Abstract
GRB7 gene encodes a multi-domain signal transduction molecule and is part of the core of the HER-2 amplicon. GRB7 is commonly co-amplified and overexpressed with HER-2 in human breast cancer. This study addresses the role of GRB7 in HER-2 positive human breast cancers resistant to HER-2 targeted therapy. HCC1954, 21MT1, and JIMT1 are basal like HER-2 positive breast cancer cell lines based on expression profiling. These three cell lines are resistant to trastuzumab and lapatinib treatment. Knockdown of GRB7 protein expression with siRNA transfection as well as lentiviral vector mediated shRNA over-expression decreased the growth of HCC1954, 21MT1, and JIMT1 cells in vitro and the growth of tumor xenografts these cells formed in animal models. When assayed by ki-67 staining and TUNEL assay, the mechanism of reduced tumor xenograft growth appeared to be distinct. Reduced proliferation and increased apoptosis were seen in 21MT1 cells, while reduced proliferation was seen in HCC1954 cells and increased apoptosis in JIMT1 cells. Phospho-proteome profiling found HER-1 tyrosine phosphorylation was reduced with GRB7 knockdown in JIMT1 cells. Immuno-blotting and immuno-precipitation experiments found HER-1 phosphorylation was reduced with GRB7 knock down in all three cell lines. HER-1 knock down via siRNA transient transfection as well as blocking HER-1 function with panitumumab decreased proliferation of all three cell lines in vitro. Our study finds that GRB7 has an essential growth promoting function which is mediated in part by HER-1 activation. The potential of HER-1 targeting in therapy resistant HER-2 positive breast cancer merits further study.
Keywords: animal models, breast cancer, signal transduction
1 |. INTRODUCTION
HER-2 gene amplification and protein over-expression is present in about 20% of breast cancer.1 Though HER-2 positive breast cancer is generally associated with poor clinical outcome, the advent of HER-2 targeted therapy has significantly improved the prognosis for patients with HER-2 positive breast cancer.2 Trastuzumab, the first humanized anti-HER-2 antibody to receive FDA approval, has significantly improved survival for patients with metastatic disease.3 For patients diagnosed with early stage disease, trastuzumab reduces the risk of recurrence and improves the overall survival in the adjuvant setting.4–6 In addition to trastuzumab, additional HER-2 targeted therapeutics have been developed that showed great promise in various clinical scenarios and received regulatory approval. These include pertuzumab and ado-trastuzumab emtansine (TDM1) and small molecule inhibitors such as lapatinib and neratinib.7–10 Amidst this remarkable success, there remains much room for improvement. In the neoadjuvant setting, about 40% of patients still have residual cancer left despite combination therapy with chemotherapy plus dual antibody with trastuzumab and pertuzumab.11,12 In the metastatic setting, metastatic HER-2 positive breast cancer remains at risk for progression notwithstanding the improvement in overall treatment efficacy.9,10
GRB7 encodes a multi-domain signal transduction molecule13 and is located in close proximity to the HER-2 gene on chromosome 17q11–12. GRB7 gene is commonly co-amplified and co-over-expressed with HER-2 in human breast cancer with HER-2 amplification and over-expression. GRB7 protein has been shown to interact with multiple signal transducing molecules. Our laboratory has previously shown that GRB7 facilitates HER-2 mediated signaling and promotes HER-2 mediated tumor formation in a xenograft model.14 Additionally, GRB7 expression has been shown to predict adverse prognosis in breast cancer.15–17 The role of GRB7 signaling in therapy resistant, HER-2 positive breast cancer cells has not been elucidated in the past however. Nor is it clear whether targeting GRB7 or GRB7 directed signaling pathway(s) can be beneficial in this patient subset. In this study, we evaluate the functional involvement of GRB7 signaling in trastuzumab and lapatinib resistant human breast cancer cell lines- HCC1954, JIMT1, and 21MT1 and explore its candidacy as a target for therapeutic intervention.
2 |. RESULTS
To investigate the growth promoting function of GRB7, we knock down the expression of GRB7 in human breast cancer cells with transient siRNA transfection. As shown in Figure 1A, successful knock down of GRB7 protein can be achieved in two breast cancer cell lines: HCC1954 and SkBr3. When the effect of GRB7 knock down in six different human HER-2 breast cancer cells on cell proliferation was examined, reduced cell proliferation was observed in all six cell lines including four cell lines that are resistant to trastuzumab: HCC1954, JIMT1, EFM192b, and BT474-TR (Figure 1B).
FIGURE 1.
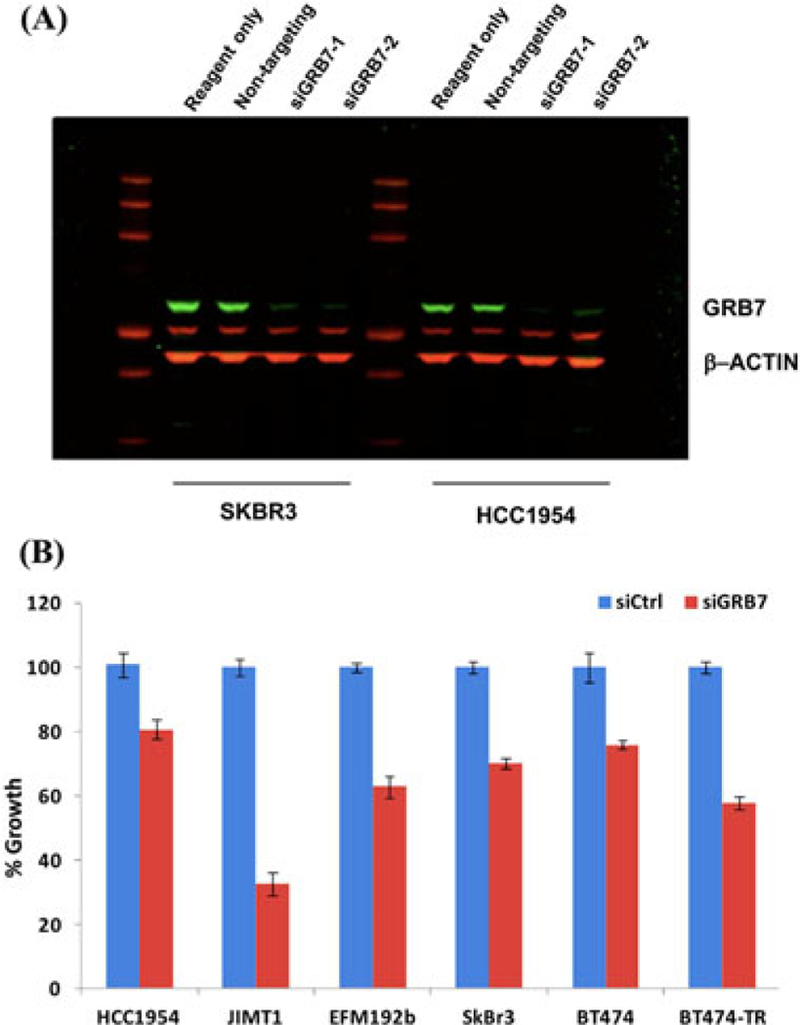
Transient knock down of GRB7 with siRNA decreased cell proliferation in multiple HER-2 positive cell lines. A, Western analysis revealed that transient knock down of GRB7 protein expression was achieved with siRNA transfection. Cell lines transfected with non-targeting siRNA served as negative controls. B, siRNA mediated transient knock down of GRB7 decreased in vitro cell growth compared to controls in six HER-2 positive human breast cancer cell lines as measured by the CellTiter Glo assay (Promega, Madison, Wl).
To examine the growth promoting function of GRB7 in HER-2 positive human breast cancer cell lines that are resistant to trastuzumab and lapatinib, we obtained stable GRB7 knock down in HCC1954, JIMT1, and 21MT1 cells via lentivirally mediated shRNA over-expression. Cells over-expressing an empty lentiviral vector without a shRNA insert serve negative controls. HCC1954, JIMT1, and 21MT1 cells are all resistant to trastuzumab with GI50 at greater than 30 μg per milliliter. HCC1954 is moderately resistant (GI50 at 0.39 μM) and JIMT1 and 21MT1 are highly resistant (GI50 at greater than 2 μM) to lapatinib treatment. As a comparison, BT474 is sensitive to both lapatinib and trastuzumab treatment with GI50 at 0.05μM and 0.12 μg per milliliter, respectively.18
As shown in Figure 2A, successful knockdown of GRB7 in each of the three cell lines was achieved as demonstrated by Western blotting. When assayed by the CellTiter Glo assay (Figure 2B, top) as well as the live cell imaging by the Incucyte (Figure 2B, bottom), all three cell lines with GRB7 knock down, maintained as polyclonal culture, showed reduced cell proliferation. The above indicate that GRB7 protein expression is required for efficient cell growth in human breast cancer cell lines that are resistant to trastuzumab and lapatinib.
FIGURE 2.
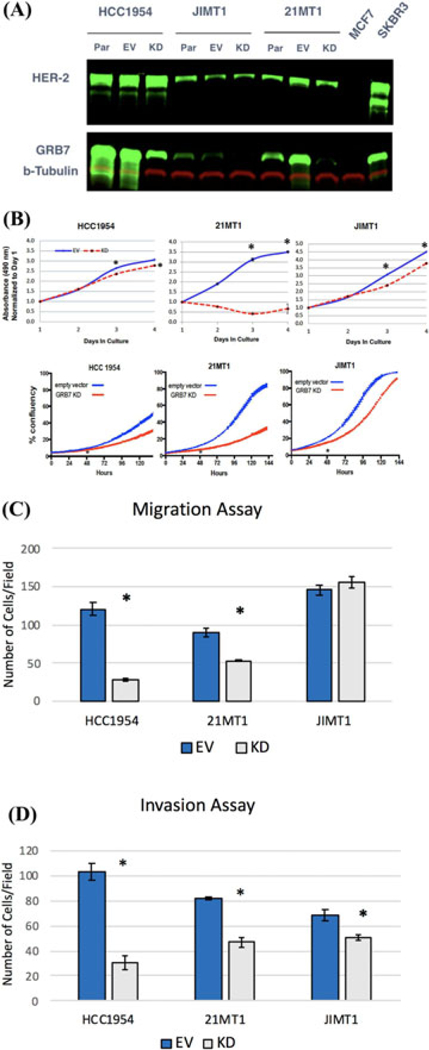
A, Western blot analysis confirmed stable GRB7 knockdown in breast cancer cell lines were achieved with lentiviral vector mediated shRNA over-expression. Par, parental cell line; EV, empty vector; KD: GRB7 knock down. B, shRNA mediated GRB7 knockdown decreased cell proliferation relative to controls in trastuzumab and lapatinib resistant cell lines as measured by the CellTiter Glo assay, Top; and Incucyte live cell imaging, Bottom. (*P < 0.05). C, Stable GRB7 knockdown decreased cell migration toward 10% FBS in HCC1954 and 21MT1 but not JIMT1 cells. (N = 4, at 100x magnification). (*P < 0.05). D, Stable GRB7 knockdown decreased cell invasion through matrigel toward 10% FBS in HCC1954, 21MT1 and JIMT1 cells. (N = 4, at 100x magnification). (*P < 0.05).
To examine the outcome of GRB7 knock down on cell motility, we performed Transwell (Figure 2C) and matrigel invasion assays (Figure 2D). GRB7 knock down decreased migration for both HCC1954 and 21MT1 cells but not JIMT1 cells. GRB7 knock down decreased invasion in all three cell lines.
To study the GRB7 function in vivo, we examined the effect of GRB7 knock down on the growth of these cell lines as tumor xenografts in immunodeficient mouse models. Between 250 thousand to a million cells were injected orthotopically into mammary fat pads of 5–6 weeks old NSG female mice. The growth of these tumor xenografts was measured with a caliper three times a week. Cells expressing an empty lentiviral vector served as negative controls. The growth rates of the tumor xenografts (Figure 3A, Top) and the final weights of the tumor xenografts (Figure 3A, Bottom) were both decreased with GRB7 knock down for all three cell lines as compared with negative controls with an empty vector infection. Taken together, these results indicate that GRB7 protein expression plays an important role for the growth of HER-2 positive breast cancer cells that are resistant to trastuzumab and lapatinib treatment both in vitro and in vivo.
FIGURE 3.
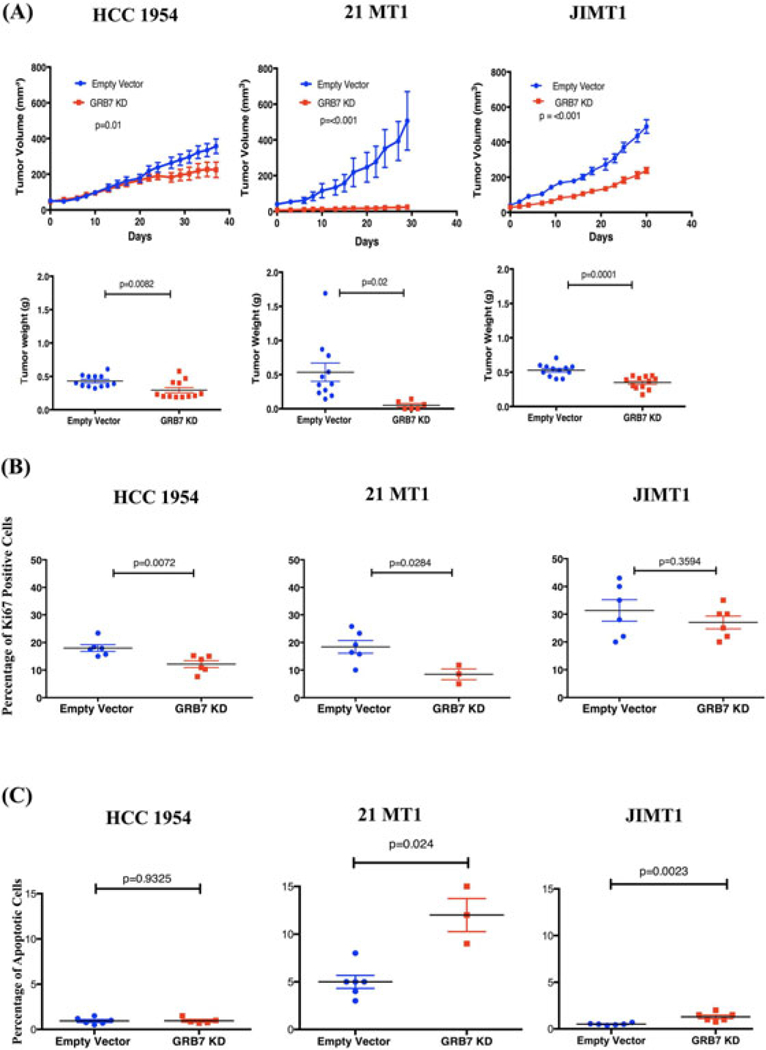
A, Knock down of GRB7 decreased the growth of tumor xenografts formed by trastuzumab and lapatinib resistant HER2 positive cell lines in immune-deficient NSG mice compared to controls and measured by volume, Top, and weight, Bottom. B, Ki-67 Staining was decreased in GRB7 knockdown xenograft tumors relative to controls in HCC1954 and 21MT1 but not in JIMT1 xenograft tumors. C, TUNEL assay showed that GRB7 knockdown increased the percentage of apoptotic cells in 21MT1 and JIMT1 but not HCC1954 xenograft tumors.
In order to further investigate the phenotypic outcome of the GRB7 knock down, we performed analysis on the tumor xenografts harvested from the animal models. We measured the cells that were Ki-67 positive (Figure 3B) as well as cells that underwent apoptosis with TUNEL assay (Figure 3C). GRB7 knock down had pleiotropic effects depending on different cellular contexts- in HCC1954 cells, GRB7 knock out was associated with a decrease in the percentage of cells that were Ki-67 positive but no change in cells undergoing apoptosis. Increased apoptosis but no change in Ki-67 cells were seen for JIMT1 cells with GRB7 knock down. In 21MT1 cells, reduction in the percentage of cells that were Ki-67 positive as well as an increase in apoptosis were seen with GRB7 knock down as compared with the control. Representative results are presented in Supplemental Figure S2.
To evaluate the effect of GRB7 knock down on signaling, we profiled several sets of phosphoproteome filters from a vendor (R&D system). We found multiple signaling molecules whose phosphorylation was altered as a result of GRB7 knock down. One common theme was reduction in the tyrosine phosphorylation of the HER-1 molecule (a representative result from the JIMT1 pair is shown in Supplemental Figure S3). We further explored this by performing the following series of experiments with lysates from HCC1954, 21MT1, and JIMT1 stably transfected with shRNA to achieve GRB7 knock down versus their respective empty vector controls.
We performed reciprocal immunoprecipitation and protein blotting experiments: we first performed immunoprecipitation with anti-phospho-tyrosine antibody followed by protein blotting with anti-HER-1, anti-HER-2 and anti-GRB7 antibody (Figure 4A). We then performed immunoprecipitation with anti-HER-1 antibody followed by immuno-blotting with anti-phospho-tyrosine antibody (Figure 4B). In both sets of experiments, we found that GRB7 knock down was associated with a decrease in the tyrosine phosphorylation of the HER-1 molecule. To confirm this, we performed Western blotting with an antibody that was specific for a phosphorylated form of HER-1 (Figure 4C). Finally, to demonstrate that the attenuation of HER-1 phosphorylation by GRB7 knockdown was not due to an off target effect, we obtained two additional stable GRB7 knock down cell populations in HCC1954 cells with two distinct shRNA containing lentiviral vectors that target GRB7 at different positions. All three GRB7 knock downs in HCC1954 cells were associated with a reduction in the HER-1 tyrosine phosphorylation (Figure 4D). Taken together the above indicates GRB7 knock down is associated with reduction in HER-1 tyrosine phosphorylation in all three therapy resistant, HER-2 positive cell lines.
FIGURE 4.
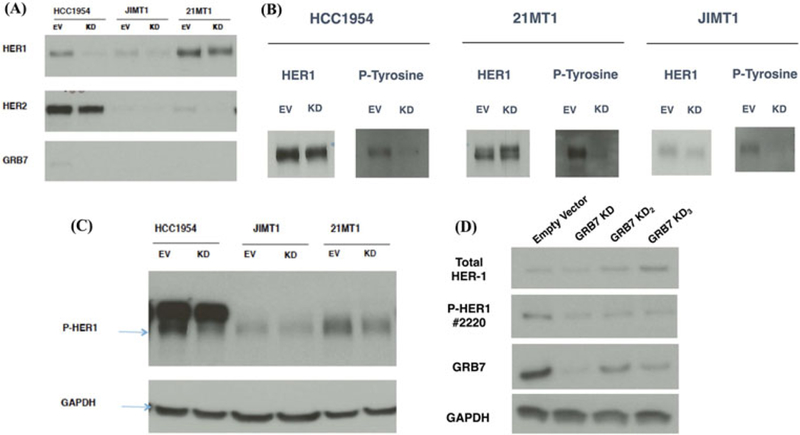
GRB7 knock down in trastuzumab and lapatinib resistant, HER-2 positive breast cancer cell lines reduced HER-1 tyrosine phosphorylation. A, Cell lysates were first immunoprecipitated with anti-phospho-tyrosine antibody (Cell Signaling Technology, #9419) followed by blotting and staining with anti-HER-1 (Cell Signaling Technology, #4267), HER-2 (Cell Signaling Technology, #2242), and GRB7 antibodies. B, Cell lysates were immunoprecipitated with anti-HER-1 antibody (Cell Signaling Technology, #8083) blotted with anti-HER-1 (Cell Signaling Technology, #4267) and anti-phospho-tyrosine antibodies (Cell Signaling Technology, #8954). C, Cell lysates were blotted with anti-phospho-HER-1 antibody (Cell Signaling Technology, #3777). EV: empty vector; KD: GRB7 knock down. D, HCC1954 cells were infected with three different shRNA targeting vectors: Clone ID: GRB7 KD = TRCN0000061385; GRB7 KD2 = TRCN0000061384; GRB7 KD3 = TRCN0000061386, (GE Healthcare, Dharmacon). Cell lysates were blotted with anti-phospho-HER-1 antibody (Cell Signaling Technology, #2220), anti-total HER-1, anti-GRB7, and anti-GAPDH antibodies
To study the effect of blocking HER-1 function in HCC1954, 21MT1, and JIMT1 cells, we first performed a transient transfection of a siRNA that targeted the HER-1 molecule with cells transfected with a non-targeting siRNA as negative controls. We demonstrated successful knock down of HER-1 protein expression by protein blotting (Figure 5A) and this was associated with a reduction of cell proliferation in all three cell lines with the CellTiter Glo assay (Figure 5B). To confirm this, we added panitumumab, a humanized anti-HER-1 antibody, at a final concentration of 100 μg per milliliter to the cell culture. We found panitumumab could decrease the proliferation in all three cell lines (Figure 5C). The above indicates that blocking HER-1 function can reduce the growth of HER-2 positive human breast cancer cell lines that are resistant to trastuzumab and lapatinib therapy.
FIGURE 5.
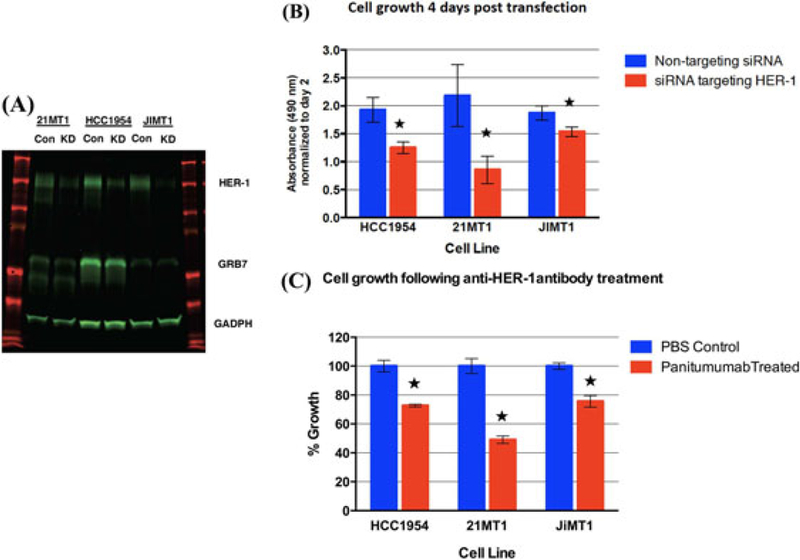
Perturbation of HER-1 signaling in trastuzumab and lapatinib resistant, HER-2 positive breast cancer cells via siRNA transient transfection and panitumumab treatment. A, Western analysis after siRNA transient transfection. Con: non-targeting control siRNA; KD: siRNA targeting HER-1. B, SiRNA mediated HER-1 knock down decreased cell proliferation. C, Panitumumab treatment decreased cell proliferation. (★ denotes P < 0.05).
3 |. DISCUSSION
Targeting of the HER-2 molecule is among the most successful stories in personalized medicine. Breast cancer with HER-2 amplification and over-expression tends to be a more aggressive disease.1 This adverse prognosis however has improved significantly with the advent of effective HER-2 targeted therapeutics. HER-2 targeted therapeutics consist of humanized HER-2 targeting monoclonal antibodies such as trastuzumab and pertuzumab, toxin antibody conjugate such as TDM1, and small molecule kinase inhibitors such as neratinib and lapatinib.19 Despite this success, there is still much room for improvement. In the pre-operative setting, aggressive chemotherapy in combination with dual antibody blockade with pertuzumab and trastuzumab achieved pathologic complete response in about 60% of the patients.11 In the advanced disease setting, TDM1 as well as chemotherapy with dual antibody blockade saw improved disease control but patients nonetheless continued to progress.9,10 In the adjuvant setting, addition of either neratinib or pertuzumab saw a more modest clinical benefit.20,21 To impact favorably the ratio of therapeutic benefits over toxicity, current efforts focus on improving the efficacy of HER-2 targeted therapy and lessening the associated toxicity as well as minimizing the toxicity of chemotherapy given concurrently.22
Multiple genes are amplified and over-expressed concurrently with HER-2 in breast cancer with chromosome 17qll-12 amplification. GRB7 gene, located in close proximity to the HER-2 gene, is one such gene. GRB7 protein contains both SH2 and PH domains and serves as an adaptor molecule that interacts with multiple signaling molecules.13,23 We and others have shown that GRB7 expression is an adverse prognostic factor for breast cancer.15–17 We have shown that GRB7 facilitates HER-2 mediated signaling and tumor formation. Over-expression of the GRB7 gene in human MCF-7 cell lines over-expressing HER-2 enhanced the growth as tumor xenografts in nude mice.14 A subsequent study showed shRNA mediated knock down of GRB7 in a trastuzumab and lapatinib sensitive, HER-2 positive human breast cancer cell line, SkBr3, reduced their growth as tumor xenografts in SCID mice.24 These data support a functionally important role of GRB7 in the growth of therapy sensitive HER-2 positive breast cancer cells.
In this study, we demonstrated that GRB7 expression is essential for the growth of three, trastuzumab and lapatinib resistant, human HER-2 positive breast cancer cell lines, both in vitro and in vivo. This observation indicates GRB7 signaling is essential for the cancer cell growth in spite of the development of resistance against HER-2 signaling blockade. Consequently, our study indicates that targeting GRB7 protein and/or GRB7 directed signaling pathways may prove to be of therapeutic benefit in the treatment of the basal type of HER-2 positive human breast cancer that has intrinsic resistance to therapy with trastuzumab and lapatinib.
The phenotypic outcome of GRB7 knock down is pleiotropic. Phospho-proteome analysis finds the changes in protein tyrosine phosphorylation are largely non-overlapping with HER-1 as an exception. This is not surprising as GRB7 protein has been shown to interact with multiple different signaling molecules. Depending on the cellular contexts and the expression of relevant signaling molecules, important cellular physiology may be modified to various extents such as cell proliferation and apoptosis with GRB7 knock down.
HER-1 protein tyrosine phosphorylation is reduced with GRB7 knock down in HCC1954, 21MT1, and JIMT1 cells. This raised the possibility that the growth promoting function of GRB7 might be mediated in part by HER-1 signaling. In support of this, we perturbed the HER-1 signaling by siRNA mediated knock down and treatment with panitumumab, a humanized anti-HER-1 antibody. Decreased cell proliferation was observed with both interventions, supporting a role of HER-1 in the growth of basal-like human HER-2 positive breast cancer cells. Of note, HCC1954, 21MT1, and JIMT1 cell lines are basal-like HER-2 positive breast cancer cell lines which demonstrated different drug resistance mechanism from luminal-like HER-2 positive cells.25
In conclusion, our study provides evidence for the functional complexity of the 17q11–12 amplification in breast cancer. Our study reveals that GRB7 signaling is essential for the growth of basal-like HER-2 positive human breast cancer cell lines and this function is mediated in part by the HER-1 signaling. Targeting HER-1 therefore may representa clinically useful therapeutic intervention for the basal-like human HER-2 positive breast cancer that has intrinsic resistance to trastuzumab and lapatinib therapy.
4 |. MATERIALS AND METHODS
4.1 |. Cell lines
HCC1954, 21MT1, and JIMT1, human HER-2 positive breast cancer cell lines were utilized because they are resistant to trastuzumab and lapatinib treatment.18 Cell lines BT474, SkBr3, and HCC1954 used in this study were obtained from ATCC. JIMTI and EFM192b were obtained from DSMZ. The establishment of resistant cell line BT474-TR was previously reported.18 Short Tandem Repeat (STR) analysis of 21MT1, JIMTI, and HCC1954 cells was performed to ascertain the identify of these cells (American Tissue Culture Collection, Manassas, VA). HCC1954 cells showed a perfect match (100%) to the HCC1954 cells available through the ATCC (CRL-2338). JIMTI cells showed a perfect match to the JIMTI cells from the DSMZ STR database. A STR profile of 21MT1 is not available from the ATCC. The 21MT1 cells used in this study generated a profile that is highly similar to their counterpart that was studied and reported in prior studies18,25 in that there is a 15/16 match (94%) with the only difference being loss of an allele in our 21MT1 cell.26 The HER-2 amplification status in 21MT1 and JIMTI cells used in this study has been independently verified by fluorescence in situ hybridization (not shown). As shown in Supplemental Figure S1 and in Ref.,18 21MT1 and JIMTI cells express relatively low HER-2 protein on Western analysis among a number of HER-2 positive human breast cancer cell lines.
4.2 |. Transient knockdown of GRB7 protein expression with siRNA transfection
The siRNA targeting GRB7 (Cat No. SI00075600, SI03083381) and AllStars negative control siRNA (Cat No. 1027280) were purchased from Qiagen Inc. (Germantown, MD). Breast cancer cells were seeded at the desired number in 96-well plates one day prior to transfection. Cells were transfected with 10 nM siRNAs using Dharmafect1 transfection regent (Dharmacon, Lafayette, CO) according to the manufacturer’s instructions.
After transfection with siRNAs for 72 h, cell viability was measured using the CellTiter-Glo® assay (Promega, Madison, Wl), a cell viability assay which uses a luciferase reaction to measure ATP levels. The percent cell growth was calculated with normalization of cell viability of groups of GRB7 siRNA (siGRB7) to AllStars Control siRNA (siCtrl). Each experiment was performed at least three times with identical results and one representative set of the experiments was presented. Both GRB7 targeting siRNAs produced similar results. The result from only one siRNA was shown (Cat No. SI00075600).
4.3 |. Stable GRB7 knockdown with lentiviral mediated shRNA over-expression
RNAi consortium (TRC) lentiviral shRNA constructs TRCN0000061384, TRCN0000061385, TRCN0000061386 were purchased from GE Healthcare (Dharmacon). As a control, a vector containing no shRNA insert was used. Breast cancer cells were seeded at the desired number in 6-well plates 1–2 days prior to infection. Infection followed manufacture’s protocol (TRC Lentiviral shRNAS—Technical Manual) with puromycin selection. Cells were assayed by Western Analysis for GRB7 expression 10–14 days following infection. All three cell lines were infected with TRCN0000061385. HCC1954 cells were infected with TRCN0000061384 (KD2) and TCRN0000061386 (KD3) for the experiment described in Figure 4D.
4.4 |. Cell proliferation was measured in an IncuCyte live cell imaging system
Each type of cells was prepared as above, seeded in 96-well plates (Corning, Costar #3595, Corning, NY) with eight replicates with a homogenous initial confluence (between 5 and 20%), loaded in the IncuCyte machine. The confluence of cells in each well was continuously measured every 1 or 2 h. Growth curves were made using the normalized confluence values of each well (setting the time zero reading as one).
4.5 |. Western blotting to monitor protein expression
Cells were lysed in buffer consisting of 20 mM Tris (pH 7.5), 150 mM NaCI, 1% Triton X-100, 5 mM EDTA, 1 mM EGTA, 10μL/mL HALT Protease & Phosphatase Inhibitor Cocktail, (ThermoFisher Scientific, Grand Island, NY) on ice for 5 min. Lysates were centrifuged at 13 000 rpm a 4°C for 5 min to remove cellular debris, separated by SDS-PAGE gel electrophoresis and transferred onto a nitrocellulose membrane. (Bio-Rad, Hercules, CA). Membranes were blocked with 5% BSA (Research Products International Corp., Mt. Prospect, IL), incubated with primary antibodies against target proteins, and then incubated with horseradish peroxidase-conjugated secondary anti-bodies (Bio-Rad). The proteins were visualized using LumiGLO ECL chemiluminescence detection system (Cell Signaling Technology, Inc., Danvers, MA). The following primary antibodies were used: anti-GRB7 (Dr. Shiuh-Wen Luoh), anti-β-actin (#4967), anti-HER-1 (#4267), anti-HER-2 (#2242), anti-phospho-EGFR (#2220 and #3777), anti-β-tubulin (#3873), anti-GAPDH (#2118) (Cell Signaling Technology, Inc.).
4.6 |. Activation status of cell signaling pathways by phospho-specific antibodies
Phosphorylation status of 43 protein kinases was analyzed in cell lysates using a membrane-based sandwich immunoassay in which captured antibodies spotted in duplicate on nitrocellulose membranes bind to specific target proteins present in the sample. Phosphorylation is detected with biotinylated phospho-specific detection antibodies and then visualized using chemiluminescent detection reagents. The signal produced is proportional to the amount of phosphorylation in the band analyzed. A representive result from the JIMT1 pair using the Proteome Profiler Human Phospho-Kinase Array Kit, R & D Systems—ARY003B (R&D Systems, Minneapolis, MN) is presented in the Supplemental Figure S3.
4.7 |. Immunoprecipitation
Cells were rinsed once with Phosphate Buffered Saline at 4°C and incubated in Lysis Buffer (20 mM Tris (pH 7.5), 150 mM NaCI, 1mM EDTA, 1mM EGTA, 1% Triton 100, and IX HALT Protease & Phosphatase Inhibitor Cocktail (Thermo Fisher Sci., Waltham, MA) on ice for 5 min, scraped and transferred to micro-centrifuge tubes. The tubes were centrifuged at 14 000×g for 5 min at 4°C and the supernatant transferred to clean tubes. The lysates were first cleared by incubating with a mouse IgG antibody-Sepharose bead conjugate (Mouse IgG, #3420, Cell Signaling Technology) for 4 h at 4°C on a rocker. The lysates were microcentrifuged for 30 s to pellet the beads and the supernatants transferred to clean tubes. The lysates were then incubated with an antibody-Sepharose bead conjugate (Phospho-Tyrosine Mouse mAb, #9419 or HER-1 Mouse mAb, #8083 both from Cell Signaling Technology) overnight at 4°C on a rocker. The lysates were microcentrifuged for 30 s to pellet the beads at 4°C. The beads were washed twice, pelleted and re-suspended in sample loading buffers for processing for Western analysis.
4.8 |. In vivo studies
All animals were used under an approved protocol of the Institutional Animal Care and Use Committee (IACUC) of Oregon Health and Science University (OHSU) and the experiments were carried out under the auspices of the Department of Comparative Medicine of OHSU. To assess the growth promoting function of GRB7 in human breast cancer cell lines in vivo, polyclonal HCC1954, 21MT1, and JIMT1 cells with GRB7 knock down or their corresponding negative control, were orthotopically injected into the mammary fat pads of female immune-deficient NSG mice. The growth rates of these tumors were measured serially with calipers, and the final tumor weights compared between GRB7 knock down and the negative control.
4.9 |. Immunohistochemistry
Xenograft tumors were harvested and fixed in 10% formalin buffer. Sections were deparaffinized followed by antigen retrieval with sodium citrate buffer and blocking of endogenous peroxidase activity with 3% H202. Slides were then incubated with primary Ab Anti-Ki-67 (Leica NCL-Ki67p) (Leica Biosystems, Buffalo Grove, IL) dilutedl:1000 in blocking buffer with 5% goat serum overnight. Slides were washed three times for 5 min each in PBS, and incubated with biotinylated goat anti-rabbit IgG diluted 1:400 for 1 h at room temperature. Following a washing in PBS, the sections were incubated with ABC, DAB, and counterstain reagents, respectively. Slide were then dehydrated and cover slipped with VECTASHIELD mount (Vector Laboratories, Burlingame, CA). Cells were enumerated from 20X fields using Leica Application Suite V4 program.
4.10 |. Apoptosis
Apoptosis was detected with terminal deoxynucleotidyltransferase-mediated dUTP Nick end labeling (TUNEL) staining using the Apo Tag kit (Millipore S7101) (MilliporeSigma, St. Louis, MO). Stained apoptotic cells were counted and percentage was determined by averaging the count in 10 random fields per slide.
4.11 |. Knock down of HER-1 with siRNA transfection in HCC 1954, 21MT1, and JIMT1 cells
Breast cancer cells were seeded at the desired number in 96-well plates (Corning Costar #3595, Corning, New York) one day prior to transfection. Cells were transfected with 0.033μM siRNA EGFR (siGENOME EGFR SMARTpool, #M003114030005, Dharmacon) or control siRNA (non-targeting siRNA control pool #2, D-001206–14-05 Dharmacon) according to manufacturer’s instructions respectively using Dharmafect 1 transfection reagent (Dharmacon). 72 h following transfection with siRNAs, cell viability was measured using the Trevigen’s (Trevigen, Gaithersburg, MD) TACS XTT Assay (TACS XTT, cat# 4891– 025-K). After substraction of the background, the absorbance of PBS-treated wells was set as 100% against which the absorbance of panitumumab-treated cells was normalized.
4.12 |. Treatment with a humanized anti- HER-1 antibody, panitumumab
Parental HCC 1954, 21MT1, and JIMT1 cells were prepared as above, seeded in 96-well plates, 8 replicates each, in a fixed volume of 75 μL with a homogenous initial confluence between 5 and 20% and incubated at 37° overnight. Panitumumab (25 μL, final concentration 100 μg/mL medium) was added to treatment wells the next day. PBS (25 μL) was added to control wells and the plates were incubated for two additional days. At the end of the incubation period, proliferation assay reagent (TACS XTT, cat# 4891– 025-K) was prepared and added as per manufacturer’s instruction. Absorbance was read with a Spectramax 384 Plus microtiter plate reader at 490 nm. The absorbance of PBS-treated wells was set as 100% and the proliferation of panitumumab-treated cells was calculated accordingly. Panitumu-mab was obtained from the OHSU pharmacy.
4.13 |. Migration/invasion assay
Assays were carried out using Corning Costar Transwell inserts (Corning #3463) and Corning BioCoat growth factor reduced Matrigel invasion chambers (Corning #354483). Breast cancer cells were grown in RPMI 1640 media with 1% FBS for 12–24 h prior to the start of the assay. Cells were counted and suspended in media with 0.1% FBS at the desired density. Inserts were placed in wells of a 24-well plates containing 750 μL of media containing 10% FBS as a chemoattractant. A 300 μL aliquot of cell suspension was added to each insert. Cells were incubated at 37°C, 5% C02 for 4–6 h for migration assays and 16–20 h for invasion assays. Cells that had migrated through the membranes were stained with crystal violet (Sigma-Aldrich #HT90132). Non-migrating cells (and Matrigel) were removed from the inside of the membranes with a cotton swab. The membranes were photographed under a microscope at 100×. Cells were counted in a minimum of three fields per membrane and averaged.
4.14 |. Statistical methods
Statistical analysis was carried out with GraphPad Prism version 6.0 unless otherwise specified (GraphPad Software, La Hoja, CA). All experiments were repeated at least three times, and the results are presented as the mean ± standard errors (SEM). Group differences were compared with an unpaired t-test. P <0.05 was considered statistically significant. A random intercept and slope model were used to measure and compare the growth rates of tumor xenografts formed by GRB7 knockdown and negative control using SAS 9.4 package, SAS Institute Inc. (Gary, NC).
Supplementary Material
ACKNOWLEDGMENTS
This work is supported in part by the VA MERIT award (1I01BX002236-01A2). The contents do not represent the views of the U.S. Department of Veterans Affairs or the United States Government. This work is dedicated to the loving memory of Dr Ed Keenan. Biostatistics support was provided by the Knight Cancer Institute Biostatistics Shared Resource at Oregon Health and Science University (NCI Cancer Center Support Grant P30 CA069533).
Funding information
VA Merit, Grant number: 1I01BX002236-01A2
Footnotes
ETHICAL APPROVAL
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. This article does not contain any studies with human participants performed by any of the authors.
CONFLICT OF INTEREST
Author Luoh served on the advisory board of Amgen. Authors Wagoner, Lai, Wang, Chin, Ramsey, Sears, and Hu declare that he/she has no conflict of interest.
SUPPORTING INFORMATION
Additional Supporting Information may be found online in the supporting information tab for this article.
REFERENCES
- 1.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235: 177–182. [DOI] [PubMed] [Google Scholar]
- 2.Slamon DJ, Godolphin W, Jones LA, et al. Studies of the HER-2/neuproto-oncogene in human breast and ovarian cancer. Science. 1989;244:707–712. [DOI] [PubMed] [Google Scholar]
- 3.Slamon DJ, Leyland-Jones B, Shak S, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–792. [DOI] [PubMed] [Google Scholar]
- 4.Slamon D, Eiermann W, Robert N, et al. Adjuvant trastuzumab in HER2-positive breast cancer. N Engl J Med. 2011;365:1273–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Romond EH, Perez EA, Bryant J, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005;353:1673–1684. [DOI] [PubMed] [Google Scholar]
- 6.Piccart-Gebhart MJ, Procter M, Leyland-Jones B, et al. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N Engl J Med. 2005;353:1659–1672. [DOI] [PubMed] [Google Scholar]
- 7.Martin M, Holmes FA, Ejlertsen B, et al. Neratinib after trastuzumab-based adjuvant therapy in HER2-positive breast cancer (ExteNET): 5-year analysis of a randomised, double-blind, placebo-controlled, phase trial. Lancet Oncol. 2017;18:1688–1700. [DOI] [PubMed] [Google Scholar]
- 8.Di Leo A, Gomez HL, Aziz Z, et al. Phase III, double-blind, randomized study comparing lapatinib plus paclitaxel with placebo plus paclitaxel as first-line treatment for metastatic breast cancer. J Clin Oncol. 2008;26: 5544–5552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baselga J, Cortes J, Kim SB, et al. Pertuzumab plus trastuzumab plus docetaxel for metastatic breast cancer. N Engl J Med. 2011;366: 109–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verma S, Miles D, Gianni L, et al. Trastuzumab emtansine for HER2-positive advanced breast cancer. N Engl J Med. 2012;367:1783–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gianni L, Pienkowski T, Im YH, et al. Efficacy and safety of neoadjuvant pertuzumab and trastuzumab in women with locally advanced, inflammatory, or early HER2-positive breast cancer (NeoSphere): a randomised multicentre, open-label, phase 2 trial. Lancet Oncol. 2012;13:25–32. [DOI] [PubMed] [Google Scholar]
- 12.Schneeweiss A, Chia S, Hickish T, et al. Pertuzumab plus trastuzumab in combination with standard neoadjuvant anthracycline-containing and anthracycline-free chemotherapy regimens in patients with HER2-positive early breast cancer: a randomized phase II cardiac safety study (TRYPHAENA). Ann Oncol. 2013;24:2278–2284. [DOI] [PubMed] [Google Scholar]
- 13.Shen TL, Guan JL. Grb7 in intracellular signaling and its role in cell regulation. Front Biosci. 2004;9:192–200. [DOI] [PubMed] [Google Scholar]
- 14.Bai T, Luoh SW. GRB-7 facilitates HER-2/Neu-mediated signal transduction and tumor formation. Carcinogenesis. 2008;29:473–479. [DOI] [PubMed] [Google Scholar]
- 15.Ramsey B, Bai T, Hanlon Newell A, et al. GRB7 protein over-expression and clinical outcome in breast cancer. Breast Cancer Res Treat. 2011;127:659–669. [DOI] [PubMed] [Google Scholar]
- 16.Giricz O, Calvo V, Pero SC, Krag DN, Sparano JA, Kenny PA. GRB7 is required for triple-negative breast cancer cell invasion and survival. Breast Cancer Res Treat. 2011;133:607–615. [DOI] [PubMed] [Google Scholar]
- 17.Nadler Y, Gonzalez AM, Camp RL, Rimm DL, Kluger HM, Kluger Y. Growth factor receptor-bound protein-7 (Grb7) as a prognostic marker and therapeutic target in breast cancer. Ann Oncol. 2010;21: 466–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu S, Hu Z, Ngamcherdtrakul W, et al. Therapeutic siRNA for drug-resistant HER2-positive breast cancer. Oncotarget. 2016;7: 14727–14741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baselga J, Coleman RE, Cortés J, Janni W. Advances in the management of HER2-positive early breast cancer. Crit Rev Oncol Hematol. 2017;119:113–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan A, Delaloge S, Holmes FA, et al. Neratinib after trastuzumab-based adjuvant therapy in patients with HER2-positive breast cancer (ExteNET): a multicentre, randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2016;17:367–377. [DOI] [PubMed] [Google Scholar]
- 21.von Minckwitz G, Procter M, de Azambuja E, et al. Adjuvant pertuzumab and trastuzumab in early HER2-Positive Breast cancer. N Engl J Med. 2017;377:122–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barroso-Sousa R, Exman P, Tolaney SM. De-escalating treatment in the adjuvant setting in HER2-positive breast cancer. Future Oncol. 2018;14:937–945. [DOI] [PubMed] [Google Scholar]
- 23.Stein D, Wu J, Fuqua SA, et al. The SH2 domain protein GRB-7 is co-amplified, overexpressed and in a tight complex with HER2 in breast cancer. Embo J. 1994;13:1331–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chu PY, Li TK, Ding ST, Lai IR, Shen TL. et al. EGF-induced Grb7 recruits and promotes Ras activity essential for the tumorigenicity of Sk-Br3 breast cancer cells. J Biol Chem. 2010;285: 29279–29285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watson SS, Dane M, Chin K, et al. Microenvironment-mediated mechanisms of resistance to HER2 inhibitors differ between HER2+ Breast cancer subtypes. Cell Syst. 2018;6:329–342. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korkola JE, Collisson EA, Heiser M, et al. Decoupling of the PI3K pathway via mutation necessitates combinatorial treatment in HER2+ Breast cancer. PLoS ONE. 2015;10:e0133219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


