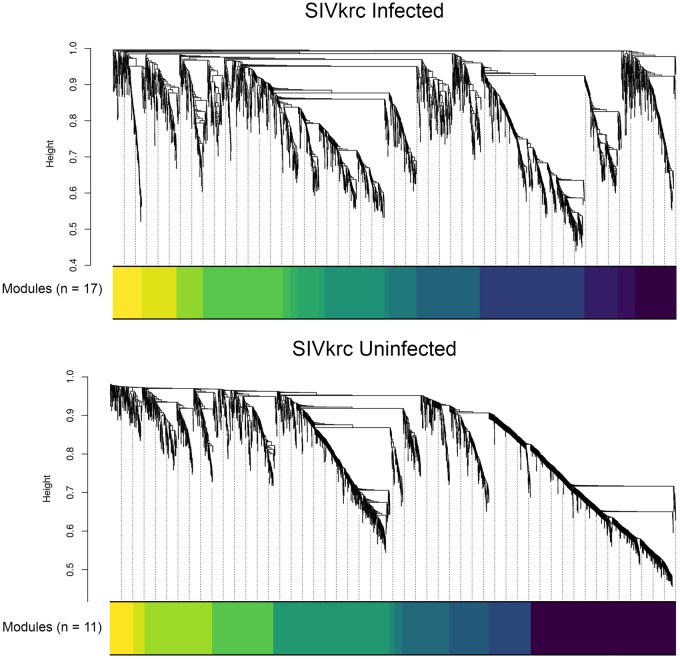
We used gene coexpression networks to identify correlated networks of genes and assess if those networks were enriched for immune-related pathways in infected individuals compared with uninfected individuals. WGCNA identified a network of 2,872 genes divided into 17 coexpression modules in infected individuals, and a network of 2,886 genes divided into 11 coexpression modules in uninfected individuals (fig. 5). GO pathway enrichment analysis found several highly enriched immune-related pathways for both infected and uninfected individuals, including immune system process, cell activation, response to external stimulus, leukocyte activation, immune response, and immune effector process, likely reflecting global patterns of immune function in the blood transcriptome. Immune-related GO pathways enriched only in infected individuals were all related to cellular immunity, including cell activation involved in immune response, leukocyte activation involved in immune response, and myeloid leukocyte activation. In contrast, the only GO pathway enriched in uninfected individuals, but not in infected individuals, was inflammatory response (supplementary table S5, Supplementary Material online).
Fig. 5.
—WGCNA identified a network of 2,872 genes divided into 17 coexpression modules in infected individuals, and a network of 2,886 genes divided into 11 coexpression modules in uninfected individuals.
Attenuated ISG and Upregulated Immunosuppressive Gene Expression in SIVkrc-Infected URC
We modeled the effects of SIVkrc infection on gene expression and identified 53 differentially expressed genes between SIVkrc-infected and -uninfected individuals, with 8 genes downregulated and 45 genes upregulated in infected individuals (supplementary table S6, Supplementary Material online). Genes downregulated in infected individuals included two immune genes, SLAMF6 (ENSG00000162739; fold change = −0.39; Q value = 0.12) and CD4 (ENSG00000010610; fold change = −1.58; Q value = 3 × 10e5). For both of these genes, there is evidence of virus-mediated downmodulation following infection. For example, for SLAMF6 (aka NTB-A), the HIV-1 accessory protein Vpu downmodulates SLAMF6 on infected T cell surfaces, thereby protecting infected cells from lysis by natural killer cells (Shah et al. 2010). Interestingly, SIVkrc lacks the Vpu accessory protein (Sakai et al. 2016). For CD4, which is the primary cell-entry receptor for both SIV/HIV, the viral accessory protein Nef downmodulates cell-surface expression of CD4 through endocytosis (Pham et al. 2014). Two GO terms were enriched in downregulated genes in infected individuals: positive regulation of biological process (Q value = 4.65E-04) and cytosolic calcium ion concentration (Q value = 4.99E-03; table 1). GO terms enriched in genes upregulated in infected individuals included broad categories related to cellular processes and biological regulation. There were no enriched GO terms related to immune response in significantly up- or down-regulated genes in infected individuals (table 1; full GO results in supplementary table S7, Supplementary Material online).
Table 1.
GO Terms Enriched in Up- and Down-Regulated Genes in SIVkrc-Infected Individuals
| Up |
Down |
||
|---|---|---|---|
| Top Enriched GO Term | Corrected GO P Value | Top Enriched GO Term | Corrected GO P Value |
| Cellular metabolic process | 8.09E-09 | Positive regulation of biological process | 4.65E-04 |
| Vesicle-mediated transport | 3.70E-07 | Positive regulation of cytosolic calcium ion concentration | 4.99E-03 |
| Regulation of catalytic activity | 7.49E-07 | ||
| Biological regulation | 8.45E-07 | ||
| Response to stimulus | 1.19E-06 | ||
| Regulation of biological process | 1.53E-06 | ||
Note.—Corrected P values are g:SCS corrected. See full GO results in supplementary table S7, Supplementary Material online.
No differentially expressed genes found in this study overlapped with ISGs upregulated during acute infection identified by Bosinger and colleagues in SMs (2009) or Jacquelin and colleagues in AGMs (2009), both cercopithecine primates. Similarly, no differentially expressed genes found in this study overlapped with the immunosuppressive genes identified in those studies. When our list of upregulated genes was searched against the Human Immunosuppressive Genes database, we identified one immunosuppressive gene, CD101 (ENSG00000134256), that was significantly upregulated in infected individuals (fold change = 0.96; Q value = 0.14).
Whole-Blood Deconvolution—CD4+ T Cell Abundance
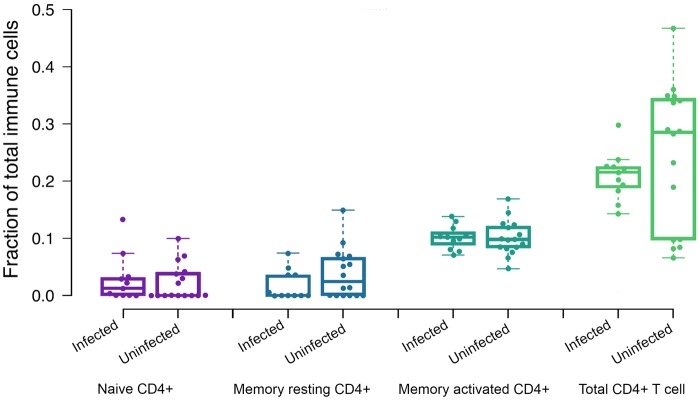
We determined the relative abundance of three CD4+ T cell subsets using computational deconvolution as implemented in CIBERSORT. The proportion of total CD4+ T cells, as a fraction of total immune cells, was 0.15, SD = 0.04. The proportions of three CD4+ T cell subsets as a fraction of total immune cells were significantly deconvoluted (P < 0.005), and abundance estimates were as follows: naïve CD4+ T cells (0.02, SD = 0.03), memory resting CD4+ T cells (0.02, SD = 0.03), and memory activated CD4+ T cells (0.10, SD = 0.02; table 2). There were no significant differences in the abundance of naïve, memory resting, memory activated, or total CD4+ T cells between infected and uninfected individuals (t[27] = 0.29, P = 0.77; t[27] = −1.4, P = 0.17; t[27] = −0.27, P = 0.78; t[27] = −1.1, P = 0.29, respectively; figure 6; full CIBERSORT results in supplementary table S8, Supplementary Material online). We also did not detect any significant differences in CD4+ T cell subsets between males and females (t[27] = −0.49, P = 0.63; table 2).
Table 2.
Relative CD4+ T Cell Subset and Total CD4+ T Cell Proportions (95% CI in Parentheses) in Infected and Uninfected Individuals, and Males and Females
| Naïve CD4+ T Cells | Memory Resting CD4+ T Cells | Memory Activated CD4+ T Cells | Total CD4+ T Cells | |
|---|---|---|---|---|
| SIVkrc infected | 0.025 (0.002–0.048) | 0.016 (0.002–0.031) | 0.1 (0.088–0.112) | 0.14 (0.118–0.167) |
| SIVkrc uninfected | 0.021 (0.007–0.036) | 0.036 (0.015–0.056) | 0.1 (0.088–0.117) | 0.16 (0.138–0.183) |
| P value | 0.78 | 0.17 | 0.79 | 0.30 |
| Males | 0.028 (0.012–0.045) | 0.023 (0.006–0.039) | 0.104 (0.092–0.116) | 0.156 (0.134–0.178) |
| Females | 0.009 (0.002–0.022) | 0.041 (0.019–0.062) | 0.095 (0.081–0.109) | 0.146 (0.0125–0.167) |
| P value | 0.20 | 0.25 | 0.41 | 0.63 |
| Total | 0.023 (0.011–0.036) | 0.028 (0.015–0.041) | 0.101 (0.092–0.111) | 0.152 (0.0136–0.169) |
Note.—P values denote no significant differences in cell proportions between infected and uninfected individuals or between males and females.
Fig. 6.
—Estimated relative proportions of CD4+ T cell subsets and total CD4+ T cells from gene expression deconvolution showing no significant difference between infected and uninfected individuals.
Discussion
Nonpathogenic SIV infection in natural hosts is not adequately defined because of the narrow set of taxa in which it has been investigated. Additionally, variation has been observed in outcomes related to infection, specifically the loss of CD4+ T cells in chimpanzees and MNDs (Keele et al. 2009; Greenwood et al. 2014), and decreased birth rate, increased infant mortality risk, and reduced longevity with findings consistent with end-stage AIDS in chimpanzees (Keele et al. 2009). Due to the small range of species in which nonpathogenic SIV infection has been studied at the molecular level, the degree to which patterns of host gene expression related to SIV infection are conserved across natural hosts is unknown. Our study helps address this gap by providing novel insights into the relationship between SIV infection and genome-wide host gene expression patterns for a primate species outside of the Cercopithecinae using high-resolution RNA-seq. The main findings from our study are that 1) genome-wide gene expression patterns in SIVkrc-infected versus -uninfected individuals support the hypothesis that SIVkrc has minimal effects on host immunoregulatory gene expression in URC, as we found no evidence for chronic immune gene activation or upregulated ISG expression in infected individuals and 2) that CD101, an immunosuppressive gene whose downregulation is related to HIV progression, was upregulated in SIVkrc-infected URC. The upregulation of an immunosuppressive gene parallels patterns observed in AGMs and SM, although each of the three host taxa shows upregulation of different immunosuppressive genes. This finding suggests that species-specific mechanisms may underlie tolerance to SIV infection in different African primate taxa. As CD101 has not been identified in previous studies (Bosinger et al. 2009; Jacquelin et al. 2009; Lederer et al. 2009), this finding expands our knowledge of natural host responses to SIV and highlights a potentially novel pathway to nonpathogenic SIV infection.
Staging of SIVkrc Infection in the Wild URC: Viral Load and Serology
Ma et al. (2013) defined AGMs as being in the acute phase of infection if they simultaneously had a high viral load and were seronegative, and in the chronic phase if they had high viral loads and were seropositive. The reasoning behind this approach is that high viral replication during acute infection generally precedes seroconversion (Fiebig et al. 2003), which can result in high viral load and seronegative results. Following this approach, we found that all of our SIVkrc-infected individuals were seropositive by HIV-2 western blot testing. This finding and our finding of no evidence for upregulated ISG expression in infected individuals strongly suggest that all infected individuals in our study were likely in the chronic phase of infection.
Shared Signatures of Gene Expression Associated with Nonpathogenic SIV Infection
We hypothesized that if SIVkrc infection is nonpathogenic in URC, then URC might share similar patterns of attenuated ISG expression in the chronic phase with AGMs and SMs. More specifically, we predicted that ISG expression would not be significantly upregulated in chronically infected URC compared with uninfected URC, as AGMs and SMs attenuate ISG expression in the chronic phase (Bosinger et al. 2009; Jacquelin et al. 2009; Lederer et al. 2009). We found evidence for attenuated ISG expression, as no ISGs were significantly upregulated in SIVkrc-infected URC compared with uninfected individuals. This result was also consistent with patterns of cytokine expression (a biomarker of immune activation) in wild AGMs, where SIV-infected individuals did not differ from uninfected individuals (Ma et al. 2013).
The results of our clustering analyses (both hierarchical clustering and PCA) also support the conclusion that SIVkrc infection is nonpathogenic in URC, because infected and uninfected individuals did not form separate clusters. Because lack of a chronic immune response characterizes nonpathogenic infection, infection status may not be a meaningful biological distinction at the level of blood transcriptome gene expression patterns. As would be expected in blood transcriptome profiles, we observed similar signatures of immune activity (enriched immune-related GO pathways) in both infected and uninfected individuals based on k-means clustering of genes, but found no evidence of elevated immune activity in infected individuals.
Evidence for the Upregulation of Immunosuppression Genes in Chronic Phase SIV Infection in Natural Hosts
Bosinger et al. (2009) and Jacquelin et al. (2009) found a small number of immunosuppressive genes in SMs and AGMs, respectively, that were upregulated upon infection and remained significantly upregulated through the end of each study (6 months, and 2 years into the chronic phase of infection, respectively). The upregulated immunosuppressive genes did not overlap between the two studies, suggesting that hosts may vary in pathways activated for immunosuppression. We hypothesized that if SIVkrc infection is nonpathogenic, URC might also show upregulation of immunosuppressive genes. We did identify an important immunosuppressive gene, CD101 (ENSG00000134256), that was significantly upregulated in infected individuals (fold change = 0.96; Q value = 0.14).
This finding is notable because CD101 has not previously been identified as an upregulated immunosuppressive gene in either AGMs or SMs even though CD101 was included in the microarrays for the previous studies, and is highly conserved across Old World monkeys. The function of CD101 suggests it could play a role in actively downregulating the acute immune response to SIV infection in natural hosts. Protein-coding variants in CD101 are associated with increased susceptibility to HIV-1 (Mackelprang et al. 2017) suggesting that it plays a role in HIV infection. Additionally, reduced expression of CD101 on mucosal CD8+ T cells in the human intestinal and pulmonary mucosa is associated with increased inflammation (Brimnes et al. 2005), demonstrating its role in immunosuppression. Perhaps most importantly, CD101 expression is strongly associated with the suppression of T regulatory cell proliferation, a hallmark of chronic immune activation that is directly associated with progression to AIDS in HIV patients (Liu et al. 1997; Giorgi et al. 2002; Deeks et al. 2004). Although these previous studies, together with our finding of CD101 upregulation in the chronic phase of SIVkrc infection, suggest that CD101 may be a component of nonpathogenic SIV infection in URC, differences between infected and uninfected individuals were relatively small and should be investigated further.
No Evidence for Decrease in CD4+ T Cell Abundance in SIVkrc Infection
In contrast to AGMs and SMs, MNDs and chimpanzees show a significant loss of memory CD4+ T cells as a consequence of SIV infection (Keele et al 2009; Greenwood et al. 2014). Because substantial variation exists between natural hosts in the effect of SIV infection on CD4+ T cell homeostasis, we were interested in whether URC would be more similar to AGMs and SMs, or MNDs and chimpanzees. We found no evidence for a decrease in abundance of any CD4+ T cell subset (naive, memory resting, or memory activated CD4+ T cell) or in total CD4+ T cell abundance in infected individuals based on deconvolution results. Given that SIVkrc and SIVmnd-1 are closely related lineages, and both SIVagm and SIVsm are distantly related, a result where URC and MNDs both showed a loss of memory CD4+ T cells in infected individuals would be consistent with a mechanism of CD4+ T cell loss driven by viral factors. Instead, our results present a more complicated picture with both host and viral factors potentially influencing the loss of activated and bystander CD4+ T cells (Doitsh and Greene 2016).
Potential Study Limitations
Our study does not perform a longitudinal assessment of the effect of SIVkrc infection on gene expression beginning in the acute phase. Because our study was conducted on a natural population, and our samples were collected cross-sectionally, we are not able to time sample collection to intentionally capture the acute response to infection. An interesting shared feature of AGMs and SMs is the robust and transient upregulation of ISGs in the acute phase of infection. Although our study shows that SIVkrc-infected URC in the chronic phase do not have upregulated ISGs, it remains unknown whether the longitudinal pattern of gene expression, beginning with acute infection, is conserved in cercopithecines or more broadly conserved across African Old World monkeys. Additionally, although it appears that natural hosts differ from humans in their ability to attenuate a chronic immune response, it may be possible that older URC individuals who have been infected for a long period of time experience some level of immunodeficiency late in life, though our data are not able to address that possibility. Although the type of repeated sampling necessary to compare to experimental infection studies will not likely be possible in natural populations, the study of host responses to SIV in natural populations nonetheless adds considerably to our understanding of variation in nonpathogenic SIV infection.
Second, we focused specifically on the effects of SIVkrc infection on host gene expression. Although we control for some of the major sources of biological and technical variation in gene expression, there are many factors, both intrinsic and extrinsic (e.g., circadian patterns and age), that influence gene regulation in natural populations. Additionally, URC are host to a myriad of infectious agents including bacteria, viruses, and eukaryotic parasites (Goldberg et al. 2008, 2009; Lauck et al. 2011, 2013; Salyer et al. 2012; Thurber et al. 2013; Bailey et al. 2014; Ghai et al. 2014; McCord et al. 2014; Sibley et al. 2014; Bailey, Lauck, Ghai, et al. 2016; Bailey, Lauck, Sibley, et al. 2016; Simons et al. 2017), all of which likely influence patterns of host gene expression. Although it is impossible to account for all of the possible factors contributing to gene expression patterns in a natural population, our study provides valuable information on the role of SIV infection in host gene expression patterns, and will inform future studies aimed at understanding coinfection dynamics.
Conclusion
We found evidence to support the hypothesis that SIVkrc infection does not strongly influence patterns of immunological gene expression in URC based on a shared pattern of attenuated ISG expression during chronic infection with AGMs and SMs.
We found no transcriptomic evidence that individuals were immunocompromised based on the observations that 1) CD4+ T cell abundance estimates did not differ between infected and uninfected individuals (despite infected individuals harboring high viral loads), 2) infected and uninfected individuals had similar enriched GO pathways in gene clusters, and 3) infected individuals did not cluster to the exclusion of uninfected individuals using multiple clustering approaches. We did find evidence for upregulation of an important immunosuppressive gene, CD101, which has not been previously reported in models of nonpathogenic SIV infection, despite both protein-coding and gene regulatory variation of CD101 being implicated in HIV infection. Based on the fundamental role of CD101 in immunosuppression, these findings may suggest evolved species-specific mechanisms for the tolerance of SIV infection in natural hosts.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This research was funded by NIH grant TW009237 as part of the joint NIH–NSF Ecology of Infectious Disease program and the UK Economic and Social Research Council, NSF BCS-1540459, National Geographic Society, NSERC, and the University of Oregon. We thank the Uganda Wildlife Authority and Uganda National Council of Science and Technology for permission to conduct this research. We are grateful to Robert Basaija, Peter Tuhairwe, Clovice Kaganzi, and Dr Dennis Twinomugisha for assistance with logistics and fieldwork. We thank Clay Small, Lucia Carbone, Suzi Fei, Dave O’Connor, and Tom Friedrich for helpful feedback on the design and analysis of the research and members of the Molecular Anthropology Group at University of Oregon for helpful feedback throughout the research process and valuable comments during manuscript preparation. Use of trade names is for identification only and does not imply endorsement by the U.S. Centers for Disease Control and Prevention (CDC). The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the CDC.
David Hyeroba, our coauthor, colleague, and friend, passed away while we were in the late stages of submitting this manuscript for publication. Dr Hyeroba was an incredible and passionate veterinarian who worked internationally to care for the health of wild and domestic animals. This manuscript would not have been possible without his expertise and skill, and his passing is an irreparable loss to our community. We dedicate this research to his memory.
Data deposition: This project has been deposited at NCBI under the accession PRJNA413051.
Literature Cited
- Anders S, Huber W.. 2010. Differential expression analysis for sequence count data. Genome Biol. 11(10):R106.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl P, Huber W.. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31(2):166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apetrei C, et al. 2011. Immunovirological analyses of chronically simian immunodeficiency virus SIVmnd-1- and SIVmnd-2-infected mandrills (Mandrillus sphinx). J Virol. 85(24):13077–13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey AL, et al. 2016. Arteriviruses, pegiviruses, and lentiviruses are common among wild African monkeys. J Virol. 90(15):6724–6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey AL, et al. 2016. Zoonotic potential of simian arteriviruses. J Virol. 90(2):630–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey AL, et al. 2014. High genetic diversity and adaptive potential of two simian hemorrhagic fever viruses in a wild primate population. PLoS One 9(3):e90714.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y.. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Stat Methodol. 57(1):289–300. [Google Scholar]
- Bosinger SE, et al. 2009. Global genomic analysis reveals rapid control of a robust innate response in SIV-infected sooty mangabeys. J Clin Invest. 119(12):3556–3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosinger SE, et al. 2013. Intact type I interferon production and IRF7 function in sooty mangabeys. PLoS Pathog. 9(8):e1003597.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brimnes J, et al. 2005. Defects in CD8+ regulatory T cells in the lamina propria of patients with inflammatory bowel disease. J Immunol. 174(9):5814–5822. [DOI] [PubMed] [Google Scholar]
- Chahroudi A, Bosinger SE, Vanderford TH, Paiardini M, Silvestri G.. 2012. Natural SIV hosts: showing AIDS the door. Science 335(6073):1188–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman CA, Struhsaker TT, Lambert JE.. 2005. Thirty years of research in Kibale National Park, Uganda, reveals a complex picture for conservation. Int J Primatol. 26(3):539–555. [Google Scholar]
- Chapman CA, et al. 2012. Protozoan parasites in group-living primates: testing the biological island hypothesis. Am J Primatol. 74(6):510–517. [DOI] [PubMed] [Google Scholar]
- Charruau P, et al. 2016. Pervasive effects of aging on gene expression in wild wolves. Mol Biol Evol. 33(8):1967–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debey-Pascher S, et al. 2011. RNA-stabilized whole blood samples but not peripheral blood mononuclear cells can be stored for prolonged time periods prior to transcriptome analysis. J Mol Diagn. 13(4):452–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks SG, et al. 2004. Immune activation set point during early HIV infection predicts subsequent CD4+ T-cell changes independent of viral load. Blood 104(4):942–947. [DOI] [PubMed] [Google Scholar]
- Doitsh G, Greene WC.. 2016. Dissecting how CD4 T cells are lost during HIV infection. Cell Host Microbe 19(3):280–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiebig EW, et al. 2003. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS 17(13):1871–1879. [DOI] [PubMed] [Google Scholar]
- Furge K, Dykema K.. 2006. PGSEA: parametric gene set enrichment analysis. R package version 1.2.
- Gao F, et al. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397(6718):436–441. [DOI] [PubMed] [Google Scholar]
- Ge SX, Son EW, Yao R. 2018. iDEP: an integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC bioinformatics.19(1):534. [DOI] [PMC free article] [PubMed]
- Ghai RR, Chapman CA, Omeja PA, Davies TJ, Goldberg TL.. 2014. Nodule worm infection in humans and wild primates in Uganda: cryptic species in a newly identified region of human transmission. PLoS Negl Trop Dis. 8(1):e2641.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgi JV, et al. 2002. Predictive value of immunologic and virologic markers after long or short duration of HIV-1 infection. J Acquir Immune Defic Syndr. 29(4):346–355. [DOI] [PubMed] [Google Scholar]
- Goldberg TL, Paige SB, Chapman CA. 2012. The Kibale EcoHealth Project: exploring connections among human health, animal health, and landscape dynamics in western Uganda. In: Aguirre AA, Ostfeld R, Daszak P, editors. New directions in conservation medicine: applied cases of ecological health. Oxford: OUP USA. p. 452–465. [Google Scholar]
- Goldberg TL, et al. 2008. Serologic evidence for novel poxvirus in endangered red Colobus monkeys, Western Uganda. Emerg Infect Dis. 14(5):801–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg TL, et al. 2009. Coinfection of Ugandan red colobus (Procolobus [Piliocolobus] rufomitratus tephrosceles) with novel, divergent delta-, lenti-, and spumaretroviruses. J Virol. 83(21):11318–11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood EJD, et al. 2014. Loss of memory CD4+ T-cells in semi-wild mandrills (Mandrillus sphinx) naturally infected with species-specific simian immunodeficiency virus SIVmnd-1. J Gen Virol. 95(Pt_1):201–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington LE, Janowski KM, Oliver JR, Zajac AJ, Weaver CT.. 2008. Memory CD4 T cells emerge from effector T-cell progenitors. Nature 452(7185):356–360. [DOI] [PubMed] [Google Scholar]
- Hirsch VM, Olmsted RA, Murphey-Corb M, Purcell RH, Johnson PR.. 1989. An African primate lentivirus (SIVsm) closely related to HIV-2. Nature 339(6223):389–392. [DOI] [PubMed] [Google Scholar]
- Jacquelin B, et al. 2009. Nonpathogenic SIV infection of African green monkeys induces a strong but rapidly controlled type I IFN response. J Clin Invest. 119(12):3544–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinska AJ, et al. 2013. Systems biology of the vervet monkey. ILAR J. 54(2):122–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandathil AJ, Sugawara S, Balagopal A.. 2016. Are T cells the only HIV-1 reservoir? Retrovirology 13(1):86.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele BF, et al. 2009. Increased mortality and AIDS-like immunopathology in wild chimpanzees infected with SIVcpz. Nature 460(7254):515–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladner JT, et al. 2016. A multicomponent animal virus isolated from mosquitoes. Cell Host Microbe 20(3):357–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S.. 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauck M, et al. 2011. Novel, divergent simian hemorrhagic fever viruses in a wild Ugandan red colobus monkey discovered using direct pyrosequencing. PLoS One 6(4):e19056.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauck M, et al. 2013. Exceptional simian hemorrhagic fever virus diversity in a wild African primate community. J Virol. 87(1):688–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederer S, et al. 2009. Transcriptional profiling in pathogenic and non-pathogenic SIV infections reveals significant distinctions in kinetics and tissue compartmentalization. PLoS Pathog. 5(2):e1000296.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhou J, White KP.. 2014. RNA-seq differential expression studies: more sequence or more replication? Bioinformatics 30(3):301–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, et al. 2017. HisgAtlas 1.0: a human immunosuppression gene database. Database (Oxford) 2017: bax094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, et al. 1997. Elevated CD38 antigen expression on CD8+ T cells is a stronger marker for the risk of chronic HIV disease progression to AIDS and death in the Multicenter AIDS Cohort Study than CD4+ cell count, soluble immune activation markers, or combinations of HLA-DR and CD38 expression. J Acquir Immune Defic Syndr Hum Retrovirol. 16(2):83–92. [DOI] [PubMed] [Google Scholar]
- Locatelli S, et al. 2008. Full molecular characterization of a simian immunodeficiency virus, SIVwrcpbt from Temminck’s red colobus (Piliocolobus badius temminckii) from Abuko Nature Reserve, The Gambia. Virology 376(1):90–100. [DOI] [PubMed] [Google Scholar]
- Locatelli S, et al. 2008. Prevalence and genetic diversity of simian immunodeficiency virus infection in wild-living red colobus monkeys (Piliocolobus badius badius) from the Taï forest, Côte d’Ivoire SIVwrc in wild-living western red colobus monkeys. Infect Genet Evol. 8(1):1–14. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15(12):550.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, et al. 2013. SIVagm infection in wild African green monkeys from South Africa: epidemiology, natural history, and evolutionary considerations. PLoS Pathog. 9(1):e1003011.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, et al. 2014. Factors associated with siman immunodeficiency virus transmission in a natural African nonhuman primate host in the wild. J Virol. 88(10):5687–5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackelprang RD, et al. 2017. Whole genome sequencing of extreme phenotypes identifies variants in CD101 and UBE2V1 associated with increased risk of sexually acquired HIV-1. PLoS Pathog. 13(11):e1006703.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCord AI, et al. 2014. Fecal microbiomes of non‐human primates in Western Uganda reveal species‐specific communities largely resistant to habitat perturbation. Am J Primatol. 76(4):347–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir KD, Gasper MA, Sundaravaradan V, Sodora DL.. 2011. SIV infection in natural hosts: resolution of immune activation during the acute-to-chronic transition phase. Microbes Infect. 13(1):14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AM, et al. 2015. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 12(5):453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiardini M, Müller-Trutwin M.. 2013. HIV-associated chronic immune activation. Immunol Rev. 254(1):78–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palesch D, et al. 2018. Sooty mangabey genome sequence provides insight into AIDS resistance in a natural SIV host. Nature 553(7686):77.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham TNQ, Lukhele S, Hajjar F, Routy JP, Cohen ÉA.. 2014. HIV Nef and Vpu protect HIV-infected CD4+ T cells from antibody-mediated cell lysis through down-modulation of CD4 and BST2. Retrovirology 11:15.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. 2008. R: a language and environment for statistical computing. Vienna (Austria: ): R Foundation for Statistical Computing. [Google Scholar]
- Reimand J, Kull M, Peterson H, Hansen J, Vilo J.. 2007. g: profiler—a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 35(Suppl 2):W193–W200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1):139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler AJ, Williams B.. 2008. Interferon-inducible antiviral effectors. Nat Rev Immunol. 8(7):559–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai Y, et al. 2016. Expression profiles of Vpx/Vpr proteins are co-related with the primate lentiviral lineage. Front Microbiol. 7:1211.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salyer SJ, Gillespie TR, Rwego IB, Chapman CA, Goldberg TL.. 2012. Epidemiology and molecular relationships of Cryptosporidium spp. in people, primates, and livestock from Western Uganda. PLoS Negl Trop Dis. 6(4):e1597.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurch NJ, et al. 2016. How many biological replicates are needed in an RNA-seq experiment and which differential expression tool should you use? RNA 22(6):839–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergushichev A. 2016. An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. bioRxiv. 10.1101/060012. [DOI]
- Shah AH, et al. 2010. Degranulation of natural killer cells following interaction with HIV-1-infected cells is hindered by downmodulation of NTB-A by Vpu. Cell Host Microbe 8(5):397–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibley SD, et al. 2014. Discovery and characterization of distinct simian pegiviruses in three wild African Old World monkey species. PLoS One 9(2):e98569.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons ND, et al. 2016. Rapid identification of major histocompatibility complex class I haplotypes using deep sequencing in an endangered Old World monkey. Conserv Genet Resour. 8(1):23–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons ND, et al. 2017. Cis‐regulatory evolution in a wild primate: infection‐associated genetic variation drives differential expression of MHC‐DQA1 in vitro. Mol Ecol. 26(17):4523–4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R.. 2003. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 100(16):9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhsaker TT. 2005. Conservation of red colobus and their habitats. Int J Primatol. 26(3):525–538. [Google Scholar]
- Subramanian A, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurber MI, et al. 2013. Co-infection and cross-species transmission of divergent Hepatocystis lineages in a wild African primate community. Int J Parasitol. 43(8):613–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Oh WK, Zhu J.. 2016. Disease-specific classification using deconvoluted whole blood gene expression. Sci Rep. 6:32976.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherall DW. 2006. The use of non-human primates in research. London: Academy of Medical Sciences. [Google Scholar]
- Weirauch MT, et al. 2014. Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158(6):1431–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson EB. 1927. Probable inference, the law of succession, and statistical inference. J Am Stat Assoc. 22(158):209–212. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.