Abstract
Paralogues pairs are more frequently observed in eels (Anguilla sp.) than in other teleosts. The paralogues often show low phylogenetic distances; however, they have been assigned to the third round of whole genome duplication (WGD), shared by all teleosts (3R), due to their conserved synteny. The apparent contradiction of low phylogenetic difference and 3R conserved synteny led us to study the duplicated gene complement of the freshwater eels. With this aim, we assembled de novo transcriptomes of two highly relevant freshwater eel species: The European (Anguilla anguilla) and the Japanese eel (Anguilla japonica). The duplicated gene complement was analysed in these transcriptomes, and in the genomes and transcriptomes of other Actinopterygii species. The study included an assessment of neutral genetic divergence (4dTv), synteny, and the phylogenetic origins and relationships of the duplicated gene complements. The analyses indicated a high accumulation of duplications (1217 paralogue pairs) among freshwater eel genes, which may have originated in a WGD event after the Elopomorpha lineage diverged from the remaining teleosts, and thus not at the 3R. However, very similar results were observed in the basal Osteoglossomorpha and Clupeocephala branches, indicating that the specific genomic regions of these paralogues may still have been under tetrasomic inheritance at the split of the teleost lineages. Therefore, two potential hypotheses may explain the results: i) The freshwater eel lineage experienced an additional WGD to 3R, and ii) Some duplicated genomic regions experienced lineage specific rediploidization after 3R in the ancestor to freshwater eels. The supporting/opposing evidence for both hypotheses is discussed.
Introduction
Large accumulations of gene duplications can originate from one single event, like a whole genome duplication (WGD) [1] or from multiple small duplication events such as small segmental duplications (SDs) [2], which are often found in tandem. Any of these duplication events may contribute to species evolution by providing raw genetic material for new phenotypic variation [1–3].
Relatively recent SDs are often found in tandem and have been found in high abundance in several organisms including yeast [4], daphnia [5], humans [2,6,7] and teleosts [8–12]. Soon after a SD, one paralogue is most commonly lost [1] possibly due to an accumulation of deleterious mutations or genetic drift [13]. In a few cases, a high abundance of SDs can persist for millions of years as seen in yeast [4], common carp [9] and humans [2,6,14]. This process has been associated with adaptation to new environments [5,15,16]. On the other hand, WGDs are presumed rare in mammals [17], but are recurrently found in amphibians and reptiles [18] and have frequently been suggested in insects [18], fungi [19], and plants [20–23]. Recent WGD events have traditionally been observed by cytological studies through the observation of additional chromosomes [22]; however, ancient WGD events are often hidden [22,24–26] by massive gene losses [27–29] and the fusion or loss of duplicated chromosomes [19,30–33]. Therefore, an ancient WGD event can only be discovered through specific analysis at a whole genome level. Consequently, discoveries of WGD events have accelerated as sequencing techniques have improved and genome-scale data has become more accessible [22,24,26]. It has been suggested that early on in the vertebrate lineage two WGDs (1R and 2R) occurred resulting in species radiation and evolution of new traits [1–3,34]. In teleosts, strong genomic evidence supports the existence of an additional WGD called the teleost specific 3rd round of WGD (3R), which occurred in the base of the teleost lineage between 350 and 320 million years ago (MYA) [35,36].
In addition to 3R, WGD events appear to be a reoccurring phenomenon in Actinopterygii even when only considering cytological evidence [37,38]. Furthermore, Inoue et al. [27] found that 70–80% of the genes originating from the 3R WGD get lost after just 60 million years. Similarly, other studies have found that in most teleosts 3–20% of the genes generated during 3R are conserved today [31]. Moreover, extensive chromosome reorganizations have been suggested in the teleost lineage associated with 3R [39,40] and after the salmonid specific 4th round of WGD (Ss4R) [41]. Therefore, it has been suggested that further discoveries of new WGDs in teleosts may increase following the development of sequencing techniques and the increase in the number of studies specifically analysing the temporal distribution and quantity of gene duplications [31]. This phenomenon of accelerating rates of WGD discoveries is currently observed in plant genomics [20–22,42].
Following a WGD event, paralogues genes will start to diverge after the recombination between duplicated genes has stopped at the transition from tetrasomic to disomic inheritance [41,43,44], also referred to as cytological rediploidization. However, after autotetraploidization tetrasomic segregation may continue due to the high similarity between the duplicated chromosomes, and thus rediploidization may be vastly delayed after a WGD event [43]. Therefore, variations in phylogenetic divergences between paralogue gene pairs originating from the same WGD event can appear in cases where a genomic region is under tetrasomic inheritance, at the time of a speciation event [43]. The resulting phylogenetic gene family trees from such event are virtually indistinguishable from gene trees where additional gene duplications have occurred [44]. In particular, in salmonids, strong evidence suggests that rediploidization after the Ss4R has been protracted in time for approximately a quarter of the genome [41,45]. In turn, this mechanism has led to several salmonid gene duplicates to not present 1:1 orthology relationships among different salmonid species, despite being created at the Ss4R [41,46,47]. A protracted pseudotetraploid period has also been suggested in teleosts after 3R [44]. In particular, the peculiar hox gene complement of the African butterfly fish (Pantodon buchholzi) is most parsimoniously explained by a hypothesis which includes protracted rediploidization for some genomic regions [44]. However, unequivocal support of protracted rediploidization beyond salmonids will require further careful phylogenomic analysis [43].
Several studies have revealed a high occurrence of duplicated genes in freshwater eels (Anguilla spp., Elopomorpha) [48–56]. While these duplicated genes often present weak conserved synteny, suggesting a 3R origin, they also present low phylogenetic divergence between paralogues, indicating that they recently started to diverge. For example, Lafont et al. [50] hypothesize that the entire genomic region containing the gene gper could have been duplicated in freshwater eels, and maybe also in other teleosts; and that the retention of duplicated genes may be higher in these eels than in other teleosts.
The occurrence of duplicated genes in freshwater eels seems to be higher than for most teleost lineages, and specifically, the remarkably high conservation of duplicated gene sequences since 3R, often hypothesized for freshwater eel genes [48–56], would be unique [57]. Owing to the fact that the availability of genetic raw material has been suggested to increase the potential of novel adaptation [42], information on the duplicated gene complement of eels may prove valuable in understanding the biology of these endangered species. Therefore, the peculiarity of the published data led us to quantify and analyse duplications in the most relevant freshwater eel species and investigate the temporal distribution of the events that created them. To this end, we assembled de novo transcriptomes of Japanese (Anguilla japonica) and European eel (Anguilla anguilla) from downloaded and newly generated Illumina RNA sequencing data, respectively. Furthermore, we performed phylogenetic reconstructions, assigned paralogue pairs to branches of the resulting species tree, and calculated fourfold synonymous third-codon transversion (4dTv) distances for each paralogue pair identified within these transcriptomes. These analyses were run on our de novo transcriptomes and on multiple other fish transcriptomes and genomes. Our analysis supports the commonly suggested hypothesis of a high abundance of paralogue pairs, unique to the freshwater eel species. However, the phylogenetic and 4dTv analyses suggest a post 3R origin, and a strong signal of synteny between the genomic environments of these paralogues opposes a hypothesis of a SD origin. Similar results were also obtained from the included Osteoglossomorpha branches and the basal Clupeocephala branch. This, in turn, suggests that the results were generated by protracted rediploidization in teleosts after the 3R. These results thus open a discussion on whether these duplicated genes are the result of a 4R WGD in a common ancestor to freshwater eels or rather have been conserved on chromosomal regions, which have experienced delayed rediploidization after the 3R.
Materials and methods
Fish husbandry
Ten immature farm European eel males (mean body mass 96.7±3.6 g ± SEM) supplied by Valenciana de Acuicultura S.A. (Puzol, Valencia, Spain) were transported to the Aquaculture Laboratory at the Universitat Politècnica de València, Spain. The fish were kept in a 200-L tank, equipped with individual recirculation systems, a temperature control system (with heaters and coolers), and aeration. The fish were gradually acclimatized to seawater (final salinity 37 ± 0.3‰), over the course of two weeks. The temperature, oxygen level and pH of rearing were 20°C, 7–8 mg/L and ~ 8.2, respectively. The tank was covered to maintain, as far as possible, a constant dark photoperiod, and the fish were starved throughout the holding period. After acclimation, the fish were sacrificed in order to collect samples of the forebrain (telencephalon, diencephalon, and olfactory bulb), pituitary, and testis tissues.
Human and animal rights
This study was carried out in strict accordance with the recommendations given in the Guide for the Care and Use of Laboratory Animals of the Spanish Royal Decree 53/2013 regarding the protection of animals used for scientific purposes (BOE 2013), and in accordance with the European Union regulations concerning the protection of experimental animals (Dir 86/609/EEC), Guidelines of the European Union (2010/63/EU). The protocol was approved by the Experimental Animal Ethics Committee from the Universitat Politècnica de València (UPV) and final permission was given by the local government (Generalitat Valenciana, Permit Number: 2014/VSC/PEA/00147). The fish were sacrificed using an overdose of anaesthesia.
RNA extraction and sequencing
High quality RNA was extracted from the forebrain, pituitary, and testis samples of one individual male eel (weight: 105.4 g, length: 38.5 cm, and eye index: 4.62), following the protocol developed by Peña-Llopis and Brugarolas [58]. The quantity and quality were tested using a bioanalyser (Agilent Technologies, USA), the samples with sufficient RNA integrity number (RIN) values (RIN > 8.2) and RNA amounts (>3 μg of total RNA) were selected. Total RNA of the three samples were shipped to the company Macrogen Korea (Seoul, South Korea). mRNA purification was carried out on these samples, using Sera-mag Magnetic Oligo (dT) Beads, followed by buffer fragmentation. Reverse transcription was followed by PCR amplification to prepare the samples for sequencing using the TruSeq stranded mRNA LT sample prep kit (Illumina, San Diego, USA). The strand information was kept in an Illumina Hiseq-4000 sequencer (Illumina, San Diego, USA). Resulting raw sequences were 101bp paired-end reads which are available at the NCBI Sequence Read Archive (SRA) under the accession no. SRP126643.
Transcriptome assemblies and genomes
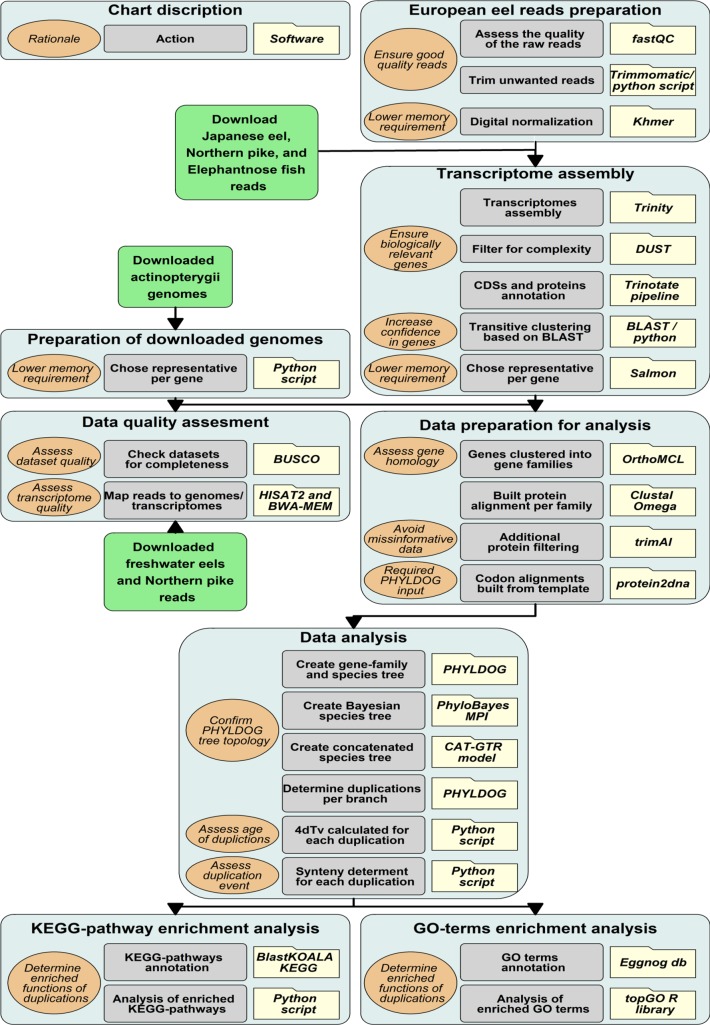
The bioinformatics methodology described below is illustrated in Fig 1. Specifically, FastQC [59] software was used to assess the quality of the raw reads generated by Macrogen. Thereafter, trimmomatic [60] was used to trim the reads, eliminating known adaptor sequences, and low quality regions. Finally, trimmed reads shorter than 50 bp were filtered out. European eel reads were digitally normalized before assembly by Khmer software [61] using a k-mer length of 25 and a coverage of 100. Furthermore, the RNA-Seq raw reads of a Japanese eel Fertilized egg (SRA, NCBI: SRR1930110), preleptocephalus (SRA, NCBI: SRR1930112), leptocephalus (SRA, NCBI: SRR1930115) and glass eel (SRA, NCBI: SRR1930117) were downloaded from NCBI. The RNA-Seq raw reads for Northern pike (Esox lucius), elephantnose fish (Gnathonemus petersii) and silver arowana (Osteoglossum bicirrhosum) were downloaded from the PhyloFish project [62]. All transcriptomes were then assembled using Trinity software [63], taking the strand orientation (for European eel) into account. Naturally produced transcripts may include intervals with a high bias for specific nucleotides (low-complexity), such transcripts may give high-scoring blast results but in fact be biologically insignificant. Therefore, the transcripts assembled were filtered according to their complexity (with a DUST score threshold of 7 and a DUST window of 64), length (with a minimum length of 500 bp), and level of expression (with a transcripts per million (TPM) threshold of 1). The DUST module from BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was used for this filtering, and Salmon software [64] was used to estimate TPM. After assembly, the coding DNA sequences (CDSs) and proteins were annotated using the Trinotate functional annotation pipeline [63]. Transcripts that share k-mers were clustered by Trinity. However, these transcripts might correspond to different transcript forms of the same gene or to closely related genes from a gene family. We split these transcripts into genes by running a transitive clustering based on a blast search. In this clustering, transcripts, which shared at least 100 bp with a minimum identity of 97%, were considered to be isoforms of the same gene. Thus, some Trinity clusters were split into several genes. For each gene, the most expressed transcript, according to the Salmon software [64], was chosen as its representative (Fig 1).
Fig 1. Methodology.
Pipeline of the bioinformatics methodology. Folders describe the software used, light grey boxes describe the action taken, light brown bubbles describes the rationale for selected actions, and light blue boxes describe the specific goal of each section. Finally, green boxes represent external data input.
Technical point
The transcriptomes of one eel was used for the European eel de novo transcriptome assembly due to the fact that transcriptome assemblies based on multiple individuals are more prone to mistake allelic variants for recent gene duplications. Despite our transitive clustering, alleles could still be present in our transcriptome. However, these would resemble very similar paralogues, and be assigned a very low 4dTv distance. Therefore, it is implausible that local density maximum of eel paralogues found at higher 4dTv values could be alleles. Additionally, since our assessment of synteny (see Synteny section) is based on the genome, the paralogue pairs from which synteny can be assessed are highly unlikely to include these potential alleles.
The Atlantic salmon (Salmo salar) genome assembled by the International Cooperation to Sequence the Atlantic Salmon Genome [41] and the Asian arowana (Scleropages formosus) genome [40] were downloaded from NCBI. The genomes of zebrafish (Danio rerio) [65], fugu (Takifugu rubripes) [66], spotted gar (Lepisosteus oculatus) [39], and platyfish (Xiphophorus maculatus) [67] were downloaded from ENSEMBL (release 87). The Northern pike genome [12] was downloaded from the Northern Pike Genome web site (Genbank accession GCA_000721915.1). For each gene in the genomes, the longest transcript was chosen as the representative. For the synteny analysis, the available European [49] and Japanese [68] eel genomes were downloaded from the ZF-Genomics and the DDBJ web site, respectively (Fig 1).
Genome and transcriptome quality assessment
In order to assess the quality of the transcriptomes and genomes, we looked for the Benchmarking set of Universal Single-Copy Orthologues (BUSCO) conserved gene set in them [69]. BUSCOs are conserved proteins which are expected to be found in complete genomes or transcriptomes. Therefore, the number of present, missing, or fragmented BUSCOs can be used as a quality control of a genome or transcriptome assembly. For this assessment, the Actinopterygii (odb9) gene set, which consists of 4584 single-copy genes that are present in at least 90% of Actinopterygii species, was used. As an additional comparison between the transcriptome and genomes of pike and eels, RNA-seq reads were mapped both to the genome and transcriptome assemblies using HISAT2 [70] and BWA-MEM [71] software, respectively (Fig 1), using default settings in both programs.
Gene families
Genes were clustered into gene families by the OrthoMCL web service [72], which uses the Markov Cluster algorithm to group homologs of all the included datasets, based on all against all BLASTP searches. Therefore, the OrthoMCL gene families were also considered gene families for this study. For each gene family, a multiple protein alignment was built. To avoid transcriptome assembly artefacts, proteins longer than 1,500 amino acids, transcripts with a DUST score higher than 7 and sequences with more than 40% gaps in the alignment, were filtered out. The software Clustal Omega [73] carried out the protein multiple alignment and trimAl [74] removed the regions with too many gaps or those difficult to align. The protein alignment was used as a template to build the codon alignment by aligning the transcript sequences against the corresponding protein using the protein2dna exonerate algorithm [75] (Fig 1).
Phylogenetic reconstruction and duplication dating
The resulting protein alignments were used by PHYLDOG [76] software to generate a species tree as well as a gene family tree corresponding to each alignment. Due to the high memory requirements of PHYLDOG, not all the gene families could be run in the same analysis, therefore 10 analyses were carried out, with 8,000 protein alignments being chosen at random for each. Once all runs were finished, we checked that the species tree topology of all the 10 species trees matched exactly. PHYLDOG uses a maximum likelihood approach to simultaneously co-estimate the species and gene family trees from all individual alignments. In order to confirm the tree topology of the PHYLDOG species tree, the species phylogeny was also reconstructed using a Bayesian approach with PhyloBayes MPI version 1.7 [77]. Furthermore, from the gene families that had one gene for each species, 100 were chosen at random to create a concatenated alignment of 43,566 amino acids. The model used was CAT-GTR and three independent MCMC chains were run for 39,872, 56,328, and 39,285 iterations (Fig 1).
PHYLDOG further tagged duplications and assigned these to specific tree branches based on the gene family trees. Between any pair of duplicated sequences, the number of transversions found in the third base of the codon was divided by the number of four-fold degenerated codons resulting in the 4dTv distance. A correction to the 4dTv was applied: ln (1–2 * distance) / -2. The 4dTv was calculated for all the duplications tagged by PHYLDOG within any gene family. These calculations are implemented by the function calculate_4dTv found in the Python scripts (S1 Material). The distribution of 4dTvs was fitted with a lognormal mixture model using the scikit-learn Gaussian Mixture class (Fig 1).
Synteny
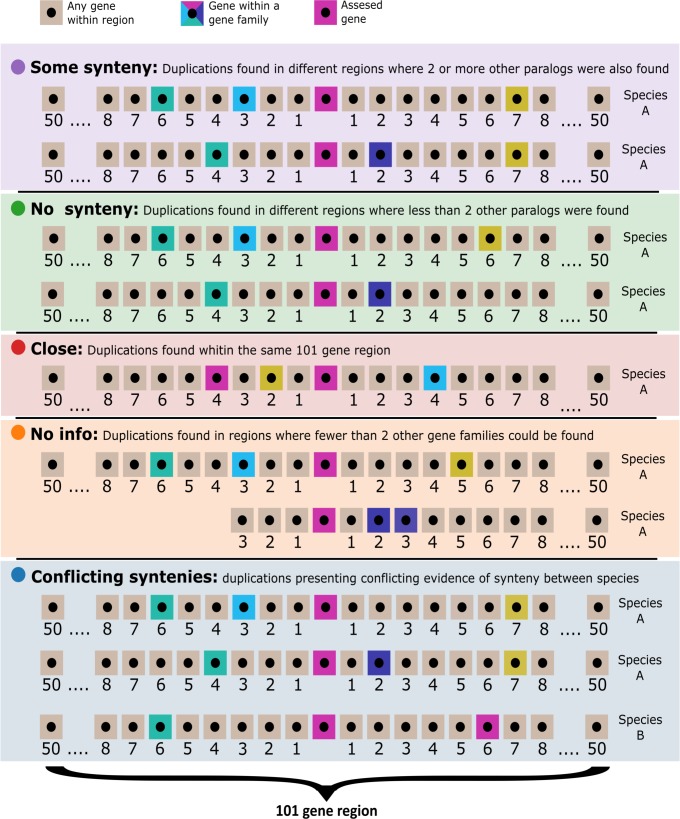
The kind of event that created each duplication was characterized by analysing the conserved synteny between the paralogues created by that duplication within a particular genome. Tandem SDs would create paralogues found close to each other in the genome, whereas the paralogues created by a WGD would be far apart, but surrounded by similar genes in each of the duplicated regions. Also, we have to consider that several phylogenetically close species can be affected by the same older duplication event. With this in mind, we categorized duplications as one of 4 classes (Fig 2): i) the paralogue genes that were found close to each other in the genome, within the 50 neighbouring genes to either side, were labelled as “close”, ii) the paralogues which were found in syntenic regions where 2 or more paralogues from other gene families were located within the 50 neighbouring genes to either side, not necessarily in the same collinear order, were labelled as “some synteny”, iii) the cases in which fewer than 2 gene families could be identified within the 50 neighbouring genes to either side, from either of the paralogues genes, were labelled as “no info”, and iv) the cases where conflicting evidence was found in the genomes of the different species affected by the duplication were labelled as “conflicting syntenies”.
Fig 2. Synteny illustration.
Visualization of the assigned synteny types: “some synteny” (), paralogues of genes found close to one duplicate are also found close to the other duplicate; “no synteny” (), less than two paralogues for other genes are found close to both paralogue duplicates; “close” (), duplicated genes are close in the genome; “no information” (), the duplicated genes are located in small scaffolds with too few gene families close by; “conflicting syntenies” (), different synteny classification found in the genomes of the different species affected by the duplication. Sand coloured boxes represent genes which have not been assigned to a gene family, pink boxes represent the gene from which synteny is being assessed; all other colour boxes represent other genes which have been assigned to a gene family.
This labelling of the duplications was carried out by the Python function “determine_if_pair_is_close_or_syntenic”and the Python class GenomeLocator, found in the scripts (S1 Material). The location of each gene in a genome was obtained by performing a BLAST search with its representative transcript against the genome (Fig 1).
Investigation of functional category enrichment
The EggNOG database has gene ontology (GO) annotations for each of its gene families [78]. To match our gene families with those from the EggNOG database, the protein sequence with least gaps per each of our families was selected and a HMMER search [79] was carried out against the EggNOG position weight matrices with an e-value threshold of 10−4. The GO annotation of the best EggNOG hit in this search was transferred to our family. The enrichment analysis was carried out using the Fisher statistic and the weight algorithm of the topGO library [80] from the Bioconductor project. The R script go_enrichment_analysis found in the scripts (S1 Material) implements this analysis. Freshwater eel transcripts were annotated using the BlastKOALA KEGG service [81] and a Fisher exact test was carried out, using the scipy implementation, to look for overrepresented KEGG pathways in the duplications assigned to the basal freshwater eel branch (Fig 1).
Results
Transcriptome assemblies
Forebrain, testis, and pituitary RNA samples, from an individual European eel, were sequenced, generating a total of 191 million Illumina reads (66, 60 and 65 million from the forebrain, testis, and pituitary, respectively), with a length of 101 bp. These reads were assembled into one de novo transcriptome, using the Trinity assembler after a digital normalization step [61] that left 75 million representative reads. The same procedure was used to generate one de novo transcriptome from Illumina RNA-sequencing reads of the Japanese eel, which was downloaded from the NCBI's Sequence Read Archive [82]. The transcriptomes of Northern pike, elephantnose fish and silver arowana were also assembled by Trinity using Illumina reads from the Phylofish database [62]. The number of unigenes (henceforth referred to as transcripts) assembled ranged from 64,857 to 78,610 (Table 1) and the number of transcript clusters ranged from 46,585 to 55,667 (henceforth referred to as genes; Table 2).
Table 1. Included transcriptomes.
| Species | N.° Reads | Q30 | Transcripts | Mean GC content (%) |
|---|---|---|---|---|
| European eel | 181,322,106 | 0.994 | 77,247 | 51.17 |
| Northern Pike | 553,710,218 | 0.989 | 68,489 | 48.05 |
| Elephantnose fish | 498,451,616 | 0.993 | 74,642 | 49.75 |
| Silver arowana | 490,649,254 | 0.992 | 78,610 | 49.18 |
| Japanese eel | 458,032,126 | 0.986 | 64,857 | 48.13 |
Metrics of included raw read datasets from European eel (Anguilla anguilla), Japanese eel (Anguilla japonica), northern Pike (Esox lucius), elephantnose fish (Gnathonemus petersi), and silver arowana (Osteoglossum bicirrhosum).
Table 2. Gene quantities far each species included.
| Species | Transcripts | Genes | Representative transcripts with predicted protein |
Gene family transcripts |
% of genes assigned to a gene family |
|---|---|---|---|---|---|
| European eel | 77,247 | 54,879 | 27,696 | 25,862 | 93.38 |
| Japanese eel | 64,857 | 46,585 | 23,780 | 23,098 | 97.13 |
| Zebrafish | 58,274 | 32,189 | 25,790 | 22,703 | 88.03 |
| Northern pike | 68,489 | 49,154 | 23,843 | 21,696 | 90.99 |
| Elephantnose fish | 74,642 | 50,455 | 24,857 | 22,036 | 88.65 |
| Spotted gar | 22,483 | 18,341 | 18,341 | 17,872 | 97.44 |
| Silver arowana | 78,610 | 55,667 | 24,938 | 21,604 | 86.63 |
| Asian arowana | 43,354 | 23,799 | 22,740 | 20,637 | 90.75 |
| Atlantic salmon | 109,584 | 55,104 | 48,593 | 42,625 | 87.72 |
| Fugu | 47,841 | 18,523 | 18,523 | 17,698 | 95.55 |
| Platyfish | 20,454 | 20,379 | 20,379 | 19,807 | 97.19 |
Quantities of included genes per included species: European eel (Anguilla anguilla), Japanese eel (Anguilla japonica), zebrafish (Danio rerio), northern pike (Esox lucius), elephantnose fish (Gnathonemus petersi), spotted gar (Lepisosteus oculatus), Asian arowana (Scleropages formosus), silver arowana (Osteoglossum bicirrhosum), Atlantic salmon (Salmo salar), fugu (Takifugu rubripes), and platyfish (Xiphophorus maculatus). “Transcripts” represents unigenes, “Genes” represents the number of transcript clusters, “Representative transcripts with predicted protein” represents the number of genes with a successful protein annotation, “Gene family transcripts” represents the representative transcripts with predicted protein with a successful gene family annotation, and “% of genes assigned to a gene family” represents the percentage of representative transcripts with predicted protein with successful gene family annotation.
Genome and transcriptome quality
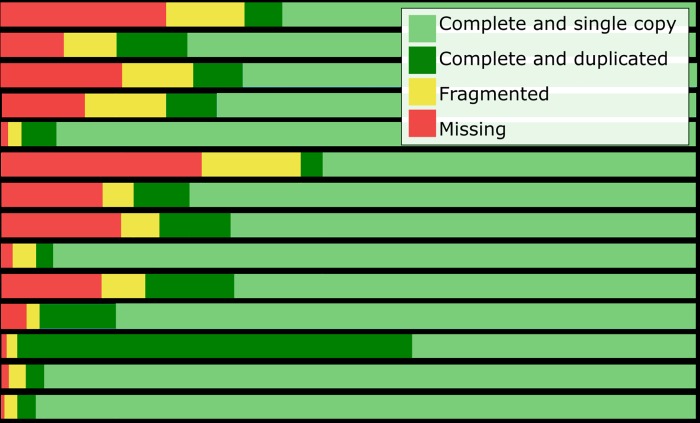
The genomes and transcriptomes considered for inclusion in the analysis were quality tested by a BUSCO assessment of completeness. In general, when available, genomes were used instead of transcriptomes, except for pike, and European eel, where the transcriptomes outperformed the genomes according to the BUSCO assessment (Fig 3). Furthermore, the Japanese eel transcriptome was preferred due to a problem with the Japanese eel genome annotation. These transcriptomes also provided a higher mapping of RNA sequencing reads compared to their corresponding genomes. The percentage of reads that mapped concordantly against the genome and the transcriptome were 65.8 and 91.9%, respectively, for European eel, 74.3 and 88.4% for Japanese eel and 44.6 and 85.8% for pike. Furthermore, previously published European eel RNA-sequencing experiments were also mapped to the available European eel genome and our de novo transcriptome. In this case, 52.2% [83], 57.9% [84], and 66.18% [85] reads mapped concordantly against the eel genome whereas 84.3% [83], 69.5% [84], and 87.32% [85] mapped against the transcriptome.
Fig 3. BUSCO analysis.
BUSCO (Benchmarking set of Universal Single-Copy Orthologues) result for included genomes and transcriptomes. The sequence of a BUSCO gene can be found complete or fragmented in each genome and it can be found once (single copy), more than once (duplicated) or not found (missing). Included genomes are: European eel (Anguilla anguilla), Japanese eel (Anguilla japonica), Asian arowana (Scleropages formosus), zebrafish (Danio rerio), northern pike (Esox lucius), spotted gar (Lepisosteus oculatus), fugu (Takifugu rubripes), platyfish (Xiphophorus maculatus) and Atlantic salmon (Salmo salar). Included transcriptomes: European eel, Japanese eel, northern pike, elephantnose fish (Gnathonemus petersii) and silver arowana (Osteoglossum bicirrhosum).
Gene families
Genes were assigned to gene families according to the gene family categorization of OrthoMCL [72]. The percentage of genes with predicted proteins assigned to a family by the OrthoMCL web service [72] ranged from 86.6% (silver arowana) to 97.4% (spotted gar; Table 2). Overall, 15,771 gene families were covered, from which 13,972 protein and codon alignments were built. These families contained between 2 and 172 genes, with 11 genes per family being the mode.
Phylogenetic reconstruction and duplication characteristics
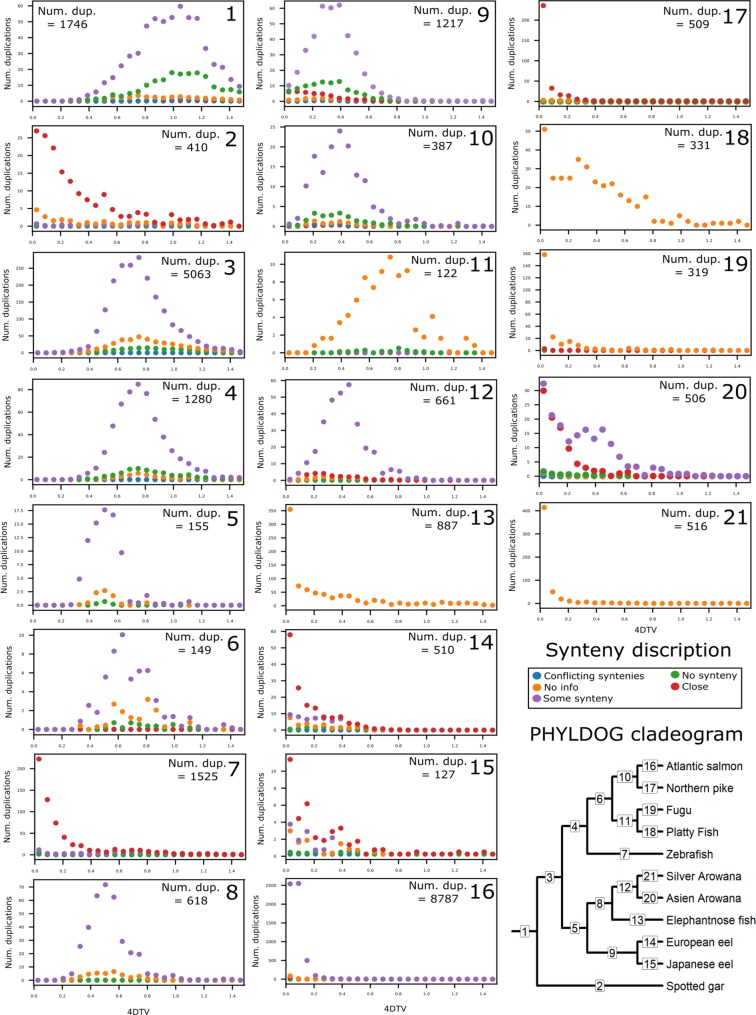
PHYLDOG software was used to tag gene duplications, create a species tree, and assign duplications to tree branches, based on gene family phylogenetic trees. Overall, trees for 10,714 gene families were created by PHYLDOG and based on the tree topology, branches in which a gene appeared to duplicate were labelled. The resulting PHYLDOG species tree matched the species tree topology created by phylobayes [77] and the resulting tree of the concatenated alignment; a cladogram of these trees is included in Fig 4. Since PHYLDOG distinguishes between gene divergence at speciation events and duplications, all genes resulting from tagged duplications are assumed to be paralogues. The assigned duplications were subsequently characterized by synteny and 4dTv distance. The 4dTv distance is used to estimate the accumulation of synonymous mutations, which can be used to estimate the time that has passed from when mutations started to accumulate. The assigned synteny classes include: “close” which indicates SDs that are a result of tandem duplications; “some synteny” which indicates a potential WGD origin (or at least a potential duplication event containing >100 genes); and “no synteny”, which supports neither a SD nor a WGD origin (Fig 2).
Fig 4. 4dTv and synteny distributions of duplications per branch of the PHYLDOG species tree.
Quantity, 4dTv and synteny distributions of duplications assigned to each branch of the PHYLDOG species tree. Each panel represents the branch with the corresponding number in the cladogram in the bottom right-hand corner. Species included in this study are: European eel (Anguilla anguilla), Japanese eel (Anguilla japonica), zebrafish (Danio rerio), northern pike (Esox lucius), spotted gar (Lepisosteus oculatus), fugu (Takifugu rubripes), platyfish (Xiphophorus maculatus), Atlantic salmon (Salmo salar), elephantnose fish (Gnathonemus petersii), Asian arowana (Scleropages formosus) and silver arowana (Osteoglossum bicirrhosum). The synteny types are the following: close (), duplicated genes are close in the genome; some synteny (), paralogues of genes found close to one duplicate are also found close to the other duplicate; no synteny (), less than two paralogeus for other genes are found close to both paralogue duplicates; no information (), the duplicated genes are located in small scaffolds with too few genes close by; conflicting syntenies (), different synteny classifications found in the genomes of the different species affected by the duplication.
PHYLDOG labelled 5,063 duplications to the basal teleost branch, after the split of the spotted gar, with a 4dTv mode of 0.75 (Fig 4, Node 3). Of the paralogues created by these duplications, 73.8% were located in regions with some synteny, 1.5% were close to each other, and 22.5% had no synteny (Fig 4, Node 3). These percentages were calculated without taking into account the duplications where no information regarding the physical location of the genes could be established. The duplications assigned to this basal teleost branch (Fig 4, Node 3) included all gene families with members in both sister clades and thus are assumed to have originated at the 3R. This branch further included hundreds of duplications found in the eels. From these duplications, 62 families had conserved 2 paralogue pairs, one of which had started to diverge at 3R and one in a common ancestor of freshwater eels after the split with osteoglossomorphs. From the paralogue pairs, which had started to diverge in a common ancestor of freshwater eels (Fig 4, Node 9), from these 62 families; 30 were located in regions with some synteny, 9 were close to each other, 11 had no synteny and for 12 pairs no information regarding the physical location of the genes could be established.
1,280 duplications were assigned to the branch basal to the included Clupeocephalan teleosts: zebrafish, fugu, platyfish, northern pike, and Atlantic salmon (Fig 4, Node 4). These duplications showed a very similar distribution with those of the 3R branch, with an overall 4dTv mode of 0.75 (Fig 4, Node 3).
The basal freshwater eel branch was assigned 1,217 duplications of which 55.3, 15.8, and 24.3% were labelled as some synteny, close and without synteny, respectively (Fig 4, Node 9). The European and Japanese eel specific branches were assigned 510 and 127 duplications, from which 32.2, and 34.7% were labelled as some synteny, 50.0 and 48.4% were labelled as close, and 17.1 and 14.7% were labelled as without synteny, respectively (Fig 4, Nodes 14 and 15).
The basal Osteoglossomorpha and the basal arowana branches were assigned 618 and 661 duplications, from which 95.7, and 76.2% were labelled as some synteny, 0.9 and 17.7% were labelled as close, and 3.5 and 5.1% were labelled as without synteny, respectively (Fig 4, Nodes 8 and 12).
The salmon and zebrafish specific branches were assigned 8,787 and 1,525 duplications, respectively, and most of these duplications seemed recent, according to their 4dTv distances. In the salmon branch, most of the duplications (87.0%) were characterized by paralogues located in syntenic regions, whereas most of the zebrafish paralogues (60.5%) were characterized as “close” (Fig 4, Nodes 16 and 7).
For all the included species the “close” paralogues (tandem SDs) tended to show low divergence according to their 4dTv, whereas the duplications found in synteny and most of the duplications without sufficient genomic location information, were more often found to have higher 4dTv distance (Fig 4).
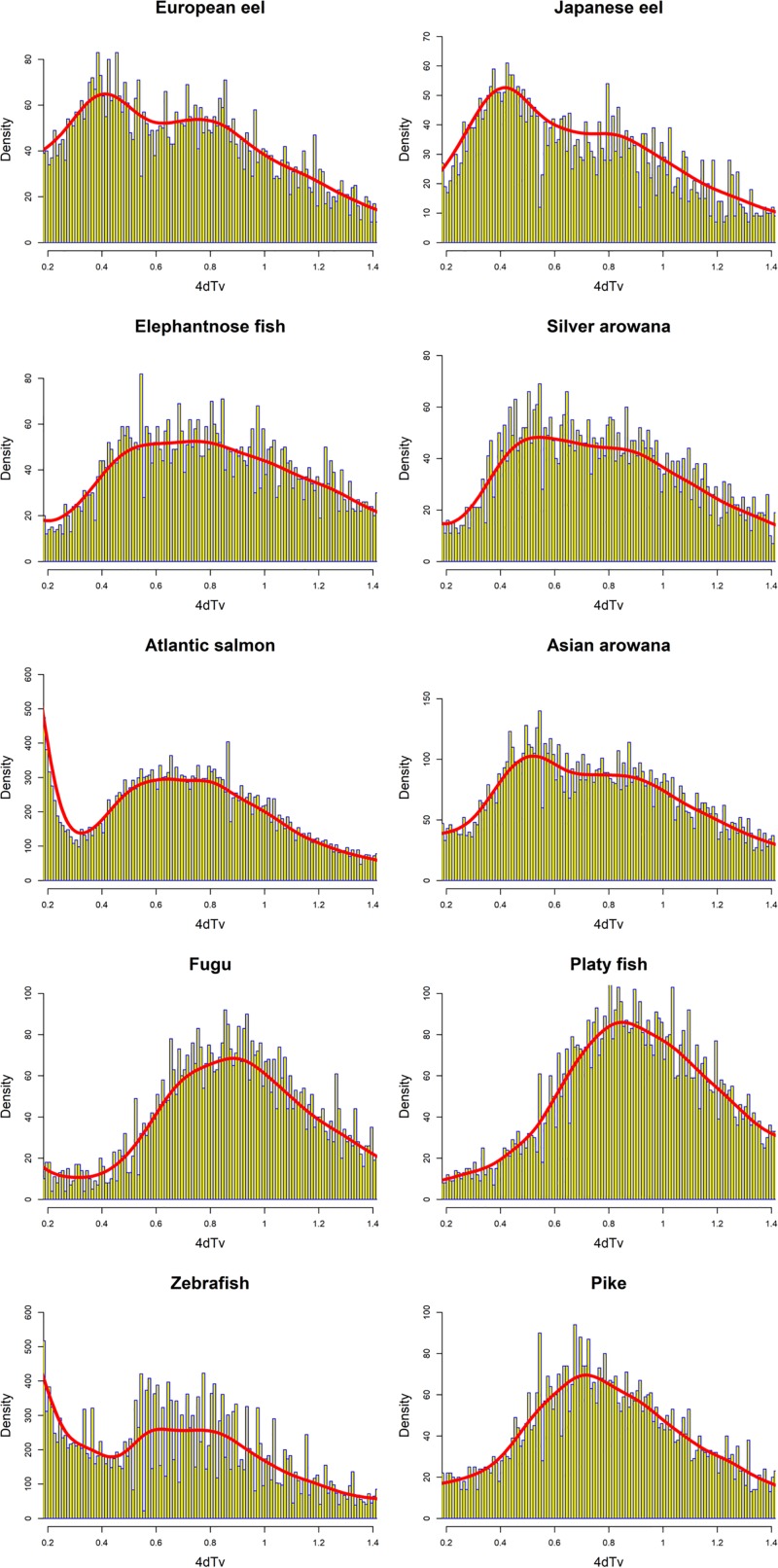
The duplications assigned to the basal freshwater eel branch showed a 4dTv mode of ~0.4 (Fig 4, Node 9). In order to investigate the relative age of all the homolog pairs found in the eels, we ran a 4dTv distance analysis independent of the PHYLDOG tree topology. In this analysis, we compared the 4dTv distribution found for European eel homologs with Japanese eel, elephantnose fish, silver arowana and Asian arowana (Fig 5). The results showed a homolog density mode at 4dTv of ~0.4 for the European and Japanese eel, and 0.5 for the speciation event that separated elephantnose fish, silver arowana, and Asian arowana from the freshwater eels (Fig 5). Furthermore, in order to obtain comparisons between older eel paralogues (>0.2 of 4dTv) and other teleosts, we produced histograms of the 4dTv distances calculated between all paralogues within each species from 0.2 to 1.4 of 4dTv (Fig 6). Additionally, a nonparametric probability density estimate was calculated, using the Gaussian mixture model and plotted on top of the histogram (Fig 6). These results show an older local density maximum (likely originating from 3R) for all teleosts ranging from 0.62 (Zebrafish) to 0.88 (Fugu) of 4dTv. Furthermore, European eel, Japanese eel, Asian arowana and possibly silver arowana showed an additional 4dTv local density maximum at 0.41, 0.42, 0.56 and 0.55 of 4dTv, respectively (Fig 6). A more recent local density maximum was seen in the Atlantic salmon distribution at 0.15 (Fig 4, Node 16).
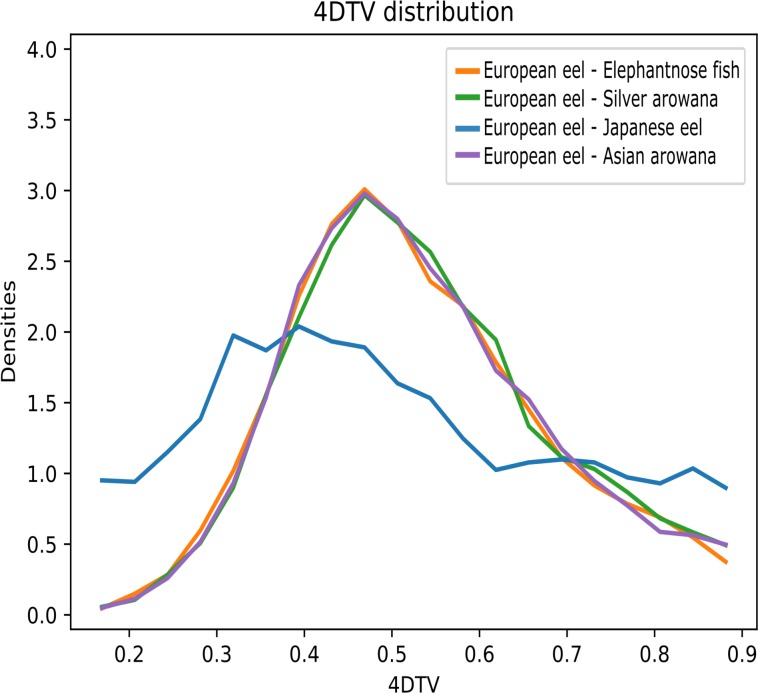
Fig 5. 4dTv distribution between European eel, elephantnose fish, and the arowanas homologs.
4dTv distribution of European eel (Anguilla anguilla) and Japanese eel homologs (), European eel and elephantnose fish (Gnathonemus petersii) homologs (), and European eel, silver arowana (Osteoglossum bicirrhosum) homologs (), and European eel and Asian arowana (Scleropages formosus) homologs ().
Fig 6. Density distribution of all 4dTv distances between teleost paralogues.
Histograms of all 4dTv distances between paralogues of the included teleosts, presented with yellow and blue bars. Furthermore, a probability density estimate curve is plotted on top of the histograms in red. Density values (y-axis) do not correspond to the density estimate. The included species are: European eel (Anguilla anguilla), Japanese eel (Anguilla japonica), zebrafish (Danio rerio), northern pike (Esox lucius), spotted gar (Lepisosteus oculatus), fugu (Takifugu rubripes), platyfish (Xiphophorus maculatus), Atlantic salmon (Salmo salar), elephantnose fish (Gnathonemus petersii), Asian arowana (Scleropages formosus) and silver arowana (Osteoglossum bicirrhosum).
Functional category enrichment
To investigate whether some functional categories were overrepresented among the paralogues assigned to the basal eel branch, two enrichment tests were carried out. First, 1,041 unique GO terms were assigned to 3,607 genes from the basal eel branch, by comparing them to the annotated EggNOG gene families. The full GO annotation can be found in S1 Table. From these terms, we performed an enrichment analysis using the topGO R library [80]. The resulting enriched GO-terms are presented in Table 3. In many cases, these terms were involved either in signalling (e.g. receptor activity, molecular transducer activity, or small GTPase mediated signal transduction), development (e.g. embryonic camera-type eye morphogenesis, gastrulation with mouth forming second, or cell migration involved in gastrulation), ion transport (e.g. anion binding, ATP hydrolysis coupled proton transport, or organic anion transmembrane transporter activity), metabolism (e.g. carbohydrate phosphorylation, ubiquitin-dependent protein catabolic process, or lipopolysaccharide biosynthetic process), or neuronal function (e.g. forebrain development, motor neuron axon guidance, or neuromast development; Table 3). Secondly, KEGG terms were assigned to 1,674 freshwater eel genes using BlastKOALA [81] and mapped onto the KEGG pathways using the KEGG Mapper tool. A Fisher test, corrected for multiple comparisons using False Discovery Rate, was used to look for enriched KEGG pathways in the basal eel branch (Table 4). Most of the KEGG pathways found to be enriched were related to the immune system, nervous system, metabolism and signal transduction. Interestingly, the most significantly enriched KEGG pathway was “Dopaminergic synapse”.
Table 3. Enriched Go-terms from the shared freshwater eel branch of Fig 4.
| Aspect | GO ID | Term | Annotated | Significant | Expected | FDR |
|---|---|---|---|---|---|---|
| Biological Process | GO:0007264 | small GTPase mediated signal transductio … | 256 | 43 | 22.97 | 0.000064 |
| Biological Process | GO:0045176 | apical protein localization | 3 | 3 | 0.27 | 0.00072 |
| Biological Process | GO:0008045 | motor neuron axon guidance | 10 | 5 | 0.9 | 0.00099 |
| Biological Process | GO:0048514 | blood vessel morphogenesis | 121 | 20 | 10.86 | 0.00257 |
| Biological Process | GO:0000132 | establishment of mitotic spindle orienta … | 8 | 4 | 0.72 | 0.00335 |
| Biological Process | GO:0048596 | embryonic camera-type eye morphogenesis | 11 | 4 | 0.99 | 0.00625 |
| Biological Process | GO:0015991 | ATP hydrolysis coupled proton transport | 20 | 6 | 1.79 | 0.00661 |
| Biological Process | GO:0008333 | endosome to lysosome transport | 2 | 2 | 0.18 | 0.00804 |
| Biological Process | GO:0015031 | protein transport | 284 | 39 | 25.48 | 0.00900 |
| Biological Process | GO:0006886 | intracellular protein transport | 156 | 19 | 14 | 0.00907 |
| Biological Process | GO:0007160 | cell-matrix adhesion | 16 | 5 | 1.44 | 0.01084 |
| Biological Process | GO:0001756 | somitogenesis | 50 | 10 | 4.49 | 0.01200 |
| Biological Process | GO:0060042 | retina morphogenesis in camera-type eye | 37 | 9 | 3.32 | 0.01887 |
| Biological Process | GO:0072358 | cardiovascular system development | 280 | 44 | 25.12 | 0.01905 |
| Biological Process | GO:0040023 | establishment of nucleus localization | 4 | 3 | 0.36 | 0.02262 |
| Biological Process | GO:0009826 | unidimensional cell growth | 3 | 2 | 0.27 | 0.02268 |
| Biological Process | GO:0030326 | embryonic limb morphogenesis | 3 | 2 | 0.27 | 0.02268 |
| Biological Process | GO:0008202 | steroid metabolic process | 34 | 3 | 3.05 | 0.02280 |
| Biological Process | GO:0007179 | transforming growth factor beta receptor … | 13 | 4 | 1.17 | 0.02382 |
| Biological Process | GO:0071840 | cellular component organization or bioge … | 846 | 81 | 75.91 | 0.02822 |
| Biological Process | GO:0048884 | neuromast development | 15 | 4 | 1.35 | 0.02854 |
| Biological Process | GO:0001569 | patterning of blood vessels | 8 | 3 | 0.72 | 0.02858 |
| Biological Process | GO:0016998 | cell wall macromolecule catabolic proces … | 8 | 3 | 0.72 | 0.02858 |
| Biological Process | GO:0046835 | carbohydrate phosphorylation | 8 | 3 | 0.72 | 0.02858 |
| Biological Process | GO:0043473 | pigmentation | 57 | 7 | 5.11 | 0.02863 |
| Biological Process | GO:0060059 | embryonic retina morphogenesis in camera … | 14 | 4 | 1.26 | 0.03104 |
| Biological Process | GO:0001702 | gastrulation with mouth forming second | 23 | 6 | 2.06 | 0.04241 |
| Biological Process | GO:0060034 | notochord cell differentiation | 6 | 3 | 0.54 | 0.04259 |
| Biological Process | GO:0061035 | regulation of cartilage development | 7 | 3 | 0.63 | 0.04260 |
| Biological Process | GO:0009103 | lipopolysaCellular Componentharide biosynthetic process | 4 | 2 | 0.36 | 0.04268 |
| Biological Process | GO:0043114 | regulation of vascular permeability | 4 | 2 | 0.36 | 0.04268 |
| Biological Process | GO:0015721 | bile acid and bile salt transport | 4 | 2 | 0.36 | 0.04268 |
| Biological Process | GO:0006511 | ubiquitin-dependent protein catabolic pr … | 91 | 14 | 8.17 | 0.04728 |
| Biological Process | GO:0030900 | forebrain development | 53 | 10 | 4.76 | 0.04832 |
| Biological Process | GO:0042074 | cell migration involved in gastrulation | 37 | 9 | 3.32 | 0.04885 |
| Cellular Component | GO:0031105 | septin complex | 6 | 4 | 0.54 | 0.00084 |
| Cellular Component | GO:0030018 | Z disc | 10 | 5 | 0.9 | 0.00099 |
| Cellular Component | GO:0031461 | cullin-RING ubiquitin ligase complex | 28 | 8 | 2.52 | 0.00555 |
| Cellular Component | GO:0008290 | F-actin capping protein complex | 2 | 2 | 0.18 | 0.00807 |
| Cellular Component | GO:0005915 | zonula adherens | 2 | 2 | 0.18 | 0.00807 |
| Cellular Component | GO:0005737 | cytoplasm | 1750 | 175 | 157.31 | 0.01734 |
| Cellular Component | GO:0005768 | endosome | 47 | 8 | 4.22 | 0.01771 |
| Cellular Component | GO:0033180 | proton-transporting V-type ATPase V1 do … | 10 | 4 | 0.9 | 0.01913 |
| Cellular Component | GO:0030424 | axon | 7 | 3 | 0.63 | 0.01920 |
| Cellular Component | GO:0000159 | protein phosphatase type 2A complex | 3 | 2 | 0.27 | 0.02275 |
| Cellular Component | GO:0005667 | transcription factor complex | 79 | 12 | 7.1 | 0.02336 |
| Cellular Component | GO:0005912 | adherens junction | 7 | 4 | 0.63 | 0.04255 |
| Cellular Component | GO:0031519 | PcG protein complex | 9 | 4 | 0.81 | 0.04258 |
| Cellular Component | GO:0005890 | sodium:potassium-exchanging ATPase compl … | 4 | 2 | 0.36 | 0.04281 |
| Cellular Component | GO:0043198 | dendritic shaft | 4 | 2 | 0.36 | 0.04281 |
| Cellular Component | GO:0005885 | Arp2/3 protein complex | 4 | 2 | 0.36 | 0.04281 |
| Cellular Component | GO:0005765 | lysosomal membrane | 16 | 4 | 1.44 | 0.04914 |
| Molecular Function | GO:0005525 | GTP binding | 251 | 43 | 22.24 | 1.6e-05 |
| Molecular Function | GO:0043168 | anion binding | 1289 | 142 | 114.24 | 0.0018 |
| Molecular Function | GO:0060089 | molecular transducer activity | 778 | 56 | 68.95 | 0.0078 |
| Molecular Function | GO:0004331 | fructose-2 6-bisphosphate 2-phosphatase … | 2 | 2 | 0.18 | 0.0078 |
| Molecular Function | GO:0045296 | cadherin binding | 2 | 2 | 0.18 | 0.0078 |
| Molecular Function | GO:0046933 | proton-transporting ATP synthase activit … | 10 | 4 | 0.89 | 0.0083 |
| Molecular Function | GO:0004702 | receptor signaling protein serine/threon … | 19 | 7 | 1.68 | 0.0112 |
| Molecular Function | GO:0008242 | omega peptidase activity | 6 | 3 | 0.53 | 0.0113 |
| Molecular Function | GO:0031683 | G-protein beta/gamma-subunit complex bin … | 6 | 3 | 0.53 | 0.0113 |
| Molecular Function | GO:0008013 | beta-catenin binding | 7 | 3 | 0.62 | 0.0185 |
| Molecular Function | GO:0016820 | hydrolase activity acting on acid anhyd … | 38 | 8 | 3.37 | 0.0219 |
| Molecular Function | GO:0004749 | ribose phosphate diphosphokinase activit … | 3 | 2 | 0.27 | 0.0221 |
| Molecular Function | GO:0008601 | protein phosphatase type 2A regulator ac … | 3 | 2 | 0.27 | 0.0221 |
| Molecular Function | GO:0003796 | lysozyme activity | 3 | 2 | 0.27 | 0.0221 |
| Molecular Function | GO:0008146 | sulfotransferase activity | 43 | 10 | 3.81 | 0.0249 |
| Molecular Function | GO:0051287 | NAD binding | 22 | 5 | 1.95 | 0.0298 |
| Molecular Function | GO:0003714 | transcription corepressor activity | 9 | 3 | 0.8 | 0.0388 |
| Molecular Function | GO:0008514 | organic anion transmembrane transporter … | 22 | 5 | 1.95 | 0.0399 |
| Molecular Function | GO:0004872 | receptor activity | 695 | 41 | 61.59 | 0.0495 |
Enriched Go-terms from the duplicated genes shared by freshwater eels. “Aspects” indicates the specific GO-term aspect of each enriched GO-term. “GO ID” indicates the identification number of each enriched GO-term. “Term “indicates the verbal description of each enriched GO-term. “Annotated” indicates the number of GO-terms which are associated with each enriched GO-term. “Significant” indicates the number of GO-terms associated to each enriched GO-term found among the duplicated genes. “Expected” indicates the number of GO-terms expected to be found linked to each enriched GO-term. “FDR” indicates the False Discovery Rate adjusted P-value from the Fisher exact test of enrichment.
Table 4. Enriched KEGG-terms from the shared freshwater eel branch of Fig 4.
| KEGG ID | Term | Annotated | Significant | Expected | FDR |
|---|---|---|---|---|---|
| 04728 | Dopaminergic synapse | 38 | 283 | 12 | 0,000001 |
| 03015 | mRNA surveillance pathway | 21 | 129 | 5 | 0,000082 |
| 04660 | T cell receptor signaling pathway | 27 | 204 | 8 | 0,000082 |
| 04071 | Sphingolipid signaling pathway | 29 | 238 | 10 | 0,000112 |
| 05142 | Chagas disease (American trypanosomiasis) | 23 | 180 | 7 | 0,000518 |
| 04659 | Th17 cell differentiation | 23 | 184 | 7 | 0,000596 |
| 05162 | Measles | 21 | 168 | 7 | 0,001190 |
| 04390 | Hippo signaling pathway | 29 | 282 | 11 | 0,001190 |
| 04658 | Th1 and Th2 cell differentiation | 19 | 148 | 6 | 0,001601 |
| 04261 | Adrenergic signaling in cardiomyocytes | 28 | 291 | 12 | 0,004378 |
| 05100 | Bacterial invasion of epithelial cells | 20 | 183 | 7 | 0,005845 |
| 05032 | Morphine addiction | 18 | 155 | 6 | 0,005845 |
| 00625 | Chloroalkane and chloroalkene degradation | 5 | 10 | 0 | 0,005845 |
| 04640 | Hematopoietic cell lineage | 12 | 79 | 3 | 0,006536 |
| 04910 | Insulin signaling pathway | 26 | 276 | 11 | 0,006551 |
| 04630 | Jak-STAT signaling pathway | 18 | 171 | 7 | 0,012920 |
| 04016 | MAPK signaling pathway—plant | 6 | 22 | 1 | 0,014553 |
| 04022 | cGMP-PKG signaling pathway | 29 | 350 | 14 | 0,015915 |
| 04917 | Prolactin signaling pathway | 15 | 133 | 5 | 0,015915 |
| 05130 | Pathogenic Escherichia coli infection | 13 | 108 | 4 | 0,019059 |
| 05418 | Fluid shear stress and atherosclerosis | 22 | 245 | 10 | 0,022738 |
| 00020 | Citrate cycle (TCA cycle) | 8 | 47 | 2 | 0,022808 |
| 04080 | Neuroactive ligand-receptor interaction | 31 | 395 | 16 | 0,023393 |
| 04151 | PI3K-Akt signaling pathway | 37 | 499 | 20 | 0,023393 |
| 04514 | Cell adhesion molecules (CAMs) | 21 | 231 | 9 | 0,023393 |
| 04391 | Hippo signaling pathway—fly | 16 | 158 | 6 | 0,034750 |
| 05340 | Primary immunodeficiency | 7 | 41 | 2 | 0,036069 |
| 04350 | TGF-beta signaling pathway | 16 | 164 | 7 | 0,038388 |
| 05133 | Pertussis | 12 | 111 | 5 | 0,045104 |
| 05152 | Tuberculosis | 21 | 247 | 10 | 0,047421 |
| 04664 | Fc epsilon RI signaling pathway | 12 | 113 | 5 | 0,048089 |
| 00510 | N-Glycan biosynthesis | 9 | 71 | 3 | 0,048943 |
| 04144 | Endocytosis | 37 | 533 | 22 | 0,048943 |
| 00350 | Tyrosine metabolism | 6 | 34 | 1 | 0,048943 |
| 04510 | Focal adhesion | 28 | 379 | 15 | 0,048943 |
Enriched KEGG-terms from the duplicated genes shared by freshwater eels. “KEGG ID” indicates the identification number of each enriched KEGG pathway. “Term”indicates the verbal description of each enriched KEGG pathway. “Annotated” indicates the number of KEGG pathways, which are associated with each enriched KEGG pathway. “Significant” indicates the number of KEGG pathways associated with each enriched KEGG pathway found among the duplicated genes. “Expected” indicates the number of KEGG pathways expected to be found associated with each enriched KEGG pathway. “FDR” indicates the False Discovery Rate adjusted P-value from the Fisher exact test of enrichment.
Discussion
The present study found more than one thousand gene families in which the gene family tree topology indicates a duplication in a common ancestor of freshwater eels sometime after the split of Elopomorpha and Osteoglossomorpha. Only phylogenetic species tree branches with previously documented WGDs (Fig 4, Nodes 1, 3, 4, and 16) and the zebrafish specific branch (Fig 4, Node 7) were assigned more duplications than the basal freshwater eel branch (Fig 4, Node 9). The vast majority of the assigned zebrafish specific duplications formed a 4dTv local density maximum at ~0 and were found “close” in the genome, thus these duplications appear to be tandem SDs, the presence of which concurs with previous studies [8,11,65].
The origin of the duplications assigned to the basal freshwater eel branch
In some cases, it has been shown that SDs could be retained at specific points in time [5,15,16]. However, most duplications assigned to the basal freshwater eel branch were detected in large syntenic blocks which opposes a hypothesis of a SD origin.
Rather the synteny results suggest that the duplications assigned to the basal freshwater eel branch originated in larger portions e.g. whole regions (large SDs), chromosomes or genomes. In particular, a WGD origin is consistent with the number of duplications observed and the 4dTv distribution (Fig 4, Node 9), which showed one distinct 4dTv density mode (4dTv ~0.4) placed along the long branch leading to the freshwater eels. Fig 6 (and Fig 5) further shows duplications which started diverging at the 3R in the eel transcriptome, as a 4dTv local density maximum of ~0.75. The notable similarity between this local density maximum and the local density maximum of the other 3R generated genes from all the included teleosts (Figs 4 and 6) suggests that these paralogues (4dTv ~ 0.75) were created by the 3R. This hypothesis is further supported by the results of the phylogenetic analysis (Fig 4, Nodes 3), which assigned hundreds of duplications, which are still present in the eel, to the 3R branch. Moreover, no modes of comparable magnitude at 4dTv ~ 0.75 can be seen in any of the post 3R branches leading to the freshwater eels (Fig 4, Nodes 5 and 9). Therefore, if the duplications assigned to the basal freshwater eel branch were created by a WGD event, and assuming instant rediploidization after the 3R, this event would be more recent and different to the 3R, and thus should be named a 4R WGD event.
However, cytological rediploidization is not always completed immediately after an autotetraploidization WGD event, as shown in the case of salmonids [43]. Therefore, the origin of the duplications assigned to the basal freshwater eel branch could also be explained by a hypothesis of lineage-specific rediploidization after the 3R. Protracted rediploidization could result in lower rates of gene losses since deleterious mutations have less time to accumulate in one paralogue and thereby create a pseudogene, which could explain the high number of paralogue pairs found in eel. This hypothesis could also explain both the PHYLDOG and 4dTv results, as paralogue genes only start to diverge after the rediploidization of their genomic region [41,43,44]. If the duplications assigned to the basal freshwater eel branch had, in fact, experienced delayed rediploidization from the 3R, the same genomic regions would have also experienced delayed rediploidization in the lineage of the remaining teleosts. Interestingly, relatively large quantities of duplications, with conserved synteny, were also assigned to the basal Clupeocephala branch and the Osteoglossomorpha branches (Fig 4, Nodes 4, 8, and 12). This observation supports the hypothesis that the duplications assigned to the basal freshwater eel branch were located in genomic regions, which experienced delayed rediploidization after the 3R. However, lineage-specific rediploidization has only been unequivocally documented in salmonids, and more studies are needed to demonstrate this process in other species. Therefore, it remains to be determined if this mechanism is a salmonid specific phenomenon. Furthermore, due to the observed 4dTv distances, the mechanisms would have protracted rediploidization for a longer time in eels than in salmonids. Moreover, the 4dTv analysis revealed very similar results for the 3R branch and the basal Clupeocephala branch (Fig 4, Nodes 3 and 4) and for the shared Osteoglossomorpha and Arowana branches, respectively (Fig 4, Nodes 8 and 12). This result supports the hypothesis that these duplications were divided due to a potential PHYLDOG artefact, explained below. Additionally, 62 gene families were found which showed a topology concurring with a 3R event followed by a 4R event, since duplication had been conserved from both events in the same gene family. From these 62 families, 30 paralogues pairs of the suspected 4R duplications were located in regions with some synteny. These trees directly oppose the hypothesis of protracted rediploidization; however, only for these 30 families. In the event of an eel 4R WGD, more such trees would be expected.
Possible PHYLDOG artefact
According to PHYLDOG, the shared teleost duplications split into two events placed in the 3R branch and the basal Clupeocephala branch (Fig 4, Nodes 3 and 4). These results could be caused by an artefact from the phylogenetic analysis. Specifically, PHYLDOG software assigns duplications to branches based on the successful identification of the daughter genes on the branches of both sister clades, thus lower genomic information (fewer or less complete genomes/transcriptomes) increases the chance of not finding a gene and thus misplacing duplications. In this case, although the number of genomes/transcriptomes are the same, the amount of genomic information is substantially different between the two daughter clades, since the genomes/transcriptomes on one side (Fig 4, from Node 5), are generally much less complete than those on the other side (Figs 3 and 4, from Node 4). In support of this hypothesis is the 4dTv analysis, in which the duplications assigned to the 3R and the basal Clupeocephala branch indicate approximately the same mode (Fig 4, Nodes 3 and 4), suggesting that they started to diverge at the same time. Therefore, a PHYLDOG artefact, in which duplications can leak down to a daughter branch which is basal to a clade containing more genomic information, is also a parsimonious explanation for most of the duplications assigned to the basal Clupeocephala branch.
Arowana results
As an unexpected result of our analysis, the included Osteoglossomorphas also appear to contain a high quantity of duplications, which likely started diverging after the split between Elopomorphas and Osteoglossomorphas. These duplications also included a high occurrence of paralogues with some conserved synteny between them. This result suggests that these genes were duplicated in larger portions e.g. whole regions (large SDs), chromosomes or genome and not by smaller SDs. When combining the basal Osteoglossomorpha and the basal arowana branches these were assigned a similar quantity of duplications as the basal freshwater eel branch. This result supports the hypothesis that some genomic regions were still under tetrasomic inheritance, from the 3R, at the time of the split between Elopomorphas and Osteoglossomorphas. However, it is also possible that the duplications were generated by a separate duplication event in a common ancestor to the included Osteoglossomorphas but have leaked into the basal arowana branch and the Asian arowana specific branch in the phylogenetic analysis due to the PHYLDOG artefact described above. The PHYLDOG artefact hypothesis is supported by the 4dTv analysis, as the 4dTv modes of these branches are very similar, and since the Elephantnose fish transcriptomes are the least complete dataset of these branches. To generate a better supported hypothesis of the origin of these duplications a study dedicated to this purpose should be conducted.
Start of divergence of the duplications assigned to the basal freshwater eel branch
In the independent 4dTv analysis, without considering phylogenetic tree topologies, the 4dTv of the homologs between the European eel and the Japanese eel, the elephantnose fish and the arowanas, showed that European eel and Japanese eel homologs have a 4dTv mode at ~ 0.4. On the other hand, the homologs between the European eel and any Osteoglossomorpha species form a 4dTv mode at ~ 0.5 (Fig 5). This result indicates that the duplications found in the freshwater eel species started diverging after the split between Elopomorphas and Osteoglossomorphas (Fig 5). Therefore, the phylogenetic reconstruction and the 4dTv distances together suggest that the duplications assigned to the basal freshwater eel branch (4dTv ~ 0.4) started diverging after the teleost specific 3R duplication event (320–350 MYA) [35,36] and after the split between eels and Osteoglossomorphas, but before the Ss4R (88–103 MYA) [86].
If the 4dTv mode observed in the basal freshwater eel branch was the result of new duplications, then these duplications would likely have originated in a common ancestor to all members of the anguillidae family, as these first appear 20–50 MYA [87]. Due to the 4dTv observed, this event could also be shared by wider Elopomorpha; however, without analysing other anguilliforms or Elopomorpha transcriptomes or genomes, this hypothesis remains speculative.
Previously published related data
In concurrence with the present study, other studies have reported data suggesting an unusually high quantity of gene duplications in eels. In the additional data included by Inoue et al. [27], the eel and zebrafish are the species with the highest percentage of duplicated genes (36.6% and 31.9%, respectively). Furthermore, an unexpectedly high number of Hox genes (73 genes) were found in the analysis of the draft eel genome [49]. In this study [49], the phylogenetic distance between Hox clusters was remarkably short, making it impossible to distinguish between the 3R “a” or “b” association of 3 out of 4 cluster pairs based on DNA sequence alone. Several other studies focusing on particular genes have likewise found paralogue pairs in eels, which are not found in other teleosts [48–56] and similarly, an unexpected short phylogenetic distance is often found between eel paralogue pairs. These results support both a 4R hypothesis and a hypothesis of a 3R origin followed by protracted rediploidization. However, many of the referenced studies also presented results of weak conserved local synteny indicating a 3R origin. These synteny results are unexpected following both a 4R hypothesis and the hypothesis of protracted rediploidization. We draw this conclusion based on the notion that the close genomic region of genes, which experienced delayed rediploidization, is highly expected to also have been under tetrasomic inheritance for an extended period [43]. Thus, these neighbouring genes should not accumulate mutations similarly to homolog regions of other teleosts, which experienced immediate rediploidization, and thus the synteny of these regions are unlikely to match.
Conclusions
The data presented in this study support the hypothesis that a remarkably high amount of paralogues pairs started to diverge in a common ancestor of the freshwater eel lineage after the split from the Osteoglossomorpha lineage. The 4dTv and phylogenetic analyses revealed a clear clustering of these paralogues in the basal freshwater eel branch with a 4dTv mode at ~0.4. The synteny of these paralogue pairs suggests they originated in large portions, most likely from a WGD event. However, the results do not unequivocally support/oppose whether i) These paralogues originated from the 3R but are located in genomic regions which have experienced protracted rediploidization; ii) These paralogues originated in a 4R WGD in a common ancestor to freshwater eels; or iii) Both i and ii have contributed to the evolution of these paralogues. The present results offer robust information on the duplicated gene complement of freshwater eels, thus providing novel insights into the peculiar biology of the critically endangered European eel. However, additional high quality genome resources of other Elopomorpha members are needed to further study the dynamics of gene duplication and conservation in early teleost evolution.
Supporting information
GO annotation from EggNOG of eel duplication gene families shared by freshwater eels. Including: “Orthogroup”, “Descriptions”, “Functional annotations”, “GO categories”, and “GO IDs”.
(CSV)
(ZIP)
Acknowledgments
We would like to thank Anna Sinclair and Mitch Fleming for language editing and helpful comments.
Data Availability
The raw RNA-sequencing reads from forebrain, pituitary, and testis samples from European eel (Anguilla anguilla) have been deposited at GenBank (http://www.ncbi.nlm.nih.gov/genbank) under accession no. SRP126643.
Funding Statement
This study received funding from the project REPRO-TEMP (AGL2013-41646-R) funded by the Spanish Ministry of Economy and Competitiveness, and from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement No 642893 (IMPRESS), which also included the predoctoral contracts of CR and JGHJ. VG has a postdoctoral grant from the Spanish Ministry of Science, Innovation and Universities (MICIU; Programa Juan de la Cierva-Incorporación; IJCI-2017-34200). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ohno S. Evolution by Gene Duplication. New York: Springer-Verlag; 1970. [Google Scholar]
- 2.Gu X, Wang Y, Gu J. Age distribution of human gene families shows significant roles of both large- and small-scale duplications in vertebrate evolution. Nat Genet. 2002;31:205–9. 10.1038/ng902 [DOI] [PubMed] [Google Scholar]
- 3.Cañestro C, Albalat R, Irimia M, Garcia-Fernàndez J. Impact of gene gains, losses and duplication modes on the origin and diversification of vertebrates. Semin Cell Dev Biol. 2013;24(2):83–94. 10.1016/j.semcdb.2012.12.008 [DOI] [PubMed] [Google Scholar]
- 4.Llorente B, Malpertuy A, Neuvéglise C, De Montigny J, Aigle M, Artiguenave F, et al. Genomic exploration of the hemiascomycetousy Yeasts: 18. comparative analysis of chromosome maps and synteny with Saccharomyces cerevisiae. FEBS Lett. 2000;487(1):101–12. [DOI] [PubMed] [Google Scholar]
- 5.Colbourne JK, Pfrender ME, Gilbert D, Thomas WK, Tucker A, Oakley TH, et al. The ecoresponsive genome of daphnia pulex. Science. 2011;331:555–62. 10.1126/science.1197761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bailey JA, Gu Z, Clark RA, Reinert K, Samonte R V, Schwartz S, et al. Recent segmental duplications in the human genome. Science. 2002;297(5583):1003–7. 10.1126/science.1072047 [DOI] [PubMed] [Google Scholar]
- 7.Vallente Samonte R, Eichler EE. Segmental duplications and the evolution of the primate genome. Nat Rev Genet. 2016;3:65–72. [DOI] [PubMed] [Google Scholar]
- 8.Blomme T, Vandepoele K, De Bodt S, Simillion C, Maere S, Van de Peer Y. The gain and loss of genes during 600 million years of vertebrate evolution. Genome Biol. 2006;7(R43):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.David L, Blum S, Feldman MW, Lavi U, Hillel J. Recent duplication of the common carp (Cyprinus carpio L.) Genome as revealed by analyses of microsatellite loci. Mol Biol Evol. 2003;20(9):1425–34. 10.1093/molbev/msg173 [DOI] [PubMed] [Google Scholar]
- 10.Jaillon O, Aury J, Brunet F, Petit J-L, Stange-Thomann N, Mauceli E, et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431(7011):946–57. 10.1038/nature03025 [DOI] [PubMed] [Google Scholar]
- 11.Lu J, Peatman E, Tang H, Lewis J, Liu Z. Profiling of gene duplication patterns of sequenced teleost genomes: evidence for rapid lineage-specific genome expansion mediated by recent tandem duplications. BMC Genomics. 2012;13(246):1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rondeau EB, Minkley DR, Leong JS, Messmer AM, Jantzen JR, Von Schalburg KR, et al. The genome and linkage map of the northern pike (Esox lucius): Conserved synteny revealed between the salmonid sister group and the neoteleostei. PLoS One. 2014;9(7):e102089 10.1371/journal.pone.0102089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Albalat R, Cañestro C. Evolution by gene loss. Nat Rev Genet. 2016;17(7):379–91. 10.1038/nrg.2016.39 [DOI] [PubMed] [Google Scholar]
- 14.Hafeez M, Shabbir M, Altaf F, Abbasi AA. Phylogenomic analysis reveals ancient segmental duplications in the human genome. Mol Phylogenet Evol. 2016;94:95–100. 10.1016/j.ympev.2015.08.019 [DOI] [PubMed] [Google Scholar]
- 15.Tautz D, Domazet-lošo T. The evolutionary origin of orphan genes. Nat Publ Gr. 2011;12(10):692–702. [DOI] [PubMed] [Google Scholar]
- 16.Chain FJJ, Feulner PGD, Panchal M, Eizaguirre C, Samonte IE, Kalbe M, et al. Extensive copy-number variation of young genes across stickleback populations. PLoS Genet. 2014;10(12):e1004830 10.1371/journal.pgen.1004830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mable BK, Leitch AR, Soltis DE, Soltis PS, Leitch IJ, Pires JC. Why polyploidy is rarer in animals than in plants’: myths and mechanisms. Biol J Linn Soc Blackwell Sci. 2004;82(82):453–66. [Google Scholar]
- 18.Otto SP, Whitton J. Polyploid incidence and evolution. Annu Rev Genet. 2000;34:401–37. 10.1146/annurev.genet.34.1.401 [DOI] [PubMed] [Google Scholar]
- 19.Albertin W, Marullo P. Polyploidy in fungi: Evolution after whole-genome duplication. Proc R Soc B Biol Sci. 2012;279(1738):2497–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463(7278):178–83. 10.1038/nature08670 [DOI] [PubMed] [Google Scholar]
- 21.Del Pozo JC, Ramirez-Parra E. Whole genome duplications in plants: An overview from Arabidopsis. J Exp Bot. 2015;66(22):6991–7003. 10.1093/jxb/erv432 [DOI] [PubMed] [Google Scholar]
- 22.Soltis DE, Visger CJ, Soltis PS. The polyploidy revolution then …and now: Stebbins revisited. Am J Bot. 2014;101(7):1057–78. 10.3732/ajb.1400178 [DOI] [PubMed] [Google Scholar]
- 23.Masterson J. Stomatal Size in Fossil Plants: Evidence for polyploidy in majority of angiosperms. Science. 1994;264(5157):421–4. 10.1126/science.264.5157.421 [DOI] [PubMed] [Google Scholar]
- 24.Parisod C, Holderegger R, Brochmann C. Evolutionary consequences of autopolyploidy. New Phytol. 2010;186(1):5–17. 10.1111/j.1469-8137.2009.03142.x [DOI] [PubMed] [Google Scholar]
- 25.Blanc G. Widespread Paleopolyploidy in Model Plant Species Inferred from Age Distributions of Duplicate Genes. Plant Cell Online. 2004;16(7):1667–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sémon M, Wolfe KH. Consequences of genome duplication. Curr Opin Genet Dev. 2007;17(6):505–12. 10.1016/j.gde.2007.09.007 [DOI] [PubMed] [Google Scholar]
- 27.Inoue J, Sato Y, Sinclair R, Tsukamoto K, Nishida M. Rapid genome reshaping by multiple-gene loss after whole-genome duplication in teleost fish suggested by mathematical modeling. Proc Natl Acad Sci. 2015;112(48):14918–23. 10.1073/pnas.1507669112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolfe KH. Yesterday’s polyploids and the mystery of diploidization. Nat Rev Genet. 2001;2(5):333–41. 10.1038/35072009 [DOI] [PubMed] [Google Scholar]
- 29.Kassahn KS, Dang VT, Wilkins SJ, Kassahn KS, Dang VT, Wilkins SJ, et al. Evolution of gene function and regulatory control after whole-genome duplication: Comparative analyses in vertebrates. Genome Res. 2009;19:1404–18. 10.1101/gr.086827.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X, Jin D, Wang Z, Guo H, Zhang L, Wang L, et al. Telomere-centric genome repatterning determines recurring chromosome number reductions during the evolution of eukaryotes. New Phytol. 2015;205(1):378–89. 10.1111/nph.12985 [DOI] [PubMed] [Google Scholar]
- 31.Glasauer SMK, Neuhauss SCF. Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol Genet Genomics. 2014;289(6):1045–60. 10.1007/s00438-014-0889-2 [DOI] [PubMed] [Google Scholar]
- 32.Chester M, Gallagher JP, Symonds VV, Cruz da Silva AV, Mavrodiev EV, Leitch AR, et al. Extensive chromosomal variation in a recently formed natural allopolyploid species, Tragopogon miscellus (Asteraceae). Proc Natl Acad Sci. 2012;109(4):1176–81. 10.1073/pnas.1112041109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gordon JL, Byrne KP, Wolfe KH. Mechanisms of chromosome number evolution in yeast. PLoS Genet. 2011;7(7): e1002190 10.1371/journal.pgen.1002190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dehal P, Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005;3(10):1700–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Christoffels A, Koh EGL, Chia JM, Brenner S, Aparicio S, Venkatesh B. Fugu genome analysis provides evidence for a whole-genome duplication early during the evolution of ray-finned fishes. Mol Biol Evol. 2004;21(6):1146–51. 10.1093/molbev/msh114 [DOI] [PubMed] [Google Scholar]
- 36.Vandepoele K, De Vos W, Taylor JS, Meyer A, Van de Peer Y. Major events in the genome evolution of vertebrates: paranome age and size differ considerably between ray-finned fishes and land vertebrates. Proc Natl Acad Sci U S A. 2004;101(6):1638–43. 10.1073/pnas.0307968100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leggatt RA, Iwama GK. Occurrence of polyploidy in the fishes. Rev Fish Biol Fish. 2003;13(3):237–46. [Google Scholar]
- 38.Comber SC LE, Smith C. Polyploidy in fishes: patterns and processes. Biol J Linn Soc. 2004;82(4):431–42. [Google Scholar]
- 39.Braasch I, Gehrke AR, Smith JJ, Kawasaki K, Manousaki T, Pasquier J, et al. Corrigendum: The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat Genet. 2016;48(6):427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bian C, Hu Y, Ravi V, Kuznetsova IS, Shen X, Mu X, et al. The Asian arowana (Scleropages formosus) genome provides new insights into the evolution of an early lineage of teleosts. Sci Rep. 2016;6:1–17. 10.1038/s41598-016-0001-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lien S, Koop BF, Sandve SR, Miller JR, Matthew P, Leong JS, et al. The Atlantic salmon genome provides insights into rediploidization. Nature. 2016;533(6020):200–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blischak PD, Mabry ME, Conant GC, Pires JC. Integrating Networks, Phylogenomics, and Population Genomics for the Study of Polyploidy. Annu Rev Ecol Evol Syst. 2018;49(1):253–78. [Google Scholar]
- 43.Robertson FM, Gundappa MK, Grammes F, Hvidsten TR, Redmond AK, Lien S, et al. Lineage-specific rediploidization is a mechanism to explain time-lags between genome duplication and evolutionary diversification. Genome Biol. 2017;18(1):1–14. 10.1186/s13059-016-1139-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin KJ, Holland PWH. Enigmatic Orthology Relationships between Hox Clusters of the African Butterfly Fish and Other Teleosts Following Ancient Whole-Genome Duplication. 2014;31(10):2592–611. 10.1093/molbev/msu202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wright JE, Johnson K, Hollister A, May B. Meiotic models to explain classical linkage, pseudolinkage, and chromosome pairing in tetraploid derivative salmonid genomes. Isozymes Curr Top Biol Med Res. 1983;10:239–60. [PubMed] [Google Scholar]
- 46.Berthelot C, Brunet F, Chalopin D, Juanchich A, Bernard M, Noël B, et al. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat Commun. 2014;5(3657). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Allendorf F, Thorgaard G. Tetraploidy and the evolution of salmonid fishes In: Evolutionary Genetics of Fishes; 1984. p. 1–53. [Google Scholar]
- 48.Dufour S, Weltzien F, Sebert M-E, Le Belle N, Vidal B, Vernier P, et al. Dopaminergic inhibition of reproduction in teleost fishes: ecophysiological and evolutionary implications. Ann N Y Acad Sci. 2005;1040:9–21. 10.1196/annals.1327.002 [DOI] [PubMed] [Google Scholar]
- 49.Henkel C V., Burgerhout E, de Wijze DL, Dirks RP, Minegishi Y, Jansen HJ, et al. Primitive duplicate hox clusters in the European eel’s genome. PLoS One. 2012;7(2):e32231 10.1371/journal.pone.0032231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lafont AG, Rousseau K, Tomkiewicz J, Dufour S. Three nuclear and two membrane estrogen receptors in basal teleosts, Anguilla sp.: Identification, evolutionary history and differential expression regulation. Gen Comp Endocrinol. 2016;235:177–91. 10.1016/j.ygcen.2015.11.021 [DOI] [PubMed] [Google Scholar]
- 51.Maugars G, Dufour S. Demonstration of the coexistence of duplicated LH receptors in teleosts, and their origin in ancestral actinopterygians. PLoS One. 2015;10(8):e0135184 10.1371/journal.pone.0135184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morini M, Pasquier J, Dirks R, Van Den Thillart G, Tomkiewicz J, Rousseau K, et al. Duplicated leptin receptors in two species of eel bring new insights into the evolution of the leptin system in vertebrates. PLoS One. 2015;10(5):1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pasqualini C, Weltzien FA, Vidal B, Baloche S, Rouget C, Gilles N, et al. Two distinct dopamine D2 receptor genes in the European eel: Molecular characterization, tissue-specific transcription, and regulation by sex steroids. Endocrinology. 2009;150(3):1377–92. 10.1210/en.2008-0578 [DOI] [PubMed] [Google Scholar]
- 54.Pasquier J, Lafont AG, Jeng SR, Morini M, Dirks R, van den Thillart G, et al. Multiple kisspeptin receptors in early osteichthyans provide new insights into the evolution of this receptor family. PLoS One. 2012;7(11):e48931 10.1371/journal.pone.0048931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rozenfeld C, Butts IAE, Tomkiewicz J, Zambonino-Infante J-L, Mazurais D. Abundance of specific mRNA transcripts impacts hatching success in European eel, Anguilla anguilla L. Comp Biochem Physiol A Mol Integr Physiol. 2016;191:59–65. 10.1016/j.cbpa.2015.09.011 [DOI] [PubMed] [Google Scholar]
- 56.Morini M, Penaranda DS, Vilchez MC, Nourizadeh-Lillabadi R, Lafont AG, Dufour S, et al. Nuclear and membrane progestin receptors in the European eel: Characterization and expression in vivo through spermatogenesis. Comp Biochem Physiol -Part A Mol Integr Physiol. 2017;207:79–92. [DOI] [PubMed] [Google Scholar]
- 57.Ravi V, Venkatesh B. The Divergent Genomes of Teleosts. Annu Rev Anim Biosci. 2018;6(1):47–68. [DOI] [PubMed] [Google Scholar]
- 58.Peña-Llopis S, Brugarolas J. Simultaneous isolation of high-quality DNA, RNA, miRNA and proteins from tissues for genomic applications. Nat Protoc. 2013;8(11):2240–55. 10.1038/nprot.2013.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Andrews S. FastQC: A quality control tool for high throughput sequence data. 2010. p. http://www.bioinformatics.babraham.ac.uk/projects/.
- 60.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–20. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Crusoe MR, Alameldin HF, Awad S, Boucher E, Caldwell A, Cartwright R, et al. The khmer software package: enabling efficient nucleotide sequence analysis. F1000 Res. 2015;4(900):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pasquier J, Cabau C, Nguyen T, Jouanno E, Severac D, Braasch I, et al. Gene evolution and gene expression after whole genome duplication in fish: the PhyloFish database. BMC Genomics. 2016;17(368):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8(8):1494–512. 10.1038/nprot.2013.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides accurate, fast, and bias-aware transcript expression estimates using dual-phase inference. bioRxiv. 2017;14(4):417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496(7446):498–503. 10.1038/nature12111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kai W, Kikuchi K, Tohari S, Chew AK, Tay A, Fujiwara A, et al. Integration of the genetic map and genome assembly of evolution in teleosts and mammals. Genome Biol Evol. 2011;3:424–42. 10.1093/gbe/evr041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schartl M, Walter RB, Shen Y, Garcia T, Catchen J, Amores A, et al. The genome of the platyfish, Xiphophorus maculatus, provides insights into evolutionary adaptation and several complex traits. Nat Genet. 2013;45(5):567–72. 10.1038/ng.2604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nomura K, Fujiwara A, Iwasaki Y, Nishiki I, Matsuura A, Ozaki A, et al. Genetic parameters and quantitative trait loci analysis associated with body size and timing at metamorphosis into glass eels in captive-bred Japanese eels (Anguilla japonica). PLoS One. 2018;13(8):1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva E V., Zdobnov EM. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31(19):3210–2. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- 70.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11(9):1650–67. 10.1038/nprot.2016.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–95. 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li L, Stoeckert CJJ, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13(9):2178–89. 10.1101/gr.1224503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7(539):1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Capella-gutiérrez S, Silla-martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25(15):1972–3. 10.1093/bioinformatics/btp348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Slater GSC, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6(31):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Boussau B, Szöll GJ, Duret L, Gouy M, Tannier E, Daubin V, et al. Genome-scale coestimation of species and gene trees. Life Sci. 2012;23:323–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lartillot N, Lepage T, Blanquart S. PhyloBayes 3: A Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics. 2009;25(17):2286–8. 10.1093/bioinformatics/btp368 [DOI] [PubMed] [Google Scholar]
- 78.Huerta-cepas J, Szklarczyk D, Forslund K, Cook H, Heller D, Walter MC, et al. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016;44:286–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Finn RD, Clements J, Eddy SR. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011;39:29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Alexa A, Rahnenfuhrer J. topGO: Enrichment analysis for gene ontology. 2016. p. R package version 2.29.0.
- 81.Kanehisa M, Sato Y, Morishima K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol. 2016;428(4):726–31. 10.1016/j.jmb.2015.11.006 [DOI] [PubMed] [Google Scholar]
- 82.Hsu HY, Chen SH, Cha YR, Tsukamoto K, Lin CY, Han YS. De novo assembly of the whole transcriptome of the wild embryo, preleptocephalus, leptocephalus, and glass eel of Anguilla japonica and deciphering the digestive and absorptive capacities during early development. PLoS One. 2015;10(9):1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Coppe A, Pujolar JM, Maes GE, Larsen PF, Hansen MM, Bernatchez L, et al. Sequencing, de novo annotation and analysis of the first Anguilla anguilla transcriptome: EeelBase opens new perspectives for the study of the critically endangered European eel. BMC Genomics. 2010;11(635):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Burgerhout E, Minegishi Y, Brittijn SA, de Wijze DL, Henkel C V., Jansen HJ, et al. Changes in ovarian gene expression profiles and plasma hormone levels in maturing European eel (Anguilla anguilla); Biomarkers for broodstock selection. Gen Comp Endocrinol. 2016;225:185–96. 10.1016/j.ygcen.2015.08.006 [DOI] [PubMed] [Google Scholar]
- 85.Ager-Wick E, Dirks RP, Burgerhout E, Nourizadeh-Lillabadi R, de Wijze DL, Spaink HP, et al. The pituitary gland of the European eel reveals massive expression of genes involved in the melanocortin system. PLoS One. 2013;8(10):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Macqueen DJ, Johnston IA, Macqueen DJ. A well-constrained estimate for the timing of the salmonid whole genome duplication reveals major decoupling from species diversification. Proc R Soc B. 2014;281(20132881):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Minegishi Y, Aoyama J, Inoue JG, Miya M. Molecular phylogeny and evolution of the freshwater eels genus Anguilla based on the whole mitochondrial genome sequences. Mol Phylogenet Evol. 2005;34:134–46. 10.1016/j.ympev.2004.09.003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
GO annotation from EggNOG of eel duplication gene families shared by freshwater eels. Including: “Orthogroup”, “Descriptions”, “Functional annotations”, “GO categories”, and “GO IDs”.
(CSV)
(ZIP)
Data Availability Statement
The raw RNA-sequencing reads from forebrain, pituitary, and testis samples from European eel (Anguilla anguilla) have been deposited at GenBank (http://www.ncbi.nlm.nih.gov/genbank) under accession no. SRP126643.