Abstract
Lc, a member of the maize (Zea mays) R/B gene family, encodes a basic helix-loop-helix transcriptional activator of the anthocyanin biosynthetic pathway. It was previously shown that translation of the Lc mRNA is repressed by a 38-codon upstream open reading frame (uORF) in the 5′ leader. In this study, we report that a potential hairpin structure near the 5′end of the Lc mRNA also represses downstream translation in the rabbit reticulocyte in vitro translation system and in transient transformation assays. Base pairing of the hairpin is important for repression because its destabilization increases translation of the uORF and the downstream ORF. However, translation of the uORF is not required for the hairpin-mediated repression. Instead, the uORF and the 5′-proximal hairpin mediate two independent levels of repression. Although the uORF represses downstream translation due to inefficient reinitiation of ribosomes that translate uORF, the hairpin inhibits ribosome loading at the 5′ end of the mRNA.
Most eukaryotic mRNAs are translated according to the ribosome scanning model (for review, see Kozak, 1999). In this model translational initiation commences with the binding of preinitiation complex (the 40S ribosomal subunit with associated factors) to the 5′ cap and the subsequent linear scanning of ribosomes to an AUG codon. When an AUG codon with favorable sequence context is encountered, the 40S subunit is joined by the 60S ribosomal subunit and polypeptide synthesis initiates. Evidence supporting this model is that sequence or structural features of 5′ leaders, including upstream AUGs and secondary structures, influence translational efficiency.
The effect of mRNA secondary structure on translation has been studied in mammalian cells by introducing synthetic hairpins into 5′ leaders (for review, see Kozak, 1999). The magnitude of the effect on translation depends on the stability and position of the hairpin. Although very stable structures within the leader (ΔG ≥ −50 kcal/mol) completely block ribosome scanning, a moderate hairpin (−30 kcal/mol) located near the 5′ end repressed translation by influencing the binding of the preinitiation complex to the mRNA (Kozak, 1986, 1989, 1998). In contrast with the usually inhibitory effects of secondary structures on translation, a −19 kcal/mol hairpin positioned 14 nucleotides downstream of an AUG codon was found to enhance translational initiation, probably by pausing ribosomes over the AUG codon, thereby favoring initiation (Kozak, 1990).
The 235-nucleotide leader of the maize (Zea mays) R gene Lc contains a 38-codon upstream open reading frame (uORF) that mediates translational repression of a downstream ORF (Fig. 1; Wang and Wessler, 1998). R genes encode myc-like transcriptional activators that control the temporal and spatial distribution of anthocyanin pigments (Ludwig et al., 1989). Although the tissue-specific expression pattern of Lc appears to be determined solely at the transcriptional level (Ludwig et al., 1989, 1990), it has been hypothesized that translational control evolved to prevent overexpression of the R protein (Damiani and Wessler, 1993).
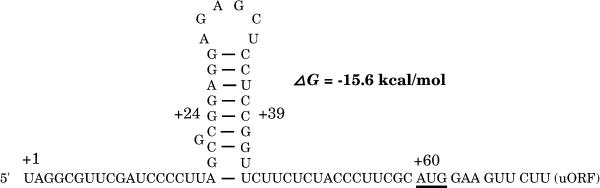
Figure 1.
A potential hairpin structure in the 5′ leader of Lc mRNA. The ΔG value of the hairpin was calculated according to Tinoco et al. (1973). Lc mRNA is numbered according to Ludwig et al. (1989) with the uORF initiation codon underlined. A G24→C substitution reduces the ΔG value from −15.6 to −5.4 kcal/mol, and a subsequent C39→G change restores the ΔG value.
In a previous study (Wang and Wessler, 1998) an in vitro assay system was utilized to visualize and quantify the 38-amino acid uORF peptide. Translation of the uORF was shown to be required for repression as an increase in uORF translation resulted in a decrease in downstream reporter gene product. Repression was unaffected by minor or major changes in the uORF coding region, suggesting that the uORF peptide itself did not mediate repression. Rather, repression is due to inefficient reinitiation of ribosomes that translate the uORF. This effect is mediated in some unknown way by the intercistronic sequence downstream of the uORF.
Here we report that translation of Lc mRNA is also repressed by a hairpin structure in the leader. Previous computer-assisted analyses indicated that the Lc leader might form a complex secondary structure with predicted ΔG value of −18 kcal/mol (Consonni et al., 1993; Damiani and Wessler, 1993). One feature of the secondary structure is a 25-nucleotide hairpin that is located 18 nucleotides from the 5′ end and has a ΔG value of −15.6 kcal/mol (Fig. 1; calculated according to Tinoco et al., 1973). The moderate stability of the hairpin and its proximity to the 5′ end suggested that it might influence translational initiation. In this study we demonstrate that base pairing within the hairpin acts in an additive manner with uORF to reduce translation of the Lc gene.
RESULTS
A Five-Nucleotide Leader Deletion Increases Downstream Reporter Gene Expression
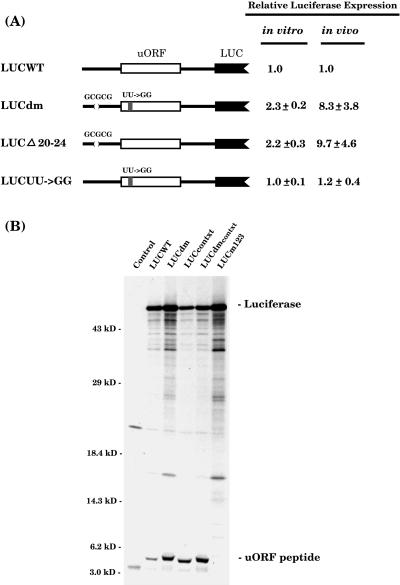
The effect of 5′-leader mutations on downstream reporter gene expression was previously assayed in the rabbit reticulocyte in vitro translation system and after bombardment into maize cells (the in vivo assay; Wang and Wessler, 1998). Construct LUCdm has two mutations: a five-nucleotide deletion near the 5′ end and a two-nucleotide substitution in the uORF. LUCdm yielded 2.3-fold more luciferase activity in vitro and 8.3-fold more activity in vivo than did the wild-type construct, LUCWT (Fig. 2A). Deletion of the nucleotides GCGCG at positions +20 to +24 is predicted to reduce stability of the potential 5′ hairpin (ΔG) from −15.6 to −5.4 kcal/mol.
Figure 2.
The effect of leader mutations on luciferase translation in vivo and in vitro. A, Constructs and their relative luciferase expression in the rabbit reticulocyte translation system (in vitro) and after bombardment into maize cells (in vivo). The parentheses symbolize the deleted five nucleotides, GCGCG, discussed in the text and a shaded box in the uORF (white box) represents the two-nucleotide substitution (UU→GG). The uORF-Luc chimeric gene was under control of the SP6 promoter for in vitro transcription. For in vivo assays, the chimeric gene was transferred to the vector pJD300, which has the cauliflower mosaic virus 35S promoter and the 3′ nopaline synthase terminator (Luehrsen et al., 1992). In both assays, the relative expression level of LUCWT was set at 1.0. Each value (±sd) in this and all subsequent figures represents the average of at least four independent assays for the in vitro data and eight bombardments for the in vivo data. B, SDS-PAGE analysis of 35S-Met-labeled in vitro translation products. RNA was not added in the control reaction. Numbers at left indicate migration of size markers in kilodaltons. The 61-kD luciferase protein and the 4.6-kD uORF peptide are indicated.
To determine whether the 5-nucleotide deletion or the 2-nucleotide uORF mutation was responsible for the increased translation, leaders containing the deletion or the uORF mutation (LUCUU→GG) were constructed and assayed. As shown in Figure 2A, whereas LUCΔ20–24 yielded 2.2-fold more luciferase activity in vitro and 9.7-fold more activity in vivo than did LUCWT, expression of LUCUU→GG was virtually identical to the wild-type construct. These results indicated that the deletion was solely responsible for the increased translation of LUCdm. This is consistent with previous results showing that minor or major changes in the amino acid sequence of the uORF had no effect on repression of the downstream ORF (Wang and Wessler, 1998).
The Leader Deletion Increases uORF Translation
RNA secondary structures such as the Lc hairpin have been shown to reduce translational initiation by interfering with ribosome loading (Kozak, 1989, 1994). If the Lc hairpin acts in this way, the weakening of the hairpin by deletion should enhance translation of the uORF and the downstream luciferase ORF.
To determine if more uORF peptide was yielded by LUCdm than by LUCWT, the products of in vitro translation of these constructs were visualized (Fig. 2B) and quantified (Table I). Although both constructs encode the approximately 5-kD uORF peptide, LUCdm yielded approximately 2-fold more peptide than LUCWT (Table I). In a similar manner, LUCdm yielded over 2-fold more luciferase protein (Fig. 2B; Table I).
Table I.
Quantitative analysis of uORF peptide and luciferase protein during in vitro translation
| RNA | Relative Amount of uORF Peptidea | Relative Amount of Luciferasea | Molar Ratio of uORF Peptide to Luciferase |
|---|---|---|---|
| LUCWT | 133 ± 10 | 100 | 1.3 ± 0.1 |
| LUCcontxt | 212 ± 17 | 59 ± 5 | 3.6 ± 0.5 |
| LUCdm | 265 ± 13 | 219 ± 12 | 1.2 ± 0.1 |
| LUCdmcontxt | 384 ± 27 | 103 ± 8 | 3.7 ± 0.3 |
| LUCWTfus | –b | 168 ± 8 | – |
| LUCdmfus | – | 327 ± 21 | – |
Values have been adjusted for the no. of labeled methionine residues in each product with the relative molar amount of luciferase in LUCWT set as 100. Each value (±sd) represents the average of at least four independent SDS-PAGE analyses.
(–), Does not apply to this RNA.
It was previously demonstrated that the sequence context of the uORF initiation codon was suboptimal for translation in the rabbit reticulocyte in vitro system (Wang and Wessler, 1998). When the uORF context was improved to match the eukaryotic consensus, the resulting construct, LUCcontxt, yielded almost 2-fold more uORF peptide, but less luciferase protein than did LUCWT (Fig. 2B; Table I). The molar ratio of uORF peptide to luciferase of LUCcontxt was approximately 3-fold higher than that of LUCWT, indicating that the uORF of LUCcontxt was more efficiently recognized by scanning ribosomes than that of LUCWT. In contrast, the 5-nucleotide deletion of LUCdm increased translation of uORF peptide and luciferase protein when compared with LUCWT (Table I). However, the molar ratio of uORF peptide to luciferase of LUCdm was virtually the same as that of LUCWT. To determine if this ratio of LUCdm could still be altered by the context change, the sequence context of the uORF initiation site of LUCdm was improved to match the eukaryotic consensus. As shown in Figure 2B and Table I, the double mutant LUCdmcontxt yielded more uORF peptide, but less luciferase than did LUCdm. This mutation led to an increase in the molar ratio of uORF peptide to luciferase when compared with LUCcontxt. Therefore, the increased translation of uORF peptide of LUCdm is not due to more efficient recognition of the uORF by scanning ribosomes.
The uORF Is Not Required for This Mode of Repression
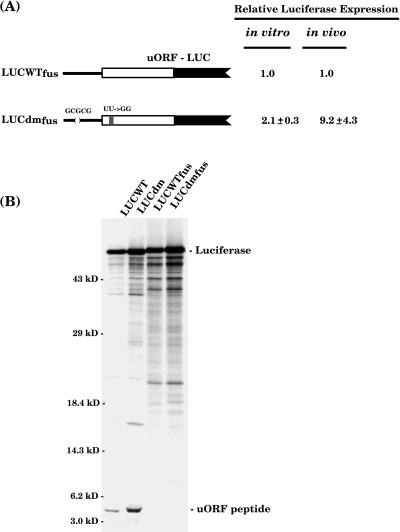
From the data presented above, the 5′-proximal hairpin of Lc appears to mediate translational repression by reducing ribosome loading. However, interpretation of these data is complicated by the presence of uORF. To determine if the 5-nucleotide deletion was sufficient to derepress reporter gene translation directly, the uORF-coding region was fused in frame to the downstream luciferase ORF. This was accomplished by deleting the intercistronic sequence along with the initiation codon of luciferase (Fig. 3A). The monocistronic constructs LUCWTfus and LUCdmfus were derived from LUCWT and LUCdm, respectively. When translated in vitro, LUCdmfus yielded 2.1-fold more luciferase activity than LUCWTfus. In addition, both constructs yielded the predicted fusion polypeptide, which should be slightly larger than the 61-kD luciferase polypeptide (Fig. 3B). In agreement with the luciferase activity measurement, LUCdmfus yielded approximately 2-fold more fusion protein than did LUCWTfus (Table I). When assayed following bombardment into maize cells, LUCdmfus yielded 9.2-fold more luciferase activity than did LUCWTfus (Fig. 3A). Therefore, the nucleotide alterations of LUCdm enhance reporter gene translation, irrespective of the presence of uORF.
Figure 3.
The effect of the leader deletion on translation does not require the presence of uORF. A, Luciferase activity measurements of the fusion constructs. The uORF-coding region (white box) was fused in frame to the luciferase ORF (black box). B, SDS-PAGE analysis of the 35S-Met-labeled fusion proteins following in vitro translation. Numbers at right indicate positions of size markers in kilodaltons.
Base Pairing of the 5′ Hairpin Is Required for Repression
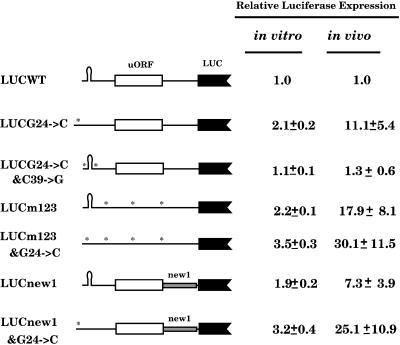
The 5-nucleotide deletion of LUCdm may increase downstream translation by disrupting the 5′ hairpin structure. To determine whether the base pairing of the 5′ hairpin correlates with repression, constructs harboring mutations that disrupt and restore the secondary structure were assayed. Of particular interest was the G at position +24 (Fig. 1). A G24→C substitution is expected to weaken the 5′ hairpin structure (from ΔG = −15.6 to −5.4 kcal/mol). As shown in Figure 4, the G24→C substitution increased downstream luciferase expression by 2.1-fold in the rabbit reticulocyte in vitro translation system and about 11-fold after bombardment into maize cells when compared with LUCWT. When the putative base pairing between nucleotides +24 and +39 was restored by a compensatory mutation (C39→G), translation was again repressed in vitro and in vivo (Fig. 4, LUCG24→C&C39→G versus LUCWT).
Figure 4.
The effect of additional mutations in the 5′ hairpin and uORF on luciferase translation. The new intercistronic sequence in LUCnew1 is represented as a shaded box. Each asterisk represents a single point mutation. The relative expression level of LUCWT was set at 1.0.
The 5′ Hairpin Acts Independently of the uORF
Taken together, the data indicate that the Lc leader mediates two levels of translational repression: one by the uORF and the other by the 5′ hairpin. Ribosomes that translate uORF reinitiate inefficiently due to some unknown feature of the intercistronic sequence. Elimination of the uORF by point mutations (LUCm123, Fig. 4) or replacement of the Lc intercistronic sequence by a random sequence (LUCnew1, Fig. 4) derepressed luciferase translation in vitro and in vivo (Wang and Wessler, 1998). In that study all of the constructs had the identical 60 nucleotides preceding uORF (including the 5′ hairpin structure).
To determine whether the inhibitory effect of the hairpin on translation is additive with the uORF-mediated repression, the G24→C mutation was introduced into LUCm123 and LUCnew1, resulting in the double mutants LUCm123&G24→C and LUCnew1&G24→C, respectively. As shown in Figure 4, the double mutant LUCm123&G24→C yielded 3.5-fold more luciferase activity than did LUCWT in vitro, whereas the construct with each single mutation (m123 or G24→C) increased luciferase translation approximately 2-fold relative to LUCWT. When the same constructs were bombarded into maize cells, the activity of LUCm123&G24→C (30.1-fold relative to LUCWT) was approximately the sum of LUCm123 (17.9-fold) and LUCG24→C (11.1-fold). The double mutant LUCnew1&G24→C also yielded more luciferase activity than LUCnew1 or LUCG24→C (Fig. 4). LUCnew1&G24→C yields 3.2-fold more luciferase activity than did LUCWT in vitro, and 25-fold more activity than LUCWT after bombardment into maize cells. The new1 mutation increased luciferase translation by 1.9-fold in vitro and 7.3-fold in vivo relative to LUCWT.
DISCUSSION
Evidence is presented that a potential hairpin in the 5′ leader of the maize Lc mRNA represses downstream translation in vitro and in maize cells. Mutations that disrupt the hairpin increased downstream reporter gene expression in the presence or absence of the uORF. When tested with dicistronic constructs, disruption of the hairpin increased translation of the uORF and the downstream ORF, suggesting that the hairpin reduced ribosome loading.
It was previously demonstrated that the 38-codon uORF in the 5′ leader of Lc mRNA represses downstream translation (Wang and Wessler, 1998). Ribosomes that translate the uORF reinitiate inefficiently due to some unknown feature of the intercistronic sequence. Inefficient reinitiation is also shown to be responsible for the uORF-mediated repression of the mammalian HER-2 oncogene (Child et al., 1999) and the Saccharomyces cerevisiae GCN4 gene (Grant and Hinnebusch, 1994). For the HER-2 uORF, inefficient reinitiation may be partially due to the short intercistronic spacing (5 nucleotides), whereas the high G + C content downstream of the GCN4 uORF4 appears to be responsible for the inefficient reinitiation. However, in both cases the exact mechanism of inefficient reinitiation is still unclear.
The data presented in this study indicate that translation of Lc mRNA is also repressed by the presence of the hairpin near the 5′ end. Evidence in support of the independence of the hairpin and uORF mechanisms is as follows. First, ribosomes that translate the uORF of LUCdm still reinitiate inefficiently (Table I). LUCWT and LUCdm yielded virtually identical molar ratios of uORF peptide to luciferase, suggesting that the ratio of scanning versus reinitiating ribosomes was not altered by the disruption of the 5′ hairpin. When the sequence context of the uORF initiation codon of LUCdm was improved to match the eukaryotic consensus, the resulting construct, LUCdmcontxt, yielded more uORF peptide, but less luciferase protein than did LUCdm. Second, the intercistronic sequence is not required for the hairpin to repress downstream translation. Deletion of the intercistronic sequence in the fusion constructs (LUCWTfus and LUCdmfus) had no effect on translational repression (Figs. 2A and 3A). Third, repression by the uORF and by the hairpin was shown to be additive. Luciferase activity could be increased by disrupting the 5′ hairpin (LUCG24→C) or by replacing the intercistronic sequence (LUCnew1) relative to LUCWT (Fig. 4). Furthermore, the luciferase activity of the double mutant, LUCnew1&G24→C, was approximately equal to the sum of the activities of LUCnew1 and LUCG24→C in vitro and in vivo.
We propose that the hairpin structure represses downstream translation by reducing ribosome loading at the 5′ end of Lc mRNA (Fig. 5). One prediction of this model is that disruption of the hairpin should increase translation of the uORF and the downstream ORF when dicistronic RNAs are assayed. This is in fact what was observed in the quantitative analysis of uORF peptide relative to luciferase protein (Table I). When translated in vitro, the wild-type dicistronic RNA (LUCWT) yielded 100 molecules of luciferase for every 133 molecules of uORF peptide. Of the 100 molecules of luciferase, it was shown previously that 60 are translated by ribosomes scanning past uORF, whereas 40 are due to reinitiation (Wang and Wessler, 1998). In other words, only 40 of 133, or 30%, of the ribosomes that translate uORF reinitiate downstream. Let us assume that deletion of the hairpin increases ribosome loading by 2-fold, thereby doubling the number of ribosomes available for translating uORF. Because LUCWT yields 133 molecules of uORF peptide, a 2-fold increase in ribosomes for LUCdm should result in approximately 266 molecules of uORF peptide. A very similar value (265) was observed for translation of uORF peptide from LUCdm (Table I). Ribosomes that scan past the uORF should also be increased by 2-fold, resulting in translation of 120 molecules of luciferase. Since 30% of the ribosomes that translate uORF reinitiate downstream, 30% of 266, or 80, molecules of luciferase would be synthesized by the reinitiating ribosomes. Thus, a 2-fold increase in ribosome loading for LUCdm would increase the synthesis of luciferase to 200 molecules (120 from leaky scanning and 80 from reinitiation). In this study 219 molecules of luciferase were translated from LUCdm (Table I).
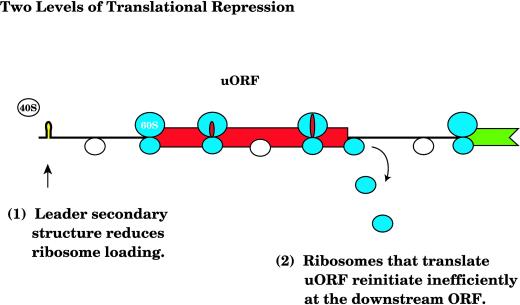
Figure 5.
Model of translational repression of Lc. The Lc uORF is represented as a red box, and the downstream ORF is represented as a green box. Scanning ribosomes (40S) are represented as white circles, and ribosomal subunits (40S and 60S) that are translating or have translated the uORF are represented as blue-shaded circles. The nascent uORF peptide is represented by red ovals and the hairpin is yellow.
Under certain circumstances sequence changes may alter mRNA stability, which can indirectly affect translation. However, it is unlikely that the observed repression or derepression in this study is due to changes in mRNA stability. First, RNAs were stable during the course of the in vitro translation reactions as assayed by RNA gel blots, and luciferase activity showed a linear increase during at least the first 30 min of incubation (data not shown). Second, several constructs, including LUCm123 and LUCnew1, were transformed into rice, and their increased expression of luciferase was shown to result directly from more efficient translation of mutant mRNAs rather than differences in mRNA stability (Wang and Wessler, 1998). Third and perhaps most significantly, the correlation between repression of luciferase expression and stability of the hairpin structure demonstrated by using the constructs LUCG24→C and LUCG24C&C39→G in the bombardment assays (Fig. 4) cannot be easily explained in terms of RNA stability changes.
Previous studies have shown that secondary structure in the 5′ leader inhibits translation by influencing the binding of 40S ribosomal subunits to the 5′ end of an mRNA (Kozak, 1986, 1989, 1994; Baim and Sherman, 1988). In mammalian cells, a hairpin of −30 kcal/mol was found to drastically inhibit translation when inserted within the first 12 nucleotides of the preproinsulin gene (Kozak, 1989). When the same −30 kcal/mol hairpin was repositioned 52 nucleotides from the 5′ end, however, it no longer inhibited translation (Kozak, 1989). This result was interpreted to mean that as long as the 40S subunit engages the mRNA at the 5′ end, the subsequent migration of the preinitiation complex could disrupt base-paired structures that occur downstream. In S. cerevisiae, a hairpin of −7.6 kcal/mol inserted near the 5′ end of the CYC1 mRNA (11 nucleotides downstream of the 5′ cap) reduced the synthesis of iso-1-cytochrome c by approximately 10-fold (Baim and Sherman, 1988).
Very few natural 5′ leaders have been studied in detail. A −10 kcal/mol hairpin present in the 5′ leader of the mammalian α-globin mRNA was shown to repress translation by approximately 2-fold in the rabbit reticulocyte translation system (Kozak, 1994). The results presented here suggest that the natural 5′ leader of the maize Lc mRNA forms a −15.6 kcal/mol hairpin structure that represses translation by approximately 2-fold in the rabbit reticulocyte in vitro translation system and approximately 10-fold after bombardment into maize cells. It is unknown at this time whether the greater magnitude of repression in maize cells may imply that plant 40S subunits are more sensitive to 5′ hairpins than mammalian ribosomes. In an alternate manner, since one mRNA must compete with others for the limited translational machinery in cells, presence of the 5′ hairpin may impose a competitive disadvantage on the mRNA and result in more repression in vivo than in vitro.
In conclusion, the results presented suggest that secondary structure is of importance in determining the translational efficiencies of mRNAs in higher plants. Even relatively weak hairpins like the one described in this study can influence translational efficiency when located in the region immediately downstream from the 5′ cap.
MATERIALS AND METHODS
Plasmid Construction
Construction of LUCWT was previously described (Wang and Wessler, 1998). In LUCWT, 186 nucleotides of the Lc 5′ leader containing the uORF and the hairpin were fused to the luciferase-coding region of vector pGEM-luc. The fortuitous mutant, LUCdm, was isolated during mutagenesis of the wild-type Lc leader in LUCWT. In the mutant leader of LUCdm, the five nucleotides from position +20 to +24 (GCGCG) were deleted, and the two nucleotides at +70 and +71 of Lc mRNA (UU) were substituted with GG (Fig. 1). LUCΔ20–24 and LUCUU→GG were constructed by mutating the wild-type Lc leader in LUCWT with the mutagenic oligonucleotides 5′-GCAAGC- TTGAGGAGAGCTCCTCCGGTTCTTCTC-3′ and 5′-CGA- AGCAATGCCCGAACTTCCATG-3′, respectively. Site-directed mutagenesis was carried out using PCR as described (Sarkar and Sommer, 1990). LUCdmcontxt was derived from LUCdm using the mutagenic oligonucleotide 5′-TCTCTACCCTTACCATGGAAGTTC-3′. For LUCWTfus and LUCdmfus, the sequence from the uORF stop codon to the initiation codon of the downstream luciferase ORF has been deleted using the mutagenic oligonucleotide 5′-CC- TTTCTTTATGTTTTTGGCGTCTTCTCTACTGATCATCAGACGATGCCTCG-3′. LUCWTfus and LUCdmfus were derived from LUCWT and LUCdm, respectively.
The mutations in constructs LUCcontxt, LUCm123, and LUCnew1 were described (Wang and Wessler, 1998). The mutagenic oligonucleotide, 5′-CTAAGCTTCCTTA-GCGCCGAGGAGAGCTCCTCCGGTTC-3′, was used to derive LUCG24→C from LUCWT, LUCm123&G24→C from LUCm123, and LUCnew1&G24→C from LUCnew1. LUCG24→C&C39→G was constructed by mutating the wild-type Lc leader in LUCWT with the mutagenic oligonucleotide 5′- CTAAGCTTCCTTAGCGCCGAGGAGAGC TCCTCGGGTTCTTCTCTACCCTT-3′.
For in vitro assays the chimeric Lc leader-luciferase gene in LUCWT or the mutant derivatives was transcribed from the SP6-derived vector, pGEM-luc (Promega, Madison, WI). For transient expression in maize (Zea mays) cells, the chimeric gene was transferred to the vector pJD300 as described (Wang and Wessler, 1998). The vector pJD300 has the cauliflower mosaic virus 35S promoter and the 3′ nopaline synthase terminator (Luehrsen et al., 1992).
In Vitro Transcription and Translation
The pGEM-luc-derived plasmids were linearized with XhoI and capped RNA was synthesized using the Riboprobe Core System-SP6 kit (Promega) according to the manufacturer. RNA samples were analyzed by RNA gel blots (Sambrook et al., 1989), probed with a 1.1-kb EcoRI fragment of the luciferase gene in pGEM-luc (Promega), and quantified by PhosphorImager scan (Molecular Dynamics, Sunnyvale, CA). Luciferase RNA (1.0 μg/μL, Promega) was included to standardize the RNA concentrations of the samples.
Each RNA was translated in the rabbit reticulocyte lysate in vitro translation system (Promega) at a final RNA concentration of 4 μg/μL. Reactions were performed at 30°C for 30 min and then frozen on dry ice. Luciferase activity increased linearly during the first 30 min of incubation (data not shown). For SDS-PAGE analysis of the in vitro translation products, 35S-Met (Amersham, Buckinghamshire, UK) was included in the translation reactions at a final concentration of 0.8 mCi/mL.
SDS-PAGE Analysis
Samples from in vitro translation reactions were fractionated on a 14% (w/v) polyacrylamide resolving gel (Sambrook et al., 1989). Gels were treated with Entensify (DuPont, Wilmington, DE) prior to autoradiography. Relative amounts of luciferase and uORF peptide were determined by using the PhosphorImager (Molecular Dynamics) and adjusted for the number of labeled Met residues in each product.
Transient Transformation Assay in Maize Cells
Maize suspension cells used for bombardment were prepared as described previously (Goff et al., 1990). Plasmid DNAs were precipitated onto 1.0-μm gold particles (60 mg/mL; Klein et al., 1989) and bombarded into maize aleurone cells with the Biolistic PDS-1000 (DuPont). Each plate of cells was cobombarded with 0.4 μg of a luciferase-containing plasmid and 0.4 μg of pAdh1CAT (also called pAI1CN [Callis et al., 1987]), which contains a chloramphenical acetyl transferase (CAT) gene under the control of the maize alcohol dehydrogenase (Adh1) promoter. After bombardment the cells were incubated on Murashige and Skoog media (Sigma, St. Louis) at 28°C for 48 h prior to enzyme assays.
Enzyme Assays
In vitro and in vivo expression levels were determined by luciferase activity assays as described (Callis et al., 1987). In vitro translation reactions were diluted 10-fold in 20 mm Tricine (N-[2-hydroxy-1,1-Bis(hydroxymethyl)ethyl]glycine), pH 7.8, and 1 μL of the diluted sample was assayed with a model 3010 Luminometer (Analytic Scientific Instruments, Alameda, CA). Luciferase activity was expressed as the number of light units detected in the first 10 s of reaction at room temperature. Relative luciferase expression was calculated by dividing the luciferase activity for each construct by the activity of LUCWT.
Bombarded maize cells were ground in 350 μL of 100 mm KPO4 (pH 7.80) and 1 mm DTT at 4°C. After centrifugation, 25 and 10 μL of the supernatant were assayed for CAT and luciferase activity, respectively. CAT activity was expressed as the ethyl acetate soluble cpm count (Sleigh, 1986). Luciferase expression levels were adjusted by the CAT activity and were expressed as the ratio of luciferase to CAT activity. The relative luciferase expression was calculated by dividing the average luciferase/CAT ratio for each construct by that of LUCWT.
ACKNOWLEDGMENT
We thank Dr. Vicki Chandler (University of Arizona) for the maize suspension cells.
Footnotes
This work was supported by the U.S. Department of Energy (grant to S.R.W.).
We dedicate this paper to the memory of Dr. Alan Jaworski.
LITERATURE CITED
- Baim SB, Sherman F. mRNA structures influencing translation in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:1591–1601. doi: 10.1128/mcb.8.4.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callis J, Fromm M, Walbot V. Introns increase gene expression in cultured maize cells. Genes Dev. 1987;1:1183–1200. doi: 10.1101/gad.1.10.1183. [DOI] [PubMed] [Google Scholar]
- Child SJ, Miller MK, Gabelle AP. Translational control by an upstream open reading frame in the HER-2/neu transcript. J Biol Chem. 1999;274:24335–24341. doi: 10.1074/jbc.274.34.24335. [DOI] [PubMed] [Google Scholar]
- Consonni G, Geuna F, Gavazzi G, Tonelli C. Molecular homology among members of the R gene family in maize. Plant J. 1993;3:335–346. doi: 10.1111/j.1365-313x.1993.tb00185.x. [DOI] [PubMed] [Google Scholar]
- Damiani J, RD, Wessler SR. An upstream reading frame represses expression of Lc, a member of the R/B family of maize transcriptional activators. Proc Natl Acad Sci USA. 1993;90:8244–8248. doi: 10.1073/pnas.90.17.8244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SA, Klein TM, Roth BA, Fromm ME, Cone KC, Radicella JP, Chandler VL. Transactivation of anthocyanin biosynthetic genes following transfer of B-regulatory genes into maize tissues. EMBO J. 1990;9:2517–2522. doi: 10.1002/j.1460-2075.1990.tb07431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CM, Hinnebusch AG. Effect of sequence context at stop codons on efficiency of reinitiation in GCN4 translational control. Mol Cell Biol. 1994;14:606–618. doi: 10.1128/mcb.14.1.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein TM, Roth BA, Fromm ME. Regulation of anthocyanin biosynthetic genes introduced into intact maize tissues by microprojectiles. Proc Natl Acad Sci USA. 1989;86:6681–6685. doi: 10.1073/pnas.86.17.6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc Natl Acad Sci USA. 1986;83:2850–2854. doi: 10.1073/pnas.83.9.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Circumstances and mechanisms of inhibition of translation by secondary structure in eukaryotic mRNAs. Mol Cell Biol. 1989;9:9134–9142. doi: 10.1128/mcb.9.11.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc Natl Acad Sci USA. 1990;87:8301–8305. doi: 10.1073/pnas.87.21.8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Features in the 5′-non-coding sequences of rabbit a and b-globin mRNAs that affect translational efficiency. J Mol Biol. 1994;235:95–110. doi: 10.1016/s0022-2836(05)80019-1. [DOI] [PubMed] [Google Scholar]
- Kozak M. Primer extension analysis of eukaryotic ribosome-mRNA complexes. Nucleic Acids Res. 1998;26:4853–4859. doi: 10.1093/nar/26.21.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- Ludwig SR, Bowen B, Beach L, Wessler SR. A regulatory gene as a novel visible marker for maize transformation. Science. 1990;247:449–450. doi: 10.1126/science.247.4941.449. [DOI] [PubMed] [Google Scholar]
- Ludwig SR, Habera LF, Dellaporta SL, Wessler SR. Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional acitvators and contains the myc-homology region. Proc Natl Acad Sci USA. 1989;86:7092–7096. doi: 10.1073/pnas.86.18.7092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luehrsen KR, de Wet JR, Walbot V. Transient expression analysis in plants using firefly luciferase reporter gene. Methods Enzymol. 1992;216:397–414. doi: 10.1016/0076-6879(92)16037-k. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sarkar G, Sommer SS. The “megaprimer” method of site-directed mutagenesis. BioTechniques. 1990;8:404–407. [PubMed] [Google Scholar]
- Sleigh MJ. A non-chromatographic assay for expression of the chloramphenicol acetyl transferase gene in eukaryotic cells. Anal Biochem. 1986;156:251–256. doi: 10.1016/0003-2697(86)90180-6. [DOI] [PubMed] [Google Scholar]
- Tinoco IJ, Borer PN, Dengler B, Levine MD, Uhlenbeck OC, Crothers DM, Gralla J. Improved estimation of secondary structure in ribonucleic acids. Nature New Biol. 1973;246:1733–1745. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Wang L, Wessler SR. Inefficient reinitiation is responsible for upstream open reading frame-mediated translational repression of the maize R gene. Plant Cell. 1998;10:1733–1745. doi: 10.1105/tpc.10.10.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]