Abstract
Extracellular vesicles such as exosomes contain several types of transcripts, including mRNAs and micro RNAs (miRNAs), and have emerged as important mediators of cell-to-cell communication. Exosome-like vesicles were identified in the ovarian follicles of several mammalian species. Although the miRNA contents have been extensively characterized, the detailed investigation of their mRNA profiles is lacking. Here, we characterize the mRNA profiles of exosome-like vesicles in ovarian follicles in a pig model. The mRNA contents of the exosome-like vesicles isolated from porcine follicular fluid were analyzed and compared with those from mural granulosa cells (MGCs) using the Illumina HiSeq platform. Bioinformatics studies suggested that the exosomal mRNAs are enriched in those encoding proteins involved in metabolic, phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K) -protein kinase B (AKT), and mitogen-activated protein kinase (MAPK) pathways. While the mRNA profile of the exosome-like vesicles resembled that of MGCs, the vesicles contained mRNAs barely detectable in MGCs. Thus, while the majority of the vesicles are likely to be secreted from MGCs, some may originate from other cell types, including theca cells and oocytes, as well as the cells of non-ovarian organs/tissues. Therefore, the mRNA profiles unveiled several novel characteristics of the exosome-like vesicles in ovarian follicles.
Introduction
Exosomes are lipid bilayer vesicles of around 40–200 nm diameter produced by most cell types [1]. These vesicles contain several bioactive materials such as proteins, lipids, micro RNAs (miRNAs), and mRNAs with properties slightly different from their originating cells [2, 3]. Exosomes mediate cell-to-cell communication by transferring these molecules to target cells, wherein the transferred molecules may affect multiple biological processes [4–6]. Exosomes produced from several organs and tissues are readily detectable in the blood stream and may be considered as potential diagnostic markers of diseases such as cancers [7–9].
The presence of exosome-like vesicles in ovarian follicular fluids was first reported in mares [10]. Exosome-like vesicles isolated from equine follicular fluid contained several proteins and miRNAs, and were taken-up by granulosa cells both in vitro and in vivo. The miRNAs detected were predicted to target several signaling pathways, including the WNT, transforming growth factor beta (TGFβ), and mitogen-activated protein kinase (MAPK) pathways [10, 11]. These pathways are involved in the regulation of ovarian functions, including folliculogenesis, luteogenesis, and steroidogenesis [12–15]. The presence of exosome-like vesicles was also reported in follicular fluids of several other mammalian species, including human, bovine, and pig [16–19]. The miRNA content in the exosome-like vesicles from human and bovine has been studied [16, 17], and these miRNAs are known to target signaling pathways similar to those targeted by equine vesicles. Therefore, these vesicles may play a critical role in the regulation of ovarian functions via miRNA transfer [20].
In general, exosomes contain many other molecules aside from miRNAs, such as mRNAs, that can be delivered and translated into proteins in recipient cells [2, 21, 22]. The exosome-mediated transfer of mRNAs was first reported by Valadi and colleagues who demonstrated that exosomes are capable of shuttling mRNAs between mast cells [2]. In addition, cancer-derived exosomes carry matrix metalloproteinase 1 (MMP1) mRNA to induce apoptosis in recipient mesothelial cells [21]. Exosomes can deliver the mRNA encoding Cre recombinase to recipient cells to perform Cre-LoxP-mediated recombination in vivo and in vitro [22]. Therefore, mRNAs are also thought to be critical mediators of exosomal functions.
The miRNA contents of follicular exosome-like vesicles have been extensively characterized, and these studies provide an insight in the function of the vesicles during folliculogenesis in mammals; however, no report has described the complete characterization of the mRNA contents of exosome-like vesicles. Therefore, in the present study, we evaluated the characteristics of transcriptomic profile of mRNA content in exosome-like vesicles from ovarian follicles using pig as a model. The presence of the exosome-like vesicles in porcine follicular fluid (pFF) has been previously reported [19]. As mural granulosa cells (MGCs) are one of the most abundant cell types in ovarian follicles, the transcriptomic profile of the MGCs was also investigated to test whether MGCs are the main producers of the follicular exosome-like vesicles.
Materials and methods
Collection of pFF and MGCs
Porcine ovaries of prepubertal gilts were collected at a commercial slaughterhouse (Tokyo Shibaura Zouki, Co., Ltd., Tokyo, Japan) and transported to our laboratory at approximately 37°C in saline. pFF was collected from antral follicles (2–5 mm in diameter), as previously reported [23]. Great care was taken to avoid contamination of the fluid with blood.
MGCs were collected from antral follicles (2–5 mm in diameter), as previously reported [19].
Isolation of exosome-like vesicles from pFF
The fraction containing exosome-like vesicles was isolated from pFF, as previously reported [19]. In brief, pFF was first centrifuged at 2,000 ×g and 4°C for 30 min, followed by another step of centrifugation at 12,000 ×g and 4°C for 45 min. The sample was filtered through a 0.22-μm membrane (Merck Millipore, Darmstadt, Germany) to remove cells and debris. After the filtration, the exosomal fraction was extracted using a Total Exosome Isolation (from serum) reagent (Life Technologies, Inc. Carlsbad, CA, USA). The samples were immediately subjected to the next procedure.
Transmission electron microscopy observations of the exosomal fraction
The exosomal fraction isolated from pFF was observed with a transmission electron microscope (JEM-1010; JEOL, Tokyo, Japan) as previously reported [19].
Western blotting analysis
Western blotting analysis was conducted as previously reported [19]. The primary antibodies used were anti-HSC70 antibody (MAB2191; Abnova, Taipei, Taiwan), anti-CD63 antibody (sc-5725; Santa Cruz Biotechnology, Texas, USA), and anti-CYCS antibody (sc-13156), and the secondary antibodies used were horseradish peroxidase conjugated anti-rat IgG antibodies (81–9520; Invitrogen) and anti-mouse IgG antibodies (115-035-044; Jackson ImmunoResearch, West Grove, PA, USA). Signals were visualized using an Immunostar LD Kit (Wako, Tokyo, Japan) and the C-DiGit Blot Scanner and Image Studio for C-DiGit (LI-COR, Lincoln, NE, USA) according to the manufacturer’s protocols.
RNA profiling using a bioanalyzer
Total RNA was extracted from the exosomal fraction and MGCs using a ReliaPrep Cell Miniprep System (Promega K.K., Tokyo, Japan). The total RNA profiles were visualized by using an Agilent 2100 bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) with Agilent RNA6000 Pico Kit (Agilent Technologies) according to the manufacturer’s protocols.
RNA sequencing with Illumina HiSeq platform
The total RNA was extracted from three biologically independent samples of the exosomal fractions and MGCs, respectively, using a ReliaPrep Cell Miniprep System (Promega K.K.). Library construction, quality control, and sequencing were performed by Filgen Inc. (Aichi, Japan). A total amount of 3 μg of RNA per sample was used as an input material for the RNA sample preparations. mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. Sequencing libraries were generated using NEBNext Ultra RNA Library Prep Kit for Illumina (NEB, USA). The library preparations were sequenced on an Illumina HiSeq platform, and 150 base paired end reads were generated. The sequenced reads (raw reads) were subjected to several quality checks. In this step, clean data (clean reads) were obtained by removing reads containing adapter and poly-N (N represents the undetermined base) as well as low quality reads from raw data. The filtering process was as follows: (1) Remove reads containing adapters, (2) remove reads containing N > 10%, and (3) remove reads containing low-quality (Q score ≤ 5) base, which was over 50% of the total base. The data have been deposited in the Data Bank of Japan (DDBJ, http://www.ddbj.nig.ac.jp, data set DRA008080).
All the downstream analyses were based on the clean data using CLC Genomics Workbench version 11.0.1 (QIAGEN K.K., Tokyo, Japan) with its default parameters. Clean reads were trimmed to remove low-quality sequence (limit = 0.05) and ambiguous nucleotides (maximal two nucleotides allowed). The proceeded reads were aligned with the Sscrofa11.1 porcine genome annotated with genes and transcripts and to generate the gene expression values in the normalized form of reads per kilobase per million mapped reads (RPKM) [24]. The criteria used to determine the detected transcripts was an average RPKM value of more than 0.1 for three replicates.
Reverse-transcription and polymerase chain reaction (RT-PCR)
The total RNA was extracted from the exosomal fraction and MGCs using a ReliaPrep Cell Miniprep System. Total RNA was reverse transcribed using a ReverTraAce qPCR Master Mix with gDNA Remover (Toyobo, Osaka, Japan), and PCR was performed using a BIOTAQ DNA polymerase (Bioline Ltd., London, UK). Samples were denatured for 2 min at 95°C and incubated for 35 cycles under the following conditions: 95°C for 30 s, 55°C for 30 s, and 72°C for 2 min, followed by the final elongation for 5 min at 72°C. The PCR products were subjected to agarose gel electrophoresis. The PCR primers used were as follows: 5’-TTTTTCGCAACGGGTTTGCC-3’ and 5’-TGTGACAGATTTTTGGTCAAGTTGT-3’ for eukaryotic translation elongation factor 1 alpha 1 (EEF1A1; NM_001003662.1); 5’-GACTCCGCCTCTCAGCTATC-3’ and 5’-GCTTGAGTGTGAGCCTTTCG-3’ for ferritin light chain (FTL; NM_001244131.1).
RNA degradation analysis
2 μg/mL RNase A (NIPPON GENE Co., Ltd., Tokyo, Japan) and 2% Triton X-100 (Sigma-Aldrich Japan K.K. Tokyo, Japan) was added into pFF, and incubated at 37°C for 20 min. As a control treatment, the equal volume of phosphate buffered saline (PBS) was added to pFF instead of the reagent and incubated at 37°C for 20 min. After the incubation, pFF was subjected into the exosome-like vesicle isolation procedure, then RNA was extracted from the exosomal fractions as described above.
The effectiveness of RNase A treatment was examined by Real-time PCR reactions. Real-time PCR reactions were performed using a THUNDERBIRD qPCR Mix (Toyobo) and an ABI Step One Plus real-time PCR system (Applied Biosystems) according to the manufacturer’s protocols. The PCR primers used were as follows: 5’-ATGCGGTGGGATCGACAAAA-3’ and 5’-AGTTTGTCCAAGACCCAGGC-3’ for EEF1A1; 5’-GAAAATGCAAAACCAGCGCG-3’ and 5’-CTTCCATAGCGTCCTGGGTT-3’ for FTL. To avoid false-positive signals, dissociation-curve analyses were performed at the end of the analyses, and the PCR products were subjected to agarose gel electrophoresis to confirm the single amplification and sizes of the products.
Data analysis
The Database for Annotation, Visualization and Integrated Discovery (DAVID) bioinformatics resources was used for pathway analysis [25, 26]. Principal component analysis (PCA) was performed using the CLC Genomics Workbench, with the data sets of porcine tissue transcriptomes downloaded from NCBI GEO (S1 Table). The downloaded raw data were subjected to the filtering procedure before using for PCA. In addition to literature, tissue specificity of transcripts was investigated with PaGenBase [27] (http://bioinf.xmu.edu.cn/PaGenBase/index.jsp) by referring to the data sets for human [28–34], as porcine data sets were unavailable.
All experiments were repeated at least three times. Statistical analyses were conducted using Microsoft Excel (Microsoft) and the program Excel-Statistics (Social Survey Research Information Co., Ltd., Tokyo, Japan). The Tukey-Kramer test was used for multiple comparisons. A P-value < 0.05 was considered statistically significant.
Results
Validation of exosome-like vesicles in the exosomal fraction isolated from pFF
To validate the detection of exosome-like vesicles in pFF, we performed the electron microscopy observation (TEM), western blotting analysis, and bioanalyzer analysis. TEM observation demonstrated that round-shaped vesicles about 100 nm in diameter were observed in the exosomal fraction (Fig 1A). With western blotting analysis, well-known exosomal makers, CD63 molecule (CD63) and heat shock protein 70 (HSC70) [35], were detected in both the exosomal fraction and MGCs. On the other hand, cytochrome C (CYCS) was readily detected in the MGCs, but it was absent in the exosomal fraction, indicating that the exosomal fraction was not contaminated with apoptotic bodies or cell debris (Fig 1B). In addition, bioanalyzer analysis demonstrated that the RNAs isolated from the exosomal fraction were enriched in small RNAs, and the peaks of ribosomal RNAs were not observed (Fig 1C) [36]. Taken together, we concluded that the exosomal fraction was enriched in exosome-like vesicles.
Fig 1. Detection of exosome-like vesicles in the exosomal fraction isolated from pFF.
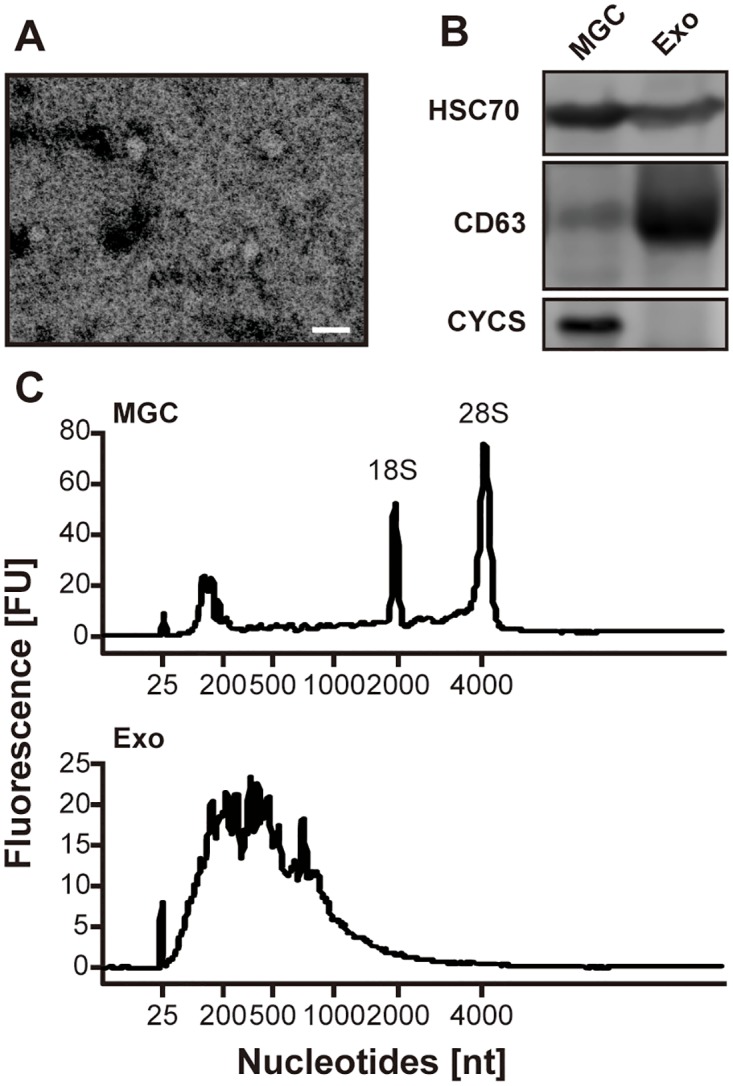
(A) Representative photograph of vesicles in the exosomal fraction isolated from pFF observed using transmission electron microscopy. The scale bar indicates 100 nm. (B) Western blotting analysis for HSC70, CD63, and CYCS. MGC, mural granulosa cell; Exo, exosomal fraction. (C) Representative electropherograms observed using bioanalyzer. FU, fluorescence intensity units; nt, nucleotides.
An overview of the RNA sequencing results
On an average, approximately 49 and 50 million reads were obtained from three independent biological replicates of the exosomal fraction and MGCs. Of those, about 33 million (67.52%) and 38 million (77.68%) paired reads were mapped to the porcine genome, respectively (S2 Table). Among the mapped fragments, approximately 7 million (43.17%) of the exosomal fraction and 12 million (62.70%) of MGCs were mapped to exons of known porcine transcripts. A total of 14,195 and 13,502 transcripts were detected in the exosomal fraction and MGCs, respectively, at an RPKM threshold of more than 0.1. Detailed information of independent samples is summarized in S2 Table.
Abundant mRNAs in the exosome-like vesicles of pFF
The top 30 annotated mRNAs in the exosomal fraction and MGCs are shown in Tables 1 and 2, respectively. EEF1A1 was the most abundant transcript in the exosomal fraction that also showed high abundance in MGCs. While the transcripts encoding ribosomal proteins (ribosomal proteins of the large subunit [RPL] and ribosomal protein of the small subunit [RPS]) were preferentially detected in the exosomal fraction, these transcripts were less abundant in MGCs. FAU, ubiquitin-like and ribosomal protein S30 fusion (FAU) and ubiquitin A-52 residue ribosomal protein fusion product 1 (UBA52) detected in the exosomal fraction also encode ribosomal protein-related products [37]. Therefore, the transcripts enriched in the follicular exosome-like vesicles seemed to encode ribosomal proteins, consistent with the results reported in human salivary exosomes [38].
Table 1. Top 30 most abundant mRNAs in exosome-like vesicles in pFF.
| Gene symbol | Ensemble ID | RPKM (Exosomal Fraction) |
RPKM (MGC) |
|---|---|---|---|
| EEF1A1 | ENSSSCG00000004489 | 5,088.21 | 4,271.57 |
| RPS27 | ENSSSCG00000006558 | 4,345.22 | 2,565.47 |
| RPL34 | ENSSSCG00000003930 | 3,173.29 | 1,406.04 |
| RPS7 | ENSSSCG00000027353 | 2,684.54 | 1,086.50 |
| RPS3 | ENSSSCG00000014855 | 2,468.42 | 935.21 |
| RPL13A | ENSSSCG00000003166 | 2,411.18 | 909.15 |
| RPS12 | ENSSSCG00000004177 | 2,384.40 | 1,187.95 |
| RPL31 | ENSSSCG00000008170 | 2,326.62 | 921.19 |
| RPS20 | ENSSSCG00000006249 | 2,321.79 | 1,211.61 |
| RPS23 | ENSSSCG00000014133 | 2,297.01 | 1,073.43 |
| RPS16 | ENSSSCG00000020817 | 2,250.29 | 721.35 |
| RPL18 | ENSSSCG00000025928 | 2,163.01 | 857.09 |
| RPS18 | ENSSSCG00000001502 | 2,123.72 | 1,140.27 |
| FTH1 | ENSSSCG00000014540 | 2,101.05 | 1,120.50 |
| UBA52 | ENSSSCG00000013907 | 2,070.60 | 1,068.80 |
| FAU | ENSSSCG00000013002 | 2,051.53 | 791.49 |
| RPL35 | ENSSSCG00000005595 | 1,924.86 | 744.56 |
| RPL34 | ENSSSCG00000009146 | 1,918.96 | 726.29 |
| RPL36 | ENSSSCG00000030010 | 1,884.65 | 861.65 |
| RPS28 | ENSSSCG00000013597 | 1,873.97 | 853.43 |
| RPLP1 | ENSSSCG00000004970 | 1,824.04 | 1,038.76 |
| RPLP2 | ENSSSCG00000012842 | 1,821.14 | 809.35 |
| RPL23A | ENSSSCG00000017768 | 1,811.04 | 1,241.32 |
| RPL13 | ENSSSCG00000024974 | 1,785.49 | 578.25 |
| RPS9 | ENSSSCG00000029785 | 1,703.30 | 565.85 |
| ACTG1 | ENSSSCG00000028355 | 1,698.27 | 1,325.06 |
| RPL17 | ENSSSCG00000029642 | 1,689.39 | 690.23 |
| FTL | ENSSSCG00000003153 | 1,644.42 | 428.01 |
| RPS25 | ENSSSCG00000015103 | 1,615.55 | 1,034.76 |
| RPL5 | ENSSSCG00000006899 | 1,612.40 | 735.82 |
Table 2. Top 30 most abundant mRNAs in porcine MGCs.
| Gene symbol | Ensemble ID | RPKM (Exosomal Fraction) |
RPKM (MGC) |
|---|---|---|---|
| COX3 | ENSSSCG00000018082 | 1,248.76 | 5,485.29 |
| COX2 | ENSSSCG00000018078 | 919.28 | 4,994.62 |
| EEF1A1 | ENSSSCG00000004489 | 5,088.21 | 4,271.57 |
| COX1 | ENSSSCG00000018075 | 980.54 | 3,863.89 |
| ATP6 | ENSSSCG00000018081 | 861.08 | 3,401.24 |
| RPS27 | ENSSSCG00000006558 | 4,345.22 | 2,565.47 |
| SERPINE2 | ENSSSCG00000016233 | 239.34 | 2,449.40 |
| INHA | ENSSSCG00000020771 | 624.58 | 2,377.75 |
| GPX3 | ENSSSCG00000017092 | 1,097.51 | 2,221.39 |
| RPS8 | ENSSSCG00000003930 | 3,173.29 | 1,406.04 |
| ACTG1 | ENSSSCG00000028355 | 1,698.27 | 1,325.06 |
| CYTB | ENSSSCG00000018094 | 277.78 | 1,284.82 |
| VIM | ENSSSCG00000011033 | 1,254.67 | 1,281.73 |
| RPL23A | ENSSSCG00000017768 | 1,811.04 | 1,241.32 |
| RPS20 | ENSSSCG00000006249 | 2,321.79 | 1,211.61 |
| RPS12 | ENSSSCG00000004177 | 2,384.40 | 1,187.95 |
| ACTB | ENSSSCG00000007585 | 1,005.93 | 1,164.69 |
| RPS18 | ENSSSCG00000001502 | 2,123.72 | 1,140.27 |
| INHBA | ENSSSCG00000021865 | 96.79 | 1,135.76 |
| FTH1 | ENSSSCG00000014540 | 2,101.05 | 1,120.50 |
| ENO1 | ENSSSCG00000022343 | 758.20 | 1,111.73 |
| RPS7 | ENSSSCG00000027353 | 2,684.54 | 1,086.50 |
| RPS23 | ENSSSCG00000014133 | 2,297.01 | 1,073.43 |
| UBA52 | ENSSSCG00000013907 | 2,070.60 | 1,068.80 |
| ND3 | ENSSSCG00000018084 | 240.18 | 1,039.76 |
| RPLP1 | ENSSSCG00000004970 | 1,824.04 | 1,038.76 |
| RPS25 | ENSSSCG00000015103 | 1,615.55 | 1,034.76 |
| CALR | ENSSSCG00000013746 | 189.64 | 1,005.75 |
| RPS29 | ENSSSCG00000005003 | 944.30 | 978.18 |
| RARRES1 | ENSSSCG00000028623 | 116.73 | 970.41 |
The other highly abundant mRNAs in the exosomal fraction were ferritin heavy chain 1 (FTH1), actin gamma 1 (ACTG1), and FTL, all of which also showed high expression in MGCs. As for MGCs, cytochrome c oxidase 3 (COX3) was the most abundant mRNA, and COX2 and COX1 were highly expressed. These COX transcripts were also detected in the exosomal fraction at a relatively high levels (RPKM of 980.54, 919.28, and 1248.76 for COX1, COX2, and COX3, respectively).
Detection of full-length mRNAs in the exosome-like vesicles
We tested whether follicular exosome-like vesicles carry full-length mRNAs. As shown in Fig 2A, the bands with the expected sizes of EEF1A1 and FTL were detected in the exosomal fraction by RT-PCR after the amplification of the regions between the first and the last exons of these transcripts. In addition, the RNA sequencing results showed high coverage of all exons of EEF1A1 and FTL transcripts (Fig 2B and 2C). Moreover, to confirm that these RNAs are confined within the vesicles, RNase A and detergent (Triton X-100) treatment of the vesicles was performed. As shown Fig 2D, the Triton X-100 with RNase A treated group exhibited significantly higher Ct value than the RNase A treated and control groups. On the other hand, there were no significant differences between the control and RNase A-treated groups. These results indicate that RNAs are confined within the vesicles and protected from exogenous RNase activity. These results suggest that the follicular exosome-like vesicles contain full-length mRNA, and therefore, the mRNAs transferred by the vesicles may be translated into proteins and affect the biological processes of recipient cells as was reported previously [2, 21].
Fig 2. Genomic view of EEF1A1 and FTL genes along with the representative RNA sequencing results of the exosomal fragments and RNA degradation assay.
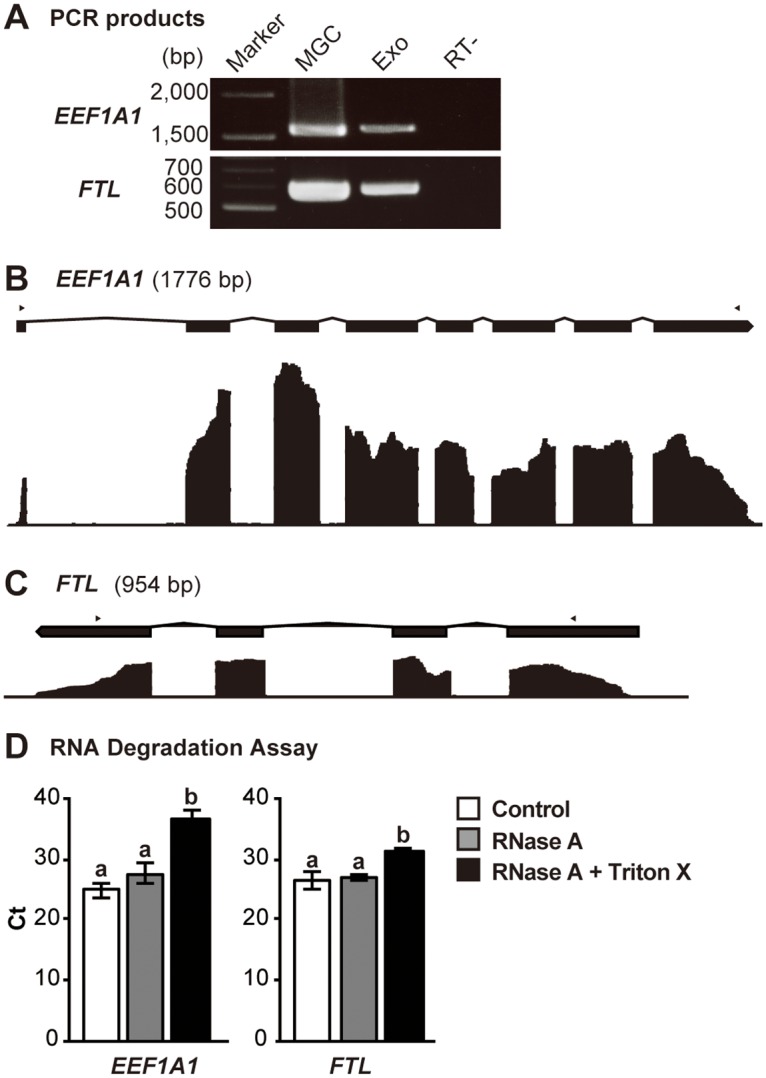
(A) RT-PCR analyses for EEF1A1 and FTL. Marker, electrophoresis marker; Exo, exosomal fraction; MGC, mural granulosa cells. (B) EEF1A1 and (C) FTL genes (upper panels) are shown in exons (black squares) and introns (polygonal lines), and the representative RNA sequencing results for each position in the genes are shown in a coverage graphs (lower panels). Arrowheads indicate positions of PCR primers used for RT-PCR shown in (A). RNA degradation assay using RNase A and Triton X-100. pFF was treated with RNase A (gray bars) with/without Triton X-100 (black bars) or PBS (control; white bars). The Ct values of total RNA extracted from the exosomal fractions were compared among these groups. Values with different letters (a and b) are significantly different (P <0.05) (n = 4).
Functional analysis of mRNAs in the exosome-like vesicles
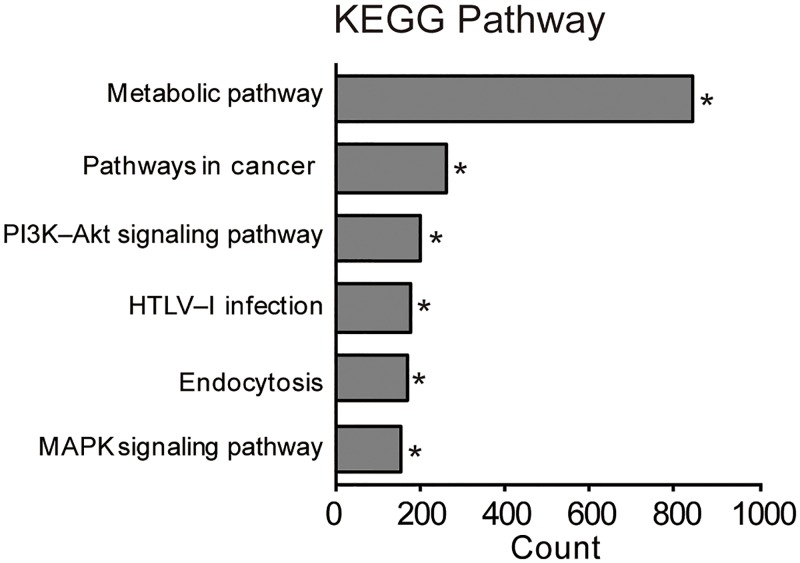
Given that follicular exosome-like vesicles contain full-length mRNAs that may be translated to affect biological processes in recipient cells, we performed the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses using DAVID (version 6.8) [25] and evaluated the potential effects of the vesicle-transferred mRNAs on recipient cells. In this analysis, 11,304 transcripts were functionally annotated with DAVID. As shown in Fig 3, the KEGG pathway analysis showed that the mRNA involved in “metabolic pathway”, “pathways in cancer”, “phosphatidylinositol-4,5-bisphosphate 3-kinase (PIK3K)-protein kinase B (AKT) signaling pathway”, “human T-lymphotropic virus (HTLV) infection”, “endocytosis”, and “MAPK signaling pathway” were enriched in the fraction. Therefore, the follicular exosome-like vesicles may affect these pathways in recipient cells.
Fig 3. KEGG pathway analysis on the mRNA profile of exosome-like vesicles.
The top six pathways are shown. For each pathway, bar plots show the counts of genes that belong to the pathway. Asterisk denotes the biological significance (P < 0.05).
Comparison of transcriptomes between the exosome-like vesicles and MGCs
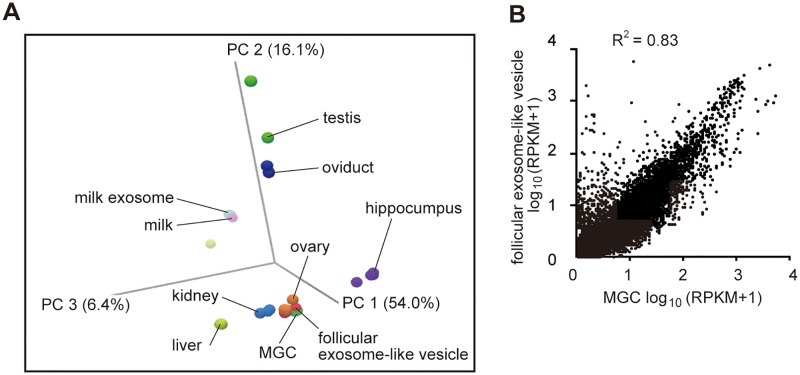
To examine the similarity of the mRNA profiles between exosome-like vesicles and MGCs, PCA was conducted (Fig 4A). Aside from the mRNA profiles of exosome-like vesicles and MGCs, those of various porcine tissues available from NCBI GEO database were used (S1 Table). As shown in Fig 3A, exosome-like vesicle samples were clustered considerably close to those of MGCs and ovary samples than to other tissue samples. Moreover, the levels of transcripts were well correlated between exosome-like vesicles and MGCs (R2 = 0.83) (Fig 4B). Therefore, the mRNAs profile of exosome-like vesicles resembled that of MGCs, and MGCs are likely to be the main source of follicular exosome-like vesicles. However, it is important to note, several transcripts were exclusively detected in the exosomal fraction by RNA sequencing (see below for detail), suggesting that some of the follicular exosome-like vesicles may be secreted from cells other than MGCs.
Fig 4. Comparisons of the mRNA profiles between exosome-like vesicles and mural granulosa cells (MGCs) in porcine follicles.
(A) Principle component analysis (PCA) of the mRNA profiles of the follicular exosome-like vesicles and MGCs together with other porcine organs (see S1 Table for detail). (B) Scatter plot comparison of mRNA profiles of follicular exosome-like vesicles and MGCs.
Unique mRNAs contained in exosome-like vesicles
Although the mRNAs profile of the exosomal fraction resembled that of MGCs, it carried mRNAs that were barely detectable in MGCs (Fig 4B). This observation suggests that the fraction may contain vesicles secreted from the cells other than MGCs. To identify the potential origin of follicular exosome-like vesicles other than MGCs, we focused on these transcripts. We used strict criteria to identify such transcripts; i.e., mRNAs from exosomal fractions with RPKM value > 10 and undetected in MGCs (RPKM < 0.1).
With this criteria, 14 mRNAs were identified as the unique transcripts in the exosomal fraction (Table 3). Some of the mRNAs are known to be specifically expressed in organs/tissues, including skeletal muscle, heart, liver, brain, and kidney [39–43]. These results suggest the possibility that a part of these exosome-like vesicles may be derived from non-ovarian organs/tissues through blood stream.
Table 3. Unique mRNAs in exosome-like vesicles undetected in MGCs.
| Gene name | Ensemble ID | Exosomal Fraction (RPKM) | High expression tissue |
|---|---|---|---|
| LOC100628118 | ENSSSCG00000025858 | 33.60 | – |
| FXYD1 | ENSSSCG00000021374 | 28.15 | skeletal muscle, heart |
| A2M | ENSSSCG00000000660 | 26.54 | liver |
| VSNL1 | ENSSSCG00000008615 | 21.48 | brain |
| COL26A1 | ENSSSCG00000007678 | 14.92 | – |
| C1QTNF5 | ENSSSCG00000024936 | 14.77 | retina |
| LCN2 | ENSSSCG00000005638 | 14.63 | trachea, bone marrow, lung |
| CLDN5 | ENSSSCG00000010123 | 13.07 | lung |
| POU5F1 | ENSSSCG00000001393 | 13.06 | colon |
| ARX | ENSSSCG00000020801 | 11.96 | ovary |
| LOC100737651 | ENSSSCG00000023783 | 11.06 | – |
| LMCD1 | ENSSSCG00000011538 | 11.00 | skeletal muscle, lung |
| ADGRA2 | ENSSSCG00000015825 | 10.72 | small intestine, thymus, colon |
| UMOD | ENSSSCG00000007859 | 10.09 | kidney |
Discussion
In this study, we performed the transcriptome analysis of the exosome-like vesicles in porcine ovarian follicles and focused on mRNA profiles. While the miRNA contents in exosome-like vesicles from ovarian follicles have been examined in detail [10, 16, 44–46], to the best of our knowledge, this is the first report to investigate the mRNA profiles of the exosome-like vesicles in ovarian follicles. The present analysis revealed that these vesicles contain various mRNAs which are enriched in those encoding ribosomal proteins. Moreover, bioinformatics analysis demonstrated that mRNA involved in the biological pathways known to important for normal follicular development were enriched in the exosome-like vesicles. While the majority of the vesicles are likely to be secreted from MGCs, some of these vesicles originated from non-ovarian organs/tissues. Therefore, the present study has unveiled several novel features of exosome-like vesicles in ovarian follicles by focusing on mRNA profiles.
The mRNAs detected in the follicular exosome-like vesicles are predicted to affect biological pathways such as metabolic, PI3K-AKT, and MAPK signaling pathways in the recipient cells. The precise control of the metabolic pathways such as lipid metabolism, glycolysis, and cholesterol biosynthesis is critical for the normal development of follicles and oocytes [47–49]. The PI3K-AKT signaling pathway is known be a critical regulator of quiescence, activation and survival of primordial follicles [50], proliferation and differentiation of granulosa and thecal cells [51], and meiotic maturation of oocytes [52]. Moreover, MAPK signaling pathway is known to be involved in steroid genesis, and oocyte maturation [13, 53, 54]. Although further studies which will test whether the transferred-mRNAs are translated into proteins in recipient cells are warranted, exosome-like vesicles may be involved in the control for the normal development of follicles and oocytes through the regulation of these pathways via mRNA transfer.
The promotive/supportive effects of exosome-like vesicles on the expansion of cumulus cells have been reported in cows and pigs [18, 19]. As normal cumulus expansion requires the activation of MAPK signaling pathway [54] and gastric cancer exosomes promote the tumor cell proliferation via MAPK signaling pathway activation [55], exosome-like vesicles may exert their effects on cumulus expansion through the transfer of mRNAs involved in the control of MAPK signaling pathway.
Some unique transcripts in exosome-like vesicles and barely detectable in MGCs were identified. A possible explanation is that the exosome-like vesicles may be secreted not only from MGCs but also from other ovarian cells such as oocytes, theca cells, ovarian interstitial cells, and non-ovarian organs/tissues. This hypothesis may be supported by the detection of CYP17A1 mRNA, known to be highly expressed in theca cells, in the exosomal fraction (RPKM = 4.23) but not in MGC samples (RPKM < 0.1). Oocyte-specific transcripts such as ZP2 and ZP3 were detected in the exosomal fraction (RPKM 0.30 and 3.22, respectively), suggesting that the oocyte-derived exosomes may exist in the follicular fluid. This possibility is supported by the presence of oocyte-derived vesicles within the perivitelline space of mouse oocytes [56]. In addition, several mRNAs known to be expressed in non-ovarian organs/tissues were detected in the exosomal fraction, suggestive of the presence of the vesicles from non-ovarian organs/tissues. Another possible origin of exosome-like vesicles is that the vesicles secreted during early periods of follicular development or gonadal stage may remain until the formation of antral follicles. This hypothesis may be supported by the detection of POU5F1 (known as OCT4) (RPKM = 13.06), known to be expressed highly at the gonadal stage, in the exosomal fraction. Further studies characterizing individual particles of follicular vesicles may clarify these possibilities.
Although the present study suggests that a part of the exosome-like vesicles present in follicular fluid may be supplied via blood from other organs/tissues, several questions remain to be answered. First, great care was taken to prevent contamination of the follicular fluid sample with blood, but the results of this study do not completely deny the possibility of contamination of our samples with blood. Second, the effects of exosome-like vesicles from other tissues on ovarian function are yet undetermined. Recent studies have showed that the adipose tissue-derived exosomes modulates insulin sensitivity in the liver and muscle tissues [57] and regulate gene expression in the liver [58]. Thus, a similar mechanism may exist between the ovary and other organ/tissues; i.e., non-ovarian organ-derived exosome may affect ovarian functions. However, further studies testing this possibility are warranted.
In summary, this study identified the characteristics of the mRNA transcriptome of exosome-like vesicles from follicular fluids of pigs. Bioinformatics analyses demonstrated that the mRNAs contained in the vesicles potentially modulate the signaling pathways involved in folliculogenesis. While most of exosome-like vesicles in the follicular fluid are likely to have originated from MGCs, our results suggest the presence of vesicles in the follicles from organs other than ovaries. Further functional investigations based on these results may help us to understand the physiological roles of exosome-transferred mRNAs in the regulation of ovarian functions.
Supporting information
(XLSX)
(XLSX)
Acknowledgments
The authors are grateful to Drs. Haruka Ito (The University of Tokyo) and Rashieda J. Hatcher (Baylor College of Medicine) for their helpful input and critical reading of the manuscript. We are grateful to Dr. Kenji Tomita (The University of Tokyo) for his technical assistance with the TEM observations.
Data Availability
Raw data used in this study have been deposited in the DNA Data Bank of Japan (DDBJ) (accession number DRA008080).
Funding Statement
This work was supported by a Grant-in-Aid for Exploratory Research from Japan Society for the Promotion of Science (JSPS) (No. 16K15052 and 17H03900 to K.S.), and a Grant-in-Aid for Research Fellowship for Young Scientists from JSPS (No. 17J00618 to Y.M.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell. 1983;33(3):967–78. Epub 1983/07/01. . [DOI] [PubMed] [Google Scholar]
- 2.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature cell biology. 2007;9(6):654–9. Epub 2007/05/09. 10.1038/ncb1596 . [DOI] [PubMed] [Google Scholar]
- 3.Ekstrom K, Valadi H, Sjostrand M, Malmhall C, Bossios A, Eldh M, et al. Characterization of mRNA and microRNA in human mast cell-derived exosomes and their transfer to other mast cells and blood CD34 progenitor cells. Journal of extracellular vesicles. 2012;1 Epub 2012/01/01. 10.3402/jev.v1i0.18389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grad LI, Yerbury JJ, Turner BJ, Guest WC, Pokrishevsky E, O’Neill MA, et al. Intercellular propagated misfolding of wild-type Cu/Zn superoxide dismutase occurs via exosome-dependent and -independent mechanisms. Proc Natl Acad Sci U S A. 2014;111(9):3620–5. Epub 2014/02/20. 10.1073/pnas.1312245111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nabet BY, Qiu Y, Shabason JE, Wu TJ, Yoon T, Kim BC, et al. Exosome RNA Unshielding Couples Stromal Activation to Pattern Recognition Receptor Signaling in Cancer. Cell. 2017;170(2):352–66.e13. Epub 2017/07/15. 10.1016/j.cell.2017.06.031 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koch S, Acebron SP, Herbst J, Hatiboglu G, Niehrs C. Post-transcriptional Wnt Signaling Governs Epididymal Sperm Maturation. Cell. 2015;163(5):1225–36. Epub 2015/11/23. 10.1016/j.cell.2015.10.029 . [DOI] [PubMed] [Google Scholar]
- 7.Liang B, Peng P, Chen S, Li L, Zhang M, Cao D, et al. Characterization and proteomic analysis of ovarian cancer-derived exosomes. J Proteomics. 2013;80:171–82. Epub 2013/01/22. 10.1016/j.jprot.2012.12.029 . [DOI] [PubMed] [Google Scholar]
- 8.Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523(7559):177–82. Epub 2015/06/25. 10.1038/nature14581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yokoi A, Yoshioka Y, Hirakawa A, Yamamoto Y, Ishikawa M, Ikeda SI, et al. A combination of circulating miRNAs for the early detection of ovarian cancer. Oncotarget. 2017;8(52):89811–23. Epub 2017/11/23. 10.18632/oncotarget.20688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.da Silveira JC, Veeramachaneni DN, Winger QA, Carnevale EM, Bouma GJ. Cell-secreted vesicles in equine ovarian follicular fluid contain miRNAs and proteins: a possible new form of cell communication within the ovarian follicle. Biol Reprod. 2012;86(3):71 Epub 2011/11/26. 10.1095/biolreprod.111.093252 . [DOI] [PubMed] [Google Scholar]
- 11.da Silveira JC, Carnevale EM, Winger QA, Bouma GJ. Regulation of ACVR1 and ID2 by cell-secreted exosomes during follicle maturation in the mare. Reprod Biol Endocrinol. 2014;12:44 Epub 2014/06/03. 10.1186/1477-7827-12-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hernandez Gifford JA. The role of WNT signaling in adult ovarian folliculogenesis. Reproduction. 2015;150(4):R137–48. Epub 2015/07/02. 10.1530/REP-14-0685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Su YQ, Wigglesworth K, Pendola FL, O’Brien MJ, Eppig JJ. Mitogen-activated protein kinase activity in cumulus cells is essential for gonadotropin-induced oocyte meiotic resumption and cumulus expansion in the mouse. Endocrinology. 2002;143(6):2221–32. Epub 2002/05/22. 10.1210/endo.143.6.8845 . [DOI] [PubMed] [Google Scholar]
- 14.Knight PG, Glister C. TGF-beta superfamily members and ovarian follicle development. Reproduction. 2006;132(2):191–206. Epub 2006/08/04. 10.1530/rep.1.01074 . [DOI] [PubMed] [Google Scholar]
- 15.Andrade GM, Meirelles FV, Perecin F, da Silveira JC. Cellular and extracellular vesicular origins of miRNAs within the bovine ovarian follicle. Reprod Domest Anim. 2017;52(6):1036–45. Epub 2017/07/12. 10.1111/rda.13021 . [DOI] [PubMed] [Google Scholar]
- 16.Sohel MM, Hoelker M, Noferesti SS, Salilew-Wondim D, Tholen E, Looft C, et al. Exosomal and Non-Exosomal Transport of Extra-Cellular microRNAs in Follicular Fluid: Implications for Bovine Oocyte Developmental Competence. PLoS One. 2013;8(11):e78505 Epub 2013/11/14. 10.1371/journal.pone.0078505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santonocito M, Vento M, Guglielmino MR, Battaglia R, Wahlgren J, Ragusa M, et al. Molecular characterization of exosomes and their microRNA cargo in human follicular fluid: bioinformatic analysis reveals that exosomal microRNAs control pathways involved in follicular maturation. Fertil Steril. 2014;102(6):1751–61.e1. Epub 2014/09/23. 10.1016/j.fertnstert.2014.08.005 . [DOI] [PubMed] [Google Scholar]
- 18.Hung WT, Hong X, Christenson LK, McGinnis LK. Extracellular Vesicles from Bovine Follicular Fluid Support Cumulus Expansion. Biol Reprod. 2015;93(5):117 Epub 2015/10/02. 10.1095/biolreprod.115.132977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsuno Y, Onuma A, Fujioka YA, Yasuhara K, Fujii W, Naito K, et al. Effects of exosome-like vesicles on cumulus expansion in pigs in vitro. J Reprod Dev. 2017;63(1):51–8. Epub 2017/02/07. 10.1262/jrd.2016-124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Di Pietro C. Exosome-mediated communication in the ovarian follicle. J Assist Reprod Genet. 2016;33(3):303–11. Epub 2016/01/28. 10.1007/s10815-016-0657-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yokoi A, Yoshioka Y, Yamamoto Y, Ishikawa M, Ikeda SI, Kato T, et al. Malignant extracellular vesicles carrying MMP1 mRNA facilitate peritoneal dissemination in ovarian cancer. Nat Commun. 2017;8:14470 Epub 2017/03/07. 10.1038/ncomms14470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zomer A, Maynard C, Verweij FJ, Kamermans A, Schafer R, Beerling E, et al. In Vivo imaging reveals extracellular vesicle-mediated phenocopying of metastatic behavior. Cell. 2015;161(5):1046–57. Epub 2015/05/23. 10.1016/j.cell.2015.04.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naito K, Fukuda Y, Toyoda Y. Effects of porcine follicular fluid on male pronucleus formation in porcine oocytes matured in vitro. Gamete Res. 1988;21(3):289–95. Epub 1988/11/01. 10.1002/mrd.1120210310 . [DOI] [PubMed] [Google Scholar]
- 24.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–8. Epub 2008/06/03. 10.1038/nmeth.1226 . [DOI] [PubMed] [Google Scholar]
- 25.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. Epub 2009/01/10. 10.1038/nprot.2008.211 . [DOI] [PubMed] [Google Scholar]
- 26.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. Epub 2008/11/27. 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan JB, Hu SC, Shi D, Cai MC, Li YB, Zou Q, et al. PaGenBase: a pattern gene database for the global and dynamic understanding of gene function. PLoS One. 2013;8(12):e80747 Epub 2013/12/07. 10.1371/journal.pone.0080747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, et al. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci U S A. 2002;99(7):4465–70. Epub 2002/03/21. 10.1073/pnas.012025199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ge X, Yamamoto S, Tsutsumi S, Midorikawa Y, Ihara S, Wang SM, et al. Interpreting expression profiles of cancers by genome-wide survey of breadth of expression in normal tissues. Genomics. 2005;86(2):127–41. Epub 2005/06/14. 10.1016/j.ygeno.2005.04.008 . [DOI] [PubMed] [Google Scholar]
- 30.Dezso Z, Nikolsky Y, Sviridov E, Shi W, Serebriyskaya T, Dosymbekov D, et al. A comprehensive functional analysis of tissue specificity of human gene expression. BMC Biol. 2008;6:49 Epub 2008/11/19. 10.1186/1741-7007-6-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krupp M, Marquardt JU, Sahin U, Galle PR, Castle J, Teufel A. RNA-Seq Atlas—a reference database for gene expression profiling in normal tissue by next-generation sequencing. Bioinformatics. 2012;28(8):1184–5. Epub 2012/02/22. 10.1093/bioinformatics/bts084 . [DOI] [PubMed] [Google Scholar]
- 32.Chen LS, Lo CF, Numann R, Cuddy M. Characterization of the human and rat phospholemman (PLM) cDNAs and localization of the human PLM gene to chromosome 19q13.1. Genomics. 1997;41(3):435–43. Epub 1997/05/01. 10.1006/geno.1997.4665 . [DOI] [PubMed] [Google Scholar]
- 33.Barbosa-Morais NL IM, Pan Q, Xiong HY, Gueroussov S, Lee LJ, Slobodeniuc V, Kutter C, Watt S, Colak R, Kim T, Misquitta-Ali CM, Wilson MD, Kim PM, Odom DT, Frey BJ, Blencowe BJ. The Evolutionary Landscape of Alternative Splicing in Veterbrate Species. Science. 2012;338(21):1587–93. [DOI] [PubMed] [Google Scholar]
- 34.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101(16):6062–7. Epub 2004/04/13. 10.1073/pnas.0400782101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lotvall J, Hill AF, Hochberg F, Buzas EI, Di Vizio D, Gardiner C, et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. Journal of extracellular vesicles. 2014;3:26913 Epub 2014/12/30. 10.3402/jev.v3.26913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lasser C, Eldh M, Lotvall J. Isolation and characterization of RNA-containing exosomes. Journal of visualized experiments: JoVE. 2012;(59):e3037 Epub 2012/01/20. 10.3791/3037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kas K, Michiels L, Merregaert J. Genomic structure and expression of the human fau gene: encoding the ribosomal protein S30 fused to a ubiquitin-like protein. Biochem Biophys Res Commun. 1992;187(2):927–33. Epub 1992/09/16. . [DOI] [PubMed] [Google Scholar]
- 38.Ogawa Y, Tsujimoto M, Yanoshita R. Next-Generation Sequencing of Protein-Coding and Long Non-protein-Coding RNAs in Two Types of Exosomes Derived from Human Whole Saliva. Biol Pharm Bull. 2016;39(9):1496–507. Epub 2016/09/02. 10.1248/bpb.b16-00297 . [DOI] [PubMed] [Google Scholar]
- 39.Pennica D, Kohr WJ, Kuang WJ, Glaister D, Aggarwal BB, Chen EY, et al. Identification of human uromodulin as the Tamm-Horsfall urinary glycoprotein. Science. 1987;236(4797):83–8. Epub 1987/04/03. . [DOI] [PubMed] [Google Scholar]
- 40.Santambrogio S, Cattaneo A, Bernascone I, Schwend T, Jovine L, Bachi A, et al. Urinary uromodulin carries an intact ZP domain generated by a conserved C-terminal proteolytic cleavage. Biochem Biophys Res Commun. 2008;370(3):410–3. Epub 2008/04/01. 10.1016/j.bbrc.2008.03.099 . [DOI] [PubMed] [Google Scholar]
- 41.Sikri KL F C, MacHugh N, Marshall RD. Localozation od Tamm-Horsfall glycoprotein in the human kidbey using immuno-fruorescence and immuno-electron microscopical techniques. Journal of Anatomy. 1981;132:597–605. [PMC free article] [PubMed] [Google Scholar]
- 42.Wei X, Xu R, Yang Z, Li Z, Liao Y, Johnson RJ, et al. Novel uromodulin mutation in familial juvenile hyperuricemic nephropathy. Am J Nephrol. 2012;36(2):114–20. Epub 2012/07/11. 10.1159/000339752 . [DOI] [PubMed] [Google Scholar]
- 43.Liu M, Chen Y, Liang Y, Liu Y, Wang S, Hou P, et al. Novel UMOD mutations in familial juvenile hyperuricemic nephropathy lead to abnormal uromodulin intracellular trafficking. Gene. 2013;531(2):363–9. Epub 2013/08/31. 10.1016/j.gene.2013.08.041 . [DOI] [PubMed] [Google Scholar]
- 44.Burns GW, Brooks KE, Spencer TE. Extracellular Vesicles Originate from the Conceptus and Uterus During Early Pregnancy in Sheep. Biol Reprod. 2016;94(3):56 Epub 2016/01/29. 10.1095/biolreprod.115.134973 . [DOI] [PubMed] [Google Scholar]
- 45.da Silveira JC, de Andrade GM, Nogueira MF, Meirelles FV, Perecin F. Involvement of miRNAs and Cell-Secreted Vesicles in Mammalian Ovarian Antral Follicle Development. Reprod Sci. 2015;22(12):1474–83. Epub 2015/03/05. 10.1177/1933719115574344 . [DOI] [PubMed] [Google Scholar]
- 46.da Silveira JC, Andrade GM, Del Collado M, Sampaio RV, Sangalli JR, Silva LA, et al. Supplementation with small-extracellular vesicles from ovarian follicular fluid during in vitro production modulates bovine embryo development. PLoS One. 2017;12(6):e0179451 Epub 2017/06/16. 10.1371/journal.pone.0179451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Collado-Fernandez E, Picton HM, Dumollard R. Metabolism throughout follicle and oocyte development in mammals. Int J Dev Biol. 2012;56(10–12):799–808. Epub 2013/02/19. 10.1387/ijdb.120140ec . [DOI] [PubMed] [Google Scholar]
- 48.Sugiura K, Pendola FL, Eppig JJ. Oocyte control of metabolic cooperativity between oocytes and companion granulosa cells: energy metabolism. Dev Biol. 2005;279(1):20–30. Epub 2005/02/15. 10.1016/j.ydbio.2004.11.027 . [DOI] [PubMed] [Google Scholar]
- 49.Su YQ, Sugiura K, Wigglesworth K, O’Brien MJ, Affourtit JP, Pangas SA, et al. Oocyte regulation of metabolic cooperativity between mouse cumulus cells and oocytes: BMP15 and GDF9 control cholesterol biosynthesis in cumulus cells. Development. 2008;135(1):111–21. Epub 2007/11/30. 10.1242/dev.009068 . [DOI] [PubMed] [Google Scholar]
- 50.John GB, Gallardo TD, Shirley LJ, Castrillon DH. Foxo3 is a PI3K-dependent molecular switch controlling the initiation of oocyte growth. Dev Biol. 2008;321(1):197–204. Epub 2008/07/08. 10.1016/j.ydbio.2008.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brown C, LaRocca J, Pietruska J, Ota M, Anderson L, Smith SD, et al. Subfertility caused by altered follicular development and oocyte growth in female mice lacking PKB alpha/Akt1. Biol Reprod. 2010;82(2):246–56. Epub 2009/10/02. 10.1095/biolreprod.109.077925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Makker A, Goel MM, Mahdi AA. PI3K/PTEN/Akt and TSC/mTOR signaling pathways, ovarian dysfunction, and infertility: an update. J Mol Endocrinol. 2014;53(3):R103–18. Epub 2014/10/15. 10.1530/JME-14-0220 . [DOI] [PubMed] [Google Scholar]
- 53.Manna PR, Stocco DM. The role of specific mitogen-activated protein kinase signaling cascades in the regulation of steroidogenesis. J Signal Transduct. 2011;2011:821615 Epub 2011/06/04. 10.1155/2011/821615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Su Y-Q, Denegre JM, Wigglesworth K, Pendola FL, O’Brien MJ, Eppig JJ. Oocyte-dependent activation of mitogen-activated protein kinase (ERK1/2) in cumulus cells is required for the maturation of the mouse oocyte–cumulus cell complex. Developmental Biology. 2003;263(1):126–38. 10.1016/s0012-1606(03)00437-8 [DOI] [PubMed] [Google Scholar]
- 55.Qu JL, Qu XJ, Zhao MF, Teng YE, Zhang Y, Hou KZ, et al. Gastric cancer exosomes promote tumour cell proliferation through PI3K/Akt and MAPK/ERK activation. Dig Liver Dis. 2009;41(12):875–80. Epub 2009/05/29. 10.1016/j.dld.2009.04.006 . [DOI] [PubMed] [Google Scholar]
- 56.Ohnami N, Nakamura A, Miyado M, Sato M, Kawano N, Yoshida K, et al. CD81 and CD9 work independently as extracellular components upon fusion of sperm and oocyte. Biol Open. 2012;1(7):640–7. Epub 2012/12/06. 10.1242/bio.20121420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ying W, Riopel M, Bandyopadhyay G, Dong Y, Birmingham A, Seo JB, et al. Adipose Tissue Macrophage-Derived Exosomal miRNAs Can Modulate In Vivo and In Vitro Insulin Sensitivity. Cell. 2017;171(2):372–84 e12. Epub 2017/09/26. 10.1016/j.cell.2017.08.035 . [DOI] [PubMed] [Google Scholar]
- 58.Thomou T, Mori MA, Dreyfuss JM, Konishi M, Sakaguchi M, Wolfrum C, et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature. 2017;542(7642):450–5. Epub 2017/02/16. 10.1038/nature21365 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
Data Availability Statement
Raw data used in this study have been deposited in the DNA Data Bank of Japan (DDBJ) (accession number DRA008080).