Abstract
DNA damage response (DDR) serves as an integrated cellular network to detect cellular stress and react by activating pathways responsible for halting cell cycle progression, stimulating DNA damage repair, and initiating apoptosis. Efficient DDR protects cells from genomic instability while defective DDR can allow DNA lesions to go unrepaired, causing permanent mutations that will affect future generations of cells and possibly cause disease conditions such as cancer. Therefore, DDR mechanisms must be tightly regulated in order to ensure organismal health and viability. One major way of DDR regulation is ubiquitination, which has been long known to control DDR protein localization, activity, and stability. The reversal of this process, deubiquitination, has more recently come to the forefront of DDR research as an important new angle in ubiquitin-mediated regulation of DDR. As such, deubiquitinases have emerged as key factors in DDR. Importantly, deubiquitinases are attractive small-molecule drug targets due to their well-defined catalytic residues that provide a promising avenue for developing new cancer therapeutics. This review focuses on the emerging roles of deubiquitinases in various DNA repair pathways.
Keywords: DNA repair, DNA Damage Response, Deubiquitination, Post-transcriptional Modification
1. Introduction
1.1. DNA damage
Eukaryotes have evolved specialized mechanisms to sense and repair unique lesion structures in the DNA induced by DNA damaging agents [1, 2]. Most lesions require specialized pathways involving sequential action of multiple proteins, whereas some lesions can be repaired directly by protein-mediated reversal. Helix-distorting DNA adducts are typically induced by ultraviolet (UV) radiation and alkylating agents and are typically repaired by the nucleotide excision repair (NER) pathway. Bases that become oxidized by reactive oxygen species are highly mutagenic as they often base pair with ‘incorrect’ bases and can be repaired by base excision repair (BER). Ionizing radiation (IR) and replication fork collapse induce highly toxic and mutagenic dsDNA breaks which can be repaired through either error-prone non-homologous end joining (NHEJ) or error-free homologous recombination (HR) [2, 3]. Interstrand crosslinking (ICL) is repaired through the Fanconi anemia (FA) pathway [4, 5]. If these DNA repair mechanisms fail and lesions persist during S phase, polymerases can still employ DNA damage tolerance (DDT) mechanisms. DDT can be achieved through translesion synthesis (TLS) to complete replication and leave lesions to be repaired later rather than undergo dangerous replication fork collapse and genome instability [1].
1.2. DNA damage response
DNA damage response (DDR) pathways operate both independently and together and must be tightly regulated in their own function. Many of these integrated DDR pathways are especially regulated by posttranslational modifications (PTMs), which are covalent modifications of amino-acid residues on target proteins that alter a variety of protein characteristics. Various PTMs are quick, reversible and dynamic, and allow for rapid responses to the cell’s changing status. They often work in a coordinated manner and are an advantageous way of regulating DDR [6, 7]. While phosphorylation has long been the most characterize and understood PTM in DDR, it is now becoming clear that ubiquitination is another major PTM in DDR.
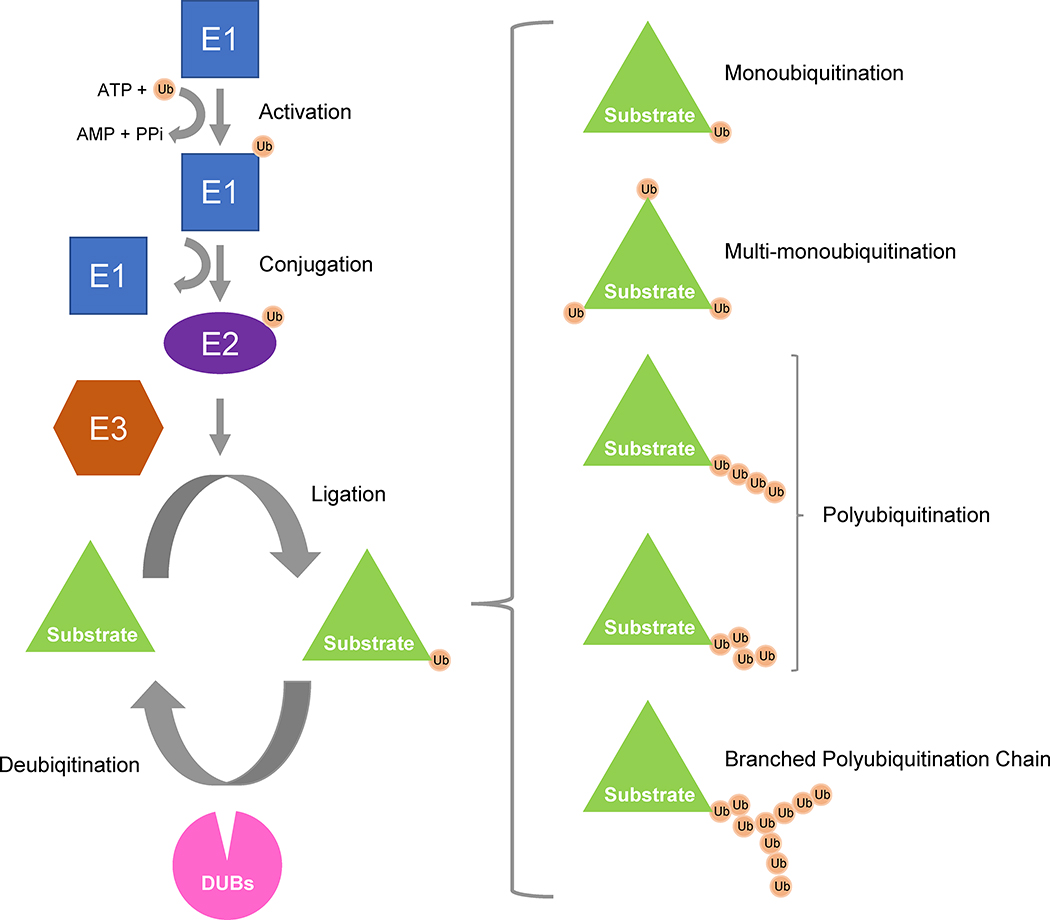
Ubiquitin is a small and highly conserved regulatory protein (8.5 kDa; 76 amino acids) that is found ubiquitously in almost all eukaryotic tissues [8, 9]. Ubiquitin’s carboxy terminal glycine residue can be covalently bonded to lysine residues in the substrate protein via an isopeptide bond. Additionally, any one of the 7 internal lysine residues of the first ubiquitin molecule (K6, K11, K27, K29, K33, K48, or K63) can be used for further linkages to secondary ubiquitin molecules, forming a polyubiquitination chain. This ubiquitination process occurs through the activity of three enzymes: E1 (ubiquitin-activating enzyme), E2 (ubiquitin-conjugating enzyme), and E3 (ubiquitin ligase). E3 ligases are mainly responsible for giving substrate specificity to the ubiquitination process [10]. The ubiquitin chains themselves vary: some substrates receive one ubiquitin molecule (monoubiquitination), one molecule in more than one location (multi-monoubiquitination), and/or chains of multiple ubiquitin moieties (polyubiquitination) which can be branched or folded in different conformations (Figure 1). Given the great diversity in the molecular nature of this covalent linkage, it is not surprising that ubiquitination can affect proteins in different ways: altering cellular localization, altering cellular activity and/or protein interactions, or signaling for catabolic degradation through the proteasome known as Ubiquitin-Proteasome System (UPS) [11–13]. More than 80% of cellular proteins are degraded by the UPS [14].
Figure 1. Ubiquitin Catalytic Process.
Ubiquitin (Ub) is first activated by ATP-dependent ubiquitin activating enzyme (E1), which results in the ubiquitin being thioester conjugated first to E1. Then the Ub is transferred from E1 to ubiquitin conjugating enzyme (E2). E2-Ub interacts with a ubiquitin ligase (E3), which then facilitates the transfer of ubiquitin to the substrate protein. Ubiquitin contains seven lysine residues that can be used to link ubiquitin molecules together. Additional ubiquitin molecules can be linked to the first one to form a polyubiquitin chain. Polyubiquitin chains linked at different positions alters the fate of the target protein. In addition, substrates can be monoubiquitinated or monoubiquitinated at multiple substrate sites (multi-monoubiquitination). Ubiquitin can be removed from the substrate by deubiquitinating enzymes (DUBs).
Like other PTMs, ubiquitination is a reversible modification. Enzymes called deubiquitinases (DUBs) can oppose the action of the E3 ligases by cleaving the isopeptide bond between the C-terminal glycine on ubiquitin and lysine residues on target proteins [9, 15]. DUBs serve several other important purposes including modulating E2 activity, editing non-proteasomal ubiquitin signals, assisting degradation machinery, ubiquitin recycling, and ubiquitin precursor processing [15, 16].
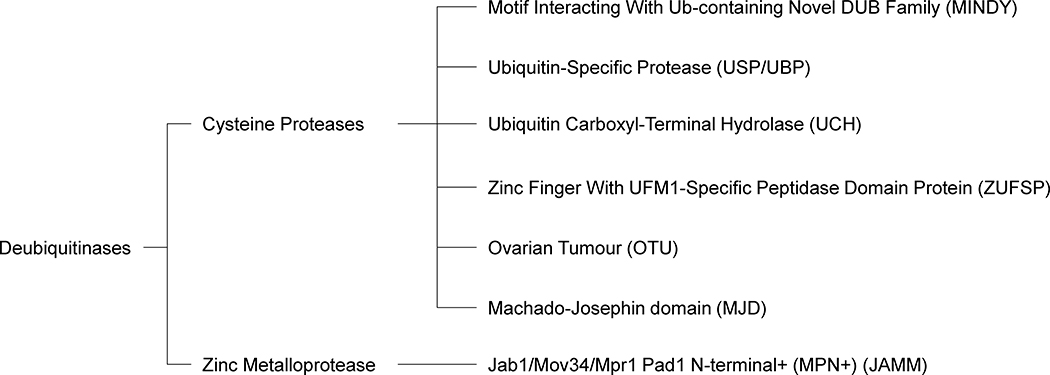
The human genome encodes approximately 100 DUBs [15, 17–19], which accounts for a major fraction of the estimated 460 proteases [20]. Seven types of DUBs are currently known and have been classified based on their active site homology (Figure 2). The DUBs of the small JAB1/MPN+/MOV34 (JAMM) family are zinc metalloproteases [21–24], the cysteine protease DUBs are the newly discovered ZUFSP/ZUP1 [25–27], and the recently discovered MIU-containing novel DUB family (MINDY) [17], ubiquitin c-terminal hydrolases (UCH) [28–30], ubiquitin specific proteases (USP, also known as UBP) [31, 32], ovarian tumor proteases (OTUs) [33, 34], and Machado-Joseph disease proteases (MJD) [35, 36].
Figure 2. Deubiquitinase Classes and Families.
Deubiquitinating enzymes (DUBs) can be classified based on their active site homology into two main classes: cysteine proteases and zinc metalloproteases. Of these two classes, there are five families of cysteine protease DUBs and one family of zinc metalloprotease DUBs. The five families of cysteine protease DUBs consist of motif interacting with Ub-containing novel DUB family (MINDY), ubiquitin-specific protease (USP/UBP), ubiquitin carboxyl-terminal hydrolase (UCH), zinc finger with UFM1-specific peptidase domain protein (ZUFSP), ovarian tumor (OTU), and Machado-Josephin domain (MJD). The one family of zinc metalloprotease DUBs consist of Jab1/Mov34/Mpr1 Pad1 N-terminal+ (MPN+) (JAMM).
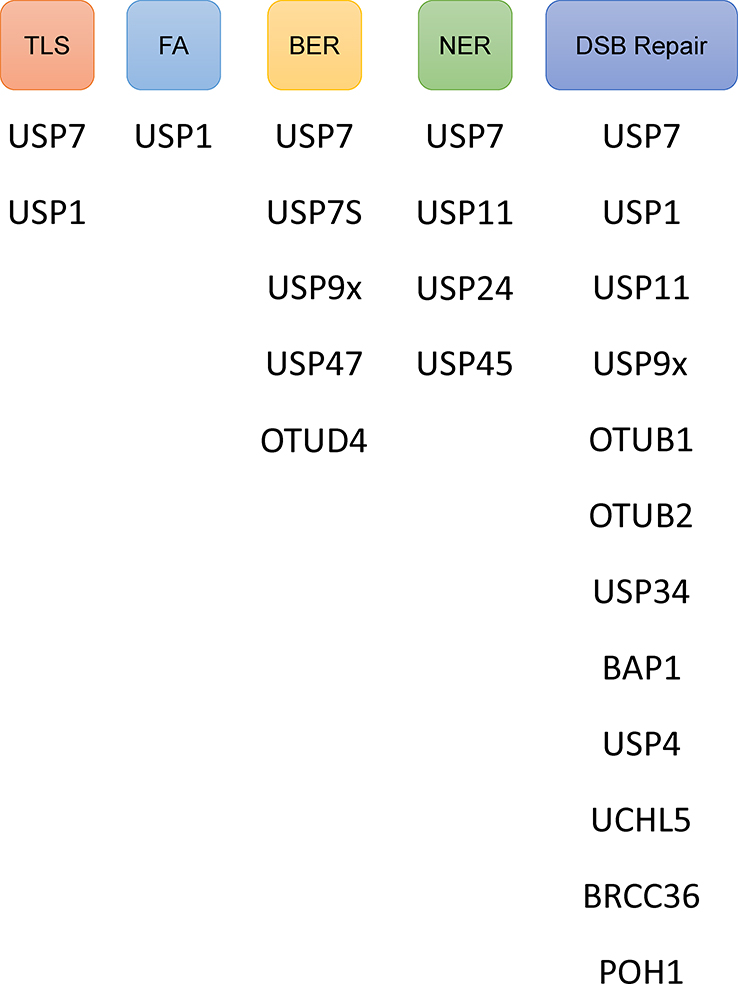
DUB activity is commonly regulated at the levels of transcription/translation and post-translational modifications which modulate intracellular abundance and localization as well as catalytic activity [37]. As ubiquitination is known to play an important role modulating and coordinating DDR, DUBs naturally follow as important factors in regulating DDR mechanisms as well (Figure 3).
Figure 3. Deubiquitinases Involved in DNA Repair Pathways.
DNA damaging agents induce unique lesions. Shown are identified DUBs involved in the DNA repair pathways. See text for explanations of their role in each particular pathway. Note that specific DUBs can be involved in multiple pathways.
2. DUBs Role in Translesion Synthesis
2.1. Translesion synthesis
Translesion synthesis involves the use of alternate, promiscuous DNA polymerases, such as Pol η, ι, κ, and Rev1, to incorporate nucleotides opposite to damaged DNA. Usually, proliferating cell nuclear antigen (PCNA) functions as a sliding clamp, a processivity factor that anchors the replisome to the DNA template. Moreover, PCNA mediates the DNA damage tolerance pathway by recruiting TLS polymerases to sites of stalled forks when monoubiquitinated [38, 39].
2.2. DUBs involved in TLS
Interestingly, the ubiquitin specific peptidase 1-USP1 associated factor 1 (USP1-UAF1) complex, which has been shown to be involved in the Fanconi anemia pathway, has also been shown to regulate TLS by deubiquitinating monoubiquitinated PCNA. The removal of ubiquitin from PCNA promotes switching from replicative polymerases to TLS polymerases. Elevated levels of PCNA monoubiquitination compete against its normal error-free DNA replication kinetics, as excessive and/or irregular recruitment of TLS polymerases to the replication fork leads to reduced fork speeds and is a contributing factor to genomic instability [38, 40, 41]. Additionally, ubiquitin specific peptidase 7 (USP7) has been implicated in the deubiquitination of RING-type E3 ubiquitin transferase RAD18 (RAD18) and Pol η to prevent their degradation via the UPS. Both RAD18 and Pol η proteins are destabilized as a consequence of the loss of USP7 resulting in compromised UV-induced PCNA monoubiquitination and Pol η recruitment to stalled replication forks, respectively [38, 42–44].
3. DUBs role in Fanconi anemia pathway
3.1. Fanconi anemia pathway
Fanconi anemia is a human genetic disorder characterized by a deficiency in the repair of DNA interstrand crosslinks, which leads to the blockage of DNA replication and transcription [45]. Repair of interstrand crosslinks can be facilitated by the FA pathway [4, 5]. The FA pathway consists of the upstream E3 ligase complex termed “the core FA complex” that is comprised of eight FA proteins (FANCA, FANCB, FANCC, FANCE, FANCF, FANCG, FANCL and FANCM) and other associated factors (FAAP100, FAAP24, FAAP20, MHF1, and MHF2) and the downstream Fanconi anemia complementation group D2-Fanconi anemia complementation group I (FANCD2-FANCI) complex [40, 45–50]. Monoubiquitinated FANCD2 is necessary for its localization to sites of DNA damage to facilitate the downstream repair of ICLs through the recruitment of DNA repair proteins to stalled sites of DNA replication. However, FANCD2 monoubiquitination alone is not sufficient for ICL repair. Over accumulation of monoubiquitinated FANCD2 can lead to impaired ICL repair [51].
3.2. USP1 is involved in FA pathway
FANCD2 deubiquitination by USP1, is also required for the FA pathway to function properly. The FA pathway depends on USP1 to maintain a proper equilibrium between monoubiquitinated and deubiquitinated FANCD2 [38]. USP1 forms a complex with UAF1 where its WD40 domain binds and stimulates the ubiquitin protease activity of USP1 to counteract the monoubiquitination of the FANCD2-FANCI heterodimer [52]. The whole cellular pool of FANCD2 is monoubiquitinated when USP1 is depleted, which may result in deregulated FANCD2 recruitment in cells to the damage sites [51]. This balancing act of FANCD2 monoubiquitination and deubiquitination is supported by the observation that upon DNA damage, USP1 is transcriptionally downregulated in order to likely facilitate FANCD2 monoubiquitination in the beginning [52]. Additionally, FANCI is required for foci formation of the core complex components. Ubiquitinated FANCI loses this activity, and therefore deubiquitination by USP1 is critical for efficient foci formation of the core complex. How this occurs is still mechanistically vague [46, 53].
More recently, the deubiquitination of the FANCD2-FANCI complex has been found to be influenced by the phosphorylation of FANCI. Specifically, phosphorylation in the FANCI S/TQ cluster may somehow enhance the FANCD2-FANCI interaction and hinder deubiquitination by USP1, as the monoubiquitination sites of FANCD2 and FANCI are buried in the FANCD2-FANCI interface and therefore inaccessible to USP1 [45]. How USP1 eventually deubiquitinates the FANCD2-FANCI complex following the completion of the DNA repair is still a question. A recent review speculates that the FANCD2-FANCI complex may somehow be removed from the damage sites after completion of repair and the DNA-free complex may be in a conformation that is subject to deubiquitination [46].
4. DUBs role in base excision repair
4.1. Base excision repair
Base excision repair (BER) is commonly used to repair small lesions in the genome, unlike NER which is the common pathway towards repairing bulky, helix-distorting lesions. For the repair of one nucleotide, the short-patch BER (SP-BER) pathway is incorporated. DNA glycosylases remove the damaged base, leaving only the sugar-phosphate backbone, also called an apurinic/apyrimidinic site (AP site). AP endonucleases such as apurinic/apyrimidinic endodeoxyribonuclease 1 (APE1) then cleave this site, leaving a 3’ hydroxyl adjacent to a 5’ deoxyribose phosphate, thereby providing a primer for DNA polymerase β (or polymerase λ in the absence of polymerase β), repairing the genome. For the repair of 2–12 nucleotides, long-patch BER (LP-BER) is incorporated. DNA synthesis is instead done by DNA Polymerase δ and Polymerase ε in conjunction with a proliferating cell nuclear antigen acting as a ‘clamp’ protein. These polymerases displace the downstream 5’ end of DNA to form a flap intermediate. The displaced strand is removed by a nuclease such as flap structurespecific endonuclease 1 (FEN1), thereby creating a ligatable substrate. Factors such as the type of lesion, the current stage of the cell cycle, and ATP concentration will affect whether the SP or LP BER pathway is taken [54].
4.2. DUBs involved in BER
DNA polymerase β is the key polymerase that plays a role in BER. Ubiquitin specific peptidase 47 (USP47) plays a role in regulating cytoplasmic levels of newly synthesized Pol β, which is targeted for degradation if nuclear levels of Pol β are sufficient for DNA repair. It is shown that knockdown of USP47 causes an increased level of ubiquitinated Pol β, decreased levels of deubiquitinated Pol β, and a deficiency in BER. This leads to accumulation of strand breaks and ultimately a decrease in cell viability. Regulation of cytoplasmic Pol β is also important since overproduction of Pol β leads to deficient repair and an increased rate of mutagenesis, both contributing to an increase in cancer susceptibility [55].
One way that DNA can be damaged is through alkylating agents such as methyl methane sulfonate. DNA alkylation can be repaired by BER as well as by oxidative demethylases. AlkB homolog 2, alpha-ketoglutarate dependent dioxygenase (ALKBH2) and AlkB homolog 3, alpha-ketoglutarate dependent dioxygenase (ALKBH3), both oxidative demethylases, remove methyl groups from double-strand DNA as well as singlestranded DNA or RNA [56, 57]. Characterization of ALKBH3-interacting proteins show that USP7 and USP9x associate with another DUB OTU deubiquitinase 4 (OTUD4) to form a protein complex that deubiquitinates ALKBH3 [58]. Knockdown of OTUD4, USP7, or USP9x have all resulted in cells being sensitized to DNA alkylation as well as reduced levels of ALKBH3. This shows how multiple DUBs play a role in the less common DDR pathways such as base repair by oxidative demethylases. It also highlights how DUBs such as USP7 can play roles in different DDR pathways [38, 58].
Interestingly, isoforms of DUBs can also play roles in DDR pathways. For example, USP7S, a S18 containing isoform of USP7, regulates the ARF-binding protein 1 (ARF-BP1, also known as Mule) E3 ligase. This enzyme stabilizes other enzymes that are involved in BER, the most important of which being that it regulates DNA polymerase β and λ. USP7S regulates Mule stability by reversing its self-ubiquitination [59].
5. DUBs role in nucleotide excision repair
5.1. Nucleotide excision repair
Many human diseases result from complex interactions between genome and environmental agents. Mutation is a frequent consequence of unrepaired DNA damage. The nucleotide excision repair (NER) pathway plays a particularly important role in the repair of environmental mutagen-induced DNA damage. NER repairs a wide variety of helix-distorting `bulky’ DNA lesions that result from damaging agents such as UV radiation, cisplatin or reactive oxygen species (ROS). It does this by cutting the damaged bases within a longer string of nucleotides out, and then resynthesizing the missing bases via the untouched template strand [60, 61].
There are two sub-pathways of NER, termed global genomic repair (GG-NER) and transcription-coupled repair (TC-NER). These pathways differ mainly in their recognition of damage. While an RNA polymerase (RNAP) blocked by a lesion initiates TC-NER, restricting recognition to the transcribed strand of active genes, GG-NER surveys the entire genome for distorting DNA lesions [3, 61]. For TC-NER, the stalled RNAP elicits the recruitment of Cockayne syndrome proteins A and B (CSA and CSB), which are both crucial in damage verification. On the other hand, GG-NER requires Xeroderma pigmentosum complementation group C (XPC) and Xeroderma pigmentosum complementation group E (XPE/DDB2) for damage recognition. XPC joins a complex with UV Excision Repair Protein RAD23 homologues and centrin 2, which scans for and recognizes lesions, and initiates NER when needed. XPC lacks the ability to recognize UV-induced lesions, which DDB2 compensates for. DDB2 can recognize these lesions and then hand the damage over to XPC to complete the NER process. TC-NER is a more complex process, yet ubiquitination plays crucial roles in both TC-NER and GG-NER [3, 60–62].
5.2. DUBs involved in GG-NER
USP24 is involved in the GG-NER pathway, specifically in the UV triggered DNA damage response. DDB2 is deubiquitinated by USP24, stabilizing the entire UV-DDB-CUL4 E3 ligase from proteasomal degradation [63]. This complex in turn ubiquitinates XPC, however it is not yet known what this linkage type and its interaction are. In addition, USP7 plays a role in regulating GG-NER. XPC, the protein which recognizes DNA damage in GG-NER, is ubiquitinated by the Cullin-RING E3 ubiquitin ligase 4 (CRL4) upon UV damage, although ubiquitination of XPC does not lead to significant proteasomal degradation. XPC can be converted back to its unmodified stage by USP7 [64]. Inhibition of USP7 increases XPC ubiquitination level, and without USP7, cells have decreased efficiency in repairing UV lesions [60, 61].
Interestingly, USP11 is another DUB that plays a part in GG-NER. USP11 knockdown in HaCaT cells increased ubiquitination of XPC as compared to control cells, suggesting that USP11 is important in maintaining NER capacity, as USP11 mediates XPC deubiquitination at the chromatin following UVB damage [65].
5.3. DUBs involved in TC-NER
In addition to being involved in the GG-NER pathway, USP7 also plays a role in TC-NER by interacting with UV stimulated scaffold protein A (UVSSA) and controlling the stability of CSB/ERCC6 and RNA polymerase II [66–68]. UVSSA is a gene that when mutated, has been found to be responsible for causing UV-sensitive syndrome disorder. USP7 is found to interact with UVSSA, likely through UVSSA recruiting USP7 along with other TC-NER factors in a UV dependent manner. For example, upon UV induced DNA damage, UVSSA can recruit USP7 to stabilize the TC-NER master organizer ERCC6 [67, 69]. Furthermore, depletion of USP7 leads to reduced recovery of RNA synthesis and destabilization of RNA polymerase II as seen in UV-sensitive syndrome cells. This suggests that USP7 and UVSSA work together to stabilize CSB and RNA Polymerase II [61, 69].
USP45 is another DUB involved in NER. UV damage repair in USP45 knockout cells is significantly impaired compared to control cell lines, suggesting USP45’s role in UV-induced damage repair. The depletion of USP45 sensitizes cells to DNA damaging agents. Specifically, the ability of cells to repair cyclobutane pyrimidine dimers (CPD), DNA lesions resulting from UV-C treatment, is greatly impaired in USP45 knockout cells. It was shown that USP45 interacts with and deubiquitinates the excision repair cross-complementation group 1 (ERCC1) subunit of the XPF-ERCC1 DNA repair nuclease which has role in the TC-NER of UV induced DNA damage [70, 71]. Evidence suggests that USP45 cleaves K48 and K63 ubiquitin linkage. Importantly, USP45 is recruited to the site of DNA damage and also controls the ability of ERCC1 to translocate to foci of DNA damage. Therefore, USP45 can affect UV damage repair by targeting ERCC1 [70].
6. DUBs role in DNA double-strand break repair pathway choice
6.1. DNA double-strand break repair
DNA double-strand breaks (DSBs) can be repaired through either error-prone non-homologous end joining (NHEJ) or error-free homologous recombination (HR) [2, 19]. Studies have shown that the appropriate choice between HR and NHEJ repair is crucial in preserving genome integrity. Ubiquitination plays a key role in the recruitment processes of these repair pathways, as such deubiquitination has been shown to influence whether the damage response favors HR or NHEJ [38, 72].
In mammalian cells, double-strand break repair begins with the recruitment and accumulation of various repair and signaling molecules by the site of the lesion. Recruitment of numerous factors to the chromatin regions surrounding DSBs requires a ubiquitination cascade [73]. The DSB sites where the molecules are recruited are commonly referred to as the ionizing radiation-induced foci (IRIF) - which protects and sequester the broken DNA-ends and modulate several cellular processes. IRIF also provide an important checkpoint signaling platform from which DNA damage-activated kinases such as ataxia telangiectasia mutated (ATM) and ataxia telangiectasia and Rad3-related protein (ATR) can globally impact transcriptional processes, cell cycle transitions, and cell fate decisions [74].
Initially, ATM and related kinases phosphorylate H2A histone family member x (γH2AX) surrounding the damage site, leading to recruitment of mediator of DNA damage checkpoint 1 (MDC1), a scaffolding protein. ATM-mediated phosphorylation of which then provides a platform for ring finger protein 8 (RNF8), a RING E3 ligase, to dock. Then, RNF8-UBC13 mediates the ubiquitination of proteins at the damage site [75, 76]. RNF8-UBC13 extends the ubiquitination signal and allows the formation of K63-linked polyubiquitin chains. Then, ring finger protein 168 (RNF168), another E3 ligase, complexes with ubiquitin-conjugating enzyme E2N (UBC13, also known as UBE2N) and recognizes the RNF8-mediated ubiquitination products and ubiquitinates H2A-type histones. A major ramification of local RNF8/RNF168-mediated ubiquitination is the recruitment of checkpoint and repair factors such as breast cancer type 1 susceptibility protein (BRCA1), receptor associated protein 80 (RAP80), tumor protein P53 binding protein 1 (53BP1), RAD51 recombinase (RAD51), and RAD18 at sites of DNA damage [73]. Evidence suggests that these proteins are in a competitive balance to promote DSB repair by HR or NHEJ through DNA-end protection or DNA-end resection. For example, the clearing of RAP80 and 53BP1 from the IRIF allows nucleases to promote DNA-end resection, which in turn allows HR to proceed [37, 77–79].
6.2. DUBs involved in DSB repair
26S proteasome-associated PAD1 homolog 1 (POH1) is a JAMM/MNP+ metalloprotease [21] that cleaves K63 chains at sites of DNA damage [80], thus limiting the IR-induced formation of K63 chains at DSB sites. It is recruited to the IRIF in a BRCA1-dependent manner, and promotes the removal of RAP80, the ubiquitin chain, and 53BP1 from the IRIF [81]. One proposed model is that POH1 degrades RAP80 and the loss of RAP80-dependent protection of the ubiquitin chain promotes the removal of ubiquitin, leading to the removal of 53BP1 from the IRIF; however, the detailed molecular mechanism remains to be elucidated. RAP80 and ubiquitin chains persist at the IRIF in POH1-depleted cells, but the ubiquitin chains are removed from the IRIF when POH1 and RAP80 are simultaneously depleted, suggesting that other DUBs but not POH1 degrade ubiquitin chains. POH1 relieves the barriers imposed by 53BP1 and RAP80 in the late stages of DDR and induces the switch from NHEJ to HR. Thus, POH1 limits excessive NHEJ repair and promotes HR repair [78, 82].
OTU domain-containing ubiquitin aldehyde-binding protein 2 (OTUB2) has been identified as a suppressor of excessive ubiquitination in the early stages of DDR. OTUB2 is shown to constitutively interact with polycomb molecule lethal(3)malignant brain tumor-like protein 1 (L3MBTL1). The ubiquitination of L3MBTL1 is required for its removal from damaged chromatin in order for 53BP1 to be recruited to the damage site. However, OTUB2 deubiquitinates L3MBTL1 and suppresses the RNF8-mediated ubiquitination of L3MBTL1, which leads to less recruitment of RAP80 and 53BP1 to DSBs and therefore increases HR. Additionally, OTUB2 suppresses K63-linked ubiquitin chain formation decreasing the recruitment of RNF168 to the DSBs. Consistent with the idea that 53BP1 and RAP80 suppress HR promoting events such as DNA-end resection and RPA foci, HR repair activity is decreased upon OTUB2 depletion [83]. In OTUB2-depleted cells, the conjugation of ubiquitin and accumulation of RNF168, 53BP1 and RAP80 at DSB sites are significantly accelerated during the beginning phases of the DDR, and total DSB repair is upregulated. However, DNA-end resection and HR are suppressed in OTUB2-depleted cells. OTUB2 enables the initiation of HR by suppressing the excessive accumulation of 53BP1 and RAP80 in an early phase of the DDR [72, 83].
BRCA1/BRCA2-containing complex subunit 36 (BRCC36) is a JAMM/MPN+ metalloprotease that cleaves the K63 chain on histone H2A [80], which may facilitate optimal recognition of RAP80. BRCC36-depletion enhances the recruitment of 53BP1 to the DSBs in RNF8-depleted cells, indicating that BRCC36 and RNF8 play opposing roles in ubiquitination-mediated DDR. Interestingly, the depletion of BRCC36 increases HR, which goes against the suppressive role of BRCC36 in RNF8-dependent DDR that should suggest that BRCC36 promotes HR. This discrepancy is probably due to inefficient accumulation of the BRCA1-A complex at DSB sites in BRCC36-depleted cells. However, the physiological role of BRCC36 in DNA repair pathway choice remains to be elucidated [72, 84].
Ubiquitin specific peptidase 11 (USP11) physically associates with H2AX and is a unique DUB for H2AX. USP11 deubiquitinates H2AX, opposing the ubiquitinating activity of the RNF8-RNF168 complex. USP11 depletion enhances the levels of ubiquitinated H2AX and is associated with prolonged 53BP1 retention and stronger ubiquitination formation at DSB sites [85]. An earlier paper identified USP11 participation in HR repair, where they proposed that the prolonged 53BP1 foci observed could be a consequence of the persistence of unrepaired DSBs or increased levels of ubiquitin-conjugation at the DSB. The increased 53BP1 retention at DSBs when USP11 is depleted could be attributed to the decrease in HR repair [86].
Ubiquitin c-terminal hydrolase L5 (UCHL5, also known as UCH37), implied through its name, contains a UCH domain [87, 88]. UCHL5 regulates DSB resection and repair by HR through protecting nuclear factor related to KappaB binding protein (NFRKB) from degradation. Specifically, UCHL5 promotes DNA-end resection and HR through regulating the stability of the NFRKB protein that is a subunit of chromatin remodeling complex INO80 [89, 90]. UCHL5 and NFRKB enhance resection by regulating the recruitment of the resection factor exonuclease 1 (EXO1) [91].
Ubiquitin specific peptidase 4 (USP4) also regulates DSB end-resection. Currently, two studies have shown USP4 positively regulates DNA-end resection in response to DNA damage, thereby promoting DSB repair by HR. USP4 interacts with the retinoblastoma binding protein 8 (CtIP, also known as RBBP8) and the MRE11RAD50-NBS1 (MRN) complex [92, 93]. Both of which have been suggested to be involved in DNA-end resection [94]. USP4 depletion has been shown to reduce the recruitment of CtIP and RAD51 to DSB sites. However, MRE11 and NBS1 recruitment to DSB sites was not affected by USP4 depletion. Additionally, in USP4-knockdown cells, DNA damage sensor replication protein A (RPA) subunit RPA2 S4/S8 phosphorylation is reduced after DNA damage [93] and RPA recruitment to DSB sites is reduced [92]. Interestingly, both studies show that USP4 counteracts its own ubiquitination by deubiquitinating itself in order to be catalytically active [92, 93].
OTU domain-containing ubiquitin aldehyde-binding protein 1 (OTUB1) has been found to negatively regulate the DNA damage-dependent ubiquitination by RNF168 and UBC13 [95]. UBC13 is a ubiquitin conjugating enzyme that heterodimerizes with ubiquitin conjugating enzyme E2 V1 (UBE2V1, also known as UEV1a2) and synthesizes K63 polyubiquitin chains at DSB sites in concert with RNF168, thereby promoting the independent recruitment of RAP80 and 53BP1 to damaged chromatin, which are critical for DNA repair [96]. Accordingly, the overexpression of OTUB1 inhibits K63 chain accumulation and 53BP1 IRIF. Interestingly, OTUB1 suppresses RNF168-dependent K63 polyubiquitination of the chromatin surrounding DSBs independently of its catalytic activity via binding to UBC13 to inhibit its E2 activity. It seems that non-catalytic roles for DUBs are possible [95–97].
Ubiquitin specific peptidase 34 (USP34) plays an important role in the recruitment of DNA damage checkpoint and mediator proteins at DSBs. Specifically, USP34 promotes RNF168-dependent functions at DSBs. USP34-depletion led to defective accumulation of BRCA1, 53BP1 and RAD18 to DSBs. Another DUB, USP7, has also been shown to deubiquitinate and stabilize RNF168 [98]; however, RNF168 is itself polyubiquitinated and destabilized upon IR treatment, which is reversed by USP34, suggesting that USP34 specifically stabilizes RNF168 when DNA becomes damaged [99].
Fascinatingly, a more recent study shows that USP7 complexes with ring finger protein 169 (RNF169) via crystal structure. RNF169 has antagonistic properties on the DSB signal transduction pathway possibly by restraining the ubiquitin-mediated DSB signaling events. Interestingly, this is the opposite of RNF168 which plays positive roles in amplifying ubiquitin-dependent DSB signals. RNF169 competes for RNF168-catalyzed ubiquitin adducts, thus limiting excessive loading of the DNA damage-mediator proteins 53BP1 and RAP80. As a result, the USP7–RNF169 axis promotes HR [100].
USP9x, discussed earlier under BER, has another well-known substrate, the BCL-2 family apoptosis regulator (MCL-1) which plays a role in DNA DSB repair [101]. Interestingly, knockdown of USP9x increases MCL-1 ubiquitination and turnover. It’s also been shown that increased USP9x expression is correlated with increased MCL-1 expression levels in these cancerous cells, and that patients with increased USP9x have poorer prognoses [102]. A recent study showed that MCL-1-depleted cells had a reduced frequency of IR-induced BRCA1, RPA, and Rad51 focus formation, decreased DNA end-resection, and decreased HR repair suggesting that MCL-1 may be important for DNA end resection. Additionally, the same study showed that there was an increase in 53BP1 and Rap1-interacting factor 1 homolog (RIF1) colocalization at DSB sites as a result of MCL-1 depletion, which blocks the recruitment of BRCA1 [101]. Therefore, if USP9x increases MCL-1 expression [102], then it could be suggested the USP9x indirectly promotes HR repair via MCL-1.
There is evidence that suggests that the USP1/UAF1 complex, previously discussed above to be involved in the FA pathway and TLS, promotes DSB repair via HR. A study in which USP1−/−, UAF1−/−/−, and USP1−/− UAF1−/−/− double-knockout DT40 cells showed hypersensitivity to camptothecin and an inhibitor of poly(ADP-ribose) polymerase (PARP), which suggests that the USP1/UAF1 complex promotes HR. Additionally, the same study disrupted the NHEJ pathway, via knockout of Ku70 (also known as XRCC6 or X-Ray repair cross complementing 6), in UAF1−/−/− cells which restored the cellular resistance to camptothecin and the PARP inhibitor suggesting that the USP1/UAF1 complex promotes HR, at least in part by suppressing NHEJ. However, the direct mechanism by which USP1/UAF1 promotes HR remains to be elucidated [103].
BRCA1 associated protein 1 (BAP1) is part of the UCH DUB family [104]. BAP1 is rapidly recruited to DSB sites, and BAP1 depletion leads to reduced BRCA1, RAD51 and RPA foci and has been suggested to decrease HR. Overexpressing BAP1 reduces ubiquitinated forms of H2A and H2AX, while depletion increases them, suggesting that BAP1 is a DUB for the ubiquitinated H2A(X). The BAP1 complex may work in unison with a protein regulator of cytokinesis 1 (PRC1) complex to promote dynamic ubiquitination/deubiquitination of H2A. A possible mechanism may be that BAP1 deubiquitinates H2A in the proximity of the DSB site to increase chromatin accessibility at the damaged region to allow DNA resection during HR. This might interfere with specific chromatin and/or histone modifications events at DSBs. Consequently, BAP1 depletion increases the amount of ubiquitinated H2A and leads to reduced HR repair and sensitivity to IR [105, 106].
6. Discussion
Cells are constantly exposed to a plethora of DNA damaging agents. DNA can endure damage from normal endogenous biochemical processes, as well as exposure to exogenous genotoxic agents. If DNA lesions are neither detected, repaired, nor removed properly from the line of transmission to daughter cells, the cells will likely become mutagenic and have a highly compromised genomic stability. Cells have evolved a DNA damage response mechanism, which is a highly integrated collection of crucial pathways that detect damage and halt cell proliferation to either repair DNA lesions or to induce apoptosis if the lesions cannot be repaired.
DDR is a crucial cellular process governing cellular health and vitality. Defective DDR is a principal characteristic of tumorigenesis, as well as cancer treatment, which attempts to induce irreparable DNA damage. Dissecting the molecular biochemical pathways that function in DDR will allow for better diagnosis of neoplastic conditions and allow for the development of personalized chemo- and immunotherapeutics that can overcome hurdles of sickening side-effects and therapeutic resistance.
Lately, there has been research focused on DUBs and their interactions with cancer stem cells (CSC), a type of cancer cell that has stem-like properties, meaning they have the ability to self-renew and differentiate into different cell types. Identifying DUBs involved in regulating CSC transcription factors and proteins could provide insight into DUB regulatory mechanisms in carcinogenesis [107]. It has been shown that p53, phosphatase and tensin homolog (PTEN), and PRC1 all play roles in CSC fate. For example, other DUBs such as USP7, USP10, USP11, ataxin-3 (ATXN3), OTU deubiquitinase 1 (OTUD1) and OTU deubiquitinase 5 (OTUD5) have been found to deubiquitinate the tumor suppressor p53 [108–116]. Furthermore, USP13 and USP18 have been shown to regulate the stabilization of PTEN, another tumor suppressor whose destabilization correlates with carcinogenesis [117, 118]. Lastly, USP7 and USP11 deubiquitinate PRC1, a protein found to contribute to cancer stemness [119–121]. It is clear that DUBs play a large role in the tightly regulated pathways that CSC use to proliferate, and learning more about these interactions would allow for the production of specific DUB-targeting therapeutics [116].
There have also been studies on the interactions between DUBs and autophagy, a process necessary for homeostasis of the cell by making sure that degradation of protein aggregates and dysfunctional organelles occurs. Without proper autophagy, disease including carcinogenesis can occur. Interestingly, ubiquitination and deubiquitination are important regulatory mechanisms of autophagy machinery as well as autophagy substrates. Thus, DUBs play a crucial role in the regulation of autophagy. One example of this is ubiquitin c-terminal hydrolase L1 (UCHL1), a DUB responsible for the accumulation of α-synuclein in the brain, a hallmark of Parkinson’s disease. It is thought that UCHL1 does this by negatively regulating the lysosomal degradation of α-synuclein. Additionally, UCHL1 has also been detected in various lung tumors but not in healthy lung tissue, implying a carcinogenic effect. Since one single DUB can play a role in different types of diseases, in this case a neurodegenerative disease and cancer, it is clear that they are an attractive therapeutic target, and that more research needs to be done on DUB regulatory mechanisms [122].
Ubiquitination has been well established as a primary means of regulating and integrating DDR pathways, and deubiquitinases have more recently gained attention as providing additional levels of DDR regulation. Mutations and deletions in DUB genes have become increasingly associated with disease conditions, including cancer, and are now being heavily explored as drug targets for cancer and other diseases [16, 77, 123]. Unlike E3 ligases, DUBs fortunately serve as attractive small-molecule drug targets due to their well-defined catalytic residues that provide a promising avenue for developing new cancer therapeutics. However, the roles of many DUBS in DDR still have not been elucidated and warrant further investigations.
Table 1.
DUBs that Regulate the DDR Pathways.
| DUB | Substrate/Target | Process | Functional Consequence | References |
|---|---|---|---|---|
| USP7 | XPC | NER | May regulate the stabilization and chromatin loading of XPC for GG-NER | [60, 61, 64] |
| UVSSA & ERCC6/CSB | NER | Regulates the stability of ERCC6/CSB for TC-NER | [61, 66–69] | |
| ALKBH2 & ALKBH3 | BER | Promotes alkylation damage resistance & regulates sensitivity to alkylating agents | [58] | |
| RNF168 & RNF169 | DSB | Stabilizes RNF168 and may regulate BRCA1 recruitment at DSB sites; HR repair increased via USP7-RNF169 axis | [98, 100] | |
| PCNA | TLS | Stabilizes RAD18 and Pol η | [42–44] | |
| USP7S | Mule/ARF-BP1 | BER | Stabilizes DNA Polymerase β and λ | [59] |
| USP24 | DDB2 | NER | Allow proper detection of UV-induced damage by stabilizing E3 ligase | [60, 61, 63] |
| USP45 | ERCC1 | NER | Promotes translocation of ERCC1 to foci of DNA damage and ability of cells to repair CPDs | [70, 71] |
| USP11 | XPC | NER | Proper damage recognition of lesions | [65] |
| H2AX | DSB | HR repair increased | [85, 86] | |
| USP9x | ALKBH2 & ALKBH3 | BER | Promotes alkylation damage resistance & regulates sensitivity to alkylated agents | [58] |
| MCL-1 | DSB | Stabilizes MCL-1 & HR repair increased; NHEJ repair decreased | [101, 102] | |
| USP47 | Polymerase β | BER | Synthesize nicked DNA | [55] |
| OTUD4 | USP7 & USP9x, ALKBH2 & ALKBH3 | BER | Promotes alkylation damage resistance via stabilization of AlkB homologues | [58] |
| OTUB1 | UBC13 | DSB | HR repair decreased | [95–97] |
| OTUB2 | L3MBTL1 & K63-linked ubiquitin chains | DSB | HR repair increased; NHEJ repair decreased | [72, 83] |
| USP34 | RNF168 | DSB | HR repair decreased | [99] |
| BAP1 | H2A | DSB | HR repair increased | [104–106] |
| UCHL5 | NFRKB | DSB | HR repair increased | [87–91] |
| USP4 | CtlP & MRN | DSB | HR repair increased | [92–94] |
| BRCC36 | K63 | DSB | HR repair increased; NHEJ repair decreased | [72, 80, 84] |
| POH1 | K63 | DSB | HR repair decreased | [21, 78, 80–82] |
| USP1 | FANCD2/FANCI | FA | Efficient foci formation at sites of DNA Damage | [40, 45–53] |
| PCNA | TLS | Promotes recruitment of TLS polymerases | [41] | |
| ??? | DSB | HR repair increased; NHEJ decreased | [103] | |
Highlights:
DUBs are involved in translesion synthesis by regulating the ubiquitination state of PCNA influencing the recruitment of TLS polymerases.
DUBs are involved in interstrand crosslink repair through regulating the ubiquitination state of the FANCD2-FANCI complex in the Fanconi anemia pathway.
DUBs are involved in base excision repair by stabilizing DNA Polymerase β as well as oxidative demethylases.
DUBs are involved in nucleotide excision repair by directly regulating the synthesis and stabilization of RNA Polymerase II and crucial damage recognition proteins.
DUBs regulate double-stranded break repair pathway choice by either promoting the recruitment or clearance of DNA-end resecting and/or DNA-end protecting proteins.
ACKNOWLEDGEMENTS
This work was supported by grants R21 ES024882 and R01 ES017784 from the National Institute of Health.
Abbreviations
- DDR
DNA damage response
- UV
Ultraviolet
- NER
nucleotide excision repair
- BER
base excision repair
- IR
ionizing radiation
- NHEJ
non-homologous end joining
- HR
homologous recombination
- ICL
interstrand crosslink
- DDT
DNA damage tolerance
- TLS
translesion synthesis
- PTM
post-translational modification
- E1
ubiquitin activating enzyme
- E2
ubiquitin conjugating enzyme
- E3
ubiquitin ligase
- UPS
ubiquitin-proteasome system
- DUB
deubiquitinating enzyme
- JAMM
JAB1/MPN+/MOV34 metalloenzyme domain DUB family
- ZUFSP
zinc finger with UFM1-specific peptidase domain protein DUB family
- MINDY
motif interacting with Ub-containing novel DUB family
- UCH
ubiquitin c-terminal hydrolase domain DUB family
- USP/UBP
ubiquitin specific protease domain DUB family
- OUT
ovarian tumor domain DUB family
- MJD
Machado-Joseph disease protein domain DUB family
- PCNA
proliferating cell nuclear antigen
- USP
ubiquitin specific peptidase
- UAF1
USP1 associated factor 1
- RAD18
RING-type E3 ubiquitin transferase RAD18
- FA
Fanconi anemia
- FANC
Fanconi anemia complementation group
- FAAP
Fanconi anemia core complex associated protein
- MHF
mph1-associated Histone-Fold protein
- SP-BER
short-path BER
- AP site
apurinic/apyrimidinic site
- APE1
apurinic/apyrimidinic endodeoxyribonuclease 1
- LP-BER
long-patch BER
- FEN1
flap structure-specific endonuclease 1
- ALKBH2
AlkB homolog 2, alpha-ketoglutarate dependent dioxygenase
- ALKBH3
AlkB homolog 3, alpha-ketoglutarate dependent dioxygenase
- OTUD
OTU domain-containing deubiquitinase protein
- ARF-BP1/Mule
ARF-binding protein 1
- ROS
reactive oxygen species
- GG-NER
global genome NER
- TC-NER
transcription coupled NER
- RNAP
RNA polymerase
- CSA
Cockayne syndrome protein A
- CSB/ERCC6
Cockayne syndrome protein B
- XPC
Xeroderma pigmentosum complementation group C
- XPE/DDB2
Xeroderma pigmentosum complementation group E
- UV-DDB-CUL4
UV-damaged DNA-binding protein-Cullin4 complex
- CRL
Cullin-RING E3 ubiquitin ligases
- UVSSA
UV stimulated scaffold protein A
- ERCC
excision repair cross-complementation group
- CPD
cyclobutane pyrimidine dimers
- DSB
DNA double-strand break
- IRIF
ionizing radiation-induced foci
- ATM
ataxia telangiectasia mutated
- ATR
ataxia telangiectasia and Rad3-related protein
- H2A
H2A Histone Family Member
- MDC1
mediator of DNA damage checkpoint 1
- RNF
ring finger protein
- UBC13/UBE2N
ubiquitin-conjugating enzyme E2N
- BRCA1
breast cancer type 1 susceptibility protein
- RAP80
receptor associated protein 80
- 53BP1
tumor protein P53 binding protein 1
- RAD51
RAD51 recombinase
- POH1
26S proteasome-associated PAD1 homolog 1
- OTUB
OTU domain-containing ubiquitin aldehyde-binding protein
- L3MBTL1
lethal(3)malignant brain tumor-like protein 1
- BRCC36
BRCA1/BRCA2-containing complex subunit 36
- UCH
ubiquitin c-terminal hydrolase
- NFRKB
nuclear factor related to KappaB binding protein
- EXO1
exonuclease 1
- CtIP/RBBBP8
retinoblastoma binding protein 8
- MRN
MRE11-RAD50-NBS1 complex
- RPA
replication protein A
- UBE2V1/UEV1a2
ubiquitin conjugating enzyme E2 V1
- RIF
Rap1-interacting factor 1 homolog
- PARP
poly(ADP-ribose) polymerase
- Ku70/XRCC6
X-Ray repair cross complementing protein 6
- BAP1
BRCA1 associated protein 1
- PRC1
protein regulator of cytokinesis 1
- CSC
cancer stem cells
- PTEN
phosphatase and tensin homolog
- ATXN3
Ataxin-3
Footnotes
Competing Interests
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ghosal G and Chen J, DNA damage tolerance: a double-edged sword guarding the genome. Transl Cancer Res, (2013). 2(3): p. 107–129.DOI: 10.3978/j.issn.2218-676X.2013.04.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoeijmakers JH, Genome maintenance mechanisms for preventing cancer. Nature, (2001). 411(6835): p. 366–74.DOI: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 3.Curtin NJ, DNA repair dysregulation from cancer driver to therapeutic target. Nat Rev Cancer, (2012). 12(12): p. 801–17.DOI: 10.1038/nrc3399. [DOI] [PubMed] [Google Scholar]
- 4.Liu T, et al. , FAN1 acts with FANCI-FANCD2 to promote DNA interstrand cross-link repair. Science, (2010). 329(5992): p. 693–6.DOI: 10.1126/science.1192656. [DOI] [PubMed] [Google Scholar]
- 5.Moldovan GL and D’Andrea AD, How the fanconi anemia pathway guards the genome. Annu Rev Genet, (2009). 43: p. 223–49.DOI: 10.1146/annurev-genet-102108-134222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dantuma NP and van Attikum H, Spatiotemporal regulation of posttranslational modifications in the DNA damage response. EMBO J, (2016). 35(1): p. 6–23.DOI: 10.15252/embj.201592595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huen MS and Chen J, The DNA damage response pathways: at the crossroad of protein modifications. Cell Res, (2008). 18(1): p. 8–16.DOI: 10.1038/cr.2007.109. [DOI] [PubMed] [Google Scholar]
- 8.Goldstein G, et al. , Isolation of a polypeptide that has lymphocyte-differentiating properties and is probably represented universally in living cells. Proc Natl Acad Sci U S A, (1975). 72(1): p. 11–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Komander D and Rape M, The ubiquitin code. Annu Rev Biochem, (2012). 81: p. 203–29.DOI: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 10.D’Arcy P, Wang X, and Linder S, Deubiquitinase inhibition as a cancer therapeutic strategy. Pharmacol Ther, (2015). 147: p. 32–54.DOI: 10.1016/j.pharmthera.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 11.Glickman MH and Ciechanover A, The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev, (2002). 82(2): p. 373–428.DOI: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 12.Mukhopadhyay D and Riezman H, Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science, (2007). 315(5809): p. 201–5.DOI: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- 13.Schnell JD and Hicke L, Non-traditional functions of ubiquitin and ubiquitin-binding proteins. J Biol Chem, (2003). 278(38): p. 35857–60.DOI: 10.1074/jbc.R300018200. [DOI] [PubMed] [Google Scholar]
- 14.Rock KL, et al. , Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell, (1994). 78(5): p. 761–771.DOI: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 15.Reyes-Turcu FE, Ventii KH, and Wilkinson KD, Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem, (2009). 78: p. 363–97.DOI: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He M, et al. , The emerging role of deubiquitinating enzymes in genomic integrity, diseases, and therapeutics. Cell Biosci, (2016). 6(1): p. 62DOI: 10.1186/s13578-016-0127-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rehman S.A. Abdul, et al. , MINDY-1 Is a Member of an Evolutionarily Conserved and Structurally Distinct New Family of Deubiquitinating Enzymes. Mol Cell, (2016). 63(1): p. 146–55.DOI: 10.1016/j.molcel.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clague MJ, et al. , Deubiquitylases from genes to organism. Physiol Rev, (2013). 93(3): p. 1289–315.DOI: 10.1152/physrev.00002.2013. [DOI] [PubMed] [Google Scholar]
- 19.Nijman SM, et al. , A genomic and functional inventory of deubiquitinating enzymes. Cell, (2005). 123(5): p. 773–86.DOI: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 20.Fortelny N, et al. , Network analyses reveal pervasive functional regulation between proteases in the human protease web. PLoS Biol, (2014). 12(5): p. e1001869DOI: 10.1371/journal.pbio.1001869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yao T and Cohen RE, A cryptic protease couples deubiquitination and degradation by the proteasome. Nature, (2002). 419(6905): p. 403–7.DOI: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- 22.Verma R, et al. , Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science, (2002). 298(5593): p. 611–5.DOI: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- 23.Maytal-Kivity V, et al. , MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem, (2002). 3: p. 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ambroggio XI, Rees DC, and Deshaies RJ, JAMM: a metalloprotease-like zinc site in the proteasome and signalosome. PLoS Biol, (2004). 2(1): p. E2DOI: 10.1371/journal.pbio.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwasna D, et al. , Discovery and Characterization of ZUFSP/ZUP1, a Distinct Deubiquitinase Class Important for Genome Stability. Mol Cell, (2018). 70(1): p. 150–164 e6DOI: 10.1016/j.molcel.2018.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coleman KE and Huang TT, In a Class of Its Own: A New Family of Deubiquitinases Promotes Genome Stability. Mol Cell, (2018). 70(1): p. 1–3.DOI: 10.1016/j.molcel.2018.03.022. [DOI] [PubMed] [Google Scholar]
- 27.Haahr P, et al. , ZUFSP Deubiquitylates K63-Linked Polyubiquitin Chains to Promote Genome Stability. Mol Cell, (2018). 70(1): p. 165–174 e6DOI: 10.1016/j.molcel.2018.02.024. [DOI] [PubMed] [Google Scholar]
- 28.Rose IA and Warms JV, An enzyme with ubiquitin carboxy-terminal esterase activity from reticulocytes. Biochemistry, (1983). 22(18): p. 4234–7. [DOI] [PubMed] [Google Scholar]
- 29.Johnston SC, et al. , Structural basis for the specificity of ubiquitin C-terminal hydrolases. EMBO J, (1999). 18(14): p. 3877–87.DOI: 10.1093/emboj/18.14.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pickart CM and Rose IA, Ubiquitin carboxyl-terminal hydrolase acts on ubiquitin carboxyl-terminal amides. J Biol Chem, (1985). 260(13): p. 7903–10. [PubMed] [Google Scholar]
- 31.Tobias JW and Varshavsky A, Cloning and functional analysis of the ubiquitin-specific protease gene UBP1 of Saccharomyces cerevisiae. J Biol Chem, (1991). 266(18): p. 12021–8. [PubMed] [Google Scholar]
- 32.Baker RT, Tobias JW, and Varshavsky A, Ubiquitin-specific proteases of Saccharomyces cerevisiae. Cloning of UBP2 and UBP3, and functional analysis of the UBP gene family. J Biol Chem, (1992). 267(32): p. 23364–75. [PubMed] [Google Scholar]
- 33.Makarova KS, Aravind L, and Koonin EV, A novel superfamily of predicted cysteine proteases from eukaryotes, viruses and Chlamydia pneumoniae. Trends Biochem Sci, (2000). 25(2): p. 50–2. [DOI] [PubMed] [Google Scholar]
- 34.Mevissen TE, et al. , OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell, (2013). 154(1): p. 169–84.DOI: 10.1016/j.cell.2013.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scheel H, Tomiuk S, and Hofmann K, Elucidation of ataxin-3 and ataxin-7 function by integrative bioinformatics. Hum Mol Genet, (2003). 12(21): p. 2845–52.DOI: 10.1093/hmg/ddg297. [DOI] [PubMed] [Google Scholar]
- 36.Burnett B, Li F, and Pittman RN, The polyglutamine neurodegenerative protein ataxin-3 binds polyubiquitylated proteins and has ubiquitin protease activity. Hum Mol Genet, (2003). 12(23): p. 3195205DOI: 10.1093/hmg/ddg344. [DOI] [PubMed] [Google Scholar]
- 37.Mevissen TET and Komander D, Mechanisms of Deubiquitinase Specificity and Regulation. Annu Rev Biochem, (2017). 86: p. 159–192.DOI: 10.1146/annurev-biochem-061516-044916. [DOI] [PubMed] [Google Scholar]
- 38.Kee Y and Huang TT, Role of Deubiquitinating Enzymes in DNA Repair. Mol Cell Biol, (2016). 36(4): p. 524–44.DOI: 10.1128/MCB.00847-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sale JE, Lehmann AR, and Woodgate R, Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat Rev Mol Cell Biol, (2012). 13(3): p. 141–52.DOI: 10.1038/nrm3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim M and Kim JM, The role of USP1 autocleavage in DNA interstrand crosslink repair. FEBS Lett, (2016). 590(3): p. 340–8.DOI: 10.1002/1873-3468.12060. [DOI] [PubMed] [Google Scholar]
- 41.Huang TT, et al. , Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol, (2006). 8(4): p. 339–47.DOI: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- 42.Kashiwaba S, et al. , USP7 Is a Suppressor of PCNA Ubiquitination and Oxidative-Stress-Induced Mutagenesis in Human Cells. Cell Rep, (2015). 13(10): p. 2072–80.DOI: 10.1016/j.celrep.2015.11.014. [DOI] [PubMed] [Google Scholar]
- 43.Qian J, et al. , USP7 modulates UV-induced PCNA monoubiquitination by regulating DNA polymerase eta stability. Oncogene, (2015). 34(36): p. 4791–6.DOI: 10.1038/onc.2014.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zlatanou A, et al. , USP7 is essential for maintaining Rad18 stability and DNA damage tolerance. Oncogene, (2016). 35(8): p. 965–76.DOI: 10.1038/onc.2015.149. [DOI] [PubMed] [Google Scholar]
- 45.Cheung RS, et al. , Ubiquitination-Linked Phosphorylation of the FANCI S/TQ Cluster Contributes to Activation of the Fanconi Anemia I/D2 Complex. Cell Rep, (2017). 19(12): p. 2432–2440.DOI: 10.1016/j.celrep.2017.05.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ishiai M, et al. , Activation of the FA pathway mediated by phosphorylation and ubiquitination. Mutat Res, (2017). 803–805: p. 89–95.DOI: 10.1016/j.mrfmmm.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 47.Kim H and D’Andrea AD, Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev, (2012). 26(13): p. 1393–408.DOI: 10.1101/gad.195248.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nijman SM, et al. , The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol Cell, (2005). 17(3): p. 331–9.DOI: 10.1016/j.molcel.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 49.van Twest S, et al. , Mechanism of Ubiquitination and Deubiquitination in the Fanconi Anemia Pathway. Mol Cell, (2017). 65(2): p. 247–259.DOI: 10.1016/j.molcel.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 50.Kennedy RD and D’Andrea AD, The Fanconi Anemia/BRCA pathway: new faces in the crowd. Genes Dev, (2005). 19(24): p. 2925–40.DOI: 10.1101/gad.1370505. [DOI] [PubMed] [Google Scholar]
- 51.Smogorzewska A, et al. , Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell, (2007). 129(2): p. 289–301.DOI: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cohn MA, et al. , A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol Cell, (2007). 28(5): p. 786–97.DOI: 10.1016/j.molcel.2007.09.031. [DOI] [PubMed] [Google Scholar]
- 53.Castella M, et al. , FANCI Regulates Recruitment of the FA Core Complex at Sites of DNA Damage Independently of FANCD2. PLoS Genet, (2015). 11(10):p.e1005563DOI: 10.1371/journal.pgen.1005563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim YJ and Wilson DM 3rd, Overview of base excision repair biochemistry. Curr Mol Pharmacol, (2012). 5(1): p. 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parsons JL, et al. , USP47 is a deubiquitylating enzyme that regulates base excision repair by controlling steady-state levels of DNA polymerase beta. Mol Cell, (2011). 41(5): p. 609–15.DOI: 10.1016/j.molcel.2011.02.016. [DOI] [PubMed] [Google Scholar]
- 56.Falnes PO, Johansen RF, and Seeberg E, AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature, (2002). 419(6903): p. 178–82.DOI: 10.1038/nature01048. [DOI] [PubMed] [Google Scholar]
- 57.Trewick SC, et al. , Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature, (2002). 419(6903): p. 174–8.DOI: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- 58.Zhao Y, et al. , Noncanonical regulation of alkylation damage resistance by the OTUD4 deubiquitinase. EMBO J, (2015). 34(12): p. 1687–703.DOI: 10.15252/embj.201490497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Khoronenkova SV and Dianov GL, USP7S-dependent inactivation of Mule regulates DNA damage signalling and repair. Nucleic Acids Res, (2013). 41(3): p. 1750–6.DOI: 10.1093/nar/gks1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chitale S and Richly H, Timing of DNA lesion recognition: Ubiquitin signaling in the NER pathway. Cell Cycle, (2017). 16(2): p. 163–171.DOI: 10.1080/15384101.2016.1261227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang L and Gong F, The emerging role of deubiquitination in nucleotide excision repair. DNA Repair (Amst), (2016). 44: p. 118–122.DOI: 10.1016/j.dnarep.2016.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chatterjee N and Walker GC, Mechanisms of DNA damage, repair, and mutagenesis. Environ Mol Mutagen, (2017). 58(5): p. 235–263.DOI: 10.1002/em.22087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang L, et al. , The deubiquitinating protein USP24 interacts with DDB2 and regulates DDB2 stability. Cell Cycle, (2012). 11(23): p. 4378–84.DOI: 10.4161/cc.22688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.He J, et al. , Ubiquitin-specific protease 7 regulates nucleotide excision repair through deubiquitinating XPC protein and preventing XPC protein from undergoing ultraviolet light-induced and VCP/p97 protein-regulated proteolysis. J Biol Chem, (2014). 289(39): p. 27278–89.DOI: 10.1074/jbc.M114.589812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shah P, et al. , Regulation of XPC deubiquitination by USP11 in repair of UV-induced DNA damage. Oncotarget, (2017). 8(57): p. 96522–96535.DOI: 10.18632/oncotarget.22105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nakazawa Y, et al. , Mutations in UVSSA cause UV-sensitive syndrome and impair RNA polymerase IIo processing in transcription-coupled nucleotide-excision repair. Nat Genet, (2012). 44(5): p. 58692DOI: 10.1038/ng.2229. [DOI] [PubMed] [Google Scholar]
- 67.Schwertman P, et al. , UV-sensitive syndrome protein UVSSA recruits USP7 to regulate transcriptioncoupled repair. Nat Genet, (2012). 44(5): p. 598–602.DOI: 10.1038/ng.2230. [DOI] [PubMed] [Google Scholar]
- 68.Zhang X, et al. , Mutations in UVSSA cause UV-sensitive syndrome and destabilize ERCC6 in transcription-coupled DNA repair. Nat Genet, (2012). 44(5): p. 593–7.DOI: 10.1038/ng.2228. [DOI] [PubMed] [Google Scholar]
- 69.Bhattacharya S, et al. , Emerging insights into HAUSP (USP7) in physiology, cancer and other diseases. Signal Transduct Target Ther, (2018). 3: p. 17DOI: 10.1038/s41392-018-0012-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Perez-Oliva AB, et al. , USP45 deubiquitylase controls ERCC1-XPF endonuclease-mediated DNA damage responses. EMBO J, (2015). 34(3): p. 326–43.DOI: 10.15252/embj.201489184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McNeil EM and Melton DW, DNA repair endonuclease ERCC1-XPF as a novel therapeutic target to overcome chemoresistance in cancer therapy. Nucleic Acids Res, (2012). 40(20): p. 9990–10004.DOI: 10.1093/nar/gks818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nakada S, Opposing roles of RNF8/RNF168 and deubiquitinating enzymes in ubiquitination-dependent DNA double-strand break response signaling and DNA-repair pathway choice. J Radiat Res, (2016). 57 Suppl 1(S1): p. i33–i40.DOI: 10.1093/jrr/rrw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Price BD and D’Andrea AD, Chromatin remodeling at DNA double-strand breaks. Cell, (2013). 152(6): p. 1344–54.DOI: 10.1016/j.cell.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thompson LH, Recognition, signaling, and repair of DNA double-strand breaks produced by ionizing radiation in mammalian cells: the molecular choreography. Mutat Res, (2012). 751(2): p. 158–246.DOI: 10.1016/j.mrrev.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 75.Wang B and Elledge SJ, Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc Natl Acad Sci U S A, (2007). 104(52): p. 20759–63.DOI: 10.1073/pnas.0710061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huen MS, et al. , RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell, (2007). 131(5): p. 901–14.DOI: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Heideker J and Wertz IE, DUBs, the regulation of cell identity and disease. Biochem J, (2015). 465(1): p. 1–26.DOI: 10.1042/BJ20140496. [DOI] [PubMed] [Google Scholar]
- 78.Kakarougkas A and Jeggo PA, DNA DSB repair pathway choice: an orchestrated handover mechanism. Br J Radiol, (2014). 87(1035): p. 20130685DOI: 10.1259/bjr.20130685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mirza-Aghazadeh-Attari M, et al. , 53BP1: A key player of DNA damage response with critical functions in cancer. DNA Repair (Amst), (2019). 73: p. 110–119.DOI: 10.1016/j.dnarep.2018.11.008. [DOI] [PubMed] [Google Scholar]
- 80.Cooper EM, et al. , K63-specific deubiquitination by two JAMM/MPN+ complexes: BRISC-associated Brcc36 and proteasomal Poh1. EMBO J, (2009). 28(6): p. 621–31.DOI: 10.1038/emboj.2009.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kakarougkas A, et al. , Co-operation of BRCA1 and POH1 relieves the barriers posed by 53BP1 and RAP80 to resection. Nucleic Acids Res, (2013). 41(22): p. 10298–311.DOI: 10.1093/nar/gkt802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Butler LR, et al. , The proteasomal de-ubiquitinating enzyme POH1 promotes the double-strand DNA break response. EMBO J, (2012). 31(19): p. 3918–34.DOI: 10.1038/emboj.2012.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kato K, et al. , Fine-tuning of DNA damage-dependent ubiquitination by OTUB2 supports the DNA repair pathway choice. Mol Cell, (2014). 53(4): p. 617–30.DOI: 10.1016/j.molcel.2014.01.030. [DOI] [PubMed] [Google Scholar]
- 84.Ng HM, et al. , The Lys63-deubiquitylating Enzyme BRCC36 Limits DNA Break Processing and Repair. J Biol Chem, (2016). 291(31): p. 16197–207.DOI: 10.1074/jbc.M116.731927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yu M, et al. , USP11 Is a Negative Regulator to gammaH2AX Ubiquitylation by RNF8/RNF168. J Biol Chem, (2016). 291(2): p. 959–67.DOI: 10.1074/jbc.M114.624478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wiltshire TD, et al. , Sensitivity to poly(ADP-ribose) polymerase (PARP) inhibition identifies ubiquitin-specific peptidase 11 (USP11) as a regulator of DNA double-strand break repair. J Biol Chem, (2010). 285(19): p. 14565–71.DOI: 10.1074/jbc.M110.104745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lam YA, et al. , Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature, (1997). 385(6618): p. 737–40.DOI: 10.1038/385737a0. [DOI] [PubMed] [Google Scholar]
- 88.Holzl H, et al. , The regulatory complex of Drosophila melanogaster 26S proteasomes. Subunit composition and localization of a deubiquitylating enzyme. J Cell Biol, (2000). 150(1): p. 119–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yao T, et al. , Distinct modes of regulation of the Uch37 deubiquitinating enzyme in the proteasome and in the Ino80 chromatin-remodeling complex. Mol Cell, (2008). 31(6): p. 909–17.DOI: 10.1016/j.molcel.2008.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gospodinov A, et al. , Mammalian Ino80 mediates double-strand break repair through its role in DNA end strand resection. Mol Cell Biol, (2011). 31(23): p. 4735–45.DOI: 10.1128/MCB.06182-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nishi R, et al. , Systematic characterization of deubiquitylating enzymes for roles in maintaining genome integrity. Nat Cell Biol, (2014). 16(10): p. 1016–26, 1–8.DOI: 10.1038/ncb3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liu H, et al. , The Deubiquitylating Enzyme USP4 Cooperates with CtIP in DNA Double-Strand Break End Resection. Cell Rep, (2015). 13(1): p. 93–107.DOI: 10.1016/j.celrep.2015.08.056. [DOI] [PubMed] [Google Scholar]
- 93.Wijnhoven P, et al. , USP4 Auto-Deubiquitylation Promotes Homologous Recombination. Mol Cell, (2015). 60(3): p. 362–73.DOI: 10.1016/j.molcel.2015.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Takeda S, et al. , Ctp1/CtIP and the MRN complex collaborate in the initial steps of homologous recombination. Mol Cell, (2007). 28(3): p. 351–2.DOI: 10.1016/j.molcel.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 95.Nakada S, et al. , Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature, (2010). 466(7309): p. 941–6.DOI: 10.1038/nature09297. [DOI] [PubMed] [Google Scholar]
- 96.Sato Y, et al. , Molecular basis of Lys-63-linked polyubiquitination inhibition by the interaction between human deubiquitinating enzyme OTUB1 and ubiquitin-conjugating enzyme UBC13. J Biol Chem, (2012). 287(31): p. 25860–8.DOI: 10.1074/jbc.M112.364752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wiener R, et al. , The mechanism of OTUB1-mediated inhibition of ubiquitination. Nature, (2012). 483(7391): p. 618–22.DOI: 10.1038/nature10911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhu Q, et al. , USP7 deubiquitinase promotes ubiquitin-dependent DNA damage signaling by stabilizing RNF168. Cell Cycle, (2015). 14(9): p. 1413–25.DOI: 10.1080/15384101.2015.1007785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sy SM, et al. , The ubiquitin specific protease USP34 promotes ubiquitin signaling at DNA double-strand breaks. Nucleic Acids Res, (2013). 41(18): p. 8572–80.DOI: 10.1093/nar/gkt622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.An L, et al. , Dual-utility NLS drives RNF169-dependent DNA damage responses. Proc Natl Acad Sci U S A, (2017). 114(14): p. E2872–E2881.DOI: 10.1073/pnas.1616602114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mattoo AR, et al. , MCL-1 Depletion Impairs DNA Double-Strand Break Repair and Reinitiation of Stalled DNA Replication Forks. Mol Cell Biol, (2017). 37(3).DOI: 10.1128/MCB.00535-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schwickart M, et al. , Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature, (2010). 463(7277): p. 103–7.DOI: 10.1038/nature08646. [DOI] [PubMed] [Google Scholar]
- 103.Murai J, et al. , The USP1/UAF1 complex promotes double-strand break repair through homologous recombination. Mol Cell Biol, (2011). 31(12): p. 2462–9.DOI: 10.1128/MCB.05058-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jensen DE, et al. , BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene, (1998). 16(9): p. 1097–112. [DOI] [PubMed] [Google Scholar]
- 105.Ismail IH, et al. , Germline mutations in BAP1 impair its function in DNA double-strand break repair. Cancer Res, (2014). 74(16): p. 4282–94.DOI: 10.1158/0008-5472.CAN-13-3109. [DOI] [PubMed] [Google Scholar]
- 106.Yu H, et al. , Tumor suppressor and deubiquitinase BAP1 promotes DNA double-strand break repair. Proc Natl Acad Sci U S A, (2014). 111(1): p. 285–90.DOI: 10.1073/pnas.1309085110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kaushal K, et al. , Deubiquitinating enzymes in cancer stem cells: functions and targeted inhibition for cancer therapy. Drug Discov Today, (2018). 23(12): p. 1974–1982.DOI: 10.1016/j.drudis.2018.05.035. [DOI] [PubMed] [Google Scholar]
- 108.Li M, et al. , A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol Cell, (2004). 13(6): p. 879–86. [DOI] [PubMed] [Google Scholar]
- 109.Yuan J, et al. , USP10 regulates p53 localization and stability by deubiquitinating p53. Cell, (2010). 140(3): p. 384–96.DOI: 10.1016/j.cell.2009.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ke JY, et al. , USP11 regulates p53 stability by deubiquitinating p53. J Zhejiang Univ Sci B, (2014). 15(12): p. 1032–8.DOI: 10.1631/jzus.B1400180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu H, et al. , The Machado-Joseph Disease Deubiquitinase Ataxin-3 Regulates the Stability and Apoptotic Function of p53. PLoS Biol, (2016). 14(11): p. e2000733DOI: 10.1371/journal.pbio.2000733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li M, et al. , Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature, (2002). 416(6881): p. 648–53.DOI: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- 113.Brooks CL and Gu W, p53 regulation by ubiquitin. FEBS Lett, (2011). 585(18): p. 2803–9.DOI: 10.1016/j.febslet.2011.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Piao S, et al. , Ovarian tumor domain-containing protein 1 deubiquitinates and stabilizes p53. Cell Signal, (2017). 33: p. 22–29.DOI: 10.1016/j.cellsig.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 115.Luo J, et al. , OTUD5 regulates p53 stability by deubiquitinating p53. PLoS One, (2013). 8(10): p. e77682DOI: 10.1371/journal.pone.0077682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Haq S, Suresh B, and Ramakrishna S, Deubiquitylating enzymes as cancer stem cell therapeutics. Biochim Biophys Acta Rev Cancer, (2018). 1869(1): p. 1–10.DOI: 10.1016/j.bbcan.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 117.Mustachio LM, et al. , The ISG15-specific protease USP18 regulates stability of PTEN. Oncotarget, (2017). 8(1): p. 3–14.DOI: 10.18632/oncotarget.13914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhang J, et al. , Deubiquitylation and stabilization of PTEN by USP13. Nat Cell Biol, (2013). 15(12): p. 1486–1494.DOI: 10.1038/ncb2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lecona E, Narendra V, and Reinberg D, Correction for Lecona et al. , “USP7 Cooperates with SCML2 To Regulate the Activity of PRC1”. Mol Cell Biol, (2017). 37(19).DOI: 10.1128/MCB.00631-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lecona E, Narendra V, and Reinberg D, USP7 cooperates with SCML2 to regulate the activity of PRC1. Mol Cell Biol, (2015). 35(7): p. 1157–68.DOI: 10.1128/MCB.01197-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Maertens GN, et al. , Ubiquitin-specific proteases 7 and 11 modulate Polycomb regulation of the INK4a tumour suppressor. EMBO J, (2010). 29(15): p. 2553–65.DOI: 10.1038/emboj.2010.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jacomin AC, Taillebourg E, and Fauvarque MO, Deubiquitinating Enzymes Related to Autophagy: New Therapeutic Opportunities? Cells, (2018). 7(8).DOI: 10.3390/cells7080112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hussain S, Zhang Y, and Galardy PJ, DUBs and cancer: the role of deubiquitinating enzymes as oncogenes, non-oncogenes and tumor suppressors. Cell Cycle, (2009). 8(11): p. 1688–97.DOI: 10.4161/cc.8.11.8739. [DOI] [PubMed] [Google Scholar]