Abstract
Escherichia coli O157:H7 is the most well-studied serotype of the enterohemorrhagic E. coli (EHEC) class of E. coli intestinal pathogens and is responsible for many outbreaks of serious food-borne illness worldwide each year. Adherence mechanisms are a critical component of its pathogenesis, persistence in natural reservoirs, and environmental contamination. E. coli O157:H7 has a highly effective virulence operon, the Locus of Enterocyte Effacement (LEE), and its encoded intimate adherence mechanism is well characterized. However, factors involved in the preceding initial attachment are not well understood. In this study, we propose a mechanism of initial adherence used by E. coli O157:H7 in vitro. We describe a bacterial protein not previously reported to be involved in adherence, Slp, and its interactions with the human host protein polymeric immunoglobulin receptor (pIgR). The human pIgR has previously been shown to act as an adherence receptor for some mucosal pathogens and is highly expressed in the intestine. Following observation of significant colocalization between E. coli O157:H7 bacteria and pIgR location on Caco-2 cells, a co-immunoprecipitation (Co-IP) assay using a human recombinant Fc-tagged pIgR protein led to the identification of this protein. Disruption of Slp expression in E. coli O157:H7, through deletion of its encoding gene slp, produced a significant adherence deficiency to Caco-2 cells at early time points associated with initial adherence. Plasmid complementation of the slp gene fully restored the wild-type phenotype. Furthermore, immunofluorescence microscopy revealed evidence that this interaction is specific to the pathogenic strains of E. coli tested and not the nonpathogenic control strain E. coli K12. Additionally, deletion of slp gene resulted in the absence of the corresponding protein band in further Co-IP assays, while the plasmid-encoded slp gene complementation of the deletion mutant strain restored the wild-type pattern. These data support the proposal that Slp directly contributes to initial adherence, with the pIgR protein as its proposed receptor.
Introduction
It has been estimated that over 700 outbreaks of foodborne illness due to Escherichia coli occurred in the United States between 2007 and 2017 [1]. Of these reported outbreaks, over 99% were due to Enterohemorrhagic E. coli (EHEC) strains, with approximately 65% belonging to the O157 serogroup [1]. EHEC infections typically result in abdominal cramping, watery or bloody diarrhea, fever, nausea, and vomiting [2]. Among these cases, a subset develop a severe complication known as hemolytic uremic syndrome (HUS), which is primarily observed in infected children under the age of five years, the elderly, and immunocompromised individuals [3]. In the U.S., E. coli O157:H7 outbreaks have highly variable rates of HUS ranging from less than 1% to greater than 15% [4, 5].
After ingestion from a contaminated source, E. coli O157:H7 uses a series of adherence mechanisms to colonize the intestine during infection. Adherence occurs in two phases: (i) initial adherence (an early, loose attachment) and (ii) intimate adherence (strong attachment) [6]. Upon infiltration of the mucin layer in the colon, the bacteria require an initial attachment in order to stay stably adjacent to the intestinal epithelial cell (IEC). The elapsed time must be sufficient for the opportunity to engage its intimate adherence mechanism before being cleared from the digestive tract [7]. E. coli O157:H7 utilizes a fairly well-characterized set of virulence factors for intimate adherence, using factors encoded in the Locus of Enterocyte Effacement (LEE) [8]. The three main factors required for intimate adherence are (i) intimin, (ii) Tir, and (iii) type three secretion system (T3SS), which are the bacterial adhesin, translocated intimin receptor, and secretion apparatus, respectively [9]. An additional receptor for intimin, the host protein nucleolin (involved in cell proliferation and growth), has also been shown to assist in intimate adherence [10, 11].
The earlier phase of initial adherence is less well defined, and it is likely that initial adherence is governed by multiple redundant mechanisms and varies between strains [6]. Several putative adhesins have been described, but the specific contribution to initial adherence is unclear. For example, long polar fimbriae (LPF) have recently been described as initial adhesins in E. coli O157:H7, but with some conflicting reports. LPF has been described in the role of translocation of EHEC across human M cells in in vitro human organ culture simulated conditions, but lpf deletion mutant strains of E. coli O157:H7 did not show any significant effects on adherence to Caco-2 cells 3 h post-infection [12]. E. coli Common Pilus (ECP) is a fimbrial adhesin associated with biofilm formation and persistent colonization of the bovine intestine [13, 14]. Deletion mutations to the ecp gene reduced adherence to Hep2 cells at up to 6 h post-infection, but no similar evidence has been shown in Caco-2 or other human intestinal cells [15, 16]. The Hemorrhagic Coli Pilus (HCP) has also been shown to contribute to adherence to human and bovine cells, and also to porcine and bovine intestinal explants [17]. An hcpA deletion in E. coli O157:H7 reduced adherence to the human intestinal epithelial cell line HT-29 in vitro after 6 h post-infection, and over-expression of hcpABC genes also resulted in increased cellular invasion, hemagglutination, biofilm formation, and motility [18]. Flagella (H antigens) are well characterized for their role in motility but have also been implicated in adherence. Flagella in E. coli O157:H7 were able to bind bovine intestinal mucus, while H-negative mutant strains showed reduced adherence to bovine intestinal tissue explants; though other studies have reported H-negative strains as being able to colonize young calves [9, 19]. Additionally, the ETEC protein EtpA, which mimics the major subunit protein flagellin, demonstrates adherence to intestinal cells in vitro through the use of recombinant protein (rEtpA) in adherence assays [20]. The exact role of flagella in EHEC initial adherence to human IECs remains unknown [21]. Many other proteins have been identified as being of interest in the study of adherence, but roles in initial adherence are either unclear, not studied in human cells, or do not yet have enough data to be conclusive [7].
A unique adherence mechanism has been described in the human mucosal pathogen Streptococcus pneumoniae, involving the human polymeric immunoglobulin receptor (pIgR). Under normal conditions, the pIgR system plays a critical role in innate mucosal immunity [22]. The human pIgR is a glycoprotein varying in size from 80–120 kDa depending on the level of glycosylation, and primarily functions as a transport for dimeric immunoglobulin A (dIgA) into the intestinal lumen [23]. In the lumen, the transported pIgR-dIgA complex is cleaved from the IEC surface and termed secretory IgA (sIgA), where it plays an important role in maintaining the balance between immune function and microflora homeostasis [24, 25]. In addition, pIgR lacking dIgA can also be transported. The unbound pIgR is also cleaved and released (free secretory component or SC), which also serves as part of innate immunity [26, 27]. Once sIgA or SC has been secreted into the intestinal lumen, a retrograde transport pathway is used to recycle the membrane-bound pIgR [28]. S. pneumoniae uses a choline-binding protein PspC as an adhesin, binding the pIgR, SC, or sIgA at domains D3 and D4 [22, 29]. PspC enhances adherence and invasion of nasopharyngeal epithelial cells through use of the pIgR recycling transport pathway [28, 30]. Although S. pneumoniae is a Gram-positive pathogen, it is a pathogen that targets mucosal epithelial cells, as does EHEC [29, 31, 32]. High expression of pIgR in the intestine presents an opportunity for other pathogens to utilize a similar mechanism for adherence [24]. A separate study in 2007 found that pigR expression was upregulated in bovine hosts when experimentally infected with E. coli O157:H7 and a non-O157 EHEC strain for 6 h (1.93 and 3.51 fold-change (FC), respectively), but not in response to non-colonizing control strains [33]. Although the cause of this behavior was not determined, the results support the question of whether a similar mucosal epithelium colonizing pathogen, such as EHEC also has the potential to use pIgR for adherence during infection.
In this study, we describe the interaction between the E. coli O157:H7 outer membrane protein Slp (carbon starvation-inducible lipoprotein) and the pIgR, and its role in initial adherence to Caco-2 cells in vitro. Utilizing immunofluorescent microscopy, we demonstrated that during initial adherence, E. coli O157:H7 had a significant correlation with its location and the location of pIgR protein on the Caco-2 cell surface. The colocalization of E. coli cells with pIgR protein was statistically significant with E. coli O157:H7 (O157-WT), but not with the nonpathogenic E. coli strain K12. Additionally, several other non-O157 pathogenic E. coli strains also showed significant colocalization patterns. These observations were further investigated using a commercially available recombinant Fc-tagged human pIgR protein (pIgR-Fc) in a co-immunoprecipitation (Co-IP) assay, leading to the identification of Slp as the interacting protein involved. A deletion mutation of E. coli O157:H7 lacking the slp gene (O157-Δslp) resulted in an elimination of the colocalization patterns observed with immunofluorescent microscopy, the absence of the band corresponding to Slp following Co-IP, and a significant adherence deficiency to Caco-2 cells. Furthermore, the O157-Δslp strain was complemented in trans with the slp gene (O157Δslp-p:slp), which restored the wild-type phenotypes seen in colocalization, Co-IP, and adherence to Caco-2 cells in vitro. Taken together, these results support the proposal of direct interaction between the E. coli O157:H7 Slp and the host pIgR and its contribution to initial adherence.
Materials and methods
Bacterial and mammalian cell growth and culture
All bacterial strains and plasmids are described in Table 1.
Table 1. Bacterial strains and plasmids.
| Strains or Plasmids | Description | Genotype |
|---|---|---|
| Strains | ||
| MG1655 | E. coli K12 nonpathogenic | O Serogroup: N/A Pathotype: N/A Shiga toxin 1 (stx1): - Shiga toxin 2 (stx2): - Intimin (eae): - slp: + |
| O157-WT EDL 932 (ATCC #43894) |
E. coli O157:H7 wild-type |
O Serogroup: O157 Pathotype: EHEC Shiga toxin 1 (stx1): + Shiga toxin 2 (stx2): + Intimin (eae): + slp: + |
| O157-Δslp |
E. coli O157:H7 Δslp |
O Serogroup: O157 Pathotype: EHEC Shiga toxin 1 (stx1): + Shiga toxin 2 (stx2): + Intimin (eae): + slp: - |
| O157Δslp-p:slp |
E. coli O157:H7 Δslp + puc18::slp |
O Serogroup: O157 Pathotype: EHEC Shiga toxin 1 (stx1): + Shiga toxin 2 (stx2): + Intimin (eae): + slp: + |
| O26 |
E. coli O26 field isolate* |
O Serogroup: O26 Pathotype: EHEC Shiga toxin 1 (stx1): + Shiga toxin 2 (stx2): + Intimin (eae): + slp: + |
| O103 | E. coli O103 field isolate* | O Serogroup: O103 Pathotype: STEC Shiga toxin 1 (stx1): + Shiga toxin 2 (stx2): + Intimin (eae): - slp: + |
| O145 | E. coli O145 field isolate* | O Serogroup: O145 Pathotype: EPEC Shiga toxin 1 (stx1): - Shiga toxin 2 (stx2): - Intimin (eae): + slp: + |
| Plasmids | ||
| pKD119 | λ Red recombinase expression plasmid | Antibiotic resistance: Tetracycline (TetR) Growth: 30°C |
| pKD3 | λ Red recombinase plasmid containing FRT-flanked chloramphenicol resistance cassette | Antibiotic resistance: Chloramphenicol (CmR) |
| pCP20 | λ Red recombinase FLP expression plasmid | Antibiotic resistance: Ampicillin (AmpR) Growth: 30°C |
| pUC18 | Complementation cloning plasmid | Antibiotic resistance: Ampicillin (AmpR) |
*Isolates were provided by Dr. Chitrita DebRoy at the E. coli Reference Center at the Pennsylvania State University, University Park, PA.
All bacterial cultures were inoculated from single colonies into Luria Bertani (LB) broth with appropriate antibiotics when applicable and grown shaking at 37°C with 5% CO2 overnight. Caco-2 cells (American Type Culture Collection (ATCC) HTB-37) were grown at 37°C with 5% CO2, in Eagle's Minimum Essential Medium (EMEM) (ATCC) plus 20% Fetal Bovine Serum (FBS) (Atlanta Biologicals) unless otherwise indicated. Caco-2 cells were seeded into tissue culture treated six well plates (Corning Life Sciences), containing glass cover slips if used for microscopy and grown to confluency.
Immunofluorescent microscopy
Mammalian cells were grown as described, rinsed once with sterile phosphate buffered saline (PBS) and fresh medium applied 2 to 4 h prior to infection. Bacterial strains were inoculated at an MOI of 20 from overnight cultures as described under ‘Bacterial and mammalian cell growth and culture’, allowed to equilibrate to 37°C for 5 min, and incubated for 0 to 6 h. After incubation, cell culture media was removed, and cells were washed gently (by tilting and swirling) with sterile PBS three times to remove unadhered bacteria. After infection, samples were fixed using 10% neutral buffered formalin (Azer Scientific) for 10 min at room temperature and rinsed with PBS three times. Fluorescent stains were applied as listed below, with two PBS rinses in between each stain; slides were stored in glycerol-based mounting medium and away from light. The pIgR protein was stained using an unconjugated PIGR rabbit IgG polyclonal antibody (Thermo Fisher Scientific) at a 1:750 dilution in PBS for 60 min, followed by a DyLight 488-conjugated secondary goat anti-Rabbit IgG (H+L) antibody (Thermo Fisher Scientific), stained at a 1:1,000 dilution in PBS for 60 min. E. coli were stained using an unconjugated E. coli goat IgG polyclonal antibody (Thermo Fisher Scientific), stained in 1:1,000 dilution for 60 min, followed by a DyLight 594-conjugated secondary donkey anti-goat IgG (H+L) Secondary Antibody (Thermo Fisher Scientific), stained at 1:1,000 dilution in PBS for 60 min. Caco-2 cell nuclei were stained using Hoechst 33342 (Thermo Fisher Scientific), stained at a 1:1,000 dilution in PBS for 5 min. Slides were washed three times in sterile PBS and mounted using glycerol based mounting media and stored away from light. Immunofluorescent images were obtained using a Keyence BZ-9000 Fluorescence Microscope (Keyence) with 40X objective lenses (Nikon) with filters for DAPI, GFP, Texas-Red, and phase contrast. Images were taken as z-stacks at 0.5 μM intervals. Covariance (the quantification of pixels with fluorescent signals in both GFP and Texas-Red channels) was measured using ImageJ (U. S. National Institutes of Health) and the JaCOP plugin [34].
Quantitative adherence
Infection of mammalian cells was done as described. After infection and incubation, samples were collected dispensing 100 μL sterile 10% triton X-100 (Sigma-Aldrich) in PBS and incubated at room temperature for 10 min until the cells detached from the well plate. A 900 μL volume of sterile PBS was added to rinse the well, and samples were collected using a sterile cell scraper for a total sample volume of 1 mL. Samples were resuspended and vortexed vigorously until no visible cell clumps remained, ten-fold serial dilutions were made by adding 100 μL of sample into 900 μL sterile PBS in succession. A 100-μL volume of each dilution was plated and grown on LB plates overnight at 37°C before counting. All adherence assays were normalized by calculating colony forming units (CFU) per inoculum to ensure accuracy, and they were performed in triplicate. To calculate adherence, bacterial counts were adjusted by dilution factor, averaged, and the percent adherence (to normalize across samples and experiments) was calculated from the number of adhered bacteria per the number of inoculated bacteria per well. The percent adherence per well was then compared to the maximum adherence count (wild-type count at 3 h post-infection) to calculate relative adherence per sample.
Where (#A) = number of adhered bacteria; (#CFU) = number of colonies counted per plate; (PDF) = plate dilution factor; (#I) = number of inoculated bacteria per well (sample); (%AI) = percent adherence of inoculum (normalization between samples); (%AM) = maximum percent adherence (%AI) of wild-type bacteria (3-h samples); (%RA) = relative percent adherence (values shown in figures). P-values were calculated using the mean %RA for the Student’s T-test and p ≤ 0.05 was considered significant [35].
Quantitative PCR (qPCR)
Quantitative real-time polymerase chain reaction (qPCR) was done using mRNA purified from bacteria adhered to eukaryotic cells as described under ‘Immunofluorescent microscopy’ (relative expression during adherence) or mRNA purified from bacteria grown in LB broth culture over time (relative expression over time). The mRNA purification was performed using the Ambion Ribopure Kit (Thermo Fisher Scientific) for total RNA recovery, Ambion MicrobEnrich Kit (Thermo Fisher Scientific) to purify prokaryotic RNA, and Ambion MicrobExpress Kit (Thermo Fisher Scientific) to purify prokaryotic mRNA. All mRNA samples were reverse-transcribed to cDNA using the iScript cDNA Synthesis kit (BioRad). The cDNA samples were then concentrated and cleaned of reverse transcription reaction components using ethanol precipitation: to each sample, 10% (of total reverse transcription reaction volume) volume of 3 M sodium acetate pH 5.2 was added; 2.5 volumes of 100% ethanol was added; sample was vortexed thoroughly and incubated at -20°C for a minimum of 6 h to form a DNA precipitate. After precipitation, samples were centrifuged (in a standard microcentrifuge) at ˃10,000xg for 30 min at 4°C; pellets were rinsed once with ice-cold 70% ethanol; pellets were then air dried and resuspended in water and assessed for purity and concentration using AD 260/280 ratios. Using cDNA as PCR template, 20 μL qPCR reactions were made using 50–500 ng of template cDNA, 10 μL Applied Biosystems SYBR Green PCR Master Mix (Thermo Fisher Scientific), and molecular-grade water up to 20 uL total volume. The qPCR thermal cycling was done using the 7500 Fast Real-Time PCR System (Applied Biosystems) using the default PCR cycling settings (hold stage: 10 min at 95°C; cycling stage (40 cycles): 15 sec at 95°C, and 1 min at 60°C) and the primers listed in Table 2.
Table 2. Primer sequences used in this study.
| Gene | Reference Sequence | Purpose | Sequence 5’-3’ |
|---|---|---|---|
| eae | NC_002695.1 | qPCR |
F:GTCGTGTCTGCTAAAACCGC R:CGGCGGAACTGGAAGTTAGT |
| gapA | NC_002695.1 | qPCR |
F:ACTTCGACAAATATGCTGGC R:CGGGATGATGTTCTGGGAA |
| slp | NC_002695.1 | qPCR |
F:GTTACCATCCTCGGCACCAT R:CAAATGCCACACCTGGATGC |
| slp | NC_002695.1 | Mutation |
F:GTGCTGCTAATGCGGATGCGACYTTCAAGGTTCAGTGTGTAGGCTGGAGCTGCTTC R:TTACTGATAGGTTAAAGAGAACCAGGCCTGTGCATTCATATGAATATCCTCCTTAG |
| slp | NC_002695.1 | Screening |
F:AGGTGCACTCATACTCAGCC R:TGCACCATAGCCGTAATCCC |
| slp | NC_002695.1 | Sequencing |
F:TCGCCTCAGAATCAGATGAAA R:ATCTGCATCTTTCGGTGGTG |
| slp | NC_002695.1 | Complementation |
F:TAAGCAAAGCTTATGGTTTTAATATTTGTTGATAAG R:TGCTTAGAATTCTTATTTGACCAGCTCAGGTGTTAC |
Relative expression was calculated using the relative expression ratio where efficiency (E) was calculated using the slope of a standard curve of ten-fold serial dilutions of DNA template, and FC was calculated using the relative expression ratio as described by Pfaffl [36].
when E = efficiency of target or reference gene, ΔCt = (Ct value of control-Ct value of sample), target = the gene of interest, ref = endogenous control gene, control = untreated sample, and sample = experimental sample [36]. A FC ≥ 2 was considered significant.
Gene deletion and complementation
E. coli O157:H7 was used to make genomic deletions using the lambda Red recombinase system described by Datsenko and Wanner [37]. Bacterial strains hosting plasmids were grown with antibiotics according to their respective resistance genes, and plasmids were purified using the Plasmid Midi Kit (Qiagen). PCR, using the mutation primers listed in Table 2, was performed using plasmid pKD3 as a template on an Eppendorf MasterCycler PCR System (Eppendorf). PCR reactions were done in 50 μL volumes: 0.25 μL Taq DNA Polymerase enzyme (5 U/μL) (Omega Bio-Tek); 10X PCR buffer (#TQ2100-00, Omega Bio-Tek) at 5 μL; 1.0 μL dNTPs (10mM) (Lucigen); 1.0 μL each of forward and reverse primers (20 μM) (IDT), and water to 50 μL total volume. All PCR cycling conditions were done as follows: 95°C for 5 min; 35 cycles of 95°C for 5 min, 55°C for 30 sec, 72°C for 3 min; followed by a final extension step of 72°C for 5 min. PCR products were run in a 1.5% agarose gel in Tris-acetate-EDTA (TAE) buffer at 150 V for 1 to 2 h to obtain clear band resolution, bands were excised from the gel, and purified using the MinElute Gel Extraction Kit (Qiagen). PCR products were transformed into electrocompetent E. coli O157:H7 containing plasmid pKD119, using volumes between 2 and 5 μL at concentrations between 100 and 500 ng/μL (as measured by Nanovue spectrophotometer). Transformed cells were incubated in 1 mL of pre-warmed 37°C SOC medium and incubated at 37°C for 1 h, and then plated on LB containing chloramphenicol. Colonies were selected for antibiotic resistance and screened by PCR. Transformants positive for antibiotic resistance and negative for screening PCR products were grown and made electrocompetent, transformed with plasmid pCP20 to excise the antibiotic resistance cassette, and screened for antibiotic resistance. Deletions were confirmed by sequencing the junction sequences flanking the deletion site, using The Pennsylvania State University’s Genomics Core Facility (University Park, PA). Plasmid complementations of deletion strains were made using pUC18 and the primers listed in Table 2. PCR was done using E. coli O157:H7 genomic DNA as a template; and PCR reactions, PCR cycling conditions, PCR product purification, and transformations were done as described.
Co-immunoprecipitation (Co-IP) and protein identification
The Co-IP assay was done using the Invitrogen Dynabeads Protein A Immunoprecipitation kit (Invitrogen). Bacterial cultures were grown from starter culture diluted into 100 mL LB broth at an OD of 0.05 and grown with shaking to prevent sediment formation for approximately 14 h at 37°C. Bacterial pellets were collected by centrifugation at 4°C at 4,000xg for 20 min and resuspended in 5mL water for sonication. Sonication was done using a hand-held sonicator (Thermo Fisher Scientific) on ice for a total of 20 min per sample, or until lysate turbidity was visibly reduced. A 5 mL volume of sonicated bacterial lysate was aliquoted into 1 mL samples. Untreated lysate was not incubated with protein; treated lysate was incubated with 10 μg of Recombinant Human Polymeric Immunoglobulin Receptor/PIgR (C-Fc) CI09 protein (Novoprotein) containing an Fc tag at the C-terminus of the protein. Protein-lysate incubation was done overnight at 4°C for 16–18 h on a rotator, and proteins collected using the Co-IP kit. Protein A coated beads from the kit were prepared for use and incubated with the lysate for 30 min at 4°C on a rotator. Beads were collected using a magnet stand and washed twice with kit wash solution. Beads pellet was then resuspended in 10 μL of kit elution solution and 10 μL of sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) buffer and incubated at 95°C for 5 min before loading into a 12% acrylamide SDS-PAGE gel. The SDS-PAGE gel was run for 2 h at 100 V at room temperature and stained with Coomasie blue (BioRad). All protein identifications were done at protein facilities using liquid chromatography and tandem mass spectrometry (LC MS/MS) analysis. A total of three analyses were completed, at The Pennsylvania State Proteomics and Mass Spectrometry Core Facility (1) and the Protein Facility of the Iowa State University Office of Biotechnology (2 and 3). The protein bands were cut from the SDS-PAGE gel and processed at the respective facility. Peptide fragmentation patterns were compared to known databases of proteins of E. coli (PSU using SEQUEST and Uniprot; ISU using Mascot or Sequest HT). Raw data were analyzed using Thermo Scientific's Proteome Discoverer Software.
Statistical analysis
All quantitative assays were performed in triplicate in independent experiments. Data are shown as the mean values ± standard deviation (SD) of the mean. P values were calculated using the two sample Student’s t test. Threshold for significance was p < 0.05 [35]. Analysis was done using XLSTAT 2018 (Addinsoft).
Results
Initial adherence timeline of E. coli O157:H7 during adherence to Caco-2 cells
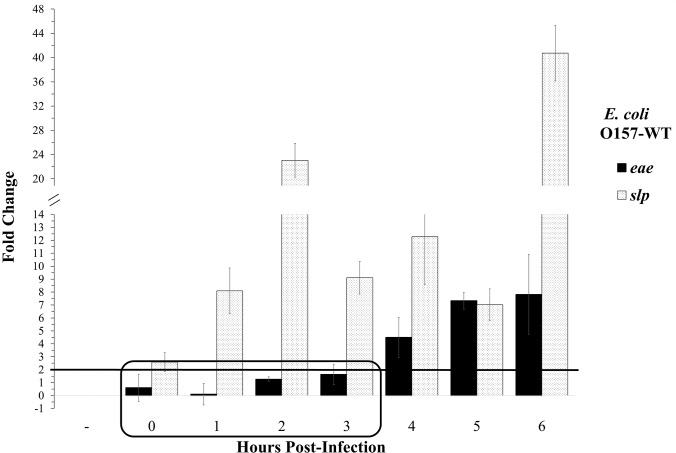
In order to effectively study the roles of different factors during attachment, a specific timeline was required to define initial adherence in this model. Using the expression of LEE-encoded genes as a benchmark for the beginning of the intimate adherence stage of attachment, significant LEE upregulation has been cited as taking place between two and 6 h post-infection in vitro. When adhered to Caco-2 cells, E. coli O157:H7 Sakai demonstrated increased production of EspA, EspB, and Tir protein production after 4.5 h post-infection [38]. During adherence to HeLa cells, LEE1 in E. coli O157:H7 Sakai has been shown to be activated by 4 h post-infection, and LEE1 and LEE5 demonstrated transient upregulation when exposed to an embryonic bovine lung cell line [39, 40, 41]. Expression of eae in an undefined E. coli O157:H7 strain demonstrated upregulation as early as 2 h after infection of HeLa cells [42]. Other studies of EPEC have shown LEE promoter or LEE-encoded gene upregulation between 4 and 6 h in several different epithelial cell lines, but the differences in the LEE operon between EPEC and EHEC strains make the relevance to E. coli O157:H7 unclear [8, 43]. Following previous work, under the specific conditions of this study the intimin-encoding eae gene was used as a benchmark for the onset of intimate adherence in this model. To study the gene expression profiles of initially attached bacteria only, the FC of eae expression in adhered E. coli O157:H7 in comparison to a non-adhered control culture was measured by qPCR using the relative expression ratio method for comparative Ct analysis [36]. The expression of eae surpassed the threshold for significance (FC ≥ 2) after 4 h and increased thereafter, consistent with previous literature (Fig 1). Thus, initial adherence in this study was defined as taking place from 0–3 h post-infection, when eae was not significantly upregulated. Additionally, the expression of slp was significantly upregulated at all time points as compared to E. coli O157:H7 grown in cell culture media alone at the same time points shown.
Fig 1. Relative expression of eae and slp during E. coli O157:H7 adherence to Caco-2 cells and the initial adherence timeline of E. coli O157:H7 in vitro.
Relative expression of eae and slp during adherence are shown as the fold-change (FC) over 6 h post-infection, as compared to E. coli O157:H7 grown in cell culture mediun alone (control). Significant upregulation is defined as FC ≥ 2.0. Threshold for significance is shown at FC = 2.0, and initial adherence timeline is shown in the boxed area.
Colocalization of pIgR with E. coli
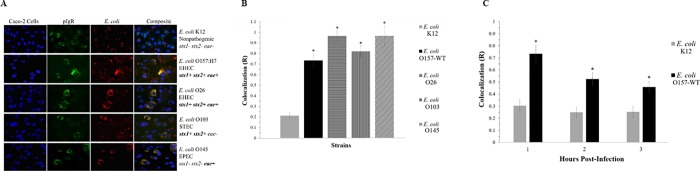
To investigate the possibility of a pIgR-mediated initial adherence mechanism in E. coli O157:H7, immunofluorescence was used to assess colocalization between the pIgR protein and adhered E. coli bacteria and quantify any correlation using covariance analysis (Fig 2A). The colocalization, shown as R, attested that there was no significant correlation between the locations of E. coli K12 bacteria and the pIgR protein; while O157-WT did exhibit a significant correlation (Fig 2B and 2C). This relationship is further detailed when observed over time, where O157-WT showed an initial peak of colocalization (R > 0.7) at one hour post-infection, and decreased over 2 and 3 h (R > 0.5 and 0.4, respectively). This pattern of early peak followed by steady decrease was consistent with the predicted pattern of an adhesin active during initial adherence, as its activity would be less necessary as the intimate adherence factors assumed adherence function at later time points. No such pattern was observed in E. coli K12. Several other non-O157 intestinal pathogenic strains of E. coli were also tested and showed significant colocalization at 2 h post-infection, indicating the possibility that an adherence mechanism is not limited to only E. coli O157:H7 (Fig 2A and 2B).
Fig 2. Colocalization of pIgR with E. coli strains during initial adherence.
(A) Colocalization of pIgR with E. coli strains at 2 h post-infection. Fluorescent signals of Caco-2 nuclei, Hoechst stain with emission at 497 nm, shown in blue; fluorescent signals of E. coli, DyLight 594 with emission at 594 nm, shown in red; fluorescent signals of pIgR protein, DyLight 488 with emission at 488 nm, shown in green; colocalization of pIgR protein with E. coli K12 and O157-WT (B) Colocalization of pIgR with E. coli strains, and (C) pIgR with E. coli K12 and O157-WT over time. Statistical significance, shown as *, was as compared to the E. coli K12 control at the same time points. P-values < 0.05 were considered significant.
Direct interaction between E. coli O157:H7 and the pIgR
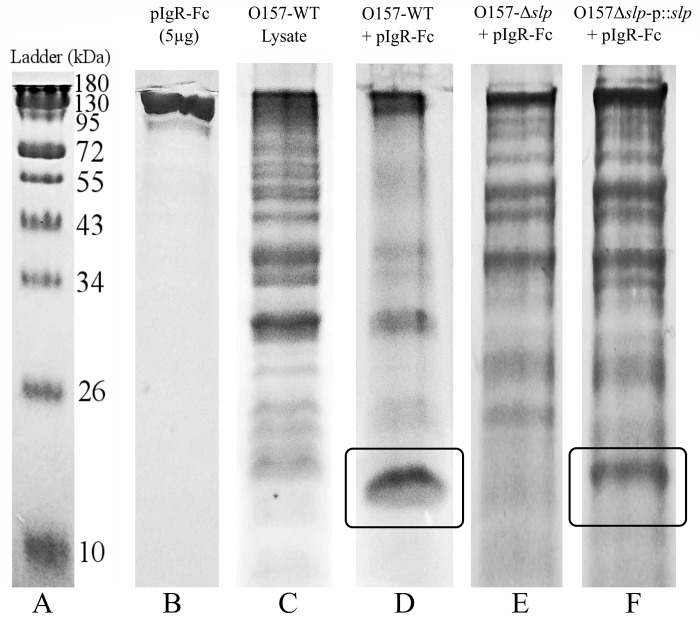
To demonstrate a direct interaction between E. coli O157:H7 and the pIgR, Co-IP assay was done using a C-terminal Fc tagged human recombinant pIgR protein (pIgR-Fc) and O157-WT proteins (whole-cell lysate was used to avoid the accidental exclusion of any proteins). Recovered proteins were run in a reducing SDS-PAGE gel (Fig 3), and bands of interest (shown in boxes) were later identified through LC MS/MS analysis (Table 3). This experiment and analysis were repeated three times, and Slp was identified in all three analyses with the highest sequence coverage. Lane D shows the presence of a concentrated band of approximately 20 kDa (enclosed in a box) in the treated sample, not visible in the untreated control sample (Lane C), indicating the likelihood of a direct binding of that bacterial protein with the pIgR-Fc (Fig 3). There were several other observable bands in the treated sample between 26–43 kDa, but they corresponded in size to the most concentrated bands present in the control, making it likely that they were non-specific and not indicative of protein binding. A direct relationship between the O157-WT protein Slp and the pIgR-Fc protein was demonstrated through disruption of the slp gene. A deletion mutation of the slp gene (O157-Δslp) eliminated the band of interest recovered with the wild-type strain (O157-WT) (Lane E), and the subsequent complementation of a plasmid-encoded slp gene (O157Δslp-p:slp) restored the wild-type phenotype (Lane F) (Fig 3).
Fig 3. Co-immunoprecipitation of E. coli O157:H7 proteins with human recombinant Fc-tagged pIgR protein.
SDS-PAGE showing the apparent sizes of proteins in the conditions used for Co-IP. The protein of ~20 kDa in size, which was co-precipitated with pIgR is enclosed in a box. The bands in the boxed areas were selected for LC MS/MS analysis.
Table 3. Protein identification using LC MS/MS.
Summary of the outer membrane proteins found to be possible identifications of the protein recovered from Co-IP. Proteins are listed in order of decreasing sequence coverage, and proteins that were identified in three of three LC MS/MS analyses are noted.
| Protein ID | Protein Description | Reference Sequence and Accession | Average % Coverage | # AAs | kDa |
|---|---|---|---|---|---|
| *Slp | Outer membrane protein Slp |
Escherichia coli O157:H7 Str. EDL933 AAG58638.1 |
54.9 | 199 | 22.2 |
| *Pal | Peptidoglycan-associated lipoprotein | Multispecies WP_001295306.1 | 53.8 | 173 | 18.8 |
| *OmpW | Outer membrane protein W | Multispecies WP_000737224.1 | 49.8 | 212 | 22.9 |
| *OmpX | Outer membrane protein X | Multispecies WP_001295296.1 | 44.7 | 171 | 18.6 |
| OmpA | Outer membrane protein A precursor |
Escherichia coli O157:H7 Str. EDL933 AIG67419.1 |
22.2 | 354 | 38.1 |
| OmpC | Outer membrane protein C precursor | Escherichia coli WP_000865552.1 | 15.4 | 367 | 40.3 |
* Proteins present in all three LC MS/MS analyses.
Slp expression over time and initial adherence in vitro
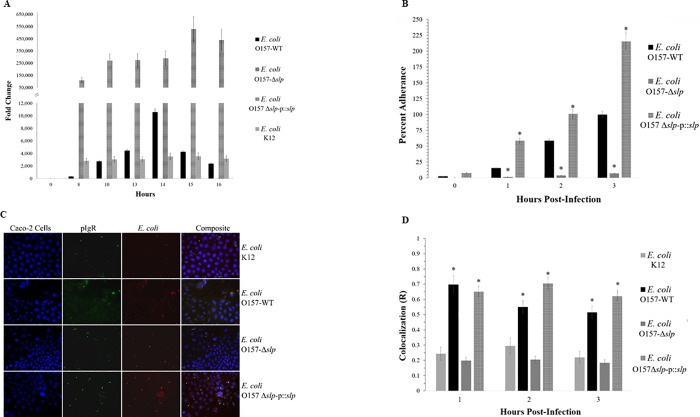
Relative expression of the slp gene showed differing patterns between E. coli K12, O157-WT, O157-Δslp, and O157Δslp-p:slp in LB broth culture over 16 h of growth, as measured by qPCR (Fig 4A). The slp gene is reported to be transcribed at a low baseline level in most conditions and upregulated during entry of bacteria into stationary-phase [44]. The O157-WT slp expression showed a transient peak at 14 h corresponding to the entry of the culture into stationary-phase growth. E. coli K12 slp expression remained relatively stable and low throughout growth, and did not show any peaks or changes at any time point. The O157-Δslp strain did not show any detectable levels of gene expression, while the O157Δslp-p:slp strain showed high levels of constitutive over-expression of slp at high levels at all time points, ranging from 10 to over 100 magnitude differences in FC as compared to E. coli K12 and O157-WT.
Fig 4. Relative slp gene expression, colocalization, and adherence in E. coli strains.
(A) Relative gene expression of slp over time in LB broth culture as measured by qPCR, when compared to expression at time 0. (B) Quantitative adherence of E. coli strains to Caco-2 cells over time, relative to the maximum number of adhered wild-type bacteria (100% adherence is wild-type at 3 h post-infection). Statistical significance, shown as *, is as compared to the O157-WT strain at the same time points. (C) Fluorescence and (D) colocalization of pIgR with E. coli O157-WT, O157-Δslp, and O157Δslp-p:slp. Statistical significance, shown as *, is as compared to the E. coli K12 control at the same time points. P-values < 0.05 were considered significant.
When measuring initial adherence to Caco-2 cells by quantifying the number of attached bacteria over time, the O157-WT presented a pattern showing a steady increase of adhered bacterial cells over 3 h (Fig 4B). These results were consistent with O157-WT bacterial cells as they were steadily coming into contact with and attaching to the Caco-2 cells over time, as the adherence increased two- to three-fold per hour (approximately 20%, 55%, and 100% at 1, 2, and 3 h post-infection, respectively). When compared to the wild-type, deletion of the slp gene resulted in a significant adherence deficiency to Caco-2 cells, with O157-Δslp adherence not reaching more than 10% at any time point. The O157-Δslp adherence increased very slightly over 3 h (from <5% to <10% at 1 and 3 h post-infection, respectively). Conversely, the O157Δslp-p:slp showed a hyper-adherent pattern, surpassing wild-type levels at all time points. The O157Δslp-p:slp adherence followed roughly the same pattern as seen in the wild-type, with adherence increasing 2 to 3-fold per h (approximately 60%, 100%, and 225% at 1, 2, and 3 h post-infection, respectively). At 3 h, the number of adhered O157Δslp-p:slp cells was more than double the wild-type.
Colocalization was measured in the O157-Δslp and O157Δslp-p:slp strains over 3 h of adherence to Caco-2 cells (Fig 4C and 4D). As in Fig 2, E. coli K12 did not show any statistically significant colocalization or any changes over time (R = 0.25, 0.28, and 0.22 at 1, 2, and 3 h post-infection). The O157-WT cells showed a significant colocalization at all timepoints, with the highest correlation (R = ~0.7) seen at 1 h post-infection, with a decrease to R = 0.55 and 0.50 at 2 and 3 h respectively. The O157-Δslp strain demonstrated adherence levels comparable to E. coli K12, with significantly diminished colocalization and no changes over time (R = approximately 0.2 at all time points). The O157Δslp-p:slp strain restored colocalization comparable to that of the wild-type strain, but colocalization did not decrease over time in the same manner (R = 0.65, 0.70, and 0.63 at 1, 2, and 3 h post-infection, respectively).
Discussion
Initial adherence of EHEC bacteria is a known key step in pathogenesis [45]. Initial adherence in EHEC strains is not yet fully understood, and it is becoming clear that mechanisms are not conserved even among EHEC strains with very similar virulence profiles, and putative adhesins have inconsistent behavior between host type [7, 46–48]. In this study, we propose an in vitro initial adherence mechanism involving the E. coli O157:H7 outer membrane protein Slp and the pIgR protein expressed on human colonic epithelial cells.
We hypothesized that the pIgR may be involved in EHEC adherence, and determined the bacterial protein binding to the human pIgR using a Co-IP assay. The Co-IP produced a distinct band of approximately 20 kDa that was significantly more concentrated when E. coli O157:H7 proteins were incubated with pIgR-Fc than with E. coli O157:H7 proteins alone. Identification using LC MS/MS led to the E. coli O157:H7 outer membrane protein Slp, a 22 kDa lipoprotein that has not been fully characterized. Its encoding gene (slp) expression is primarily known to be upregulated in pure culture during entry into stationary-phase growth or during carbon starvation [44]. Its function is generally accepted to be involved with membrane stability during stationary-phase growth, but additional alternative functions have also been proposed [49–51]. There has also been evidence of increased slp gene expression during biofilm formation, though the significance of these findings is unclear [52]. The slp is located on a genomic acid fitness island (AFI) along with several other defined and undefined genes and operons [53, 54]. The slp gene and AFI are present in all E. coli, but there are significant differences between strains. In E. coli K12, the AFI is approximately 14 kb, whereas the E. coli O157:H7 AFI is 23 kb due to the insertion of an O-island sequence [54–57]. These differences in sequence, combined with differences in transcription regulation, provide insight into possible reasons why E. coli K12 does not show a colocalization phenotype with pIgR despite having the slp gene. As shown in Fig 4A, the E. coli K12 slp gene is not expressed under the same culture conditions as E. coli O157:H7, suggesting that slp expression is managed by a different mechanism in E. coli K12 than in E. coli O157:H7 [54].
It is notable that Slp contributes to initial adherence and is located on an acid fitness island, because pH is a potent signaling factor for virulence and adherence gene expression, and the acidic pH encountered by E. coli in the stomach results in a gene expression profile fit for initial adherence [14]. Acid resistance response regulators downregulate the expression of LEE-encoded effectors not required during the early phases of attachment (i.e. eae and other intimate adherence related genes), and upregulate genes related to motility and initial adherence [14, 55, 58, 59]. Given that one of the primary acid resistance response transcription regulators GadE is known to be highly involved in the regulation of AFI gene expression where slp is located, it follows that pH dependent regulation may play a key role in expression of initial adhesins [56, 59]. Given the complexity and sensitivity of acid-responsive regulatory mechanisms, further study is required to fully understand these systems and how their gene products, such as Slp, affect pathogenesis in vivo.
The role of the pIgR in pathogenesis is a known mechanism for other pathogens, but it had not been demonstrated in E. coli adherence. EHEC infection can affect pigR expression; however, the effects of EHEC on the pIgR system do not explain the results reported [60–63]. In order to upregulate pigR expression as early as 0 to 3 h post-infection, exposure of the host cell to immunogenic signals such as LPS or Stx would have to be immediate, prolonged and stable; which is unlikely to occur in the variable intestinal environment [61]. Additionally, there is no evidence to suggest that the pIgR has any mechanism of location specificity, and any inflammatory signals only affect the rate at which pIgR reaches the membrane without any effect on its location [24, 64, 65].
E. coli O157:H7 and other EHEC strains pose a large public health burden worldwide, and the need for effective treatments or interventions is urgent [3]. Interventions are aimed at preventing transmission from the colonized bovine host at either the pre- or post-harvest stage, by treating or preventing EHEC colonization before slaughter, or reducing or eliminating contamination post-harvest [66]. To date, interventions such as vaccines and post-harvest sanitation have produced limited effects, but the identification of an outer membrane protein highly conserved among E. coli may provide a potential new target [66–68]. Bacterial lipoproteins are a broad range of proteins and have been known to be involved with many virulence functions, including adherence, and other genes with low levels of sequence homology have been identified in other Gram-negative bacteria or enteric pathogens, which suggests the possibility of a similar role among some limited classes of bacteria [50, 69]. In S. pneumoniae, the cell signaling pathway responsible for internalization is also used by other pathogens of diverse nature such as Staphylococcus aureus, Neisseria meningitidis, and Listeria monocytogenes [28], suggesting the possibility that this adherence mechanism might be more widespread than previously known [28, 70, 71].
Acknowledgments
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture (USDA). USDA is an equal opportunity provider and employer.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was completely funded by the Research Assistant Fund of SK. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. There was no additional external funding received for this study.
References
- 1.U.S. Centers for Disease Control and Prevention. National Outbreak Reporting System (NORS). https://wwwn.cdc.gov/norsdashboard/. Accessed 1 Dec 2018.
- 2.Hunt JM. Shiga toxin-producing Escherichia coli (STEC). Clin Lab Med. Elsevier Ltd; 2010; 30: 21–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mohawk KL, O’Brien AD. Mouse models of Escherichia coli O157:H7 infection and Shiga toxin. J Biomed Biotechnol. 2011;2011:258185 10.1155/2011/258185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rangel JM, Sparling PH, Crowe C, Griffin PM, Swerdlow DL. O157:H7 Outbreaks in the US 1982–2002. Emerg Infect Dis. 2005; 11: 603–609. 10.3201/eid1104.040739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gould LH, Demma L, Jones TF, Hurd S, Vugia DJ, Smith K, et al. Hemolytic uremic syndrome and death in persons with Escherichia coli O157:H7 infection, foodborne diseases active surveillance network sites, 2000–2006. Clin Infect Dis. 2009; 49: 1480–1485. 10.1086/644621 [DOI] [PubMed] [Google Scholar]
- 6.Croxen M a Finlay BB. Molecular mechanisms of Escherichia coli pathogenicity. Nat Rev Microbiol. 2010; 8: 26–38. 10.1038/nrmicro2265 [DOI] [PubMed] [Google Scholar]
- 7.Bardiau M, Szalo M, Mainil JG. Initial adherence of EPEC, EHEC and VTEC to host cells. Vet Res [Internet]. 2010. [cited 2014 Oct 22];41(5):57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmidt MA. LEEways: tales of EPEC, ATEC and EHEC. Cell Microbiol. 2010; 12: 1544–1552. 10.1111/j.1462-5822.2010.01518.x [DOI] [PubMed] [Google Scholar]
- 9.Farfan MJ, Torres AG. Molecular mechanisms that mediate colonization of Shiga toxin-producing Escherichia coli strains. Infect Immun. 2012; 80: 903–913. 10.1128/IAI.05907-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sinclair JF, O’Brien AD. Intimin types α, β, and γ bind to nucleolin with equivalent affinity but lower avidity than to the translocated intimin receptor. J Biol Chem. 2004; 279: 33751–33758. 10.1074/jbc.M401616200 [DOI] [PubMed] [Google Scholar]
- 11.Sinclair JF, O’Brien AD. Cell surface-localized nucleolin is a eukaryotic receptor for the adhesin intimin-gamma of enterohemorrhagic Escherichia coli O157:H7. J Biol Chem. 2002; 277: 2876–2885. 10.1074/jbc.M110230200 [DOI] [PubMed] [Google Scholar]
- 12.Cordonnier C, Etienne-Mesmin L, Thévenot J, Rougeron A, Rénier S, Chassaing B, et al. Enterohemorrhagic Escherichia coli pathogenesis: Role of long polar fimbriae in Peyer’s patches interactions. Sci Rep. 2017. March 20;7:44655 10.1038/srep44655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farfan MJ, Cantero L, Vergara A, Vidal R, Torres AG. The long polar fimbriae of STEC O157:H7 induce expression of pro-inflammatory markers by intestinal epithelial cells. Vet Immunol Immunopathol. 2013; 152: 126–131. 10.1016/j.vetimm.2012.09.017 [DOI] [PubMed] [Google Scholar]
- 14.Barnett Foster D. Modulation of the enterohemorrhagic E. coli virulence program through the human gastrointestinal tract. Virulence. 2013; 4: 315–323. 10.4161/viru.24318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rendón M a, Saldaña Z, Erdem AL, Monteiro-Neto V, Vázquez A, Kaper JB, et al. Commensal and pathogenic Escherichia coli use a common pilus adherence factor for epithelial cell colonization. Proc Natl Acad Sci U S A. 2007; 104: 10637–10642. 10.1073/pnas.0704104104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McWilliams BD, Torres AG. Enterohemorrhagic Escherichia coli adhesins. Microbiol Spectr. 2014. June;2(3). 10.1128/microbiolspec.EHEC-0003-2013 Review. [DOI] [PubMed] [Google Scholar]
- 17.Xicohtencatl-Cortes J, Monteiro-Neto V, Saldaña Z, Ledesma MA, Puente JL, Girón JA. The type 4 pili of enterohemorrhagic Escherichia coli O157:H7 are multipurpose structures with pathogenic attributes. J Bacteriol. 2009; 191: 411–421. 10.1128/JB.01306-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xicohtencatl-Cortes J, Monteiro-Neto V, Saldaña Z, Ledesma MA, Puente JL, Girón JA. The type 4 pili of enterohemorrhagic Escherichia coli O157:H7 are multipurpose structures with pathogenic attributes. J Bacteriol. 2009; 91: 411–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma VK, Bearson SMD, Bearson BL. Evaluation of the effects of sdiA, a luxR homologue, on adherence and motility of Escherichia coli O157:H7. Microbiology. 2010; 156: 1303–1312. 10.1099/mic.0.034330-0 [DOI] [PubMed] [Google Scholar]
- 20.Roy K, Hilliard GM, Hamilton DJ, Luo J, Marguerite M, Fleckenstein JM, et al. Enterotoxigenic Escherichia coli EtpA mediates adhesion between flagella and host cells. Nature. 2009; 457: 594–598. 10.1038/nature07568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giron J, Torres G, Freer E, Kaper JB. The flagella of enteropathic Escherichia coli mediate adherence to epithelial cells. Mol Micro. 2002; 44: 361–379. [DOI] [PubMed] [Google Scholar]
- 22.Hammerschmidt S, Tillig MP, Wolff S, Vaerman J-P, Chhatwal GS. Species-specific binding of human secretory component to SpsA protein of Streptococcus pneumoniae via a hexapeptide motif. Mol Microbiol. 2002; 36: 726–736. [DOI] [PubMed] [Google Scholar]
- 23.Mostov KE, Blobel G. A transmembrane precursor of secretory component. J Biol Chem. 1982; 257: 11816–11821. [PubMed] [Google Scholar]
- 24.Johansen F-E, Kaetzel CS. Regulation of the polymeric immunoglobulin receptor and IgA transport: new advances in environmental factors that stimulate pIgR expression and its role in mucosal immunity. Mucosal Immunol. 2011;4: 598–602. 10.1038/mi.2011.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernandez MI, Pedron T, Tournebize R, Olivo-Marin JC, Sansonetti PJ, Phalipon A. Anti-inflammatory role for intracellular dimeric immunoglobulin A by neutralization of lipopolysaccharide in epithelial cells. Immunity. 2003; 18: 739–749. [DOI] [PubMed] [Google Scholar]
- 26.Asano M, Komiyama K. Polymeric immunoglobulin receptor. J Oral Sci. 2011; 53: 147–156. [DOI] [PubMed] [Google Scholar]
- 27.Kaetzel CS. The polymeric immunoglobulin receptor: bridging innate and adaptive immune responses at mucosal surfaces. Immunol Rev. 2005; 206: 83–99. 10.1111/j.0105-2896.2005.00278.x [DOI] [PubMed] [Google Scholar]
- 28.Agarwal V, Asmat TM, Dierdorf NI, Hauck CR, Hammerschmidt S. Polymeric immunoglobulin receptor-mediated invasion of Streptococcus pneumoniae into host cells requires a coordinate signaling of SRC family of protein-tyrosine kinases, ERK, and c-Jun N-terminal kinase. J Biol Chem. 2010; 285: 35615–35623. 10.1074/jbc.M110.172999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu L, Lamm ME, Li H, Corthesy B, Zhang J-R. The human polymeric immunoglobulin receptor binds to Streptococcus pneumoniae via domains 3 and 4. J Biol Chem. 2003; 278: 48178–48187. 10.1074/jbc.M306906200 [DOI] [PubMed] [Google Scholar]
- 30.Zhang J, Mostov KE, Lamm ME, Nanno M, Shimida S, Ohwaki M, et al. The polymeric immunoglobulin receptor translocates pneumococci across human nasopharyngeal epithelial cells. Cell. 2000; 102: 827–837. [DOI] [PubMed] [Google Scholar]
- 31.Dave S, Carmicle S, Hammerschmidt S, Pangburn MK, McDaniel LS. Dual roles of PspC, a surface protein of Streptococcus pneumoniae, in binding human secretory IgA and factor H. J Immunol. 2004; 173: 471–477. 10.4049/jimmunol.173.1.471 [DOI] [PubMed] [Google Scholar]
- 32.Kouki A, Haataja S, Loimaranta V, Pulliainen AT, Nilsson UJ, Finne J. Identification of a novel streptococcal adhesin P (SadP) protein recognizing galactosyl-α1-4-galactose-containing glycoconjugates: convergent evolution of bacterial pathogens to binding of the same host receptor. J Biol Chem. 2011; 286: 38854–38864. 10.1074/jbc.M111.260992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J, Hovde CJ. Expression profiles of bovine genes in the rectoanal junction mucosa during colonization with Escherichia coli O157:H7. Appl Environ Microbiol. 2007; 73: 2380–2385. 10.1128/AEM.02262-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bolte S, Cordelières FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006; 224: 213–232. 10.1111/j.1365-2818.2006.01706.x [DOI] [PubMed] [Google Scholar]
- 35.Mcdonald JH, Dunn KW. Statistical tests for measures of colocalization in biological microscopy. J Microsc. 2013; 252: 295–302. 10.1111/jmi.12093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001. May 1;29(9):e45 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Datsenko K a Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000; 97: 6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abe H, Tatsuno I, Tobe T, Okutani A, Sasakawa C. Bicarbonate ion stimulates the expression of locus of enterocyte effacement-encoded genes in enterohemorrhagic Escherichia coli O157:H7. Infect Immun. 2002; 70: 3500–3509. 10.1128/IAI.70.7.3500-3509.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alsharif G, Ahmad S, Islam MS, Shah R, Busby SJ, Krachler AM. Host attachment and fluid shear are integrated into a mechanical signal regulating virulence in Escherichia coli O157:H7. Proc Natl Acad Sci U S A.2015; 112: 5503–5508. 10.1073/pnas.1422986112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crane JK, Byrd IW, Boedeker EC. Virulence inhibition by zinc in Shiga-toxigenic Escherichia coli. Infect Immun. 2011; 79: 1696–1705. 10.1128/IAI.01099-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roe AJ, Naylor SW, Spears KJ, Yull HM, Dransfield T, Oxford M, et al. Co-ordinate single-cell expression of LEE4- and LEE5-encoded proteins of Escherichia coli O157:H7. Mol Microbiol. 2004; 54: 337–352. 10.1111/j.1365-2958.2004.04277.x [DOI] [PubMed] [Google Scholar]
- 42.Yang B, Feng L, Wang F, Wang L. Enterohemorrhagic Escherichia coli senses low biotin status in the large intestine for colonization and infection. Nat Commun. 2015. March 20;6:6592 10.1038/ncomms7592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yona-Nadler C. Integration host factor (IHF) mediates repression of flagella in enteropathogenic and enterohaemorrhagic Escherichia coli. Microbiol. 2003; 149: 877–884. [DOI] [PubMed] [Google Scholar]
- 44.Alexander DM, St John AC. Characterization of the carbon starvation-inducible and stationary phase-inducible gene sip encoding an outer membrane lipoprotein in Escherichia coli. Mol Microbiol.1994; 11: 1059–1071. [DOI] [PubMed] [Google Scholar]
- 45.Melton-celsa A, Mohawk K, Teel L, Brien AO. Pathogenesis of Shiga-toxin producing Escherichia coli. Curr Top Microbiol Immunol. 2012; 357: 67–103. 10.1007/82_2011_176 [DOI] [PubMed] [Google Scholar]
- 46.Kalita A, Hu J, Torres AG. Recent advances in adherence and invasion of pathogenic Escherichia coli. Curr Opin Infect Dis. 2014; 27: 459–464. 10.1097/QCO.0000000000000092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Szalo IM, Goffaux F, Pirson V, Piérard D, Ball H, Mainil J. Presence in bovine enteropathogenic (EPEC) and enterohaemorrhagic (EHEC) Escherichia coli of genes encoding for putative adhesins of human EHEC strains. Res Microbiol. 2002; 153: 653–658. [DOI] [PubMed] [Google Scholar]
- 48.Tatsuno I, Horie M, Abe H, Makino K, Shinagawa H, Taguchi H. toxB gene on pO157 of enterohemorrhagic Escherichia coli O157:H7 is required for full epithelial cell adherence phenotype. Infect Immun. 2001; 69: 6660–6669. 10.1128/IAI.69.11.6660-6669.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mates AK, Sayed AK, Foster JW. Products of the Escherichia coli acid fitness island attenuate metabolite stress at extremely low pH and mediate a cell density-dependent acid resistance. J Bacteriol. 2007; 189: 2759–2768. 10.1128/JB.01490-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Price GP, John ACS. Purification and analysis of expression of the stationary phase-inducible Slp lipoprotein in Escherichia coli: role of the Mar system. FEMS Microbiol Lett. 2000; 193: 51–56. 10.1111/j.1574-6968.2000.tb09401.x [DOI] [PubMed] [Google Scholar]
- 51.Wu VCH, Qiu X, de los Reyes BG, Lin CS, Pan Y. Application of cranberry concentrate (Vaccinium macrocarpon) to control Escherichia coli O157:H7 in ground beef and its antimicrobial mechanism related to the downregulated slp, hdeA and cfa. Food Microbiol. 2009; 26: 32–38. 10.1016/j.fm.2008.07.014 [DOI] [PubMed] [Google Scholar]
- 52.Schembri MA, Kjaergaard K, Klemm P. Global gene expression in Escherichia coli biofilms. Mol Microbiol. 2003; 48: 253–267. [DOI] [PubMed] [Google Scholar]
- 53.Zhao B, Houry WA, Johnson JN, Ahrendt E, Braun JE. Acid stress response in enteropathogenic gammaproteobacteria: an aptitude for survival. Biochem Cell Biol. 2010; 88: 301–314. 10.1139/o09-182 [DOI] [PubMed] [Google Scholar]
- 54.Carter MQ, Louie JW, Fagerquist CK, Sultan O, Miller WG, Mandrell RE. Evolutionary silence of the acid chaperone protein HdeB in enterohemorrhagic Escherichia coli O157: H7. Appl Environ Microbiol. 2012; 78: 1004–1014. 10.1128/AEM.07033-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tramonti A, De Canio M, De Biase D. GadX/GadW-dependent regulation of the Escherichia coli acid fitness island: Transcriptional control at the gadY-gadW divergent promoters and identification of four novel 42 bp GadX/GadW-specific binding sites. Mol Microbiol. 2008; 70: 965–982. 10.1111/j.1365-2958.2008.06458.x [DOI] [PubMed] [Google Scholar]
- 56.Hommais F, Krin E, Coppée JY, Lacroix C, Yeramian E, Danchin A, et al. GadE (YhiE): a novel activator involved in the response to acid environment in Escherichia coli. Microbiol. 2004; 150: 61–72. [DOI] [PubMed] [Google Scholar]
- 57.Zhao B, Houry WA. Acid stress response in enteropathogenic gammaproteobacteria: an aptitude for survival. Biochem Cell Biol. 2010; 88: 301–314. 10.1139/o09-182 [DOI] [PubMed] [Google Scholar]
- 58.Morgan JK, Carroll RK, Harro CM, Vendura KW, Shaw LN, Riordan JT. Global regulator of virulence A (GrvA) coordinates expression of discrete pathogenic mechanisms in enterohemorrhagic Escherichia coli through interactions with GadW-GadE. J Bacteriol. 2015; 198: 394–409. 10.1128/JB.00556-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Foster JW. Escherichia coli acid resistance: tales of an amateur acidophile. Nat Rev Microbiol. 2004; 2: 898–907. 10.1038/nrmicro1021 [DOI] [PubMed] [Google Scholar]
- 60.Croxen MA, Law RJ, Scholz R, Keeney KM, Wlodarska M, Finlay BB. Recent advances in understanding enteric pathogenic Escherichia coli. Clin Microbiol Rev. 2013; 26: 822–880. 10.1128/CMR.00022-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schneeman TA., Bruno MEC, Schjerven H, Johansen, Chady L, Kaetzel CS, et al. Regulation of the polymeric Ig receptor by signaling through TLRs 3 and 4: linking innate and adaptive immune responses. J Immunol. 2005; 175: 376–384. 10.4049/jimmunol.175.1.376 [DOI] [PubMed] [Google Scholar]
- 62.Bruno MEC, Frantz AL, Rogier EW, Johansen F-E, Kaetzel CS. Regulation of the polymeric immunoglobulin receptor by the classical and alternative NF-κB pathways in intestinal epithelial cells. Mucosal Immunol. 2011; 4: 468–478. 10.1038/mi.2011.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Karpman D, Ståhl A-L. Enterohemorrhagic Escherichia coli pathogenesis and the host response. Microbiol Spectr. 2014; 2: 1–15. [DOI] [PubMed] [Google Scholar]
- 64.Phalipon A, Corthésy B. Novel functions of the polymeric Ig receptor: well beyond transport of immunoglobulins. Trends Immunol. 2003; 24: 55–58. [DOI] [PubMed] [Google Scholar]
- 65.Cardone MH, Smith BL, Mennitt PA, Mochly-Rosen D, Silver RB, Mostov KE. Signal transduction by the polymeric immunoglobulin receptor suggests a role in regulation of receptor transcytosis. J Cell Biol. 1996; 133: 997–1005. 10.1083/jcb.133.5.997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Callaway T, Carr M. Diet, Escherichia coli O157: H7, and cattle: a review after 10 years. Curr Issues Mole. Biol. 2009; 1: 67–80. [PubMed] [Google Scholar]
- 67.Naylor SW, Nart P, Sales J, Flockhart A, Gally DL, Low JC. Impact of the direct application of therapeutic agents to the terminal recta of experimentally colonized calves on Escherichia coli O157:H7 shedding. Appl Environ Microbiol. 2007; 73: 1493–1500. 10.1128/AEM.01736-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arthur TM, Brichta-Harhay DM, Bosilevac JM, Kalchayanand N, Shackelford SD, Wheeler TL, et al. Super shedding of Escherichia coli O157:H7 by cattle and the impact on beef carcass contamination. Meat Sci. 2010; 86: 32–37. 10.1016/j.meatsci.2010.04.019 [DOI] [PubMed] [Google Scholar]
- 69.Kovacs-Simon A, Titball RW, Michell SL. Lipoproteins of bacterial pathogens. Infect Immun. 2011; 79: 548–561. 10.1128/IAI.00682-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Almeida MT, Mesquita FS, Cruz R, Osório H, Custódio R, Brito C, et al. Src-dependent tyrosine phosphorylation of non-muscle myosin heavy chain-IIA restricts Listeria monocytogenes cellular infection. J Biol Chem. 2015; 290: 8383–8395. 10.1074/jbc.M114.591313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Agerer F, Michel A, Ohlsen K, Hauck CR. Integrin-mediated invasion of Staphylococcus aureus into human cells requires Src family protein-tyrosine kinases. J Biol Chem. 2003; 278: 42524–42531. 10.1074/jbc.M302096200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.