Abstract
Current understanding of chromosome folding largely relies on chromosome conformation capture (3C)-based experiments, where chromosomal interactions are detected as ligation products after chromatin crosslinking. To measure chromosome structure in vivo, quantitatively and without crosslinking and ligation, we implemented a modified version of DamID named DamC, which combines DNA-methylation based detection of chromosomal interactions with next-generation sequencing and biophysical modelling of methylation kinetics. DamC performed in mouse embryonic stem cells provides the first in vivo validation of the existence of topologically associating domains (TADs), CTCF loops and confirms 3C-based measurements of the scaling of contact probabilities. Combining DamC with transposon-mediated genomic engineering shows that new loops can be formed between ectopic and endogenous CTCF sites, which redistributes physical interactions within TADs. DamC provides the first crosslinking- and ligation-free demonstration of the existence of key structural features of chromosomes and provides novel insights into how chromosome structure within TADs can be manipulated.
Introduction
Characterizing chromosome folding is fundamental to better understand gene expression and how it possibly constrains genome evolution. Chromosome conformation capture (3C) methods, and notably its high-throughput sequencing-based derivatives such as Hi-C, 5C and 4C1, have significantly contributed to current understanding of genome architecture revealing that chromosome folding is driven by at least two independent mechanisms. On the one hand, the mutually exclusive associations between transcriptionally active or inactive loci generate the so-called A and B compartments2. On the other hand, chromatin loops are formed between regulatory sequences and between convergent CTCF binding sites, the latter through cooperative action between cohesin and the DNA-binding protein CTCF3. The interplay between compartmentalization and CTCF-cohesin looping results in complex hierarchies of folding domains4,5, among which topologically associating domains (TADs)6–8 stand out as preferential functional units9. The involvement of CTCF in loop formation has been demonstrated using global depletion experiments10,11, as well as targeted deletions and inversions of CTCF sites leading to loss of looping interactions12–14. The underlying mechanisms are however still incompletely understood. An influential hypothesis is that CTCF-mediated interactions occur as cohesin extrudes chromatin loops until it is blocked by CTCF bound to DNA in a defined orientation15. According to this hypothesis, ectopic insertion of CTCF sites could result in newly established loops onto endogenous CTCF sites, depending on their mutual orientation. Whether this actually occurs and how it modifies interactions within TADs has however not been demonstrated.
In 3C, detection of spatial proximity relies on formaldehyde crosslinking followed by digestion and ligation of crosslinked chromatin1. Crosslinking and ligation are sources of potential experimental bias, raising the question of whether structures detected by 3C methods actually exist in living cells16–19. 3C crosslinking frequencies are assumed to be proportional to absolute chromosomal contact probabilities and used to build mechanistic physical models of chromosome folding20,21, including the loop-extrusion model14,15,22,23. However, formal proof of this assumption is missing. Independent techniques such as DNA fluorescence in situ hybridization (DNA FISH)6,24, genome architecture mapping (GAM)25, native 3C26 and split-pool recognition of interactions by tag extension (SPRITE)27 have also detected loops, TADs and compartments. Nevertheless, these methods still involve substantial biochemical manipulation of cells, and employ either crosslinking or ligation.
An alternative approach to study chromosomal contacts without crosslinking and ligation is recruiting an ectopic DNA modifying enzyme to specific genomic locations, and detecting chemically modified DNA at sequences that physically interact with the recruitment sites. Three previous studies have provided proof of principle for such an approach using a modified version of DamID26,28,29. In DamID, the bacterial DNA adenine methyltransferase Dam is fused to a DNA-binding protein resulting in adenine methylation within GATC motifs in the neighborhood of the protein DNA binding sites30. Methylated GATCs (GmATC) are specifically digested by the DpnI restriction enzyme, allowing to determine DNA binding locations of the fusion protein after normalization for nonspecific methylation by freely diffusing Dam. Methylation at distal chromosomal sites interacting with the viewpoint in 3D can also be observed26,28,29 if interaction-specific methylation is significantly higher than nonspecific methylation. However, previous studies detected methylated DNA with semi-quantitative PCR readouts and analyzed interactions of one viewpoint with a limited number of restriction sites, similar to early 3C experiments31. This and the lack of formal schemes to convert methylation states into contact probabilities have prevented these versions of DamID from reaching the resolution and throughput needed to detect TAD boundaries and CTCF loops. Thus to date no crosslinking- and ligation-free method is available to study chromosome interactions in the context of contacts made by all other surrounding genomic sequences. Remarkably, evidence that CTCF-associated loops exist is based exclusively on crosslinking methods.
Here we present DamC, a new modified version of DamID coupled to physical modelling of DNA methylation kinetics. In DamC, Dam is recruited to ectopically inserted Tet operators (TetOs) through fusion to the reverse tetracycline receptor (rTetR). Methylated DNA is detected by high-throughput sequencing allowing the identification of chromosomal contacts at high genomic resolution across hundreds of kilobases around viewpoints. Modelling of this process shows that experimental output in DamC is proportional to chromosomal contact probabilities, providing a theoretical framework for the interpretation of data.
Using DamC we provide the first crosslinking- and ligation-free validation of structures identified by 3C methods. By comparing DamC with 4C-seq and Hi-C at hundreds of genomic locations in mouse embryonic stem cells (mESC), we confirm the existence of TADs and CTCF loops. We also show that the scaling of contact probabilities measured in DamC is the same as in 4C and Hi-C, providing evidence in favor of current interpretations of 3C-based data in terms of physical models of chromosome folding. We additionally demonstrate that ectopic insertion of CTCF sites can lead to the formation of new loops with endogenous CTCF-bound sequences and alter sub-TAD contacts. This shows that chromosome structure can be manipulated by inserting short ectopic sequences that rewire interactions within TADs.
Results
DamC: Methylation-based detection of chromosomal contacts in vivo
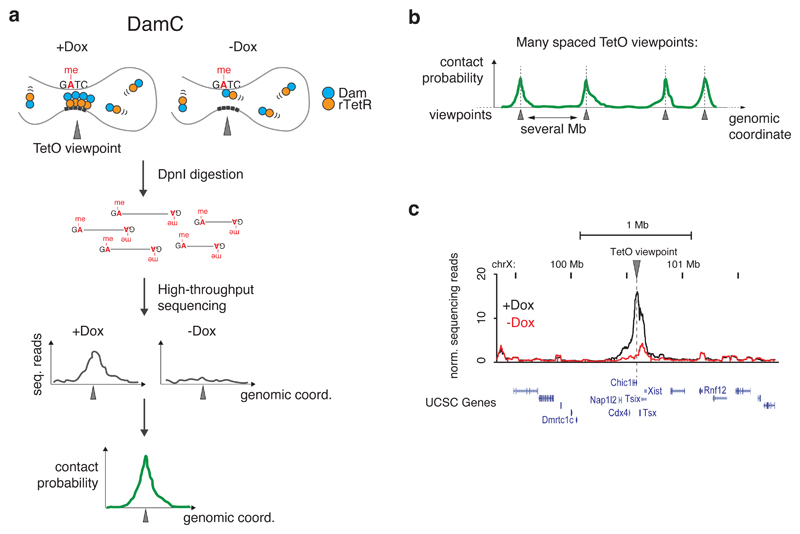
Based on the results of previous studies26,28,29, we reasoned that fusing Dam to rTetR and inserting an array of TetOs in the genome would ensure targeted, inducible recruitment of large numbers of Dam molecules to a specific genomic viewpoint in the presence of doxycycline (Dox) (Figure 1a, left). In the absence of Dox, rTetR-Dam would not bind to the viewpoint, allowing accurate estimation of nonspecific methylation (Figure 1a, right) and precise background correction. Coupled to high-throughput sequencing, this strategy could provide 4C-like, ‘one vs. all’ profiles32 of contact probabilities from the TetO viewpoint (Figure 1a) across large genomic distances and at high genomic resolution (one GATC every ~250bp on average). Inserting multiple TetO arrays separated by large genomic distances would allow interrogating chromosomal interactions in parallel from many viewpoints (Figure 1b). We refer to this method as ‘DamC’.
Figure 1. DamC: methylation-based measurement of chromosomal interactions.
a. Scheme of DamC experiments. In the presence of doxycycline (+Dox), rTetR-Dam binds to a genomic viewpoint through a TetO array and methylates adenines in GATC sites that contact the viewpoint. In the absence of doxycycline (-Dox), only nonspecific methylation by freely diffusing rTetR-Dam occurs. Methylated GATCs can be detected by digestion of genomic DNA with DpnI and next-generation sequencing of the restriction sites. Correction for nonspecific methylation allows extracting contact probabilities with the TetO viewpoint. b. Insertion of multiple TetO arrays spaced by several Mb allows detecting interaction from single viewpoints in parallel. c. Proof-of-principle experiment showing increased methylation in cis following the recruitment of rTetR-Dam to an array of 256 TetO sites in the 5’UTR of the Chic1 gene in the presence of Dox (black), compared to the -Dox control where rTetR-Dam is not recruited (red).
To test this approach, we employed female mESCs carrying an array of 256 TetOs at the 3’ end of the Chic1 gene within the X inactivation center33. We transfected an X0 subclone of these cells with an rTetR-Dam expression plasmid and measured methylation after 24 hours34. Quantification of the methylated GmATCs by high-throughput sequencing revealed significantly higher methylation upon Dox induction compared to the uninduced control over approximately 300kb around the TetO viewpoint (Figure 1c). Thus, targeted recruitment of Dam leads to increased methylation in cis over long genomic distances, consistent with previous observations using semi-quantitative methods for the detection of methylation26,28,29. Since methylation is determined by the interplay between methyltransferase activity and passive demethylation during DNA replication, we reasoned that it should be possible to model this process and derive chromosomal contact probabilities from sequencing readouts.
DamC enrichment is proportional to chromosomal contact probabilities
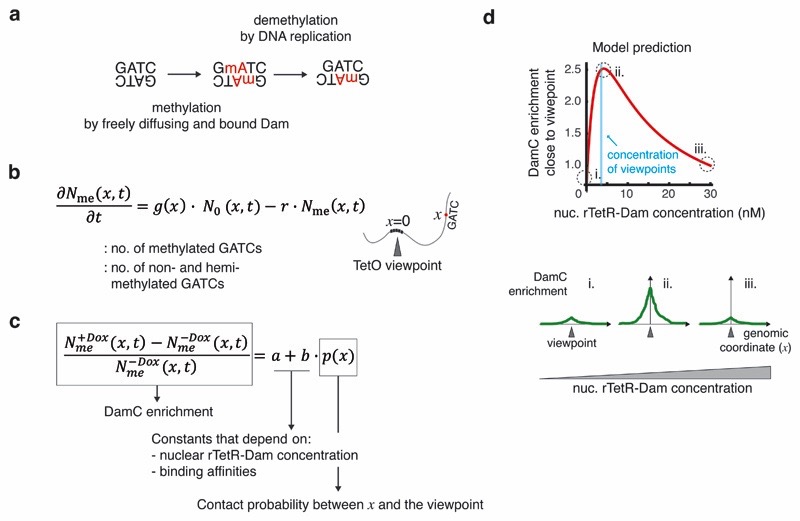
The methylation level of a single GmATC is determined by a dynamic interplay between methylation (by freely diffusing or TetO-bound Dam) and passive demethylation by DNA replication, when a fully methylated GmATC becomes two hemi-methylated sites that are essentially not detected in DamID35 (Figure 2a). To identify experimental quantities that are directly proportional to chromosomal contact probability, we generated a physical model describing the time evolution of methylation at an arbitrary genomic distance from the TetO viewpoint (Supplementary Note 1) through rate equations (Figure 2b) which take into account that methylation by TetO-bound Dam only occurs in the presence of Dox (Figure 1a). Methylation rates are allowed to depend on local biases (e.g. chromatin accessibility or mappability). Under the assumptions that methylation is faster than demethylation35 and the duration of an experiment, we found that contact probabilities between the GATC site and the TetO viewpoint are directly proportional to a measurable quantity. This quantity, which we refer to as ‘DamC enrichment’, is simply the relative difference between methylation levels in the presence and absence of Dox (Figure 2c). Thus, DamC can directly measure chromosomal contact probabilities.
Figure 2. Physical model of methylation dynamics.
a. Unmethylated GATCs can be methylated by either freely diffusing or TetO-bound rTetR-Dam with rate g, and partially demethylated during DNA replication with rate r. Partially demethylated GATCs are inefficiently cut by DpnI and do not contribute to the DamC experiment. b. Model of methylation dynamics. The time evolution of the number of methylated and non- and hemi-methylated GATCs located at genomic distance x from a TetO viewpoint is described in terms of ordinary differential equations governed by rates g and r. c. The DamC enrichment at a generic location x is independent of time and proportional to the contact probability between x and the viewpoint. Proportionality constants a and b depend on the nuclear rTetR-Dam concentration and the binding affinities to TetO and nonspecific genomic sites. d. Model prediction using example parameter values (rTetR-TetO affinity = 5 nM, nonspecific affinity = 80 nM, 600 TetO insertions corresponding to ~2 nM in a nuclear volume of ~490 fl, and contact probability of 0.5 corresponding to an interaction occurring in half of the cell population). The behavior of the curve is conserved across a wide range of physiologically relevant parameter values (see Supplementary Figure 1b).
For a given contact probability between the GATC site and the TetO viewpoint, the model predicts that DamC enrichment depends on: 1) the nuclear rTetR-Dam concentration, 2) the rTetR-Dam binding affinity for the TetO array, and 3) the average nonspecific binding affinity of rTetR-Dam for endogenous genomic sites (Figure 2c). DamC enrichment does not depend on local methylation biases and therefore should not be affected by differential accessibility or mappability, provided that interactions with the TetO viewpoint can increase methylation at the GATC site (i.e. local methylation is not saturated in the absence of Dox). In real experiments, where binding affinities are fixed, the main determinant of DamC enrichment is the nuclear concentration of rTetR-Dam. In particular, DamC enrichment should be maximal when rTetR-Dam concentration is around the nuclear concentration of TetO viewpoints (Supplementary Figure 1a), and negligible when the concentration is very high or very low (Figure 2d). This does not depend on the particular values of the affinity constants (Supplementary Figure 1b) and implies that maximal DamC enrichment occurs at different Dam concentrations depending on the number of viewpoints. Thus, modeling predicts that accurate control of rTetR-Dam nuclear concentrations is needed to perform DamC with optimal signal-to-noise ratio.
DamC from hundreds of genomic viewpoints validates model predictions
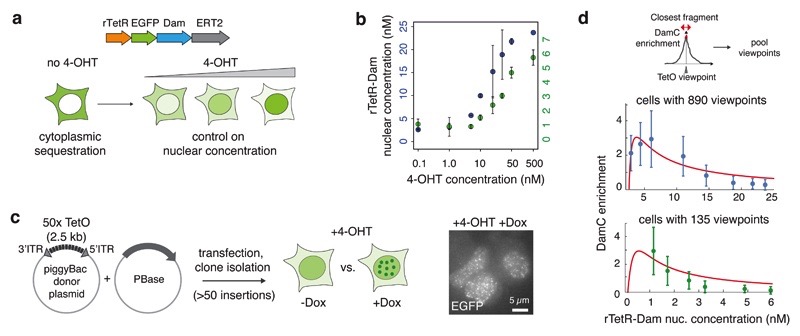
To test model predictions and measure chromosomal interactions using DamC, we established mESCs allowing to control the rTetR-Dam nuclear concentration. We first created a stable cell line expressing rTetR fused with enhanced green fluorescent protein (EGFP), Dam, and the mutant estrogen ligand-binding domain ERT2. ERT2 ensures cytoplasmic localization of the fusion protein in the absence of 4-hydroxy-tamoxifen (4-OHT), preventing constitutive GATC methylation. It also enables to control its nuclear level by changing 4-OHT concentrations in the culture medium (Figure 3a) as confirmed by increasingly nuclear accumulation of EGFP upon increasing 4-OHT doses (Supplementary Figure 2a).
Figure 3. An inducible mESC line to perform DamC and test the model predictions.
a. mESCs expressing rTetR-Dam-EGFP-ERT2 allow control of the nuclear concentration of the fusion protein by changing the amount of 4-hydroxy-tamoxifen (4-OHT) in the culture medium. b. Nuclear concentration of the rTetR-Dam fusion protein as a function of 4-OHT concentration in the polyclonal population with 890 insertions (blue) and in the subclone with 135 insertions (green). Number of protein copies per nucleus were determined using mass spectrometry on nuclear extracts and divided by the average nuclear volume (~490 fl) as determined using DAPI staining (see Supplementary Figure 2). Error bars are s.d. of two biological replicates (independent cell cultures). c. Random integration of large numbers of 50x TetO platforms using the piggyBac transposon. Accumulation of EGFP signal to nuclear foci in the presence of Dox (right: max. intensity projection over 10 Z planes) indicates binding of rTetR-Dam to the arrays and allows selecting clones with large numbers of insertions. d) Quantification of DamC experiments as a function of rTetR-Dam concentration in cells with 890 (upper panel, blue) and 135 (lower panel, green) TetO viewpoints. Blue data points, mean and s.d. from over the 100 TetO viewpoints with highest enrichment. Green data points, mean and s.d. from over 130 TetO viewpoints (5 viewpoints were excluded due to absence of DamC signal). Red line, model fit to the experimental data.
To measure chromosomal interactions in a wide variety of randomly selected genomic contexts in parallel, we further inserted arrays of 50 TetOs (each spanning approx. 2.7kb) using the piggyBac transposon system36 (Figure 3c). This resulted in clonal mESC lines carrying at least 100 TetO arrays, judging from EGFP accumulation in nuclear foci in the presence of 4-OHT and Dox (Figure 3c). We further selected one polyclonal population carrying a total of 890 TetO array insertions and one clonal line with 135 insertions (Supplementary Table 1 and 2), as determined by mapping piggyBac insertion locations (Methods). To quantitatively measure rTetR-Dam nuclear concentrations as a function of 4-OHT concentration, we analyzed nuclear protein extracts using mass spectrometry. Combining the proteomic ruler strategy with parallel reaction monitoring (PRM) (Methods, Supplementary Figure 2b-c and Supplementary Table 3) we estimated nuclear rTetR-Dam concentrations to vary gradually between approximately 3 and 25 nM, and 1 and 6 nM in the polyclonal and pure clonal lines, respectively, when increasing the 4-OHT concentration from 0.1 to 500 nM (Figure 3b).
We performed DamC after treating cells overnight with different doses of 4-OHT in the presence and in the absence of Dox. Experiments were performed using a custom next-generation sequencing library preparation protocol that includes unique molecular identifiers (UMI) and increases the coverage of methylated GATC sites genome-wide, thus maximizing proportionality between methylation levels and sequencing readout (Supplementary Figure 2d-e, Methods). We quantified DamC enrichment in the immediate vicinity of TetO viewpoints, and plotted it as a function of rTetR-Dam concentration quantified by mass spectrometry (Figure 3d). For the polyclonal line we considered the 100 insertions with highest signal-to-noise ratios, corresponding to the most abundant insertions. In the pure subclone, all insertions showed similar enrichment levels except five, possibly as a consequence of recombination of the TetO array or high levels of transcription at the insertion point preventing TetO binding. These insertions were discarded from analysis and their coordinates are provided in the Methods section.
Consistent with model predictions, in the polyclonal mESC line maximum enrichment occurs at ~3 nM corresponding to ~860 viewpoints (Figure 3d, upper panel). Model fitting returned an estimate of 0.4 nM for the specific rTetR-TetO binding constant, in the range of in vitro measurements37, and 17nM for the average non-specific binding constant. Again in line with the model, enrichment in the clonal line carrying ~7-fold less viewpoints (130) was compatible with maximal enrichment occurring at ~7-fold lower rTetR-Dam nuclear concentration (0.4 nM). These results provide a validation of the DamC model and support the interpretation of the DamC enrichment in terms of contact probabilities. They additionally highlight that in our experimental system, maximal DamC enrichment in cells with ~100 insertions is observed in a range of rTetR-Dam nuclear concentrations corresponding to 0.1-1 nM 4-OHT (Supplementary Figure 2f). In the following analysis, reads from these two conditions were pooled to maximize read coverage.
DamC reveals the existence of TADs and loops in vivo
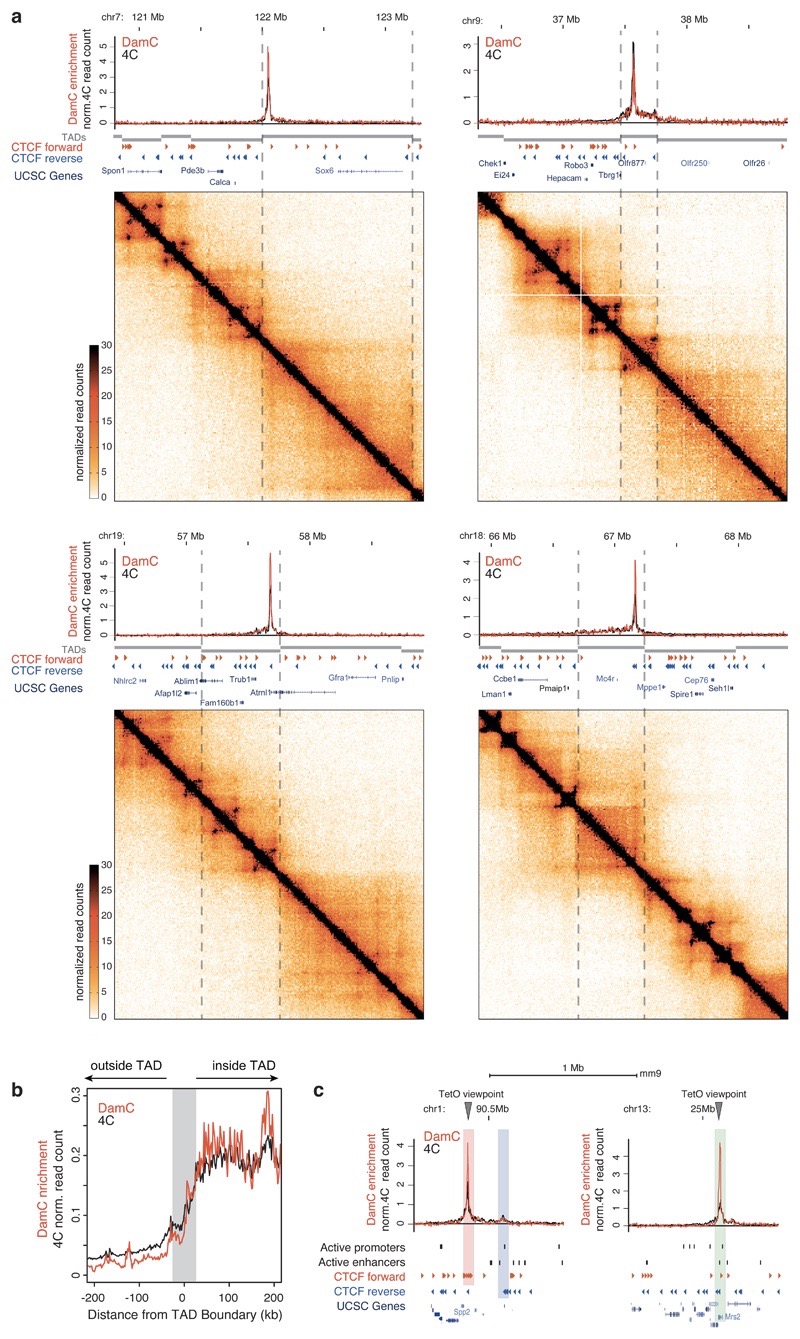
Under optimal 4-OHT concentrations (0.1-1 nM pooled), zooming into individual TetO viewpoints in the clonal line with 130 insertions revealed significant DamC enrichment over hundreds of kilobases around each viewpoint (Fig. 4a). Since biological replicates were highly correlated (Supplementary Figure 3a), we analyzed merged data. DamC enrichment profiles showed remarkable agreement with 4C performed using the same TetO arrays as viewpoints and DpnII as primary restriction enzyme (Figure 4a and Methods). DamC enrichment was systematically concentrated within TAD boundaries detected in Hi-C (Figure 4a) and steeply decayed across TAD boundaries by roughly a factor two, in excellent agreement with 4C (Figure 4b). Only a minor fraction of TetO insertions occurred in close proximity (<1kb) to either an active regulatory element or a CTCF site (Supplementary Figure 3b). Also in these cases, DamC enrichment profiles highly overlapped with 4C (Figure 4c) and revealed looping interactions between endogenous convergent CTCF sites (Figure 4c, left), which were confirmed using the partner CTCF sites as reciprocal viewpoint in 4C (Supplementary Figure 3c). The targeted TetO insertion at the 3’ end of Chic133 (Figure 1c) allowed measuring chromosomal interactions within the the well-characterized Tsix TAD6,38. In accordance with 4C, DamC recapitulated the previously observed CTCF-mediated interactions between Chic1, Linx and Xite/Tsix6, as well as the boundaries of the Tsix TAD (Supplementary Figure 3d). Additional DamC and 4C profiles are plotted in Supplementary Figure 4 and bedGraph tracks are available online (Data Accessibility section).
Figure 4. DamC confirms the existence of TAD boundaries and quantitatively correlates with 4C and Hi-C.
a. Four representative DamC and 4C interaction from the same piggyBac-TetO viewpoints, aligned with Hi-C experiments performed in the same cell line. Dashed lines mark TAD boundaries in mESC detected using CaTCH9. Hi-C data were binned at 10 kb resolution. DamC was performed using 0.1 and 1 nM 4-OHT (pooled). Data from two biological replicates were pooled for DamC, 4C and Hi-C. b.Aggregated plot over 130 TetO viewpoints showing DamC and 4C data aligned to TAD boundaries identified using CaTCH9. Gray shading: +/- 40 kb uncertainty on boundary definition9. c. Interaction profiles from viewpoints located <1kb from a CTCF site (left) belonging to a cluster of forward sites (red shading) interacting with reverse CTCF sites (blue shading) and <1kb from the active promoter of the Mrs2 gene (right), highlighted in the green shaded area.
We next investigated whether despite evident global similarities, DamC and 4C showed local differences. We defined a deviation score measuring differences between DamC and 4C interaction profiles within windows of 20 DpnI/II restriction fragments (5kb on average) (Supplementary Figure 3e). Most dissimilar windows were enriched in active chromatin marks (Supplementary Figure 3f), although local differences between DamC and 4C within these regions are relatively mild (Supplementary Figure 3e). We reasoned that local discrepancies might be due to the fact that the methylation signal highly correlates with chromatin accessibility measured by DNase I sensitivity (Supplementary Figure 3g). Correction by nonspecific methylation generally normalizes for chromatin accessibility in the DamC enrichment, unless GATC sites are highly methylated in the absence of Dox preventing further increases when interacting with the TetO viewpoint in +Dox conditions. However, only 0.05% of GATC sites within DNase I hypersensitive regions were saturated (Methods), and masking DNase I hypersensitive sites did not increase the overall similarity between DamC and 4C profiles (Supplementary Figure 3h). Thus local differences between the two techniques are not due to saturated methylation levels, and are possibly due to experimental factors not described by the DamC model and thus not accounted for in the calculation of DamC enrichment.
In summary, crosslinking- and ligation-free measurements of contact probabilities using DamC quantitatively agree with 4C, confirm the existence of TAD boundaries and show that crosslinking and ligation do not significantly distort the detection of chromosomal interactions.
piggyBac-TetO insertions do not perturb chromosome structure
Next, we set off to understand whether insertion of TetO/piggyBac cassettes themselves could perturb local chromosome structure. We compared TetO insertion sites with the corresponding WT loci in Hi-C experiments (Supplementary Figure 5a) using a modified version of deviation score defined in Supplementary Figure 3d describing differences in virtual 4C profiles extracted from Hi-C data (Supplementary Figure 5b). Deviation scores between WT cells and cells carrying TetO arrays were similar to those between Hi-C replicates at random genomic locations, and significantly smaller than those between different WT loci (Supplementary Figure 5b). Finally, 4C profiles obtained with and without TetO viewpoints were indistinguishable (Supplementary Figure 5c). Thus, piggyBac-mediated insertion of TetO arrays does not lead to measurable perturbations on chromosome structure.
In vivo detection and manipulation of CTCF-mediated interactions
Loops between convergent CTCF sites are a defining feature of chromosome architecture. However, it is unclear whether new loops can be established between endogenous and ectopically inserted CTCF sites. Early 3C observations suggested that ectopic sequences containing CTCF sites can change the surrounding chromosomal interactions39,40; however, experimental resolution in 3C did not allow to resolve single CTCF loops, and inserted sequences contained additional regulatory regions. Since piggyBac-TetO constructs alone do not perturb chromosome structure, we further engineered them to insert ectopic CTCF sites in the genome and detect resulting structural modifications without confounding effects.
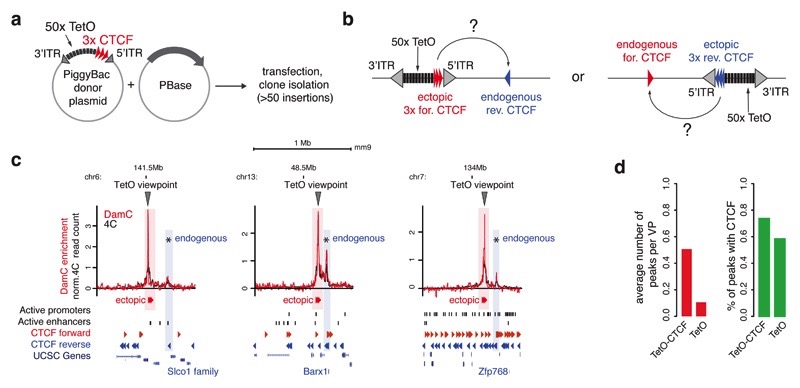
Starting from the founder rTetR-GFP-Dam-ERT2 mESC line described in Figure 3a, we randomly introduced modified piggyBac cassettes where the TetO array is flanked by three CTCF sites oriented outwards (Figure 5a). To test if ectopically inserted CTCF sites could establish loops with endogenous CTCF sites (Figure 5b), we selected one clone carrying 91 insertions, for which we could map insertion positions and genomic orientations (Supplementary Table 4), and performed 4C and DamC with 0.1-1 nM 4-OHT.
Figure 5. DamC-based detection of CTCF loops.
a. Modified piggyBac strategy to insert TetO viewpoints flanked by three CTCF sites oriented outwards. b. The TetO-CTCF cassette can insert in the genome in both directions and lead to the formation of interactions with either forward or reverse endogenous CTCF sites. c. Three representative interaction profiles obtained using DamC and 4C from TetO-CTCF viewpoints. Asterisks indicate interactions identified by PeakC that overlap with CTCF sites. Shaded boxes indicate the overlap with the genomic positions of endogenous and ectopic CTCF locations. d. Left, average number of peaks per viewpoint detected by PeakC at least 20kb away from the viewpoint, in cells with TetO-CTCF or TetO-only insertions. Viewpoints landing within 1kb from an endogenous CTCF site were excluded. Right, percentage of peaks containing a CTCF motif that is bound based on ChIP-seq data11.
Interaction profiles from TetO-CTCF viewpoints displayed prominent distal peaks (Figure 5c) detected by both DamC and 4C. We used the PeakC algorithm, developed to analyze 4C profiles41, to identify distal preferential interactions. Using stringent thresholds (Methods) and excluding viewpoints within 1kb from an endogenous CTCF site (Supplementary Figure 3b and Supplementary Figure 6a), we detected 38 specific interactions separated by at least 20 kb from single TetO-CTCF viewpoints (~0.5 distal peaks per insertion site on average, Supplementary Figure 6b). Of those, 74% contained one or more bound CTCF sites based on ChIP-seq datasets in mESC11, predominantly (79%) convergent with the ectopic CTCF insertion (Figure 5d). As a comparison, in the cell line harboring TetO viewpoints without CTCF we detected only 0.1 peaks per insertion site (Supplementary Figure 6b), of which 58% contained one or more bound CTCF sites (Figure 5d). These correspond to endogenous CTCF loops since in virtually all these cases, the TetO was located between 1 and 20 kb away from an endogenous CTCF. Thus, peaks in the TetO-CTCF line are likely to coincide with new loops established by ectopic CTCF sites. Insertions without distal peaks predominantly correspond to TetO-CTCF cassettes integrated either in CTCF ‘deserts’, or conversely close (<30kb) to the nearest endogenous convergent CTCF site and in regions with many endogenous CTCF sites (Supplementary Figure 6c), resulting in short-distance loops that are difficult to distinguish in 4C and DamC profiles. Additional TetO-CTCF DamC and 4C profiles are plotted in Supplementary Figure 7 and bedGraph tracks are available online (Data Accessibility section).
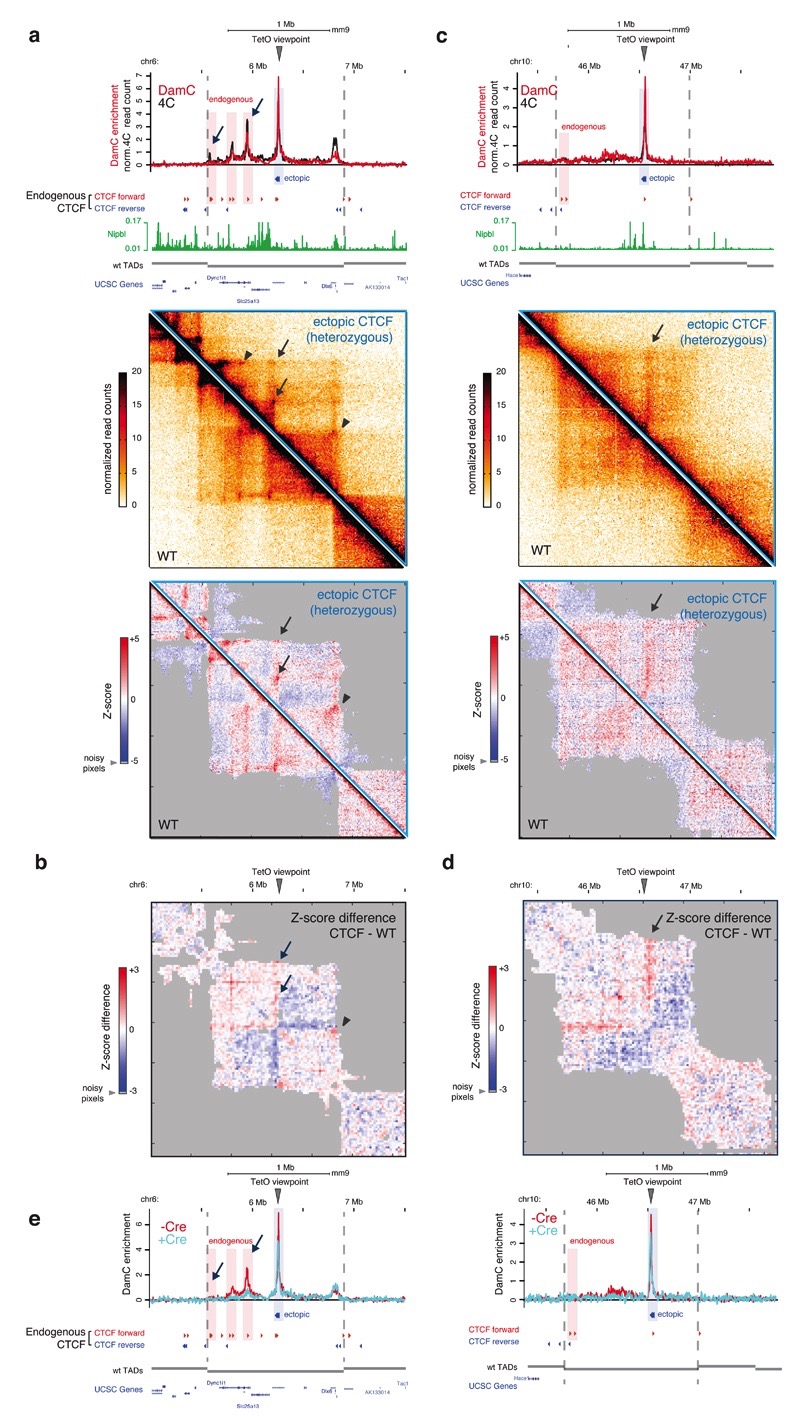
We then performed Hi-C in the TetO-CTCF line and compared it to the data obtained from TetO-only mESCs (see Figure 4a), where insertion locations are different. Since TetO-CTCF insertions are heterozygous, the Hi-C readout is confounded by the presence of a WT allele. Nevertheless, in a fraction of insertions showing prominent distal CTCF peaks in 4C and DamC, we could detect the formation of new structures in Hi-C and notably new loops (Figure 6a, arrows and Supplementary Figure 6d) leading to increased partitioning of interactions within TADs and the appearance of sub-TAD boundaries (Figure 6b). Ectopic CTCF insertion also reinforced pre-existing interactions between convergently oriented sites (Figure 6a, arrowheads), possibly by bringing them closer by effect of the new loops. Even insertions without prominent distal CTCF peaks could be associated with new structures (Figure 6c) reminiscent of stripes predicted by the loop extrusion model15 and recently observed in Hi-C data at endogenous locations42. Consistent with the loop extrusion model interpretation, the stripe shown in Figure 6c occurred at a location where the three ectopic CTCF sites landed close to a cluster of Nipbl sites, and far from the nearest convergent CTCF sites (~800 kb). Formation of an ectopic CTCF-associated stripe also resulted in modifications of intra-TAD chromosomal interactions (Figure 6d).
Figure 6. Ectopic CTCF insertion leads to the formation of new loops and stripes.
a. TetO-CTCF insertion site giving rise to ectopic loops with convergently oriented endogenous CTCF sites. Top, interaction profiles measured with DamC and 4C are overlaid with the position of CTCF ChIP-seq sites from ref.11 and Nipbl ChIP-seq data from ref.42. Middle, Hi-C data from the ESC lines carrying wither the heterozygous TetO-CTCF insertion or two wild-type alleles. Bottom, distance-normalized Z-scores, highlighting interactions that are either enriched (red) or depleted (blue) compared to the expected interaction frequency. Arrows, interactions between convergent CTCF sites that are established upon CTCF insertion. Arrowheads, pre-existing interaction that are strengthened after CTFC insertion. Hi-C data are binned at 10 kb resolution. Data from two biological replicates (independent cell cultures) were pooled for DamC, 4C and Hi-C. b) Z-score difference between heterozygous CTCF and wild-type cells showing increased partitioning of interactions inside the TAD. Hi-C data were binned at 20kb. Shaded areas correspond to ‘noisy’ interactions that did not satisfy a quality control filter based on their correlations with immediate nearest neighbors (see Methods). c. Same as panel a for an insertion on chromosome 10, occurring in proximity to an isolated cluster of Nipbl binding and giving rise to a stripe-like interaction pattern. d. Z-score differences for the locus shown in panel c. e. DamC interaction profiles from the same viewpoints as in panel a and c, before and after Cre-mediated excision of ectopically inserted CTCF sites (but not of the piggyBac cassette).
Finally, to formally prove that new structures are induced by ectopic CTCF binding sites (rather than the piggyBac-TetO cassette), we removed the three CTCF sites by Cre-assisted recombination using two flanking LoxP sites (Supplementary Figure 6e). DamC performed in one mESC clone where CTCF sites had been excised at both loci shown in Figure 6a-b (Supplementary Figure 6f) revealed that removal of CTCF sites led to loss of distal interactions (Figure 6e).
In summary, DamC identifies chromatin loops formed through specific long-range chromatin interactions. Additionally, our data shows that ectopically inserted CTCF sites can establish new loops with endogenous CTCF sites and stripes, leading to modified partitioning of interactions within TADs.
Quantitative properties of chromosome folding in vivo
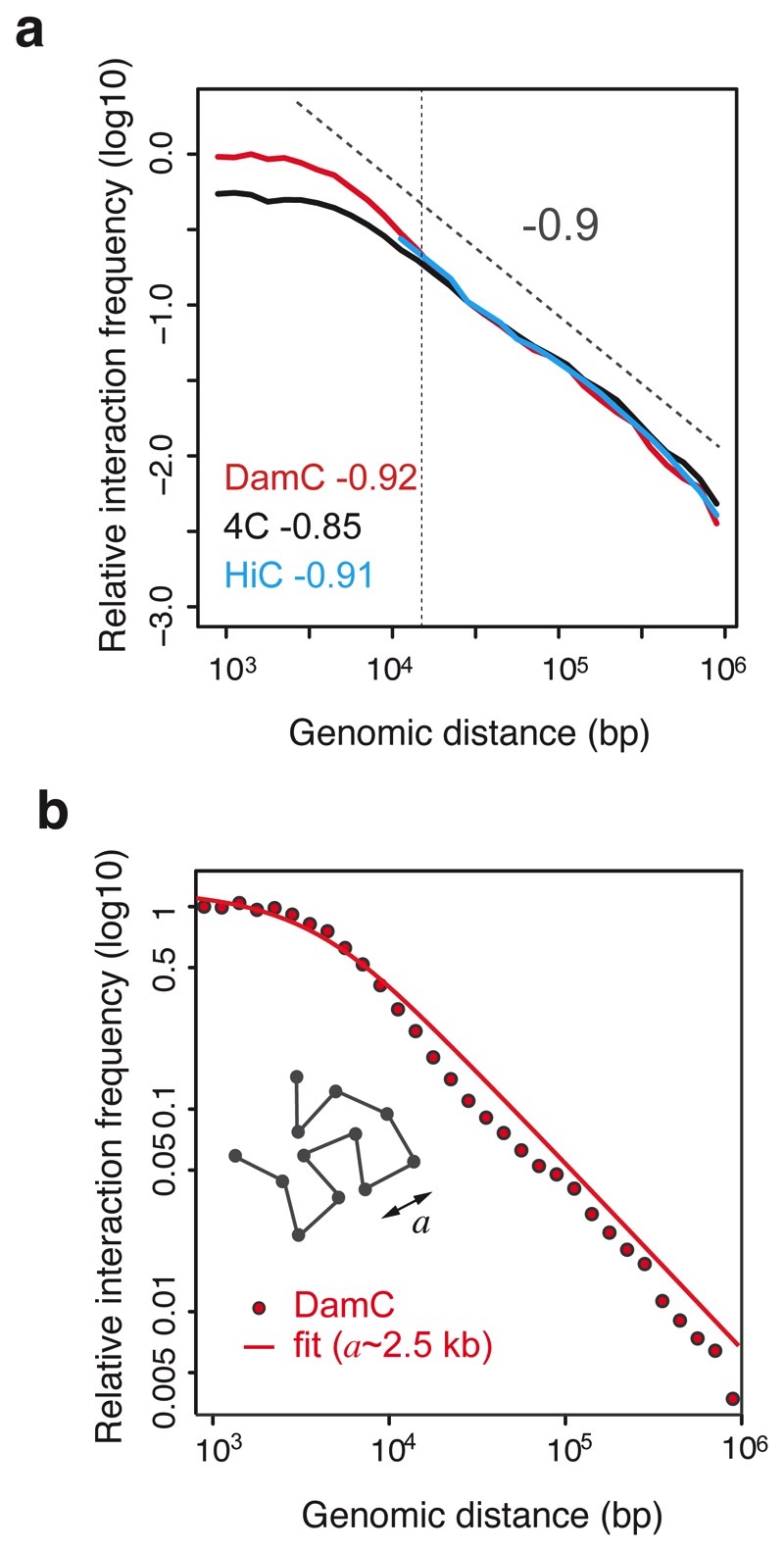
Given the high similarity between DamC and 4C both with and without CTCF sites at the viewpoint, we next asked whether DamC and 3C-based techniques measured the same scaling of interaction probabilities. We pooled all viewpoints from TetO-only and TetO-CTCF lines and plotted the data as a function of genomic distance from the viewpoints. For distances between 15 kb and 1 Mb, fitting both DamC and 4C with a power law resulted in decay exponents around 0.9, in excellent agreement with Hi-C from the same cells and viewpoints (Figure 7a), and in accordance with previous measurements in similar genomic ranges14,43.
Figure 7. Scaling analysis of contact probabilities in vivo.
a. Scaling of contact probabilities measured in DamC, 4C and Hi-C from all 130 TetO and 91 TetO-CTCF viewpoints. Power-law fitting was performed between 15kb and 1Mb. b. Best fit of scaling measured by DamC with a polymer model with persistence length a (See Supplementary Note 1). The best value of a extracted from the fit is ~2.5 kb.
Below ~10 kb, both DamC and 4C showed a gentler decay as recently observed in Hi-C experiments on yeast chromosomes44. In Ref.44 this was attributed to crosslinking artefacts but DamC, showing the same behavior, argues against this explanation. The leveling-off of contact probabilities at short genomic distances can be explained in terms of a simple coarse-grained polymer model with a persistence length of ~2.5 kb (Figure 7b, Supplementary Note 1). We cannot formally rule out alternative explanations such as experimental factors not accounted by the DamC model and thus not normalized in the calculation of enrichment. One such scenario could be that the spacing between GATC sites imposes an effective capture range of few kb, consistent with micrococcal nuclease-based Micro-C experiments showing that yeast chromatin is flexible at lower scales45. However, in the absence of Micro-C measurements on mammalian chromatin, we can safely assume that DamC provides an upper limit to the persistence length of chromosomes in vivo of approximately 2.5kb.
Discussion
In this work we provide the first in vivo, high resolution, systematic measurements of chromatin contacts that do not require crosslinking nor ligation using DamC. An essential feature of this method is that its experimental output is directly proportional to contact probabilities. This is supported by rigorous modeling of methylation kinetics (Figure 2), providing a rational basis to quantitatively interpret sequencing results. Importantly, DamC confirms that contact frequencies drop across TAD boundaries approximately by a factor 2, in accordance with 4C (Figure 4b) and previous estimations based on Hi-C15,46. Such modest decrease raises the question of how TAD boundaries can functionally insulate enhancers and promoters from a biophysical point of view, although they might represent an optimal compromise between enriching and depleting interactions between regulatory sequences within and across boundaries, respectively9.
DamC detects chromosomal contacts on short spatial distances, since GATC motifs can only be methylated if Dam directly binds DNA. We estimate a detection range of <10nm, given that the expected physical size of the rTetR-EGFP-Dam-ERT2 fusion protein does not exceed 3 nm47. Decreases in interaction frequencies at TAD boundaries, as well as increases due to CTCF loops, therefore closely match what a promoter would ‘experience’ through its bound protein complexes. Interestingly DamC also picks up ‘non-specific’ interactions due to random collisions within the chromatin fiber to the same extent as 4C and Hi-C (Figure 4a, Supplementary Figure 4). Thus, random collisions do occur in vivo, despite not being detected in crosslinking-free analysis of chromosome folding using native 3C26.
Scaling of crosslinking probabilities measured in Hi-C data are at the core of physical models developed to explain chromosome folding and infer its mechanistic determinants15,38,48–50, including the highly influential loop extrusion model. Importantly, DamC confirms scaling exponents measured in 4C and Hi-C (Figure 7). Since DamC enrichment is proportional to actual short-range contact probabilities, our measurements provide strong evidence in favor of chromosome folding models based on Hi-C. Scaling analysis at short genomic distances additionally suggests that mouse chromosomes might have a persistence length of approximately 2.5kb, corresponding to ~40 nm assuming a linear density of ~60 bp/nm38.
The finding that loops can be established de novo upon insertion of CTCF binding sites and can be detected in vivo (Figure 5-6) confirms earlier reports39,40 and argue that chromosome structure at the TAD level can be manipulated in a ‘gain of function’ manner by adding new structures. New structures formed upon ectopic insertion of three CTCF sites can significantly modify intra-TAD interactions and can result in the formation of new boundaries within pre-existing TADs (Figure 6b and 6d). Remarkably, we could only detect newly formed interactions within pre-existing TAD boundaries, possibly due to the fact that TAD boundaries are particularly enriched in clusters of CTCF sites7,9 providing efficient barriers to loop extrusion.
A limitation of DamC is that it requires genetic manipulation to insert genomic viewpoints and to stably express rTetR-Dam-ERT2 allowing accurate control of nuclear Dam concentration. This prevents DamC in its current form to be considered as an alternative to 3C-based methods in routine experimentation. However DamC can be performed by transiently nucleofecting cells with a Dam-TetR expression plasmid, which ensures low expression levels (Figure 1c), and future implementations based on TAL effector proteins (similar to TALE-ID26) or catalytically inactivated Cas9 could overcome the need for targeted insertion of TetO arrays.
The current TetR-based implementation of DamC might nevertheless be beneficial in situations where 4C cannot be used, notably to detect chromosomal interactions in a tissue-specific context by expressing the rTetR-Dam fusion under a tissue-specific promoter51 and starting from small numbers of cells52. Contrary to 3C methods where one ligation event per allele can be retrieved at most, in the course of a DamC experiment (~18 hours) several GATCs might be contacted by a TetO viewpoint depending on the temporal dynamics of chromosome structure. Based on our previous measurements of the dynamics of the TetO array at the Chic1 locus53 as well as recent data from other chromosomal locations54,55, several contacts might be created and disassembled in 18 hours. If n GATC sites are methylated in this time window, DamC would in principle require n times less cells than 4C to build similar contact profiles. In this manuscript we analysed ~10 thousand cell equivalents per 4C and DamC experiment, but scaling down cell numbers in DamC will be an interesting future development.
In summary, by coupling a methylation-based readout with physical modeling, DamC enables systematic and quantitative crosslinking- and ligation-free measurements of chromatin interaction frequencies. Our experiments provide an orthogonal validation of 3C-based findings, including TADs and endogenous as well as ectopically induced CTCF loops, and demonstrate that 3C methods do not significantly distort the detection of chromosomal interactions.
Methods
Physical modeling
Detailed descriptions of the physical model of methylation kinetics in DamC, as well as of the polymer model with persistence length are available as a separate file (Supplementary Note 1).
Cell culture and sample collection
All cell lines are based on feeder-independent PGK12.1 female mouse embryonic stem cells (mESC), kindly provided by Edith Heard’s laboratory. The founder cell line in our study is an X0 sub-clone of the PGKT2 clone described in (Masui et. al. 2011), carrying the insertion of a 256x TetO array within the 3’UTR of the Chic1 gene on chromosome X and the additional deletion of the Linx promoter6 Cells were cultured on gelatin-coated culture plates in Dulbecco Modified Eagle’s medium (Sigma) in the presence of 15% foetal calf serum (Eurobio Abcys), 100 µM β-mercaptoethanol, 20 U/ml leukemia inhibitory factor (Miltenyi Biotec, premium grade) in 8% CO2 at 37°C. Cells were tested for mycoplasma contamination once a month and no contamination was detected. After insertion of the rTetR-Dam vector (see below), cells were cultured in the presence of 250 µg/mL hygromycin. To induce nuclear translocation of the rTetR-Dam fusion protein to the nuclei, mESC were trypsinized and directly seeded in culture medium containing 4-hydroxy-tamoxifen (4-OHT) at the concentrations indicated in the main text for 18 hours. Binding of the Dam fusion protein to the TetO arrays was induced by simultaneously adding 2.5 μg/ml doxycycline (Dox).
Generation of cell lines expressing rTetR-Dam and carrying random insertions of TetO arrays
The rTetR-EGFP-Dam-ERt2 construct was cloned into a pBroad3 backbone (Invivogen) carrying a mouse Rosa26 promoter. We used a modified rTetR based on the rtTA-M2 transactivator in Ref.58, which has substantially decreased affinity for the Tet operator in the absence of Dox. The construct was randomly integrated in the PGKT2 X0 subclone by co-transfecting 5x105 cells with 3 μg pBROAD3-rTetR-ICP22-EGFP-EcoDam-Ert2 and 0.2 μg of pcDNA3.1hygro plasmid using Lipofectamin 2000 (Thermo Fisher Scientific). After 10 days of hygromycin selection (250 μg/ml), one clone (#94.1) expressing low levels of EGFP was selected and expanded for subsequent experiments. To obtain large numbers of viewpoints for DamC experiments, stable random integrations of arrays of Tet operator (TetO) sites were introduced in the #94.1 mESC clone using the piggyBac transposon system. A mouse codon optimized version of the piggyBac transposase36 was cloned in frame with the red fluorescent protein tagRFPt (Evrogen) into a pBroad3 vector using Gibson assembly cloning (pBroad3_hyPBase_IRES_tagRFPt). 5x105 cells were co-transfected with 0.2ug of pBroad3_hyPBase_IRES_tagRFPt and 1µg of a piggyBac donor vector containing an array of 50 TetO binding sites using Lipofectamin 2000 (Thermo Fisher Scientific). Cells with high levels of RFP were FACS sorted two days after transfection and seeded at three serial 10x dilutions in 10-cm dishes to ensure optimal density for colony picking. To identify clones with high numbers of TetO integration sites, cells were screened for large numbers of nuclear EGFP accumulation foci using live-cell imaging (see below) in the presence of 500nM 4-OHT and 2.5ug/ml Dox. One polyclonal population (#94.1_2.7) and one subclone (#94.1_2.7_pureclone3) was further expanded.
To introduce CTCF binding sites flanking the TetO viewpoints, the piggyBac donor vector was modified as follows. Three CTCF binding motifs (TGGCCAGCAGGGGGCGCTG, CGGCCAGCAGGTGGCGCCA and CGACCACCAGGGGGCGCTG) were selected based on high CTCF occupancy in ChIP-seq experiments11 and cloned into the piggyBac donor vector in an outwards direction with respect to the TetO array, including 100bp of their surrounding endogenous genomic sequence (chr8:13461990-13462089, chr1:34275307-34275419 and chr4:132806684-132806807, respectively). The three CTCF binding motifs were flanked by two LoxP sites for CRE assisted recombination. 5x105 #94.1 were co-transfected with 0.2ug of pBroad3_hyPBase_IRES_rfp and 1µg of the modified piggyBac donor vector using Lipofectamin 2000. Cells with high levels of RFP were FACS sorted two days after transfection and seeded at three serial 10x dilutions in 10-cm dishes for colony picking. Clones with >50 of integration sites were identified through accumulation of EGFP at nuclear TetO foci in the presence of 500nM 4-OHT and 2.5ug/ml dox. One clone (#94.1_216_C3) was further selected for analyis.
Transient transfection
To transiently express rTetR-Dam for the proof-of-principle experiment in Figure 1d, the PKGT2 X0 subclone was transiently transfected with pBroad3-rTetR-EGFP-Dam-ERt2 using the Amaxa 4D-Nucleofector X-Unit and the P3 Primary Cell 4D-Nucleofector X Kit (Lonza). 5x106 cells were resuspended in 100 µl transfection solution (82ul primary solution, 18ul supplement 1, 2μg pBroad3-rTetR-EGFP-Dam-ERt2) and transferred in a single Nucleocuvette (Lonza). Nucleofection was done using the protocol CG109. Transfected cells were directly seeded in pre-warmed 37°C culture medium containing 10nM 4-OHT +/- 2.5 μg/ml Dox. Genomic DNA was collected 18 hours after transfection. Sequencing libraries were prepared as previously described34,59.
Mapping of piggyBac insertion sites
2µg of genomic DNA were fragmented to an average of 500bp by sonication (Covaris S220, duty cycle: 5%, peak power: 175W, duration: 25sec). End-repair, A-tailing and ligation of full-length barcoded Illumina adapters were performed using the TruSeq DNA PCR-free kit (Illumina) according to the manufacturer guidelines with the exception that large DNA fragments were not removed. 750ng of libraries for each sample were pooled together, and fragments of interest were captured using biotinylated probes against the the piggyBac inverted terminal repeats (ITRs) sequence and the xGen Hybridisation Capture kit (IDT) according to the manufacturer protocol (probes concentration of 2.25pmol/µl). Following the capture, libraries were amplified for 12 cycles using the Kapa Hi-fi polymerase and the following primers: 5’-AATGATACGGCGACCACCGAGAT, 5’-CAAGCAGAAGACGGCATACGAGA. Final libraries were purified using AMPure XP beads (1:1 ratio), quality controlled and sequenced on the NextSeq500 platform (paired-end 300 cycles mid-output) for a total of 8x108 paired-end reads per sample on average.
Capture probe sequences are as follows:
ITR3-1 [Btn]ATCTATAACAAGAAAATATATATATAATAAGTTATCACGTAAGTAGAACATGAAATAACAATATAATTATCGTATGAGTTAAATCTTAAAAGTCACGTAAAAGATAATCATGCGTCATTT,
ITR3-2 [Btn]TCCAAGCGGCGACTGAGATGTCCTAAATGCACAGCGACGGATTCGCGCTATTTAGAAAGAGAGAGCAATATTTCAAGAATGCATGCGTCAATTTTACGCAGACTATCTTTCTAGGGTTAA,
ITR5-1[Btn]TTAACCCTAGAAAGATAATCATATTGTGACGTACGTTAAAGATAATCATGCGTAAAATTGACGCATGTGTTTTATCGGTCTGTATATCGAGGTTTATTTATTAATTTGAA,
ITR5-2 [Btn]ATTAAGTTTTATTATATTTACACTTACATACTAATAATAAATTCAACAAACAATTTATTTATGTTTATTTATTTATTAAAAAAAAACAAAA ACTCAAAATTTCTTCTATAAAGTAACAAA.
Genotyping of CTCF integration sites by PCR
We designed primers binding to endogenous genomic DNA sequence outside the piggyBac 3’ ITR based on the genomic position of mapped piggyBac insertion sites. We then amplified the junction between the ITR and the genome using Phusion High-Fidelity DNA Polymerase (Thermo Scientific) with one genomic primer and a T7 promoter primer (5’TAATACGACTCACTATAGGG3’) flanking the piggyBac CTCF integration cassette (see Supplementary Figure 4d). PCR products were purified and Sanger sequenced. For the verification of CTCF integrations shown in Figure 4 and Supplementary Figure 4e on chromosome 6 and chromosome 10, the following genomic primers were used: Ch6_flxCTCF_11F (5’AGGCATTCTGTCCAACTGGT3’) and Chr10_flxCTCF_13F (5’TGTTGAGCATCTATCACATTCCTTA3’).
Excision of CTCF sites using Cre recombinase
In order to excise ectopically inserted CTCF sites from the #94.1_216_C3 clone, 5x105 cells were transfected with 0.5 μg of pIC-Cre60 using Lipofectamine 2000 (Thermo Fisher Scientific). After 4 days under G418 selection (300 ug/ml), single colonies were expanded and genotyped following the procedure described above.
Live-cell Imaging
Gridded-glass-bottom dishes (Mattek) were coated with 2 ug/ml recombinant mouse E-cadherin (R&D Systems, 748-EC) in PBS at 4°C overnight. 5x105 cells were seeded in full medium one day before imaging supplemented with 4-OHT and Dox as indicated above. Cells were imaged with a Nikon Eclipse Ti-E inverted widefield microscope (Perfect Focus System with real time drift correction for live cell imaging) operating in TIRF mode using a CFI APO TIRF 100x/1.49 oil objective (Nikon). A 488nm, 200mW Toptica iBEAM SMART laser was used as excitation source. Cells were maintained at a constant temperature of 37°C and 8% CO2 within an incubation box. Images were collected with an Evolve™ 512 Delta EMCCD high speed Camerang using Visiview (Visitron). Background subtraction (150-pixel rolling ball radius) and maximum intensity projections were performed in ImageJ.
Nuclear volume measurements
3x106 Cells from the #94.1_2.7 mESC clone were cultured in gelatin-coated 6-well plates in full medium, dissociated 5 minutes at room temperature with Accutase (GIBCO), then centrifuged 4 minutes at 950 rpm and resuspended in 500 µl of culture medium. 25-µl droplets of cell suspension were spotted on coverslips previously coated with poly-L-lysine, let adsorb on ice for 5 minutes and washed gently once with 1X PBS. Cells were then permeabilized on ice for 5 min in 1X PBS and 0.5%Triton X-100, and coverslips were stored in 70% EtOH at -20C. Nuclei were counterstained with 0.2 mg/ml DAPI and Z-stack images were acquired using a Zeiss Z-1 microscope equipped with a 40x oil immersion (NA=1.3) (voxel size 0.227x0.227x0.73 µm). Z-stacks were then deconvolved using the Huygens software (20 iterations of the CMLE algorithm). To segment individual nuclei, we binarized DAPI images based on a single intensity threshold based on the fact that image histograms of all Z-stacks were bimodal (threshold = 7000 in 32-bit images). The volumes of binary 3D objects was then calculated using the 3DObjectCounter plugin in FIJI/ImageJ, excluding objects on the edges of each Z-stack.
Preparation of nuclear extracts
Cell nuclei were extracted as previously described61. Briefly, 107 mES cells were seeded in ES medium (see above) supplemented with the appropriate concentration of 4-OHT on a gelatin coated 15 cm2 dish. The next day, cells were harvested using trypsin and washed twice in ice cold PBS. Next, cells were carefully resuspended in 500μl ice cold Buffer A1 (10mM HEPES pH7.9, 10mM KCl, 1.5mM MgCl2, 0.34M sucrose, 10% glycerol, 0.1% Triton-X 100, 1mM DTT, 1mM PMSF) to obtain nuclei. After incubating for 5 minutes on ice, extracted nuclei were washed twice with buffer A1.
Mass spectrometry
Nuclear extracts were dissolved in 400 µL 50mM HEPES pH 8.5 in 8.3M guanidine hydrochloride. All the samples were heated at 95°C for 5 min, sonicated using Bioruptor® sonication device and supplemented with 5mM TCEP and 10mM CAA. To reduce sample complexity, lysates were diluted to 6M guanidine hydrochloride and transferred onto 100kDa molecular weight cut-off Amiconultra-0.5 centrifugal filter units. Samples were concentrated for 2 x 15 minutes at 14kG followed by refill of the filter with 6M guanidine hydrochloride in 50mM HEPES pH 8.5 and 3 x 45 minutes at 14kG followed by refill of the filter with 1M guanidine hydrochloride in 50mM HEPES pH 8.5. For digestion 10µg of Lys-C (Wako Chemicals) and 10µg of trypsin (Thermo Fisher) were added to each sample and incubated over night at 37°C. In the morning additional 10µg of trypsin was added, incubated for 3h and acidified using TFA.
To estimate nuclear proteins copy numbers samples were desalted using SEP-PAK (Waters) and subjected to high pH offline fractionation on a YMC Triart C18 0.5 x250 mm column (YMC Europe GmbH) using the Agilent 1100 system (Agilent Technologies). 96 fractions were collected for each experiment and concatenated into 48 fractions as previously described62. For each LC-MS analysis, approximately 1 µg of peptides were loaded onto a PepMap 100 C18 2 cm trap (Thermo Fisher) using the Proxeon NanoLC-1000 system (Thermo Fisher). On-line peptide separation was performed on the 15 cm EASY-Spray C18 column (ES801, Thermo Fisher) by applying a linear gradient of increasing ACN concentration at a flow rate of 150 nL/min. An Orbitrap Fusion Tribrid (Thermo Fisher) or an Orbitrap Fusion LUMOS Tribrid (Thermo Fisher) mass spectrometers were operated in a data-dependent mode. The top 10 most intense precursor ions from the Orbitrap survey scan were selected for higher-energy collisional dissociation fragmentation and analyzed using the ion-trap.
Mass spectrometry data processing
Maxquant version 1.5.3.8 was used to search raw mass spectrometry data using default settings63,64 against the mouse protein sequences from Uniprot database (release 2017-04). The label free quantification (LFQ) algorithm was used for quantification. The protein groups table were loaded to Perseus software65 (version 1.5.0.0) filtered for potential contaminants and reverse hits. Protein copy numbers per cell were calculated using the Protein ruler plugin of Perseus by standardization to the total histone MS signal56 (Wisniewski et al., 2014). The LFQ values were normalized using same normalization for all samples. To estimate cytoplasmic contamination “GOCC slim name” annotations provided in Perseus were used. Exclusively cytoplasmic proteins were defined as being associated with GOCC terms “cytoplasm” or “cytosol” and not associated with terms “nucleus”, “nuclear”, “nucleoplasm” and “nucleosome”. Exclusively nuclear proteins were defined as being associated with GOCC terms “nucleus”, “nuclear”, “nucleoplasm” and “nucleosome” and not associated with terms “cytoplasm” or “cytosol”. The cytoplasmic contamination was estimated using a ratio of summed LFQ intensity between exclusively cytoplasmic proteins and exclusively nuclear proteins in samples with and without nuclear extraction.
Parallel reaction monitoring (PRM) data acquisition and analysis
To select peptides for PRM assays, the rTetR-Dam-EGFP-ERT2 construct was enriched using magnetic ChromoTek's GFP-Trap beads and analyzed using shotgun data-dependent acquisition LC-MS/MS on an Orbitrap Fusion Lumos platform as decribed above. For PRM analysis the resolution of the orbitrap was set to 240k FWHM (at 200 m/z), the fill time was set to 1000 ms and ion isolation window was set to 0.7 Th. For LC-MS analysis of samples derived from a polyclon carrying 890 TetO array insertions, approximately 1 µg of peptides were loaded onto a PepMap 100 C18 2 cm trap (Thermo Fisher) using the Proxeon NanoLC-1000 system (Thermo Fisher). On-line peptide separation was performed on the 15 cm EASY-Spray C18 column (ES801, Thermo Fisher) by applying a linear gradient of increasing ACN concentration at a flow rate of 150 nL/min. Whereas for LC-MS analysis of samples derived from a polyclon carrying 100 TetO array insertions, approximately 1 µg of peptides were on-line separated on a 50 cm µPACTM cartridge (PharmaFluidics) by applying a linear gradient of increasing ACN concentration at a flow rate of 300 nL/min using the Proxeon NanoLC-1000 system (Thermo Fisher).The acquired PRM data were processed using Skyline 4.13566. The transition selection was systematically verified and adjusted when necessary to ensure that no co-eluting contaminant distorted quantification based on traces co-elution (retention time) and the correlation between the relative intensities of the endogenous fragment ion traces, and their counterparts from the library. As a loading control the mean of total MS1 signal was estimated using RawMeat v2.0b1007.
DamC library preparation
DamC experiments are based on a newly developed DamID-seq NGS library preparation protocol to maximize the proportionality between methylation levels and sequencing readout (Supplementary Figure 2c). One crucial issue in the calculation of enrichment as in Figure 2c is that small fluctuations in -Dox methylation in the denominator can be amplified into large fluctuations in enrichment levels. GATC sites must therefore be equally and robustly represented in the DamID sequencing library irrespective of their methylation level. From this perspective, the principal limitation of the original DamID protocol59 for our present application was its dependence on the genomic distance between two GmATC sites, resulting in large adaptor-ligated molecules and as a consequence in a strong bias towards densely methylated regions. In our optimized protocol, GmATC sites are sequenced independently of the neighboring GATC methylation status resulting in a ~30% increase in GmATC coverage at equivalent sequencing/read depth (Supplementary Figure 2e). In addition, we introduced unique molecular identifiers (UMIs) allowing a precise enrichment quantification after excluding PCR duplicates from the sequencing data.
Overall, the DamC library construction protocol can be divided in 3 parts: 1) ligation of UMI adapters with a “one-tube” strategy, 2) integration of the second sequencing adapter, followed by 3) a final PCR amplification. Briefly, 3x106 cells were harvested using trypsin after 18 hours of induction with tamoxifen +/- doxycyclin. Genomic DNA was extracted using the Qiagen blood and tissue kit adding 250U of RNaseA in step 1. Genomic DNA was eluted in 80ul ddH2O. DNA concentration was measured using the Qbit DNA Broad Range kit. Genomic DNA (350ng input) was treated with Shrimp Alkaline Phosphatase treatment (NEB, 1U), followed by DpnI digestion (ThermoFisher Scientific, 10U), A-tailing (0.6mM final dATP, 5U Klenow exo-, ThermoFisher Scientific), and UMI adapters ligation (30U T4 DNA ligase, PEG4000, ThermoFisher Scientific) performed within the same tube and buffer (Tango 1X, ThermoFisher Scientific) by heat inactivating each enzymatic step followed by adjustment with the reagents required for the next step. UMI adapters were made by annealing the following oligos: 5’-AATGATACGGCGACCACCGAGATCTACACNNNNNNNNACACTCTTTCCCTACACGACGCTCTTCCGATC*T and 5’-pGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGT. Ligation reactions were treated with Exonuclease I (20U, ThermoFisher Scientific) then purified using AMPureXP beads (1:0.8 ratio, Agencourt) and the second sequencing adapter (5’ TGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNNNN*N 3’, IDT) was tagged using heat denaturation and second strand synthesis (5U T4 DNA Polymerase, ThermoFisher Scientific). The tagging reaction was purified using AMPure XP beads (1:1 ratio) followed by a final library amplification (12 cycles) using 1U of Phusion polymerase, 2μl 10 μM DAM_UMIindex_PCR (5’ AATGATACGGCGACCACCGAGATCTACA*C 3’), and 2μl 10μM NEBnext indexed primer (NEB). Final libraries were purified AMPure XP beads (1:1 ratio) and QCed using Bioanalyser and Qbit. DamC libraries were sequenced on a NextSeq500 (75 cycles single-end) with a custom sequencing protocol (dark cycles at the start of read1 to "skip" the remaining DpnI site TC sequence). Samples index were determined using index1 read, and UMI sequence using index2 read. Detailed number of total and valid reads can be found in Supplementary Table 5.
4C-seq
4C sample preparation was performed as previously described67. Briefly, 107 cells were cross-linked in 2% formaldehyde for 10 minutes and quenched with glycine (final concentration 0.125M). Cells were lysed in 150 mM NaCl/50 mM Tris-HCl (pH 7.5)/5 mM EDTA/0.5% NP-40/1% Triton X-100. The first digest was performed with 200 U DpnII (NEB), followed by ligation at 16° C with 50 U T4 DNA ligase (Roche) in 7 mL. Ligated samples were de-crosslinked with Proteinase K (0.05 ug/uL) at 65° C, purified, and digested with 50 U Csp6I (Thermo Fisher Scientific) each, followed by ligation with 100 U T4 DNA ligase in 14 mL and purification. Resulting products were directly used as PCR template for genomic dedicated 4C viewpoints. Primers for PCR were designed using guidelines described previously67. We obtained the following read counts: #94.1_2.7 cell line (135 TetO insertions only), 5.7x106 valid reads in total (+Dox, two replicates). For the #94.1_216_C3 line (TetO-CTCF), 3.5x106 valid reads on average per sample; for the experiments shown in Supplementary Figure 3c and 4c, we obtained an average of 3.2x106 reads per sample. Detailed number of total and valid reads can be found in Supplementary Table 5.
In vitro Cas9 digestion of 4C templates
In order to detect chromosomal interactions directly from the genome integrated TetO platform, viewpoint primers were designed to amplify directly from the DpnII fragments contained in the TetO sequence. The 2.7 kb TetO platform contains a total of 50x contiguous repeats of the same TetO DpnI/II viewpoint. To prevent PCR amplification and sequencing of TetO repeats due to tandem ligation of two or more TetO DpnII fragments in a given 4C circle, an in vitro Cas9 digestion was performed on the 4C templates. Cas9 was targeted into the TetO repeats in between viewpoint primers using a single gRNA. In vitro transcribed gRNA template was obtained using the Megashortscript T7 transcription kit (Invitrogen). gRNA was purified with 4 × AMPure purification (Agencourt). Purified Cas9 protein was kindly provided by N. Geijsen. Cas9 was pre-incubated with the sgRNA for 30 min at 37 °C. Subsequently, 4C template DNA was added to the pre-incubated gRNA-Cas9 complex and incubated for 3–6 h at 37 °C for digestion. Cas9 was inactivated by incubating at 70 °C for 5 min.
Hi-C library preparation
6x106 mESC were harvested and diluted in 1x PBS to final 1x106 cells/ml, then crosslinked with 1% formaldehyde and quenched with 0,125M glycine for 5 min at RT. After two 1x PBS washes, cells pellets were obtained by centrifugation, snap frozen and stored at -80°C. Pellets were thawed on ice and resuspended in 500 ul lysis buffer (10 nM Tris-HCl pH8.0, 10 nM NaCl, 0.2%NP40, 1x Roche protease inhibitors) and left 30 min on ice. Cells were then pelleted by centrifugation (954 x g, 5 min, 4°C), washed once with 300 ul 1x NEB2 buffer and nuclei were extracted by 1 h incubation at 37°C in 190 ul 0.5%SDS 1xNEB2 buffer. SDS was neutralized by diluting the sample with 400 ul NEB2 buffer and adding 10% Triton X-100. After 15 min of incubation at 37°C, nuclei were pelleted, washed once in PBS and resuspended in 300 ul NEB2 buffer. 400U of MboI (NEB, 25 000 units/ml) were added and incubated at 37°C overnight. The next day, nuclei were pelleted again, resuspended in 200 ul fresh NEB2 buffer and additional 200U of MboI were added for two more hours before heat inactivation at 65°C for 15 min. 43 ul of end-repair mix (1.5 μL of 10 mM dCTP; 1.5 μL of 10mM dGTP; 1.5 μL of 10 mM dTTP; 37.5 μL of 0.4 mM Biotin-11-dATP (Invitrogen) and 1 μL of 50U/μL DNA Polymerase I Large Klenow fragment (NEB) were added to the nuclear suspension, incubated at 37°C for 45 min and heat inactivated at 65°C for 15 min. The end repair mix was exchanged with 1.2 ml of ligation mix (120μL of 10X T4 DNA Ligase Buffer; 100 μL of 10% Triton X-100; 6 μL of 20 mg/mL BSA; 969μL of H2O) plus 5 ul of T4 ligase (NEB, 2000 units/ml) and ligation was performed at 16°C overnight. Nuclei were reconstituted in 200 ul fresh NEB2 buffer followed by RNA digestion in 0.5 mg/ml RNAse A for 10 min at 37°C. Samples were de-crosslinked with Proteinase K at 65°C overnight and DNA was purified using phenol/chloroform. 2 ug of DNA sample were sonicated using Diagenode Bioruptor Pico. MyOne Streptavidin T1 (Life Technologies # 65601) magnetic beads were used to capture biotinylated DNA followed by A-tailing. Adapter ligation was performed according to NEB Next Ultra DNA Library prep kit instructions. Two independent PCR reactions with multiplex oligos for Illumina sequencing were performed and pooled for the final PCR clean-up by magnetic AMPure bead (Beckman Coulter) purification. The final libraries were eluted in nuclease-free water, QCed by Bioanalyzer and Qubit. HiC libraries were sequenced on a Illumina Nextseq500 platform (2x42bp paired end). We obtained an average of 3.5x108 valid reads per sample (TetO-only and TetO-CTCF cells, -Dox, two biological replicates each). Detailed number of total and valid reads can be found in Supplementary Table 5.
Sequencing data processing and data analysis
DamC analysis
All samples were aligned to mouse mm9 using qAlign (QuasR package68) using default parameters. PCR duplicates were removed using a custom script. Briefly, reads were considered PCR duplicates if they map to the same genomic location and have the same 8-bp UMI sequence. We quantified the number of reads mapped to each GATC that could be uniquely mapped using qCount (QuasR package68). The query object we used in qCount was a GRanges object containing the uniquely mappable 76-mers GATC loci in the genome shifted upstream (plus strand) or downstream (minus strand) by 5 base-pairs (3 dark cycles + GA, see the ‘Bulk DamID-seq Library preparation’ paragraph in the Methods section). Each sample was then normalized to a common library size of 10M reads and a pseudo-count of 0.2 was added. Prior to calculating DamC enrichments, a running average over 21 restriction fragments was performed and the mean value was assigned to the central GATC. Enrichment was then calculated as in Figure 2c: E=([+Dox]-[-Dox])/[-Dox] where [+Dox] and [-Dox] are the normalized and running-averaged number of reads in the presence and absence of Dox, respectively. We defined the DamC signal to be saturated if it satisfies the following criteria: 1) it belongs to the top highest 25% genome wide both in +Dox and -Dox samples, and 2) the ratio between +Dox and -Dox methylation is close to 0.5, i.e. belongs to the [0.45, 0.55] quantile of all ratios genome wide. Coordinates of excluded viewpoints in the clonal cell line with TetO integrations are: chr6:25758950, chr8:26653938, chr8:96714938, chr11:33429300, and chr11:51411650.
4C analysis
Mapping of 4C reads was performed as described for DamC, with the exception of UMI de-duplication since 4C libraries did not include UMIs and quantification was done by counting the reads mapped exactly to the GATC sites. The two restriction fragments immediately flanking the piggyBac-TetO cassette were excluded from subsequent analyses.
Hi-C Analysis
Hi-C data were analysed using HiC-Pro version 2.7.10 with --very-sensitive --end-to-end --reorder option. Briefly, reads pairs were mapped to the mouse genome (build mm9). Chimeric reads were recovered after recognition of the ligation site. Only unique valid pairs were kept. Contact maps at a given binning size were then generated after dividing the genome into equally sized bins and applying iterative correction70 on binned data.
Fit of scaling plots
Average normalized Hi-C counts, DamC enrichment or 4C counts were calculated for all pairs of loci separated by logarithmically binned distance intervals. The binning size in logarithmic scale (base 10) was 0.1. Curves were fitted in log-log scale using the lm function in R.
Fitting the DamC model to DamC experiments as a function of 4-OHT concentration
The DamC enrichment depends on the rTetR-TetO specific and nonspecific dissociation constants, the concentration of TetO and the nuclear rTetR-Dam concentration (Supplementary Note 1). In addition, it depends on the actual contact probability between the genomic location where it is calculated and the TetO viewpoint. In Figure 3d we calculated the DamC enrichment at the closest fragments to the 100 TetO viewpoint with higher signal to noise ratio in the polyclonal line. We assumed that the contact probability between the TetO array and the closest fragment is ~1, and fitted the model to the experimental data using the other parameters with the NonlinearModelFit function in Mathematica. The constraints that the dissociation constants and the concentration of TetO are positive were imposed. The goodness of the fit was evaluated using the adjusted R2 (0.73). In the clonal line, we assumed that the specific dissociation rTetR-TetO does not change compared to the polyclonal line and by setting the concentration of viewpoints to 135 per cell, we fitted the nonspecific dissociation constant using the NonlinearModelFit function in Mathematica. Model fitting resulted in an estimate of 5nM for the average non-specific binding constant accounting for rTetR and Dam interactions with GATC sites genome-wide. The goodness of the fit was evaluated using the adjusted R2 (0.68).
ChromHMM
In order to assign chromatin states, we used the ChromHMM software57 with four states. We used histone modifications as in Supplementary Table 6. The four states correspond to active (enriched in H3K36me3, H3K27ac, H3K4me1 and H3K9ac), poised (H3K36me3, H3K27ac, H3K4me1, H3K9ac and H3K27me3), inert (no enrichment) and heterochromatic (H3K9me3) states.
Deviation scores
Given a set of restriction fragments (or genomic bins) {xi} belonging to a window [a,b], the deviation score is defined as
Where f and g are data vectors (e.g. DamC enrichment, 4C or virtual 4C counts) and < >[a,b] represents the average in the window [a,b]. If two profiles are identical in the window [a,b], then the deviation score is zero; increasing deviation from zero indicates increasing dissimilarity.
PiggyBac-TetO integration site mapping
Paired-end reads (see ‘Mapping of piggyBac insertion sites’ above) were trimmed to 50 bp using a custom script. Read1 and Read2 were mapped separately to the piggyBac-TetO sequence using QuasR (qAlign). Only hybrid pairs with one of the reads mapping to array were kept. The second reads from hybrid pairs were mapped to the mouse genome (build mm9) using QuasR (qAlign). Reads were then piled up in 25 bp windows using csaw (windowCounts function). Integration sites can be identified because they correspond to local high read coverage. Local coverage was calculated by resizing all non-zero 25-bp windows up to 225 bp (expanding by 100 bp upstream and downstream). Overlapping windows were then merged using reduce (from GenomicRanges) thus resulting in a set of windows {wi}. The size distribution of wi is multimodal, and only wi from the second mode on were kept. For each wi we estimated the coverage ci as the number of non-zero 25-bp windows. Only wi’s where the coverage were higher than 16 were considered. The exact position of the integration sites were then identified with the center of wi.
Determination of the orientation of TetO-CTFC insertions
In order to determine the orientation of ectopically inserted TetO-CTCF sites, we exploited the fact that the three CTCF sites are oriented within the piggyBac casette in the 3’ ITR - 5’ ITR direction. If the genomic position of the 5’ ITR is upstream of the 3’ ITR, then CTCF sites are in the reverse orientation (- strand), and vice versa. To determine the relative orientation of the 3’ and 5’ ITRs in the genome, we used only reads that run through the junction between the ITRs and the genome. More precisely, we extracted reads that contain an exact match to 30bps of the ITRs (3’ and 5’ ITR separately), trimmed the ITR sequence and mapped the reads to the mouse genome using qAlign (from QuasR). We quantified the reads at single bp resolution using scanBam. Only integration sites where both 5’ and 3’ ITRs are mapped are kept. This resulted in 91 integration sites (Supplementary Table 4).
Z-score analysis of Hi-C data
In order to identify and exclude ‘noisy’ interactions in Hi-C maps we used a custom algorithm named ‘Neighborhood Coefficient of Variation’ (van Bemmel et al., under revision). Since the chromatin fiber behaves as a polymer, the contact probability of a given pair of genomic loci i and j, is correlate to that of fragments i+N and j+N if N is smaller (or on the order of) than the persistence length of the chromatin fiber. Hence, a given pixel in a Hi-C map can be defined as noisy if its numerical value is too different from those corresponding to neighboring interaction frequencies. To operatively assess the similarity between neighboring interactions, we calculated the coefficient of variation (CV) within a 10x10 pixel square centered on every interaction and discarded all pixels whose CV is larger than a certain threshold. Given that the distribution of the coefficient of variation of Hi-C samples in this study is multimodal with the first component terminating around CV=0.6, we set the CV threshold to 0.6. Discarded interactions appear as grey pixels in the differential Hi-C maps. For differential analysis between TetO-CTCF and wt samples, we calculated the difference between distance-normalized Z-scores calculated for each individual map71. The Z-score is defined as (obs-exp)/stdev where (obs) is the Hi-C signal for a given interaction and (exp) and (stdev) are the genome-wide average and standard deviation of Hi-C signals at the genomic distance separating the two loci.
4C peak calling
In order to call specific interactions in 4C profiles, we used the peakC package41 using the following parameters: qWr = 2.5 and minDist = 20000. peakC was applied to 2 replicates of 4C profiles at single fragment resolution. Peak regions were then extended 1kb upstream and downstream. Overlapping peaks were merged.
Supplementary Material
Acknowledgments
This work is dedicated to the memory of Maxime Dahan. Research in the Giorgetti lab is funded by the Novartis Foundation and the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No 759366 ‘BioMeTre’). The Kind lab was funded by the ERC (grant agreement No 678423 ‘EpiID’) and EMBO (LTF 1214-2016 to I). RSG acknowledges support from the European Union’s Horizon 2020 research and innovation program under the Marie Sk1odowska-Curie grant agreement no. 705354 and an EMBO Long-Term fellowship ALTF 1086-2015. We would like to thank Peter Cron for cloning TetO-piggyBac plasmids, Sirisha Aluri and Stéphane Thiry for assistance with high-throughput sequencing, Michael Stadler for help with bioinformatics analysis, Stefan Grzybek and Hans-Rudolf Hotz for server supports, Edith Heard and Rafael Galupa (Institut Curie, PSL Research University, CNRS UMR3215, INSERM U934, Paris, France) for kindly providing PGK cells. We are grateful to Dirk Schuebeler and Rafael Galupa for critically reading the manuscript and Geoffrey Fudenberg for useful comments on scaling behavior. We acknowledge The ENCODE Project Consortium and in particular the Ren and Hardison laboratories for ChIP-Seq data sets in ESC.
Footnotes
Code availability
The custom-made codes used to analyze the data can be obtained upon request to L. Giorgetti.
Data availability
The sequencing data from this study, including bedgraph files for the visualization of DamC and 4C profiles from all the samples described in the manuscript are available at the NCBI Gene Expression Omnibus, with accession code GEO GSE128017. A UCSC session containing all the DamC and 4C tracks used can be found at https://genome.ucsc.edu/s/zhan/DamC_publication_2019. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE72 partner repository with the dataset identifier PXD013507. Source data for Figure 1, 3-7 and Supplementary Figure 1-3, 5 and 6 are available online.
Author contributions
JR generated cell lines and performed DamC experiments. YZ wrote the model with assistance from GT and analyzed the data. CV performed 4C in WdL’s lab. MK assisted with cell culture and DamC library preparation and performed Hi-C experiments. IG and JK helped with experimental design and data analysis. VI performed mass spectrometry experiments and analysis. TP provided constructs for initial experiments and discussed the data. RSG provided CTCF site sequences and tested CTCF binding in preliminary experiments. EM contributed designing the initial experiments. SAS developed the DamC library preparation protocol and performed piggyBac insertion mapping experiments. LG designed the study and wrote the paper with JR and YZ and input from all the authors.
Competing Interests Statement
The authors declare no competing financial interests.
References
- 1.Denker A, de Laat W. The second decade of 3C technologies: detailed insights into nuclear organization. Genes Dev. 2016;30:1357–1382. doi: 10.1101/gad.281964.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lieberman-Aiden E, et al. Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rao SSP, et al. A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Norton HK, et al. Detecting hierarchical genome folding with network modularity. Nat Methods. 2018;15:119–122. doi: 10.1038/nmeth.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fraser J, et al. Hierarchical folding and reorganization of chromosomes are linked to transcriptional changes in cellular differentiation. Mol Syst Biol. 2015;11:852–852. doi: 10.15252/msb.20156492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nora EP, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sexton T, et al. Three-Dimensional Folding and Functional Organization Principles of the Drosophila Genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 9.Zhan Y, et al. Reciprocal insulation analysis of Hi-C data shows that TADs represent a functionally but not structurally privileged scale in the hierarchical folding of chromosomes. Genome Res. 2017;27:479–490. doi: 10.1101/gr.212803.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zuin J, et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci. 2014;111:996–1001. doi: 10.1073/pnas.1317788111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nora EP, et al. Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell. 2017;169:930–944.e22. doi: 10.1016/j.cell.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Wit E, et al. CTCF Binding Polarity Determines Chromatin Looping. Mol Cell. 2015;60:676–684. doi: 10.1016/j.molcel.2015.09.023. [DOI] [PubMed] [Google Scholar]
- 13.Guo Y, et al. CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell. 2015;162:900–910. doi: 10.1016/j.cell.2015.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanborn AL, et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci. 2015;112:E6456–E6465. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fudenberg G, et al. Formation of Chromosomal Domains by Loop Extrusion. Cell Rep. 2016;15:2038–2049. doi: 10.1016/j.celrep.2016.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gavrilov A, Razin SV, Cavalli G. In vivo formaldehyde cross-linking: it is time for black box analysis. Brief Funct Genomics. 2015;14:163–165. doi: 10.1093/bfgp/elu037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gavrilov AA, et al. Disclosure of a structural milieu for the proximity ligation reveals the elusive nature of an active chromatin hub. Nucleic Acids Res. 2013;41:3563–3575. doi: 10.1093/nar/gkt067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williamson I, et al. Spatial genome organization: contrasting views from chromosome conformation capture and fluorescence in situ hybridization. Genes Dev. 2014;28:2778–2791. doi: 10.1101/gad.251694.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Belmont AS. Large-scale chromatin organization: the good, the surprising, and the still perplexing. Curr Opin Cell Biol. 2014;26:69–78. doi: 10.1016/j.ceb.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fudenberg G, Mirny LA. Higher-order chromatin structure: bridging physics and biology. Curr Opin Genet Dev. 2012;22:115–124. doi: 10.1016/j.gde.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tiana G, Giorgetti L. Integrating experiment, theory and simulation to determine the structure and dynamics of mammalian chromosomes. Curr Opin Struct Biol. 2018;49:11–17. doi: 10.1016/j.sbi.2017.10.016. [DOI] [PubMed] [Google Scholar]
- 22.Alipour E, Marko JF. Self-organization of domain structures by DNA-loop-extruding enzymes. Nucleic Acids Res. 2012;40:11202–11212. doi: 10.1093/nar/gks925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nichols MH, Corces VG. A CTCF Code for 3D Genome Architecture. Cell. 2015;162:703–705. doi: 10.1016/j.cell.2015.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang S, et al. Spatial organization of chromatin domains and compartments in single chromosomes. Science. 2016;353:598–602. doi: 10.1126/science.aaf8084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beagrie RA, et al. Complex multi-enhancer contacts captured by genome architecture mapping. Nature. 2017;543:519–524. doi: 10.1038/nature21411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brant L, et al. Exploiting native forces to capture chromosome conformation in mammalian cell nuclei. Mol Syst Biol. 2016;12:891. doi: 10.15252/msb.20167311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quinodoz SA, et al. Higher-Order Inter-chromosomal Hubs Shape 3D Genome Organization in the Nucleus. Cell. 2018;174:744–757.e24. doi: 10.1016/j.cell.2018.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lebrun E, Fourel G, Defossez P-A, Gilson E. A Methyltransferase Targeting Assay Reveals Silencer-Telomere Interactions in Budding Yeast. Mol Cell Biol. 2003;23:1498–1508. doi: 10.1128/MCB.23.5.1498-1508.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cléard F, Moshkin Y, Karch F, Maeda RK. Probing long-distance regulatory interactions in the Drosophila melanogaster bithorax complex using Dam identification. Nat Genet. 2006;38:931–935. doi: 10.1038/ng1833. [DOI] [PubMed] [Google Scholar]
- 30.van Steensel B, Henikoff S. Identification of in vivo DNA targets of chromatin proteins using tethered Dam methyltransferase. Nat Biotechnol. 2000;18:424–428. doi: 10.1038/74487. [DOI] [PubMed] [Google Scholar]
- 31.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing Chromosome Conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 32.van de Werken HJG, et al. Robust 4C-seq data analysis to screen for regulatory DNA interactions. Nat Methods. 2012;9:969–972. doi: 10.1038/nmeth.2173. [DOI] [PubMed] [Google Scholar]
- 33.Masui O, et al. Live-Cell Chromosome Dynamics and Outcome of X Chromosome Pairing Events during ES Cell Differentiation. Cell. 2011;145:447–458. doi: 10.1016/j.cell.2011.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peric-Hupkes D, et al. Molecular Maps of the Reorganization of Genome-Nuclear Lamina Interactions during Differentiation. Mol Cell. 2010;38:603–613. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kind J, et al. Single-Cell Dynamics of Genome-Nuclear Lamina Interactions. Cell. 2013;153:178–192. doi: 10.1016/j.cell.2013.02.028. [DOI] [PubMed] [Google Scholar]
- 36.Cadiñanos J, Bradley A. Generation of an inducible and optimized piggyBac transposon system†. Nucleic Acids Res. 2007;35:e87. doi: 10.1093/nar/gkm446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kamionka A, Bogdanska-Urbaniak J, Scholz O, Hillen W. Two mutations in the tetracycline repressor change the inducer anhydrotetracycline to a corepressor. Nucleic Acids Res. 2004;32:842–847. doi: 10.1093/nar/gkh200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giorgetti L, et al. Predictive Polymer Modeling Reveals Coupled Fluctuations in Chromosome Conformation and Transcription. Cell. 2014;157:950–963. doi: 10.1016/j.cell.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hou C, Zhao H, Tanimoto K, Dean A. CTCF-dependent enhancer-blocking by alternative chromatin loop formation. Proc Natl Acad Sci U S A. 2008;105:20398–20403. doi: 10.1073/pnas.0808506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rawat P, Jalan M, Sadhu A, Kanaujia A, Srivastava M. Chromatin Domain Organization of the TCRb Locus and Its Perturbation by Ectopic CTCF Binding. Mol Cell Biol. 2017;37:e00557–16. doi: 10.1128/MCB.00557-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Geeven G, Teunissen H, de Laat W, de Wit E. peakC: a flexible, non-parametric peak calling package for 4C and Capture-C data. Nucleic Acids Res. doi: 10.1093/nar/gky443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vian L, et al. The Energetics and Physiological Impact of Cohesin Extrusion. Cell. 2018;173:1165–1178.e20. doi: 10.1016/j.cell.2018.03.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bonev B, et al. Multiscale 3D Genome Rewiring during Mouse Neural Development. Cell. 2017;171:557–572.e24. doi: 10.1016/j.cell.2017.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scolari VF, Mercy G, Koszul R, Lesne A, Mozziconacci J. Kinetic Signature of Cooperativity in the Irreversible Collapse of a Polymer. Phys Rev Lett. 2018;121 doi: 10.1103/PhysRevLett.121.057801. 057801. [DOI] [PubMed] [Google Scholar]
- 45.Hsieh T-HS, et al. Mapping Nucleosome Resolution Chromosome Folding in Yeast by Micro-C. Cell. 2015;162:108–119. doi: 10.1016/j.cell.2015.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dekker J, Mirny L. The 3D Genome as Moderator of Chromosomal Communication. Cell. 2016;164:1110–1121. doi: 10.1016/j.cell.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Erickson HP. Size and Shape of Protein Molecules at the Nanometer Level Determined by Sedimentation, Gel Filtration, and Electron Microscopy. Biol Proced Online. 2009;11:32. doi: 10.1007/s12575-009-9008-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brackley CA, et al. Predicting the three-dimensional folding of cis-regulatory regions in mammalian genomes using bioinformatic data and polymer models. Genome Biol. 2016;17:59. doi: 10.1186/s13059-016-0909-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kalhor R, Tjong H, Jayathilaka N, Alber F, Chen L. Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nat Biotechnol. 2012;30:90–98. doi: 10.1038/nbt.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosa A, Everaers R. Structure and Dynamics of Interphase Chromosomes. PLOS Comput Biol. 2008;4:e1000153. doi: 10.1371/journal.pcbi.1000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.La Fortezza M, et al. DamID profiling of dynamic Polycomb-binding sites in Drosophila imaginal disc development and tumorigenesis. Epigenetics Chromatin. 2018;11:27. doi: 10.1186/s13072-018-0196-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tosti L, et al. Mapping transcription factor occupancy using minimal numbers of cells in vitro and in vivo. Genome Res. 2018 doi: 10.1101/gr.227124.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tiana G, et al. Structural Fluctuations of the Chromatin Fiber within Topologically Associating Domains. Biophys J. 2016;110:1234–1245. doi: 10.1016/j.bpj.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gu B, et al. Transcription-coupled changes in nuclear mobility of mammalian cis-regulatory elements. Science. 2018;359:1050–1055. doi: 10.1126/science.aao3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Germier T, et al. Real-Time Imaging of a Single Gene Reveals Transcription-Initiated Local Confinement. Biophys J. 2017;113:1383–1394. doi: 10.1016/j.bpj.2017.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wiśniewski JR, Hein MY, Cox J, Mann M. A “Proteomic Ruler” for Protein Copy Number and Concentration Estimation without Spike-in Standards. Mol Cell Proteomics. 2014;13:3497–3506. doi: 10.1074/mcp.M113.037309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Urlinger S, et al. Exploring the sequence space for tetracycline-dependent transcriptional activators: Novel mutations yield expanded range and sensitivity. Proc Natl Acad Sci. 2000;97:7963–7968. doi: 10.1073/pnas.130192197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vogel MJ, Peric-Hupkes D, van Steensel B. Detection of in vivo protein-DNA interactions using DamID in mammalian cells. Nat Protoc. 2007;2:1467–1478. doi: 10.1038/nprot.2007.148. [DOI] [PubMed] [Google Scholar]
- 60.Gu H, Zou Y-R, Rajewsky K. Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre-loxP-mediated gene targeting. Cell. 1993;73:1155–1164. doi: 10.1016/0092-8674(93)90644-6. [DOI] [PubMed] [Google Scholar]
- 61.Sanulli S, et al. Jarid2 Methylation via the PRC2 Complex Regulates H3K27me3 Deposition during Cell Differentiation. Mol Cell. 2015;57:769–783. doi: 10.1016/j.molcel.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Y, et al. Reversed-phase chromatography with multiple fraction concatenation strategy for proteome profiling of human MCF10A cells. PROTEOMICS. 2011;11:2019–2026. doi: 10.1002/pmic.201000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 64.Cox J, et al. Accurate Proteome-wide Label-free Quantification by Delayed Normalization and Maximal Peptide Ratio Extraction, Termed MaxLFQ. Mol Cell Proteomics. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tyanova S, et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 66.MacLean B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Splinter E, de Wit E, van de Werken HJG, Klous P, de Laat W. Determining long-range chromatin interactions for selected genomic sites using 4C-seq technology: From fixation to computation. Methods. 2012;58:221–230. doi: 10.1016/j.ymeth.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 68.Gaidatzis D, Lerch A, Hahne F, Stadler MB. QuasR: quantification and annotation of short reads in R. Bioinformatics. 2015;31:1130–1132. doi: 10.1093/bioinformatics/btu781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Servant N, et al. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015;16:259. doi: 10.1186/s13059-015-0831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Imakaev M, et al. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat Methods. 2012;9:999–1003. doi: 10.1038/nmeth.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sanyal A, Lajoie B, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Perez-Riverol Y, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47(D1):D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.