Abstract
Objective:
The purpose of this study is to evaluate the role mitochondrial inheritance plays in primary open-angle glaucoma (POAG) characteristics in African Americans.
Methods:
POAG cases from the L1c2 and L1b mitochondrial haplogroups were compared in a retrospective case-case study. Twenty-six pairs of self-identified African American POAG cases from L1c2 and L1b mitochondrial haplogroups matched on age (mean [SD] = 71.2 [9.6] and 71.3 [9.6] years, respectively; p = 0.97), sex (21 female and 5 male pairs), and family history of glaucoma (positive in 15/26 [58%] pairs) were included.
Results:
L1c2 subjects displayed higher vertical cup-to-disc ratio (0.75 [0.12] and 0.67 [0.16], respectively; p = 0.01, Bonferroni-corrected p = 0.08), worse pattern standard deviation on visual field (VF) testing (5.5 [3.5] and 3.5 [2.7]; p = 0.005, Bonferroni-corrected p = 0.02), and more severe glaucoma based on American Glaucoma Society staging criteria (p = 0.04, Bonferroni-corrected p = 0.32) compared to L1b subjects. L1c2 also trended towards worse mean deviation on VF compared to L1b (−8.2 [7.6] and −5.8 [6.8], respectively, p = 0.17). Best corrected visual acuity, central corneal thickness, maximum intraocular pressure (IOP), and cataract severity were comparable between L1c2 and L1b haplogroups (p ≥ 0.49), as was retinal nerve fiber layer thickness on optical coherence tomography (75.1 [14.1] and 75.1 [13.0]; p = 0.99).
Conclusion:
Results demonstrated worse glaucomatous cupping and more severe VF loss in the L1c2 compared to the L1b haplogroup despite comparable IOP. Findings implicate mitochondrial inheritance as a factor affecting POAG severity and may ultimately contribute to stratifying POAG patients into phenotypically and genotypically distinct subgroups.
Keywords: Glaucoma, Primary open-angle glaucoma, African Americans, mitochondrial genetics, mitochondrial haplogroups
Introduction
Primary open angle glaucoma (POAG) is a phenotypically complex condition of progressive optic neuropathy characterized by optic nerve excavation and a corresponding pattern of visual field loss. African Americans are four times more likely than Caucasians to develop POAG, with the disease presenting earlier, progressing more rapidly, and exhibiting more resistance to treatment compared to its presentation in Caucasians [1–3]. Glaucomatous optic neuropathy shares many similarities with those present in primary mitochondrial diseases, and a growing body of literature suggests that mitochondrial function plays a role in POAG pathogenesis [4]. Due to high-energy demands, both the optic nerve and retinal ganglion cells are highly susceptible to impairments of mitochondrial distribution and respiratory capacity [5–8]. Consistent with mitochondrial inheritance, a maternal family history of glaucoma is also more prevalent in patients with POAG [9].
The mitochondrial genome is classified into haplogroups based upon single nucleotide polymorphisms (SNPs) that have accumulated over time [10]. The Primary Open Angle African American Glaucoma Genetics (POAAGG) study, a genome wide association study aimed at elucidating novel genetic variants impacting POAG risk, previously identified mitochondrial haplogroups and variants associated with differential POAG risks [11]. Pooled sequencing of mitochondrial variants in the POAAGG population implicated the L1c2 haplogroup as conferring increased risk of developing POAG, with odds ratio (OR) > 1.4 between cases and controls [12]. This finding echoed published literature implicating select subclades of the L haplogroup in elevating POAG susceptibility in Saudi Arabian and North African populations [13].
However, apart from an exploratory study published by our group, no study has examined whether clinical presentations of POAG differed between mitochondrial haplogroups. To bridge this knowledge gap, this study compared POAG severity between 26 age, sex, and family history-matched pairs from the risk-conferring L1c2 haplogroup and the phylogenetically similar but equivalent-risk L1b haplogroup (OR = 1.0). The L1c2 haplogroup is defined in part by missense mutations m.6150G>A (V83I; OR = 1.8) and m.6253C>T (M117T; OR 1.6) in the cytochrome c oxidase subunit 1 (MT-CO1) gene [12]. An exploratory study by our group supported an association between haplogroup L1c2 and increased POAG severity [14]. Utilizing a rigorously matched POAAGG cohort, this study replicated previous findings and confirmed worse glaucomatous cupping and visual field (VF) deficits in the L1c2 haplogroup despite equivalent intraocular pressures (IOP), implicating an association between POAG severity and mitochondrial inheritance.
Methods
This study is a retrospective case-case comparison between POAG subjects from the L1c2 and L1b mitochondrial haplogroups. These subjects are a subset of those enrolled in the POAAGG study, whose recruitment criteria were detailed previously [15]. Briefly, POAAGG subjects who self-identified as African American were recruited from the clinical practices of the Department of Ophthalmology at the University of Pennsylvania and affiliates. At the time of enrollment, all POAG cases were evaluated and diagnosed by fellowship-trained glaucoma specialists based on an open anterior chamber angle, characteristic optic nerve findings, and corresponding VF changes. In addition, electronic medical records of all POAG cases included in this study were reviewed by two fellowship-trained glaucoma specialists (QNC and VMA) and confirmed to have POAG. This study was approved by the University of Pennsylvania Institutional Review Board (IRB) and adhered to the tenets of the Declaration of Helsinki. Informed consent was obtained.
Haplogroup Classification
Microarray genotyping (Illumina Multi-Ethnic Genotyping Array) of the POAAGG cohort was performed. Genotypes from the 1,293 mitochondrial positions were extracted using Genome studio software. After standard quality control measures, polymorphisms at six positions on the mitochondrial DNA (mtDNA) (2352, 2768, 6150, 6253, 6480, 6548) were used to infer haplogroup classifications based on the Phylotree (build 17), MITOMAP, and MITOMASTER databases [10,16]. Subjects were classified as L1c2 based on the presence of variants m.6150G>A and m.6253T>C. L1b is classified by the presence of variants m.2352T>C, m.2678A>G, m.6548C>T, and the absence of variants at the other positions of interest. The L1b haplogroup was chosen for comparison based upon its close phylogenetic association to the L1c2 haplogroup and the lack of risk-associated variants. Haplogroup classifications were confirmed by Haplogrep2 software using genotype data from 913 variable mtDNA positions [17].
Patient Selection and Matching
At the time of this study, 87 POAG subjects out of the ~2500 POAG subjects in the POAAGG cohort belonged to the L1c2 haplogroup. Forty-eight subjects were excluded due to < 2 ophthalmology visits and/or missing data within 1 year of enrollment date. In order to minimize bias from confounders when comparing POAG characteristics, the remaining L1c2 subjects were individually matched with L1b based on sex, age +/− 2 years, and family history of glaucoma. To maintain age-matching, the phenotypic dataset was also reviewed to ensure all parameters included in the analysis were recorded within 1 year of the age at comparison. During extended clinical data review, 13 L1c2 subjects did not have an exact L1b match, resulting in a final cohort consisted of 26 matched pairs (52 total subjects).
Phenotypic Data Review
The following ocular parameters from the electronic medical records of clinic visits were collected after matching and consisted of: best corrected visual acuity (BCVA), central corneal thickness (CCT), maximum recorded IOP, and vertical cup-to-disc ratio (CDR) based on clinical assessment from glaucoma specialists. Optic nerve optical coherence tomography (OCT) and VF testing results were reviewed to extract mean retinal nerve fiber layer (RNFL) thickness, mean deviation (MD) and pattern standard deviation (PSD). OCTs with < 6/10 signal strength, VFs with > 20% fixation loss and >30% false positive/negative rates were excluded. Visual fields were tested using both Humphrey and Octopus visual field systems. A single eye from L1b was missing BCVA data, both eyes from a single L1c2 subject did not have maximum IOP recorded, and both eyes from 3 L1b subjects were missing CCT information. Seven L1b and 7 L1c2 subjects did not receive optic nerve OCT imaging within 1 year of age at comparison.
A glaucoma specialist (QNC) assigned glaucoma severity (mild, moderate, or severe) to each subject based on the American Glaucoma Society (AGS) staging guidelines. All individuals in data extraction and analysis were masked to haplogroup status.
Statistical Analysis
Generalized linear regression models were used to compare POAG ocular measurements between haplogroups. General estimating equation was utilized to account for the correlation between eyes in these ocular measurements. Lens status and cataract severity were incorporated in the regression model to control for any effect of media opacity in VF testing. In these analyses, if either one of a haplogroup pair had missing data from both eyes, both subjects were excluded from analysis. All statistical analyses were performed using the SAS software version 9.4. The two-sided p-values were reported without correcting for multiple comparisons, and if p-value<0.05, the conservative Bonferroni-corrected p-value was also calculated to account for all 8 multiple comparisons of glaucoma associated ocular characteristics reported in Table 2.
Table 2.
Glaucoma associated ocular characteristics of L1b and L1c2 subjects.
| POAG Characteristics | L1b with POAG* (n = 52 eyes) |
L1c2 with POAG** (n = 52 eyes) |
P value (Bonferroni- corrected P value) |
|---|---|---|---|
| CCT, mean (SD), μm | 529.7 (48.9) | 538.0 (40.8) | 0.49 |
| CDR, mean (SD) | 0.67 (0.16) | 0.75 (0.12) | 0.01 (0.08) |
| Maximum IOP, mean (SD), mmHg | 23.4 (7.0) | 22.9 (6.5) | 0.78 |
| OCT RNFL, mean (SD), μm | 75.1 (13.0) | 75.1 (14.1) | 0.99 |
| BCVA, mean (SD), logMAR | 0.14 (0.26) | 0.11 (0.20) | 0.52 |
| MD, mean (SD), dB | −5.8 (6.8) | −8.2 (7.6) | 0.17 |
| PSD, mean (SD), dB | 3.5 (2.7) | 5.5 (3.5) | 0.005 (0.02)+ |
Abbreviations: CCT, central corneal thickness; SD, standard deviation; CDR, vertical cup-to-disc ratio; IOP, intraocular pressure; OCT RNFL, mean retinal layer fiber layer thickness on optical coherence tomography; MD, mean deviation on visual field testing; PSD, pattern standard deviation on visual field testing.
A single eye was missing BCVA, and 3 L1b subjects (6 eyes) was missing CCT.
Maximum IOP was missing from both eyes of a single L1c2 subject.
Results are statistically significant after Bonferroni correction
Results
The 26 L1c2 and L1b subject pairs (52 POAG cases total) consisted of 21 females and 5 males in each group, of which 15 pairs (58%) reported a positive family history of glaucoma (Table 1). The mean (standard deviation [SD]) age was 71.2 (9.6) years for the L1c2 group and 71.3 (9.6) years for the L1b group (p = 0.97). The mean (SD) age difference between haplogroup pairs was 7.3 (8.6) months. Although the majority of the 30 subjects with a family history of glaucoma reported maternal inheritance, the difference was not statistically significant (p = 0.17).
Table 1.
Demographic characteristics of the age-, sex-, and family history-matched POAG cases from mitochondrial haplogroups L1b and L1c2.
| Demographic Characteristics | L1b with POAG (n = 26) |
L1c2 with POAG (n = 26) |
p value | |
|---|---|---|---|---|
| Age, mean (SD), years | 71.3 (9.6) | 71.2 (9.6) | 0.97 | |
| Sex, No. (%) | Female | 21 (81%) | 21 (81%) | 1.00 |
| Male | 5 (19%) | 5 (19%) | ||
|
Family History of Glaucoma, No. (%) |
No | 11 (42%) | 11 (42%) | 1.00 |
| Yes | 15 (58%) | 15 (58%) | ||
|
Maternal Family History of Glaucoma among those with positive family history of glaucoma, No. (%) |
No | 2 (14%) | 2 (14%) | 0.17 |
| Yes | 8 (53%) | 12 (80%) | ||
| Unknown | 5 (33%) | 1 (7%) | ||
|
Lens/Cataract Status+, No. (%) |
Pseudophakia | 11 (22%) | 14 (28%) | 0.84 |
| Cataract grades 0 – 1+ | 16 (32%) | 16 (32%) | ||
| Cataract grades 2 – 3+ | 23 (46%) | 20 (40%) | ||
Abbreviation: SD, standard deviation.
No subject had 4+ cataract severity.
L1c2 subjects displayed higher CDR (0.75 [0.12]) compared to L1b subjects (0.67 [0.16]; p = 0.01; Bonferroni-corrected p=0.08, Figure 1). The L1c2 group also exhibited worse PSD on VF testing compared to the L1b group (5.5 [3.5] dB and 3.5 [2.7] dB, respectively; p = 0.005, Bonferroni-corrected p=0.04). The majority of matched pairs received VF testing using the same system (i.e. both Humphrey vs. both Octopus): 17 pairs were tested using the Humphrey VF system, 2 pairs using the Octopus VF system, and 7 pairs using a mix of both. After excluding the 7 pairs who were not tested on the same type of VF system, the L1c2 group still exhibited worse PSD compared to the L1b group (5.4 [3.5] dB and 3.1[2.7] dB, respectively; p = 0.002, Bonferroni-corrected p=0.02). While L1c2 trended towards worse MD compared to L1b (−8.2 [7.6] dB versus −5.8 [6.8] dB, respectively), differences were not statistically significant (p = 0.17). RNFL thicknesses were comparable between groups (p = 0.99; Table 2). BCVA, CCT, IOP, and lens status/cataract severity were comparable between haplogroups (p ≥ 0.49; Table 2). Accounting for cataract severity and pseudophakic status in the regression model did not reveal a significant effect of media opacity on VF testing between group comparisons.
Figure 1:
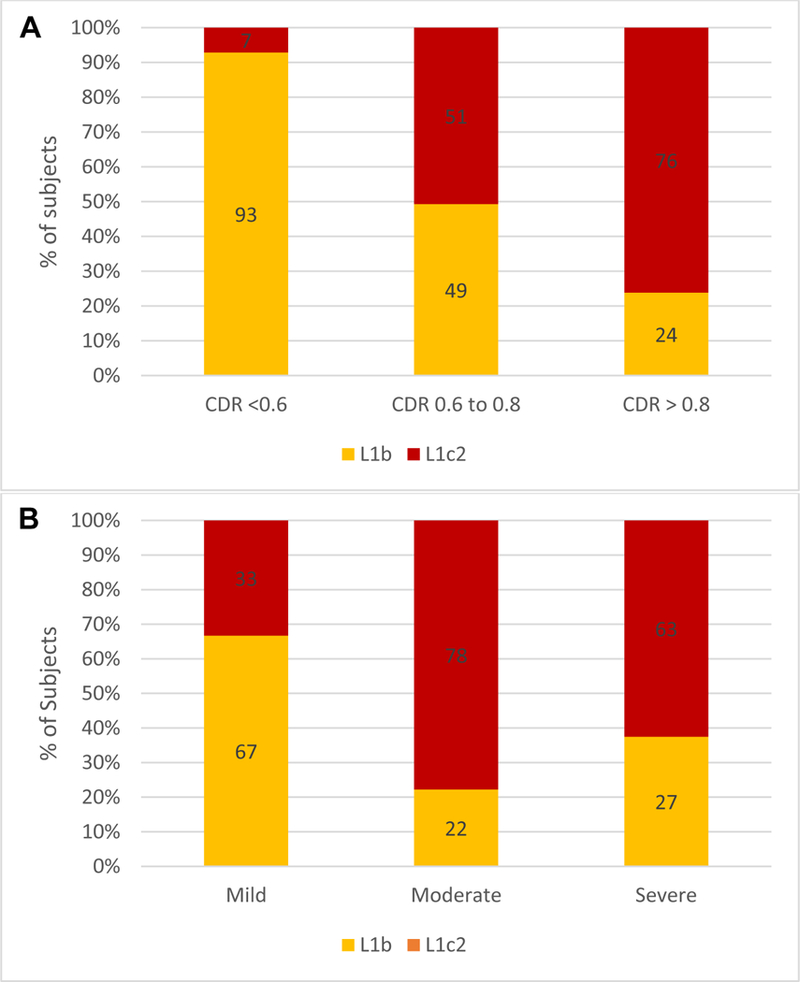
(A) Distribution of vertical cup-to-disc ratio (CDR): the L1c2 haplogroup trended towards more severe glaucomatous cupping compared to L1b (P = 0.01, Bonferroni-corrected P = 0.08). (B) Distribution of glaucoma severity based on American Glaucoma Society staging criteria: the L1c2 haplogroup trended towards more severe glaucoma compared to L1b (P = 0.04, Bonferroni-corrected P = 0.32).
A single grader (QNC) blinded to haplogroup status assigned glaucoma severity based on AGS criteria. The L1c2 haplogroup was associated with more severe disease compared to the L1b group (P=0.04; Bonferroni-corrected p = 0.32, Figure 1B). Nine (35%) L1c2 subjects had mild glaucoma, 7 (27%) had moderate glaucoma, and 10 (38%) had severe glaucoma. In comparison, the majority (18 subjects, 69%) of L1b subjects were classified as mild, 2 (8%) were classified as moderate, and 6 (23%) were classified as severe.
Discussion
In this study, ocular parameters were compared between age, sex, and family history matched African American POAG subjects from the risk-conferring mitochondrial haplogroup L1c2 and the risk-equivalent haplogroup L1b. The L1c2 haplogroup was found to have worse optic neuropathy and more severe glaucomatous VF loss despite comparable enrollment and maximum recorded IOPs. Consistent with mitochondrial inheritance and previously reported data from our group, a trend towards higher self-reported maternal family history of glaucoma in L1c2 subjects was observed [9,14].
These findings echo those of a prior exploratory cohort (n=29 pairs), which concluded that the L1c2 haplogroup was associated with higher CDR and worse VF parameters despite comparable IOP [14]. Compared to the exploratory cohort, the strength of our study lies in the application of stringent age-matching criteria with respect to all phenotypic data, including VFs and OCTs. The L1b haplogroup was chosen for comparison based upon its close phylogenetic association to the L1c2 haplogroup and the lack of risk-associated MT-CO1 variants. Each subject was strictly matched on age, sex, and family history of glaucoma. Disease severity was additionally standardized by a fellowship-trained glaucoma specialist based upon a widely-utilized staging criteria put forth by the AGS. The fact that our study demonstrated similar results to the exploratory cohort is confirmatory. The previous study also demonstrated a strong association with males in the L1c2 haplogroup and glaucoma severity. Our current cohort only had 5 males, and is underpowered for any subset analyses based on sex. To the best of our knowledge, studies from our group are the first to associate the mitochondrial L1c2 haplogroup with greater POAG severity, whether in an African American or some other population.
As a consequence of our matching study design, which is tailored to minimize impact from possible confounders, the study is limited by a small sample size. The power calculation indicates that the study has 80% power to detect an effect size of 0.46 and 90% power to detect an effect size of 0.54 for these ocular parameters of POAG. The small sample size may have attributed to the fact that the CDR and glaucoma severity comparisons did not survive Bonferroni corrections. The small sample size is partly due to the low prevalence of the L1c2 haplogroup in the POAAGG cohort (~4%) and is partially a result of excluding several pairs from analysis due to the lack of matching data. 48 L1c2 subjects were initially excluded as they had no data due to missed ophthalmology visits, which may introduce ascertainment bias, as it is possible that disease severity may affect adherence to care and patient visits. For our analysis, both members of the pair were excluded if one member was missing data to allow rigorous phenotypic evaluation and AGS severity grading. Unpaired regression including pairs with partially missing data showed no difference in RNFL thickness between groups. Other limitations include using both Humphrey and Octopus visual fields, and that cataract grading was not standardized across all providers. However, these differences were not found to have significantly affected the results.
In light of our findings, it is worth discussing which inherited variants associated with the L1c2 haplogroup may be implicated in POAG pathogenesis. The L1c2 haplogroup is defined in part by the missense mtDNA variants Val83Ile and Met117Thr in the N-terminal region of MT-CO1. We have previously identified these MT-CO1 mutations as conferring increased risk of POAG in the POAAGG study cohort, with OR of 1.8 and 1.6 between cases and controls, respectively [12]. MT-CO1 encodes cytochrome oxidase c subunit, a key component of the catalytic core of the respiratory chain enzyme cytochrome c oxidase (Complex IV). The optic nerve and retinal ganglion cells have been shown to be highly vulnerable to impaired cellular respiration as a result of high energy demands [5–8]. Decreased Complex IV activity has also been directly associated with increased reactive oxygen species generation and decreased cellular energy synthesis, both of which have been linked with glaucomatous optic neuropathy [18].
In summary, our study confirmed an association between mitochondrial inheritance and POAG severity, and suggests that mitochondrial haplogroup designation may assist in POAG stratification. Future directions will include a larger sample size and a longer follow-up duration to allow for glaucoma progression analysis, evaluation for specific patterns of VF defects, and assessment for signs and symptoms of systemic mitochondrial disturbances, if present. Finally, targeted functional studies of MT-CO1 variants will elucidate pathogenic mechanisms and may direct future diagnostic and therapeutic strategies for POAG.
Acknowledgments
Financial Support: This work was supported by NIH-NEI RO1EY023557 and the Department of Ophthalmology at the Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA. Funds were also provided by the F.M. Kirby Foundation, Research to Prevent Blindness, The Paul and Evanina Bell Mackall Foundation Trust, and the National Eye Institute, National Institutes of Health, Department of Health and Human Services, under eyeGENE™ contract Nos. HHSN260220700001C and HHSN263201200001C. Dr. Cui is supported by NIH-NEI 5K12EY015398 (PI: Maguire, Maureen). The sponsor or funding organization had no role in research design or execution.
Footnotes
No conflicting commercial relationship exists for any author.
REFERENCES
- 1.Leske MC. The epidemiology of open-angle glaucoma: a review. Am J Epidemiol. 1983;118(2):166–191. [DOI] [PubMed] [Google Scholar]
- 2.Tielsch JM, Sommer A, Katz J, Royall RM, Quigley HA, Javitt J. Racial variations in the prevalence of primary open-angle glaucoma. The Baltimore Eye Survey. JAMA. 1991;266(3):369–374. [PubMed] [Google Scholar]
- 3.Tielsch JM, Katz J, Sommer A, Quigley HA, Javitt JC. Family history and risk of primary open angle glaucoma. The Baltimore Eye Survey. Arch Ophthalmol. 1994;112(1):69–73. [DOI] [PubMed] [Google Scholar]
- 4.Lascaratos G, Garway-Heath DF, Willoughby CE, Chau KY, Schapira AH. Mitochondrial dysfunction in glaucoma: understanding genetic influences. Mitochondrion. 2012;12(2):202–212. [DOI] [PubMed] [Google Scholar]
- 5.Osborne NN, del Olmo-Aguado S. Maintenance of retinal ganglion cell mitochondrial functions as a neuroprotective strategy in glaucoma. Curr Opin Pharmacol. 2013;13(1):16–22. [DOI] [PubMed] [Google Scholar]
- 6.Lee S, Van Bergen NJ, Kong GY, et al. Mitochondrial dysfunction in glaucoma and emerging bioenergetic therapies. Exp Eye Res. 2011;93(2):204–212. [DOI] [PubMed] [Google Scholar]
- 7.Bristow EA, Griffiths PG, Andrews RM, Johnson MA, Turnbull DM. The distribution of mitochondrial activity in relation to optic nerve structure. Arch Ophthalmol. 2002;120(6):791–796. [DOI] [PubMed] [Google Scholar]
- 8.Carelli V, Ross-Cisneros FN, Sadun AA. Mitochondrial dysfunction as a cause of optic neuropathies. Prog Retin Eye Res. 2004;23(1):53–89. [DOI] [PubMed] [Google Scholar]
- 9.Shin DH, Becker B, Kolker AE. Family history in primary open-angle glaucoma. Arch Ophthalmol. 1977;95(4):598–600. [DOI] [PubMed] [Google Scholar]
- 10.van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat. 2009;30(2):E386–394. [DOI] [PubMed] [Google Scholar]
- 11.Collins DW, Gudiseva HV, Trachtman BT, et al. Mitochondrial sequence variation in African-American primary open-angle glaucoma patients. PLoS One. 2013;8(10):e76627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins DW, Gudiseva HV, Trachtman B, et al. Association of primary open-angle glaucoma with mitochondrial variants and haplogroups common in African Americans. Mol Vis. 2016;22:454–471. [PMC free article] [PubMed] [Google Scholar]
- 13.Abu-Amero KK, Gonzalez AM, Osman EA, Larruga JM, Cabrera VM, Al-Obeidan SA. Mitochondrial DNA lineages of African origin confer susceptibility to primary open-angle glaucoma in Saudi patients. Mol Vis. 2011;17:1468–1472. [PMC free article] [PubMed] [Google Scholar]
- 14.Collins DW, Gudiseva HV, Chavali VR, et al. The MT-CO1 V83I polymorphism is a risk factor for primary open-angle glaucoma in African American men. Invest Ophthalmol Vis Sci. 2018;59(5):1751–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charlson ES, Sankar PS, Miller-Ellis E, et al. The primary open-angle african american glaucoma genetics study: baseline demographics. Ophthalmology. 2015;122(4):711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lott MT, Leipzig JN, Derbeneva O, et al. mtDNA Variation and Analysis Using Mitomap and Mitomaster. Curr Protoc Bioinformatics. 2013;44:1 2321–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weissensteiner H, Pacher D, Kloss-Brandstatter A, et al. HaploGrep 2: mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucleic Acids Res. 2016;44(W1):W58–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Srinivasan S, Avadhani NG. Cytochrome c oxidase dysfunction in oxidative stress. Free Radic Biol Med. 2012;53(6):1252–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]


