To the Editor:
Regulatory T (Treg) cells have been subject of many debates and controversies. Discrepancies exist in regard to Treg-cell development, their recognition of antigen, specificity of immune regulation, requirement for T-cell receptor (TCR) activation, and regulatory mechanisms.1,2 We previously used somatic cell nuclear transfer (SCNT) to generate a naturally occurring Treg (nTreg)-cell model, T138.3 Particularly, we found that the TCR strength of T138 contradicted findings in wild-type (WT) mice, prompting us to hypothesize that Treg cells are heterogeneous with distinctive subsets. With our unbiased SCNT approach, we generated another Treg-cell model (T143) using FoxP3GFP-expressing CD4+ T cells from the spleen of NOD-FoxP3GFP-Rag+/− mice as described previously.3–5
We identified the V(D)J rearrangements underlying T143 as Trav5–4-J40 and Trbv15-J2–3-C2 (see Fig E1, A, in this article’s Online Repository at www.jacionline.org). Thymic development of T143-Rag1−/− revealed a significant increase in CD4 single-positive (CD4SP) cells (~12% WT vs ~39% T143) and a significant decrease in CD4-CD8 double-positive (DP) cells when compared with WT NOD mice (~77% WT vs ~34% T143) (Fig 1, A). Interestingly, CD4 expression levels in DP cells were lower in T143 than in WT. Strikingly, we identified thymic FoxP3+ T cells in CD4SP cells of T143-Rag−/− (Fig 1, B; ~6% WT vs 0.3% T143). In the spleen, we found approximately 10% FoxP3+ CD4+ T cells in T143-Rag1−/− compared with approximately 17% in WT (Fig E1, B). Given that we had previously identified a fate-determining role for nTreg cell–derived TCRβ, we tested whether TCRβ of T143 contributes to lineage choice. We found that 2 copies of T143 TCRβ skewed T-cell development similarly as seen in T143-Rag1−/− (lower frequency of DP, higher frequency of CD4SP, and lower CD4 levels on DP), while a single copy had negligible impact on lineage choice (Fig E1, C).
FIG E1.
TCR of aTreg-cell T143 model, and splenic Treg cell. A, Identification of the V(D)J rearrangements for TCR-α (upper) and TCR-β (lower) chains in aTreg-cell model T143. B, Representative FACS plots (left) and scatter plot (right) showing the presence and frequencies of splenic FoxP3+ T cells in WT NOD and T143-Rag1−/− mice. C, Representative flow cytometric analysis of the impact of T143-derived TCRβ on thymic development in mice possessing a single copy (middle) or 2 copies of TCRβ (right). FACS, Fluorescence-activated cell sorting. Error bars are mean ± SEM. ***P < .001.
FIG 1.
Development and differentiation of SCNT aTreg-cell model T143. FACS plots and scatter dot plots of thymic development (A) and Treg-cell frequencies (B) in thymus. TCR strength determined as expression of Nur77 (C) or CD5 (D) on indicated cells. Differentiation of FoxP3− CD4+ T cells from indicated mice into Treg cells (E) or TH1 (F) in vitro. FACS, Fluorescence-activated cell sorting; MFI, mean fluorescence intensity; ns, not significant. Error bars = mean ± SEM. ***P < .001.
By determining expression levels of Nur77 and CD5, we previously showed that FoxP3+ nTreg and FoxP3− pre-nTreg cells possess comparable TCR strength, thus arguing against agonist-dependent development.3,6 We determined TCR strength in T143, and found significantly higher Nur77 and CD5 levels in FoxP3+ CD4+ T cells than in their FoxP3− counterpart, suggesting higher TCR strength (Fig 1, C and D). Hence, we refer to T143 as agonist-induced Treg (aTreg) cell.
We previously showed that FoxP3− CD4+ pre-nTreg cells carrying the same TCR as nTreg cells were poised toward FoxP3 expression and away from TH1 differentiation. Thus, we determined whether FoxP3− CD4+ T cells carrying the same TCR as aTreg cells were poised similarly. FoxP3− CD4+ T cells from T143 were significantly better in differentiating into FoxP3+ T cells than from WT (~62% T143 vs 52% WT) (Fig 1, E). However, we observed no significant difference in their ability to differentiate into TH1 (Fig 1, F; ~48% WT vs ~52% T143). For consistency, we refer to FoxP3− CD4+ T cells from aTreg cells as pre-aTreg cells.
We previously found that CD8+ T cells expressing nTreg cell–derived TCRβ had reduced capabilities to express FoxP3 upon induction. We observed similar results for CD8+ T cells expressing aTreg cell–derived TCRβ (see Fig E2, A, in this article’s Online Repository at www.jacionline.org; ~48% WT vs ~8% T143), whereas we found no difference in their capability to differentiate into IFN-γ–secreting cells (~76% WT vs ~79% T143, Fig E2, B).
FIG E2.
Characterization of CD8+ T cells plasticity. A, CD8+ T cells from WT NOD and T143-ββ mice were purified and cultured with anti-CD3 and anti–CD28-coated beads in the presence of IL-2, TGFβ, and anti–IFN-γ to induce FoxP3 expression. Cells were analyzed after 4 days in culture. Representative flow cytometric analysis (left) and scatter plot (right) are shown. B, CD8+ T cells from WT NOD and T143-ββ mice were purified and cultured with IL-2, IL-12, and anti–IL-4 to induce TH1 differentiation. Cells were analyzed after 4 days in culture. Representative flow cytometric analysis (left) and scatter plot (right) are shown. n.s., Not significant. Error bars are expressed as mean ± SEM; ***P < .001.
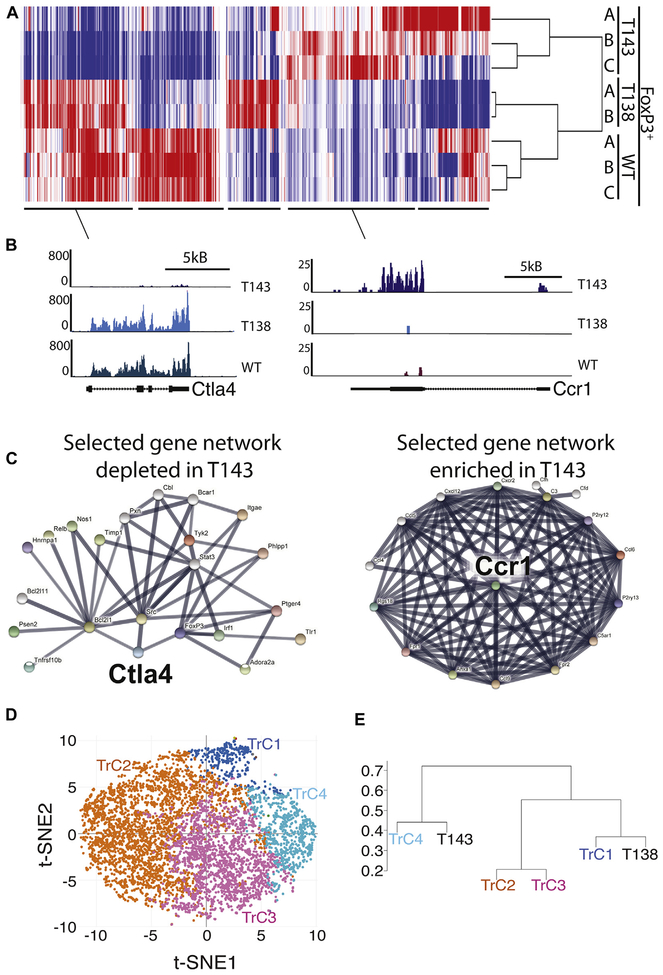
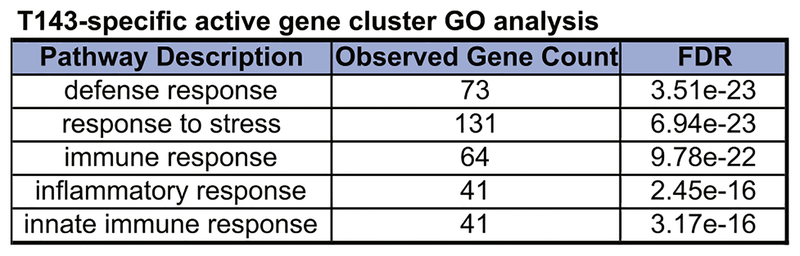
The development, frequencies, and plasticity of T143 were quite distinct from those in our previously reported T138, and urged us to compare transcriptional programs of both. We performed RNA-Seq on FoxP3+ CD4+ T cells from T143, T138, and WT. Genome-wide hierarchical clustering revealed differentially expressed genes in FoxP3+ T cells from T138, T143, and the bulk from WT (Fig 2, A). We identified a gene cluster that was specifically activated (eg, Ccr1), and one that was specifically repressed in aTreg cells (eg, Ctla4) (Fig 2, A and B). Next, we used Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) to determine specific protein-protein interaction network in these differentially expressing gene clusters.7 Ccr1 forms an interconnected gene network with many aTreg cell–specific active genes, whereas Ctla4 forms a distinctive repressive gene network absent in aTreg cells (Fig 2, C). GO term analysis of T143-specific active gene network showed enrichment in immune functions (see Fig E3 in this article’s Online Repository at www.jacionline.org). These data demonstrated that aTreg cells execute transcription programs unlike nTreg cells.
FIG 2.
Transcriptomes of nTreg, aTreg, and WT cells show distinct subsets. Hierarchical clustering of RNA-Seq data from T143, T138, and WT CD4+ Treg cells (A). T143-specific repressed (Ctla4) or active (Ccr1) gene clusters (B) and protein-protein interaction gene network (C). PCA and t-SNE analyses revealed potentially 4 Treg-cell clusters (TrC) in WT Treg cells (D). Hierarchical clustering shows T138 clusters with TrC1, and T143 with TrC4 (E). PCA, Principal-component analysis; t-SNE, t-distributed Stochastic Neighbor Embedding.
FIG E3.
GO analysis of T143-specific active gene cluster. T143-specific active genes are enriched for immune functions.
Our data on T138 and T143 demonstrated that nTreg and aTreg cells are 2 distinct thymic-derived Treg-cell subsets.3 To determine heterogeneity within the WT Treg-cell pool, we performed single-cell RNA-seq (scRNA-Seq) analysis. Principal-component analysis was performed and visualized as scatter plot using t-distributed Stochastic Neighbor Embedding. Our analysis revealed 4 potential Treg-cell clusters (TrC1–4) within WT Treg cells (Fig 2, D). To determine whether our nTreg-cell and aTreg-cell models align with any of these 4 TrCs, we averaged our previous RNA-Seq data from T143 and T138 and compared it with TrC1–4. Strikingly, we found that nTreg-cell model T138 clustered closely with TrC1, whereas aTreg-cell model T143 clustered with TrC4, suggesting that each cluster represents a transcriptionally different Treg-cell subset (Fig 2, E).
We used SCNT and scRNA-Seq to demonstrate that nTreg and aTreg cells are 2 distinct thymic Treg-cell subsets. Despite the lower relative frequency of thymic Treg cells in aTreg cells3 (1.1% in T138, and 0.3% in T143), the absolute numbers of thymic Treg cells were comparable, and the overall frequencies in T138 and T143 were higher than in current existing transgenic Treg-cell models. Although we currently lack a complete understanding of lymphocyte receptor diversification especially in gnathostomata,8 our data clearly demonstrate that TCRs have a profound impact on T-cell development and linage specification, emphasizing the need to generate additional novel SCNT models on pure backgrounds.
We demonstrated that aTreg cells develop in an agonist-dependent manner, whereas nTreg cells develop in an agonist-independent manner. Thus, we hypothesize that the transcription factor AIRE (autoimmune regulator) might support the development of some Treg cells, such as aTreg cells, but not of nTreg cells. This would explain why Treg cells exist—albeit in reduced numbers—in Aire-deficient mice and humans (referred to as autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy), yet autoimmune diseases occur.
In line with our hypothesis that WT Treg cells are heterogeneous, our scRNA-Seq analysis revealed potentially 4 Treg-cell clusters. Strikingly, we identified 2 clusters, TrC1 and TrC4, that closely resemble our nTreg-cell and aTreg-cell models, respectively. So far, several mechanisms had been identified by which Treg cells might regulate effector cells, such as through the expression of Ctla4. Our RNA-Seq data showed that T143 expressed significantly less Ctla4 than did T138, which supports the hypothesis that different Treg-cell subsets might execute different immune regulatory function. This suggests that blocking antibodies against CTLA4 used in cancer immunotherapy might only target certain Treg-cell subsets.9 Clearly, more SCNT-derived Treg-cell models are needed to determine common features of Treg cells within a subset, and to unambiguously distinguish Treg-cell subsets.
Using SCNT we can deconstruct the Treg-cell pool and generate accurate Treg-cell models that enable us to determine the differences in development, TCR strength, signaling, plasticity, and especially their functions. The existence of different Treg-cell subsets could explain the plentitude of controversial data reported on Treg cells. Importantly, novel insights into the biology of Treg-cell subsets might provide new opportunities for the treatments of autoimmune diseases and cancer, and to optimize vaccination strategies.
METHODS
Mice and SCNT
B6D2F1 females were generated in-house by breeding C57BL/6 with DBA/2 mice for the use of oocyte donors. NOD-FoxP3GFP-Rag+/− mice were used to isolate donor T cells for SCNT. The SCNT procedure was performed as previously described.E1,E2 Briefly, oocytes from superovulated B6D2F1 female were enucleated, and donor cells were transferred into enucleated oocytes. Embryonic stem cells were derived from resulting SCNT blastocysts and used to generate chimeric mice. NOD-T143-Rag−/− mice were obtained through breeding with NOD-Rag−/− mice, and various genotypes were used for analysis as indicated. All mice experiments were approved by the Institutional Animal Care and Use Committee at the Scripps Research Institute.
TCR identification
The TCR of T143 was identified as previously described.E1 Briefly, RNA was isolated from spleenocytes of SCNT mice (Macherey-Nagel, Bethlehem, Pa). 5′-RACE was performed, and results were analyzed using the Ensembl Genome Browser (www.ensembl.org) and the international ImMunoGeneTics information system (www.imgt.org).
Fluorescence-activated cell sorting and cell sorting
Single-cell suspensions of thymus, spleen, or lymph nodes were prepared and red blood cells were lysed. Cells were incubated with various combinations of the following antibodies: CD3, CD4, CD5, CD8, CD19, FoxP3, Nur77, IFN-γ (eBioscience, San Diego, Calif, and Tonbo, San Diego, Calif). Samples were collected using FACS LSRII (BD, San Jose, Calif) or sorted using FACSAria (BD), and data were analyzed with FlowJo software (Tree Star, Ashland, Ore). Differences between groups were analyzed using 2-tailed t test (Graphpad Prism software, La Jolla, Calif).
Nur-77 in vitro assay
T Cells were purified from spleen of WT NOD and T143-Rag−/− mice and activated with anti-CD3/28 beads (Dynabeads, Invitrogen, Carlsbad, Calif). Cells were intracellularly stained by Nur77-PE 5 to 6 hours after activation (eBioscience). Nur-77 levels were determined using flow cytometry and normalized to unstimulated FoxP3− CD4+ T cells from NOD.
Bulk RNA-Seq and data analysis
RNAs from indicated populations were isolated from the spleen of either WT NOD or indicated SCNT mice using Nucleospin RNA XS kit and treated with DNase according to manufacturer’s protocol (Macherey-Nagel). RNA was processed into RNA-Seq libraries using Truseq Stranded mRNA-Seq sample prep kit (Illumina, San Diego, Calif) following manufacturer’s instructions. Briefly, polyA-mRNA was enriched using oligo-dT magnetic beads. Next, RNA was fragmented and reverse-transcribed, and cDNA was end-repaired, adenylated, adapter-ligated, and PCR amplified. RNA-Seq libraries were then sequenced on Illumina HiSeq2500 at single-end 50-bp (base pair), resulting in 25 million to 30 million reads per library. Sequencing reads were aligned to the mouse genome (mm10, MGSCv38) using STAR (v2.2.0c).E3 The RNA-Seq data have been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) and are accessible through GEO Series accession number GSE97850. Gene expression was quantified using aligned reads to exons of RefSeq transcripts using HOMER.E4 Differential gene expression was determined with edgeRE5 and StringDB.E6
Single-cell RNA-Seq and data analysis
Spleenic FoxP3+ CD4+ T cells from NOD-FoxP3GFP mice were purified using flow cytometry, washed, and diluted to 1000 cells/μL in 0.04% BSA/PBS. Chromium single-cell 3′ RNA-Seq library generation was carried according to 10X Genomics protocolsE7 (Pleasanton, Calif). Single-cell suspension was combined with reverse transcriptase master mixes and loaded onto the chromium single-cell 39 chip together with single-cell 3’ gel beads and partition oil for GEM generation on the chromium controller. GEMs were then incubated for 2 hours at 55°C for reverse transcription reaction and then cDNA was purified using Silane DynaBeads and SPRISelect beads. cDNA was then amplified for 12 cycles and sheared using Covaris. Fragmented cDNA was then end-repaired, A-tailed, and adapter-ligated before index PCR for 10 cycles. Final single-cell libraries were sequenced on the Illumina NextSeq500. sc-RNA-Seq data were analyzed using the 10x Genomics Chromium platform. For comparative analysis, expression counts from sc-RNA clusters from the pool of FoxP3+ CD4+ T cells were averaged per gene and normalized in units of tag per million to match normalization of bulk expression counts from nTreg-cell T138 and aTreg-cell T143 models before clustering. A representative set of 231 genes for clustering was generated from the intersection of differentially expressed genes in 4 Treg-cell clusters, and differentially expressed genes from our nTreg-cell T138 and aTreg-cell T143 models. To overcome the scarcity of the count matrix from single-cell subsets, gene expressions were standardized to mean of 0 and SD of 1 across all sets (4 clusters TrC1–4 and 2 models, T138 and T143). Hierarchical clustering of sets was performed using Pearson correlation as distance measure with average linkage criterion.
T-cell differentiation
T-cell differentiation experiments were performed as described previously.E8 Briefly, FoxP3- CD4+ T cells were purified and activated using anti-CD3/CD28 beads (Dynabeads, Invitrogen). For TH1 differentiation, 1 × 105 T cells were stimulated in the presence of IL-2 (20 U/mL; eBioscience), IL-12 (20 ng/mL; Tonbo), and anti–IL-4 (10 μg/mL; Tonbo). For Treg-cell differentiation, cells were stimulated in the presence of IL-2 (100 U/mL; eBioscience), TGF-β (5 ng/mL; Tonbo), and anti–IFN-γ (10 μg/mL; Tonbo). After 4 days, T cells were stimulated with ionomycin (500 ng/mL) and phorbol 12-myristate 13-acetate (50 ng/mL) in the presence of monensin (eBioscience) for 4 hours before intracellular staining. FoxP3 staining was performed using FoxP3/transcription factor staining kit (Tonbo).
Acknowledgments
We thank David Nemazee, Changchun Xiao, Amanda Gavin, and Michael Petrascheck for fruitful discussions and critical reading of the manuscript. We thank the FACS core, and the animal resources department at The Scripps Research Institute.
Research reported in this publication was supported by The Scripps Research Institute start up fund (O.K.) and the National Cancer Institute of the National Institutes of Health under award number T32CA009370 (E.K.).
Disclosure of potential conflict of interest: M. Sabouri-Ghomi is an employee of Fate Therapeutics. K. O. Lui receives grants from Research Grants Council of Hong Kong (grant nos. 24110515 and 14111916), Health and Medical Research Fund (grant nos. 03140346 and 04152566), and the Croucher Foundation. The rest of the authors declare that they have no relevant conflicts of interest.
REFERENCES
- 1.Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol 2012;30:531–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shevach EM, Thornton AM. tTregs, pTregs, and iTregs: similarities and differences. Immunol Rev 2014;259:88–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ku M, Chang SE, Hernandez J, Abadejos JR, Sabouri-Ghomi M, Muenchmeier NJ, et al. Nuclear transfer nTreg model reveals fate-determining TCR-beta and novel peripheral nTreg precursors. Proc Natl Acad Sci U S A 2016;113: E2316–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirak O, Frickel EM, Grotenbreg GM, Suh H, Jaenisch R, Ploegh HL. Transnuclear mice with pre-defined T cell receptor specificities against Toxoplasma gondii obtained via SCNT. J Vis Exp 2010;e2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kirak O, Frickel EM, Grotenbreg GM, Suh H, Jaenisch R, Ploegh HL. Transnuclear mice with predefined T cell receptor specificities against Toxoplasma gondii obtained via SCNT. Science 2010;328:243–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moran AE, Holzapfel KL, Xing Y, Cunningham NR, Maltzman JS, Punt J, et al. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J Exp Med 2011;208:1279–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43:D447–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pancer Z, Cooper MD. The evolution of adaptive immunity. Annu Rev Immunol 2006;24:497–518. [DOI] [PubMed] [Google Scholar]
- 9.Du X, Tang F, Liu M, Su J, Zhang Y, et al. A reappraisal CTLA-4 checkpoint blockade in cancer immunotherapy. Cell Res 2018;28:416–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- E1.Kirak O, Frickel EM, Grotenbreg GM, Suh H, Jaenisch R, Ploegh HL. Transnuclear mice with pre-defined T cell receptor specificities against Toxoplasma gondii obtained via SCNT. J Vis Exp 2010;e2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E2.Kirak O, Frickel EM, Grotenbreg GM, Suh H, Jaenisch R, Ploegh HL. Transnuclear mice with predefined T cell receptor specificities against Toxoplasma gondii obtained via SCNT. Science 2010;328:243–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E3.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E4.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010;38:576–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E5.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E6.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43:D447–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E7.Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun 2017;8: 14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E8.Ku M, Chang SE, Hernandez J, Abadejos JR, Sabouri-Ghomi M, Muenchmeier NJ, et al. Nuclear transfer nTreg model reveals fate-determining TCR-beta and novel peripheral nTreg precursors. Proc Natl Acad Sci U S A 2016;113:E2316–25. [DOI] [PMC free article] [PubMed] [Google Scholar]