Abstract
Objective.
To determine the precision of new and established methods for estimating duration of HIV infection.
Design.
A retrospective analysis of HIV testing results from serial samples in commercially-available panels, taking advantage of extensive testing previously conducted on 53 seroconverters.
Methods.
We initially investigated four methods for estimating infection timing: 1) “Fiebig stages” based on test results from a single specimen; 2) an updated “4th gen” method similar to Fiebig stages but using antigen/antibody tests in place of the p24 antigen test; 3) modeling of “viral ramp-up” dynamics using quantitative HIV-1 viral load data from antibody-negative specimens; and 4) using detailed clinical testing history to define a plausible interval and best estimate of infection time. We then investigated a “two-step method” using data from both methods 3 and 4, allowing for test results to have come from specimens collected on different days.
Results.
Fiebig and “4th gen” staging method estimates of time since detectable viremia had similar and modest correlation with observed data. Correlation of estimates from both new methods (3 and 4), and from a combination of these two (“2-step method”) was markedly improved and variability significantly reduced when compared with Fiebig estimates on the same specimens.
Conclusions.
The new “two-step” method more accurately estimates timing of infection and is intended to be generalizable to more situations in clinical medicine, research, and surveillance than previous methods. An online tool is now available that enables researchers/clinicians to input data related to method 4, and generate estimated dates of detectable infection.
INTRODUCTION
Biomarkers that change over time are often used to assess the chronicity or duration of an illness. When physicians “stage” cases of most infectious diseases (e.g., syphilis, hepatitis B, or Epstein-Barr virus infections), they use serologic assays to assess infection duration. Knowing if a case has very early or later-stage infection can help a physician interpret patient symptoms, inform the prognosis, and affect treatment and public health management. Unfortunately, while serologic staging is conceptually appealing, staging systems often provide very limited information in practice. One major issue is that staging systems are developed using specific assays, and thus require that those assays be used to yield valid estimates of infection duration for individuals. Even more important, test results often come from visits that are weeks or months apart, making it challenging to estimate infection timing. To assign a specific stage directly from a set of test results, one must also be looking at tests from specimens collected during the time serologies are evolving. This means infection staging can only be done during a brief window of time.
HIV infection staging has been approached in this manner, using a serologic staging rubric developed by Fiebig, et. al.1 Originally proposed in 2003, this system describes five discrete, sequential stages of early infection with different HIV test result patterns. These stages are referenced in nearly all discussions related to acute HIV infection.2 Unfortunately, the window of acute HIV infection is brief, and discrepant results indicating acute infection are rarely seen in clinical practice. Also, three of the five key assays used to define Fiebig stages—the HIV p24 antigen ELISA, HIV IgG antibody ELISA and HIV antibody Western blots—are classes of HIV test that are no longer available or commonly used. The types of tests available today (“4th generation” antigen/antibody HIV assays, IgM or IgG-antibody rapid tests, HIV-½ differentiation assays, and quantitative viral loads) were not part of the Fiebig analysis. Recently, Ananworanich and colleagues3 proposed a simplified system of three “4th gen stages” in which an antigen/antibody assay would function in place of the p24 Ag ELISA. To date, neither the staging system proposed by the Fiebig nor that of Anorworanich have been independently validated as a predictor of HIV infection timing.
The shortcomings of traditional serologic staging are germane in cases of HIV seroconversion or acute HIV infection, (i.e. after detectable HIV viremia but before the antibodies to HIV infection are present in detectable levels). In these situations there is urgency to identify HIV infection related to important prevention and clinical benefits. For HIV prevention, the risk of HIV transmission to exposed infants and sexual partners is many times greater for index patients who are acutely or recently infected—and different interventions are thus often recommended for infants4 and for sexual partners5 determined to have been exposed to acute or recent HIV infection.6, 7 Clinically, ART initiated in the first two months after infection has been associated with reduced seeding of HIV reservoirs8, 9 and better HIV specific immune responses.10-12 In randomized trials comparing immediate versus delayed ART for patients with recent (<3–6 month) HIV infection, immediate ART has resulted in improved HIV control and increased CD4 cell levels,13 and reduced long-term malignancy risk14 when compared to ART initiated after a delay. The sense of clinical urgency around acute HIV infections often drastically affects the way that diagnosing emergency room physicians, HIV testing counselors, and clinics respond to a new diagnosis of HIV—leading to urgent prioritization of partner management strategies and triggering rapid care navigation and immediate ART (prior to meeting a primary provider, and without waiting for a drug resistance result).15
Accurate, individual-level information on HIV infection timing is key to interpreting many research studies of HIV transmission, early disease pathogenesis, ART, and diagnostics. Further, the concept of infection time is central to global surveillance programs, which measure HIV incidence in part by classifying individuals in population surveys as being infected less than some average duration of time, as recently reviewed in Murphy et al.16
For prospective studies, as well as those using stored specimens and historical test results from many years ago, updated strategies for staging infection are needed. In the fourteen years since the Fiebig staging method was first published, investigators at the U.S. Centers for Disease Control and Prevention (CDC)17-19 and others3, 20-22 have reported additional experiments using seroconversion panels that have much more precisely described the observed distribution of test conversion times for many more assays. The CDC studies clearly estimate the median and range of times that elapse between individuals’ first detectable HIV viremia and their conversion on nearly all other HIV tests.23
We hypothesized that these test-specific “diagnostic delays” could be used to infer a range of plausible dates of first detectable viremia from any testing history—even when same-sample “acute infection staging” was not possible, and that this method should provide equally accurate estimates for historical or modern assays. We also hypothesized that quantitative viral load information would be as useful or more accurate than p24 or antigen/antibody assay information in assessing the timing of acute HIV infections detected prior to antibody seroconversion. In this paper, we describe methods based on these principles; demonstrate their performance and compare them with older methods (i.e. Fiebig and 4th gen staging models); and provide guidance including links to online tools for improved estimation of HIV infection duration.
METHODS
HIV test results including qualitative and quantitative HIV-1 viral load were obtained through testing of commercially available seroconversion panels from 53 individuals documented to have seroconverted to detectable HIV infection while participating in programs for repeat blood plasma donation. We sought to identify and access all available data from plasma donor panels in which all or most of the Fiebig-era tests, as well as more modern tests, had been performed and results were available across a variety of timepoints. This included panels from CDC and a variety of commercial panels used in the original Fiebig analysis and other subsequent analyses. We selected panels for the analysis in which the last specimen where no RNA target was detected “Target not detected” (TND) or “negative”) or with an HIV viral load quantified <100 copies/mL on any assay, was followed within 7 days by at least one specimen with >100 copies/mL HIV RNA on any assay. HIV test results for each timepoint were analyzed including results that were publicly available for each specimen panel,24, 25 and additional results of re-testing of plasma aliquots by a variety of HIV diagnostic assays as these became available over time; more detail on these methods have been reported elsewhere.17-19, 23 As most of the panels used were commercially available, archival HIV testing data were available on each company’s website24, 25 and these were also included in relevant analyses.
In theory, the ideal reference standard for an infection dating scheme would be the infectious event itself; however, as no assay can measure the infectious event, there is a need to agree on an assay-based reference standard. We call this the date of detectable infection (DDI), defined operationally as a hypothetical date on which “ramp-up viremia” first exceeded 1 copy/mL of HIV RNA in plasma, or DDI1, likely several days after exposure.16 To estimate the actual date of infection, the average estimated duration of the eclipse phase23 can be added to the estimate of days since DDI1 for a specific subject. In any analysis, the threshold for DDI can be set to a value appropriate for viral detection during the ramp-up phase. For this paper, we used a modified reference standard for the specimen sets we examined, defining the date of detectable infection as the date at which viremia exceeded 100 copies/mL (the DDI100). This particular threshold was chosen for consistency with previous papers1 and to account for variability in assay detection limits in our retrospective, pooled dataset.
For each individual seroconverter’s specimen set, a date of detectable infection (DDI100) was estimated as occurring between the last bleed at which HIV RNA was either TND or detected with <100 copies/mL, and the first bleed at which HIV RNA was detected ≥ 100 copies/mL. Within this (maximum 7 day) window, the exact date of crossing the 100-copy threshold was estimated for each seroconverter by linear interpolation of log10-viral load graphed vs. time. For this calculation, TND values were assigned half the detection limit of the most sensitive negative RNA assay used.
We initially investigated four methods (detailed in the Supplemental appendix) for estimating infection time: 1) using Fiebig stages;1 2) using the “4th gen” modified staging system;3 3) using quantitative viral load results from antibody-negative specimens utilizing a linear mixed effects regression model of “viral ramp-up” dynamics with a random slope and intercept; and 4) using the details in the clinical testing history, including negative and positive assay results to define a plausible interval and best estimate of infection time. Examples of this testing history-based estimation procedure are illustrated in Figure 1, and more detail is available in Grebe et al. (forthcoming). We explored whether methods 3 or 4 performed at least as well as the widely-accepted Fiebig method, with greater flexibility and applicability to today’s currently available assays. After initial results indicated suitable performance of the viral ramp-up method (method 3), we lastly investigated a “two-step method” for evaluating the complete testing history. The two-step method was designed to incorporate information for people identified after viral ramp-up was complete and the antibody response had developed, which is the most common real-life application of these methods. The two-step method calculated infection date estimates on viral load information (method 3) when it was available on an antibody negative specimen, but otherwise used the rest of the subject’s testing history in the calculation using method 4.
Figure 1. The testing history method.
The panels illustrate how an estimated date of detectable infection (EDDI), and a “plausible interval” for this estimate, can be inferred from HIV testing histories that include the dates and qualitative results on each HIV assay used. Testing history timelines are shown for three representative HIV seroconverters, with hypothetical dates of tests inputted and date of detectable infection (DDI) outputs shown along the x-axis as calendar days. Triangles represent HIV tests performed (solid=positive; empty=negative). The “diagnostic delay” from first detectable infection to initial positivity has been determined experimentally for most assays (see Table S1). For any result, the diagnostic delay for the assay used can be subtracted from the test date, to give an earlier date which is interpreted differently depending on whether the result is now negative or positive. If the result is negative, we infer that the patient’s actual DDI probably occurred sometime after that earlier date (else we would expect the assay result to now already be positive). If the result is positive, we infer that the DDI had probably already occurred by the earlier date (else, the result would still likely be negative). In the figures, white bars show ranges for the EDDI that are inconsistent with the particular result. Taken across all results in the history, these define the “earliest-and latest-plausible dates of detectable infection” (EP-DDI and LP-DDI). The “plausible interval” for the EDDI is bounded by these dates as shown by the black bar. The best estimate of the EDDI from the qualitative HIV testing history is then the midpoint of this plausible interval (shown by the circle). The method applies to discrepant qualitative HIV test results obtained on the same day (e.g., “antibody negative/RNA positive” (Panel A), but also applies when discrepant HIV results were obtained on two (Panel B) or multiple (Panel C) encounters: information from multiple test encounters may narrow the plausible interval and hence improve precision of the EDDI. Additional abbreviations: quant = quantitative; VL = viral load; RT = rapid test; WB = Western blot; Full = fully reactive assay.
Statistical analysis.
All analyses presented in this paper concern only the specimen panels with a documented date of first detectable viremia at a 100-copy threshold (with results going from <100 to >100 copies RNA within less than 7 days). The DDI100 was determined for each seroconverter panel. The time in days since the DDI100 could then be directly calculated for each specimen in that panel.
For primary analyses and for sensitivity analyses in which assumptions and included data varied, the performance of each prediction model was assessed by comparing model-estimated time with the gold standard for DDI100. Prediction errors were assessed by the root mean square error when comparing to a gold standard. Specifically, if Xi represents the gold standard estimate of the infection time for person i, and Yi is the corresponding estimate derived by a method under consideration, then the overall root mean square deviation exhibited is
The standard errors were computed using bootstrap resampling. When comparing the accuracy of two methods which give estimates Yi and Zi for each patient, we computed the statistic RY-RZ and conducted hypothesis tests using the bootstrap confidence interval. We also used the intraclass correlation as a measure of agreement.
RESULTS
Data from 206 specimens (n) and 31 individual seroconverters (N) were obtained and considered eligible for inclusion in the final analyses, based on an interval ≤7 days between the last TND and first detectable result on a quantitative viral load. Of these, 14 individual series/99 specimens (N/n = 14/99) were represented in the 2003 Fiebig manuscript analysis (“Fiebig panels”) while 30 individual series/107 specimens (N/n = 30/107) were represented in the CDC papers from which most of the offsets in Table S1 were derived (the “CDC panels”). Intervals between last specimen with HIV RNA <100 and first specimen with HIV RNA >100 ranged from 1 to 7 days with a mean of 3.5 days. The duration from DDI100 for specimens in the analysis ranged from 1 – 43 days. To preliminarily validate individual methods, the panel data used for original model development were excluded; i.e. data from Fiebig panels were excluded from the Fiebig method validation, and data from CDC panels were excluded from the testing history method validations. After this validation, we performed a head-to-head comparison of the different candidate methods using the entire specimen set.
In the initial method validation, Fiebig method estimates of time since DDI100 for observations with Fiebig stage I-V on CDC panels (N/n = 14/92) had a modest correlation with observed times in the original Fiebig data, with an intraclass correlation (ICC) of 0.671 (95% CI 0.542, 0.770) and a root mean square (RMS) error of 13.15 (95% CI 10.77, 15.87). The distribution of times associated with each Fiebig stage (including all panels for the head-to-head comparison) are shown in Table 1: times were generally very close to those expected in stages 1–4, with average error less than 7 days at each of these stages. Estimates of infection time for Fiebig stage V were much shorter than expected, perhaps due to truncation of sample collection in the plasma donor population. Estimates for the three “4th gen” stages gave similarly small errors (ICC of 0.779 and RMS error of 4.2). The Fiebig and 4th gen staging methods performed similarly in the final comparison using all staged specimens for each method, as shown in Table 2 and illustrated in Figure 2(A and B).
Table 1.
Performance of previously published Fiebig and 4th gen staging systems
| Model and stage | Specimens | Predicted and observed times after first viremia (DDI100) for each stage | |||
|---|---|---|---|---|---|
| Predicted time | Distribution of actual times observed | Error in model estimates | |||
| N/n | Median | [25th,75th] IQR | RMS difference (predicted-observed) |
||
| Fiebig (after Fiebig, et al., 2003)2 | |||||
| Stage 1 RNA+ p24− 3G− | 30/62 | 9 days | 9 days | [8,12] days | 3.5 days |
| Stage 2 RNA+ p24+ 3G− | 29/55 | 15 | 16 | [14,19] | 3.1 |
| Stage 3 RNA+ p24+ 3G+ 2G/WB− | 15/27 | 19 | 20 | [20,23] | 4.5 |
| Stage 4 RNA+ WB indeterminate | 10/18 | 23 | 27 | [22,30] | 6.8 |
| Stage 5 RNA+ WB+ (p31−) | 8/54 | 61 | 35 | [30,41] | 26.2 |
| Stage 1-2* RNA+ 3G− | 31/117 | 12 | 13 | [9,16] | 7.7 |
| Stage 1-3* RNA + Ab rapid test− | 31/144 | 13 | 14 | [10,20] | 7.8 |
| Stage 1-4* RNA + unspecified Ab− | 31/162 | 17 | 15 | [11,21] | 10.5 |
| 4th Gen (after Ananworanich, et al. 2013)1 | |||||
| Stage 1 RNA+ 4G− 3G− | 30/64 | 9 | 9 | [8,12] | 3.4 |
| Stage 2 RNA+ 4G+ 3G− | 28/53 | 19 | 16 | [14,19] | 4.0 |
| Stage 3 RNA+ 4G+ 3G+ WB indeterminate | 18/45 | 22 | 23 | [21,27] | 5.4 |
Abbreviations: 3G = 3rd Generation (IgM-sensitive antibody assays); 4G = 4th Generation (p24 antigen/antibody assays); RMS = root mean square; IQR = interquartile range; DDI100 = days since detectable infection on a 100-copy viral load; N = unique individuals; n = specimens from distinct timepoints
Because it is often impossible to determine the precise Fiebig stage, examples are also given for the infection times seen for combinations of stages which can sometimes be inferred from ‘incomplete’ staging information
Table 2.
Performance of newer proposed prediction methods, as compared to complete Fiebig staging as a benchmark method
| Method compared to Fiebig (reference standard) | Specimens | New method performance | Fiebig performance (benchmark) |
Method comparison |
||
|---|---|---|---|---|---|---|
| N5/n6 | ICC1 | RMS Difference2 |
ICC | RMS Difference2 | P value3 | |
| 4th gen staging system | 31/162 | 0.779 | 4.2 | 0.756 | 4.2 | 0.96 |
| Quantitative RNA viral load model (Ab4- specimens only) | ||||||
| IgM-sensitive Ab test (−) specimens only | 14/41 | 0.899 | 2.2 | 0.591 | 3.5 | <0.001 |
| Any Ab test (−) specimens | 14/51 | 0.877 | 2.6 | 0.754 | 3.4 | 0.21 |
| Testing history only (quantitative viral load not entered) | 31/161 | 0.744 | 4.4 | 0.758 | 4.2 | 0.09 |
| Testing history, including quantitative RNA viral load | 31/157 | 0.834 | 3.5 days | 0.759 | 4.1 days | 0.002 |
ICC = intraclass correlation of model predictions with observed infection duration
RMS = root mean square – difference between predicted and observed data was calculated as a measure of estimation error
P values shown indicate whether the estimation error (RMS difference) is statistically different with the newer model, as compared to with the benchmark model
Ab = Antibody
N = unique individuals
n = specimens from distinct timepoints
Figure 2. Estimates obtained using alternative staging methods, plotted versus time since infection (days since DDI100).
In each figure, infection times predicted by each method (y axis) are compared with observed times (x axis) for 206 specimens from 31 seroconverters frequently donating blood plasma (all available data from all panels, including those used in model development and those used in model validation). Infection times are given as days since first viremia detected at the 100 RNA-copy/mL threshold (DDI100); timing of first viremia was estimated within <3.5 days. Black circles represent specimens that were negative on the Western blot, while specimens with indeterminate Western blots are shown with white (unfilled) circles. Plots (A) and (B) show the distributions of infection times predicted by Fiebig2 and 4th gen3 staging methods, respectively. Plot (C) shows the distribution of times for the seroconversion testing history method only, “step 2” of the 2-step method described in Figure 1, using the diagnostic delays in Table S1 for specimens with discrepant results occurring on the same day. Plot (D) shows the distribution of times for the 2-step method. While the testing history method can take all results into account, the plot again shows results limited to tests performed on the same day. In all four plots results were used from five assay types: viral load, p24 antigen ELISA, IgG-only, IgM/IgG antibody immunoassays and Western blot. In Plot (D), for antibody positive specimens only the qualitative results on the five assay types were used; for antibody negative specimens the method also used quantitative viral load information.
The two new methods developed for this paper were the acute viremia “step 1” model—applicable to antibody-negative specimens only—and the more generalizable “step 2” model computing a plausible interval and EDDI from all positive and negative results in the testing history. As shown in Table 2 and Figure 2C, both of these new methods performed better than complete Fiebig staging in head-to-head comparison using the same test result information and specimens. For the step 1 acute viremia model and for the combined, 2-step procedure (Figure 2D), correlation of estimates with actual infection time was markedly improved and errors significantly reduced when compared with Fiebig estimates on the same specimens (Table 2). By itself, the step 2 model gave performance that was similar to that of Fiebig and 4G staging when applied to discrepant results on same-day testing (see Table 2, and Figure 2C). However, the goal of including the new method was to improve flexibility over more traditional “staging” approaches—making it possible to use real world results, including results on newer assays and from multiple test dates during seroconversion. We therefore tested the sensitivity of the step 2 model to these situations in a series of scenarios where more or less information was included in the testing history model. As shown in Figure 3 and Table 3, the inclusion of information from two separate dates actually improved precision of estimates when compared with using the same system on same-day discrepant results. A scenario using newer assays (e,g, an antigen/antibody immunoassay, followed by the Bio-Rad Geenius™ HIV-½ Supplemental Assay) also gave significantly better performance than one using older tests (e.g. antibody only, IgM-sensitive immunoassay, and the Bio-Rad GS HIV-1 Western blot) on the same specimens (Table 3).
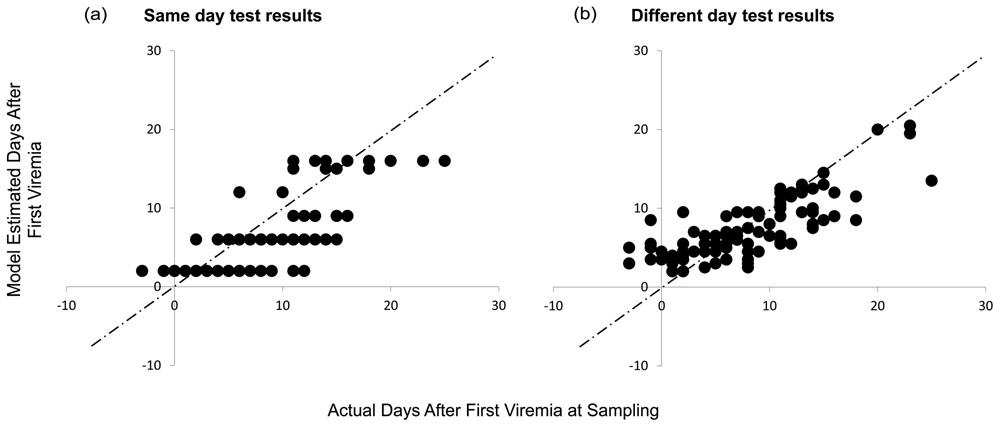
Figure 3. Estimates of infection duration obtained using discrepant, same-day qualitative test results (A) or using test results obtained from the same subjects on two different days (B).
Estimates of time since first viremia detectable at 100 RNA copies/mL (DDI100) are shown on the y axis for alternative scenarios of all panel observations using the testing history method, versus observed times on the x axis. In the first same-day scenario (Panel A), time was calculated based on results occurring on that day. In the serial testing scenario (Panel B), the data from that date were ignored and instead, test results from the visit prior to and after that visit were used to generate the DDI100. In both scenarios results were used from five assay types: viral load, p24 antigen ELISA, IgM-sensitive and recombinant IgG-sensitive antibody immunoassays, and Western blot, and analysis was limited to n=104 specimens from N=30 individuals with results for all 5 assay types.
Table 3.
Sensitivity of “testing history” method estimates to the type of testing history information available
| Staging scenarios compared | Information used by the “testing history method” | Specimens | Model performance: predicted vs. observed infection duration |
||
|---|---|---|---|---|---|
| Results included at each observation1 |
Observations included | N/n | ICC | RMS difference2 | |
| “Fiebig-like”: same-day tests, vs.results from different days | RNA (qualitative), p24, 3G, 2G, WB | Same day | 30/104 | 0.698 | 4.1 |
| RNA (qualitative), p24, 3G, 2G, WB | Two days (two bleeds apart) | 30/104 | 0.715 | 3.7* | |
| Newer Assays | RNA (qualitative), 3G, WB | Same day | 14/64 | 0.739 | 4.3 |
| RNA (qualitative), 4G, Geenius | Same day | 14/64 | 0.698 | 5.3* | |
statistically significant difference in prediction accuracy between model scenarios at a level of p<0.05.
Abbreviations: ICC = intraclass correlation; RMS = root mean square; 3G = 3rd Generation (IgM-sensitive antibody assays); 2G = 2nd Generation (Recombinant IgG-sensitive antibody assays); 4G = 4th Generation (p24 antigen/antibody assays); WB = Western blot; N = unique individuals; n = specimens from different timepoints
For these analyses the testing history was assumed not to include the quantitative viral load.
RMS = root mean square – difference between predicted and observed data was calculated as a measure of estimation error
DISCUSSION
In this paper we demonstrate a novel method for estimating the timing of HIV infection that is more accurate than prior methods and much more likely to be applicable in the real world. Performance differences favoring the new method were generally small but statistically significant; whether or not such differences are meaningful in practice will depend on the clinical or research question being asked. We furthermore showed that estimates by the new method were robust to variation in the types of HIV test used, and were equally accurate whether tests were included from specimens on the same day or from specimens taken on different days during seroconversion. The new system may thus overcome key limitations of previous methods. Up to now, acute infection staging has relied on successfully obtaining and testing an acute infection sample, and has required results from multiple different tests, some of which are no longer available. By using results from any two tests, even if obtained from specimens collected on different days, the new method is intended to be generalizable to many more situations in clinical medicine, research and surveillance.
One of the key features distinguishing the current method is its use of information on acute viremia (early viral load) when this is available. We found that calculating the duration of an acute infection based on the acute viral load was more accurate than when the viral load result was treated as a qualitative (“positive”) result alongside other results to determine a “stage”. Importantly, this ability to back-calculate infection time is only applicable during the “ramp-up” phase of initial viremia—thus, an acute viral load should only be used to estimate infection time if it appears to be from the ramp-up period, e.g. it is known to be from a day when there was a negative antibody test. “Step 1” of the 2-step method is therefore to determine whether the acute viral load-based estimate can be used. If not, infection duration is estimated using the remaining complete testing history, considering all qualitative results, including qualitative interpretation of viral load results.
In addition to showing some advantages of the new method, we also independently validated the performance of previous staging systems. This exercise provides a basis for standardizing results across studies or programs that used older methods and those that will use the new method. It also provides reassurance that many acute infection studies that used Fiebig or 4th gen staging data may not need to be reanalyzed, as the accuracy advantage to the new method was relatively small (on the order of half a week). For some types of studies—such as acute HIV intervention studies, where timing in first 1–2 weeks of acute viremia may be critical—the error inherent in Fiebig and 4th gen systems may warrant focused re-analyses.
In other research settings (e.g., vaccine trials), or when making decisions about clinical acute infection treatment, the enhanced flexibility of assessing timing without an antibody-negative but viremic sample may be the more important feature of the new proposed system. In many of these situations, the interval defined by a seroconversion testing history may be many months or even years—or a last negative HIV test may not actually be documented. In these cases, it is possible that additional tests that measure the slow maturation of the HIV antibody response will provide more information. At present, these “tests for recent infection” (e.g., the LAg Avidity assay and others) are recommended only for use in population surveys26 and not for use in estimating individual-level infection timing. If appropriate criteria could be developed to incorporate these data to improve individual timing estimates, this could further expand opportunities for improved clinical and public health management of newly diagnosed HIV infections.
It is important to note that some patient or viral characteristics (e.g. hepatitis C co-infection,27, 28 viral type,29 use of HIV pre-exposure prophylaxis (PrEP),30, 31 or very early treatment during acute HIV infection32 can delay antibody seroconversion and/or viral ramp up in an individual, leading to unexpected (and inaccurate) results from any of the methods described here. Further research is needed into the impact of these increasingly common factors on antibody and viral kinetics, particularly as they affect results of HIV diagnostic or incidence assays.
We have provided expanded guidance as well as updated diagnostic delay tables and tools for computing EDDIs and plausible intervals online, at https://tools.incidence-estimation.org/idt/. The tool facilitates calculation of an EDDI and plausible interval for almost any individual with at least one negative and one positive HIV test result, with the interval bound by the EP-DDI and LP-DDI. For many applications, the EP-DDI and LP-DDI may be more useful than the EDDI itself: examples might include determining subject eligibility for an acute intervention, talking to a patient about partners who may have been exposed during the plausible interval, or selecting specimens for inclusion in a research study. Researchers with access to cohorts with both multiple HIV test results and independent estimates of a plausible date of infection may pursue validation of our methods compared to their gold standard, understanding that there are known differences between estimated DDI and date of infection. Note that names for assays are sometimes regional; readers unsure about assays with the same or similar name are advised to refer to original source publications for clarification of assays and estimated diagnostic delays, available within the tool. More details about the analytical framework underlying this tool are available in Grebe et al. (forthcoming). This publicly available tool should be useful to researchers, clinicians or public health program staff wishing to use the methods described here to explore how to estimate DDI information from seroconversion testing histories; and will be updated as methods are further developed based on additional research.
Supplementary Material
Supplementary Table S1. Diagnostic delays used as model inputs for determining plausible interval and estimated infection times.
ACKNOWLEDGEMENTS
The Consortium for the Evaluation and Performance of HIV Incidence Assays (CEPHIA) comprises: Oliver Laeyendecker, Thomas Quinn, David Burns (National Institutes of Health); Alex Welte, Eduard Grebe, Reshma Kassanjee, David Matten, Hilmarié Brand, Trust Chibawara (South African Centre for Epidemiological Modelling and Analysis); Gary Murphy, Elaine McKinney, Jake Hall (Public Health England); Michael Busch, Sheila Keating, Mila Lebedeva, Dylan Hampton (Vitalant Research Institute); Christopher Pilcher, Shelley Facente, Kara Marson; (University of California, San Francisco); Susan Little (University of California, San Diego); Anita Sands (World Health Organization); Tim Hallett (Imperial College London); Sherry Michele Owen, Bharat Parekh, Connie Sexton (Centers for Disease Control and Prevention); Matthew Price, Anatoli Kamali (International AIDS Vaccine Initiative); Lisa Loeb (The Options Study – University of California, San Francisco); Jeffrey Martin, Steven G Deeks, Rebecca Hoh (The SCOPE Study – University of California, San Francisco); Zelinda Bartolomei, Natalia Cerqueira (The AMPLIAR Cohort – University of São Paulo); Breno Santos, Kellin Zabtoski, Rita de Cassia Alves Lira (The AMPLIAR Cohort – Grupo Hospital Conceição); Rosa Dea Sperhacke, Leonardo R Motta, Machline Paganella (The AMPLIAR Cohort – Universidade Caxias Do Sul); Esper Kallas, Helena Tomiyama, Claudia Tomiyama, Priscilla Costa, Maria A Nunes, Gisele Reis, Mariana M Sauer, Natalia Cerqueira, Zelinda Nakagawa, Lilian Ferrari, Ana P Amaral, Karine Milani (The São Paulo Cohort – University of São Paulo, Brazil); Salim S Abdool Karim, Quarraisha Abdool Karim, Thumbi Ndungu, Nelisile Majola, Natasha Samsunder (CAPRISA, University of Kwazulu-Natal); Denise Naniche (The GAMA Study – Barcelona Centre for International Health Research); Inácio Mandomando, Eusebio V Macete (The GAMA Study – Fundacao Manhica); Jorge Sanchez, Javier Lama (SABES Cohort – Asociación Civil Impacta Salud y Educación (IMPACTA)); Ann Duerr (The Fred Hutchinson Cancer Research Center); Maria R Capobianchi (National Institute for Infectious Diseases “L. Spallanzani”, Rome); Barbara Suligoi (Istituto Superiore di Sanità, Rome); Susan Stramer (American Red Cross); Phillip Williamson (Creative Testing Solutions / Blood Systems Research Institute); Marion Vermeulen (South African National Blood Service); and Ester Sabino (Hemocentro do Sao Paolo). CEPHIA was supported by grants from the Bill and Melinda Gates Foundation (OPP1017716, OPP1062806 and OPP1115799). Additional support for analysis was provided by a grant from the US National Institutes of Health (R34 MH096606).
Footnotes
Disclaimer: The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention (CDC). Use of trade names is for identification purposes only and does not constitute endorsement by the CDC or the US Department of Health and Human Services.
REFERENCES
- 1.Fiebig EW, Wright DJ, Rawal BD, Garrett PE, Schumacher RT, Peddada L, et al. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS. 2003;17(13):1871–9. [DOI] [PubMed] [Google Scholar]
- 2.Cohen MS, Gay CL, Busch MP, Hecht FM. The detection of acute HIV infection. The Journal of Infectious Diseases. 2010;202 Suppl 2:S270–7. [DOI] [PubMed] [Google Scholar]
- 3.Ananworanich J, Fletcher JL, Pinyakorn S, van Griensven F, Vandergeeten C, Schuetz A, et al. A novel acute HIV infection staging system based on 4th generation immunoassay. Retrovirology. 2013;10:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Panel on Treatment of Pregnant Women with HIV Infection and Prevention of Perinatal Transmission. Recommendations for Use of Antiretroviral Drugs in Transmission in the United States; 2018. [Google Scholar]
- 5.Priddy FH, Pilcher CD, Moore RH, Tambe P, Park MN, Fiscus SA, et al. Detection of acute HIV infections in an urban HIV counseling and testing population in the United States. Journal of Acquired Immune Deficiency Syndromes. 2007;44(2):196–202. [DOI] [PubMed] [Google Scholar]
- 6.Rutstein SE, Ananworanich J, Fidler S, Johnson C, Sanders EJ, Sued O, et al. Clinical and public health implications of acute and early HIV detection and treatment: a scoping review. Journal of the International AIDS Society. 2017;20(1):21579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Green N, Hoenigl M, Chaillon A, Anderson CM, Kosakovsky Pond SL, Smith DM, et al. Partner services in adults with acute and early HIV infection. AIDS. 2017;31(2):287–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takata H, Buranapraditkun S, Kessing C, Fletcher JL, Muir R, Tardif V, et al. Delayed differentiation of potent effector CD8(+) T cells reducing viremia and reservoir seeding in acute HIV infection. Science Translational Medicine. 2017;9(377). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li JZ, Etemad B, Ahmed H, Aga E, Bosch RJ, Mellors JW, et al. The size of the expressed HIV reservoir predicts timing of viral rebound after treatment interruption. AIDS. 2016;30(3):343–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosenberg ES, Altfeld M, Poon SH, Phillips MN, Wilkes BM, Eldridge RL, et al. Immune control of HIV-1 after early treatment of acute infection. Nature. 2000;407(6803):523–6. [DOI] [PubMed] [Google Scholar]
- 11.Herout S, Mandorfer M, Breitenecker F, Reiberger T, Grabmeier-Pfistershammer K, Rieger A, et al. Impact of Early Initiation of Antiretroviral Therapy in Patients with Acute HIV Infection in Vienna, Austria. PloS One. 2016;11(4):e0152910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fidler S, Porter K, Ewings F, Frater J, Ramjee G, Cooper D, et al. Short-course antiretroviral therapy in primary HIV infection. The New England Journal of Medicine. 2013;368(3):207–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grinsztejn B, Hosseinipour MC, Ribaudo HJ, Swindells S, Eron J, Chen YQ, et al. Effects of early versus delayed initiation of antiretroviral treatment on clinical outcomes of HIV-1 infection: results from the phase 3 HPTN 052 randomised controlled trial. The Lancet Infectious Diseases. 2014;14(4):281–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lundgren JD, Babiker AG, Gordin F, Emery S, Grund B, Sharma S, et al. Initiation of Antiretroviral Therapy in Early Asymptomatic HIV Infection. The New England Journal of Medicine. 2015;373(9):795–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pilcher CD, Ospina-Norvell C, Dasgupta A, Jones D, Hartogensis W, Torres S, et al. The Effect of Same-Day Observed Initiation of Antiretroviral Therapy on HIV Viral Load and Treatment Outcomes in a U.S. Public Health Setting. Journal of Acquired Immune Deficiency Syndromes. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murphy G, Pilcher CD, Keating SM, Kassanjee R, Facente SN, Welte A, et al. Moving towards a reliable HIV incidence test - current status, resources available, future directions and challenges ahead. Epidemiology and Infection. 2016:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masciotra S, McDougal JS, Feldman J, Sprinkle P, Wesolowski L, Owen SM. Evaluation of an alternative HIV diagnostic algorithm using specimens from seroconversion panels and persons with established HIV infections. Journal of Clinical Virology. 2011;52 Suppl 1:S17–22. [DOI] [PubMed] [Google Scholar]
- 18.Masciotra S, Luo W, Youngpairoj AS, Kennedy MS, Wells S, Ambrose K, et al. Performance of the Alere Determine HIV-½ Ag/Ab Combo Rapid Test with specimens from HIV-1 seroconverters from the US and HIV-2 infected individuals from Ivory Coast. Journal of Clinical Virology. 2013;58 Suppl 1:e54–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Owen SM, Yang C, Spira T, Ou CY, Pau CP, Parekh BS, et al. Alternative algorithms for human immunodeficiency virus infection diagnosis using tests that are licensed in the United States. Journal of Clinical Microbiology. 2008;46(5):1588–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.da Motta LR, Vanni AC, Kato SK, Borges LG, Sperhacke RD, Ribeiro RM, et al. Evaluation of five simple rapid HIV assays for potential use in the Brazilian national HIV testing algorithm. Journal of Virological Methods. 2013;194(1–2):132–7. [DOI] [PubMed] [Google Scholar]
- 21.Eller L, Manak M, Malia J, De Souza M, Shikuku K, Lueer C, et al. Reduction of HIV Window Period by 4th Gen HIV Combination Tests. Conference on Retroviruses and Opportunistic Infections (CROI); Mar 3–6, 2013; Atlanta, GA. [Google Scholar]
- 22.Perry KR, Ramskill S, Eglin RP, Barbara JA, Parry JV. Improvement in the performance of HIV screening kits. Transfusion Medicine. 2008;18(4):228–40. [DOI] [PubMed] [Google Scholar]
- 23.Delaney KP, Hanson DL, Masciotra S, Ethridge SF, Wesolowski L, Owen SM. Time Until Emergence of HIV Test Reactivity Following Infection With HIV-1: Implications for Interpreting Test Results and Retesting After Exposure. Clinical Infectious Diseases. 2017;64(1):53–9. [DOI] [PubMed] [Google Scholar]
- 24.ZeptoMetrix. HIV Seroconversion Panels 2016. [Available from: http://www.zeptometrix.com/store/quality-control-panels/seroconversion-panels/hiv/.
- 25.SeraCare. AccuVert Seroconversion Panels 2016. [Available from: https://www.seracare.com/products/validation-and-lot-qualification-materials/seroconversion-panels/.
- 26.UNAIDS/WHO Working Group on Global HIV/AIDS and STI Surveillance. When and How to Use Assays for Recent Infection to Estimate HIV Incidence at a Population Level. Geneva; 2011. [Google Scholar]
- 27.Kuhar DT, Henderson DK, Struble KA, Heneine W, Thomas V, Cheever LW, et al. Updated US Public Health Service guidelines for the management of occupational exposures to human immunodeficiency virus and recommendations for postexposure prophylaxis. Infection Control and Hospital Epidemiology. 2013;34(9):875–92. [DOI] [PubMed] [Google Scholar]
- 28.Moyer VA US Preventive Services Task Force. Screening for hepatitis C virus infection in adults: U.S. Preventive Services Task Force recommendation statement. Annals of Internal Medicine. 2013;159(5):349–57. [DOI] [PubMed] [Google Scholar]
- 29.Delaney KP, Wesolowski LG, Owen SM. The Evolution of HIV Testing Continues. Sexually Transmitted Diseases. 2017;44(12):747–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Donnell D, Ramos E, Celum C, Baeten J, Dragavon J, Tappero J, et al. The effect of oral preexposure prophylaxis on the progression of HIV-1 seroconversion. AIDS. 2017;31(14):2007–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cohen SE, Sachdev D, Lee SA, Scheer S, Bacon O, Chen MJ, et al. Acquisition of tenofovir-susceptible, emtricitabine-resistant HIV despite high adherence to daily pre-exposure prophylaxis: a case report. The Lancet HIV. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henrich TJ, Hatano H, Bacon O, Hogan LE, Rutishauser R, Hill A, et al. HIV-1 persistence following extremely early initiation of antiretroviral therapy (ART) during acute HIV-1 infection: An observational study. PLoS Medicine. 2017;14(11):e1002417. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1. Diagnostic delays used as model inputs for determining plausible interval and estimated infection times.