Abstract
Starch-branching enzymes (SBE) break the α-1,4 linkage of starch, re-attaching the chain to a glucan chain by an α-1,6 bond, altering starch structure. SBEs also facilitate starch accumulation by increasing the number of non-reducing ends on the growing chain. In maize (Zea mays), three isoforms of SBE have been identified. To examine the function of the SBEIIa isoform, a reverse genetics polymerase chain reaction-based screen was used to identify a mutant line segregating for a Mutator transposon within Sbe2a. To locate the insertion within the second exon of Sbe2a, the genomic sequence of Sbe2a containing the promoter and 5′ end was isolated and sequenced. Plants homozygous for sbe2a::Mu have undetectable levels of Sbe2a transcripts and SBEIIa in their leaves. Characterization of leaf starch from sbe2a::Mu mutants shows reduced branching similar to yet more extreme than that seen in kernels lacking SBEIIb activity. Characterization of endosperm starch from sbe2a::Mu mutants shows branching that is indistinguishable from wild-type controls. These mutant plants have a visible phenotype resembling accelerated senescence, which was correlated with the Mutator insertion within Sbe2a. This correlation suggests a specific role for SBEIIa in leaves, which may be necessary for normal plant development.
Starch biosynthesis occurs through the action of four enzymes: ADP-Glc pyrophosphorylase (EC 2.7.7.27), starch synthase (EC2.4.1.21), starch-branching enzyme (EC 2.4.1.18), and debranching enzyme (EC 2.4.1.41) (for reviews, see Smith et al., 1997; Myers et al., 2000). The resulting starch may be classified into two types, transitory and reserve, defined by their utilization by plants. Transitory starch accumulates in photosynthetic tissues during light periods to provide a source of photo-assimilates for metabolic sinks at night. Reserve starch accumulates in storage organs, such as cereal endosperm, where it is used later by the germinating embryo. Both transitory starch and reserve starch are composed of two polysaccharides, amylose and amylopectin (Takeda et al., 1988). The differences in the structures of these two polysaccharides result in differences in their chemical and physical properties. Amylose is essentially a linear α-1,4-linked glucan chain of approximately 1,000 residues, whereas amylopectin has numerous branch points that are formed by α-1,6 linkages joining linear chains of an average of 21 residues in length. Starch-branching enzymes (SBE) catalyze the formation of these branch points by breaking α-1,4 linkages and re-attaching the reducing ends of the glucan chains by α-1,6 bonds. Introduction of branches into the glucan chain increases the number of non-reducing ends, thereby facilitating starch synthesis. In this way, SBEs can affect both the structure and quantity of the starch produced.
Three isoforms of SBEs, SBEI, SBEIIa, and SBEIIb, have been identified in leaves and/or kernels of maize (Zea mays; Boyer and Preiss, 1978; Dang and Boyer, 1988). SBEIIa and SBEIIb have been shown to share several biochemical properties (Guan and Preiss, 1993; Takeda et al., 1993) and their cDNA sequences share high homology (Gao et al., 1997), however they differ in terms of their expression pattern (Gao et al., 1996). SBEIIa and SBEIIb have similar Km values for amylose, and both tend to produce shorter constituent chains than SBEI when reacted with amylose in vitro. The cDNA sequences of Sbe2a and Sbe2b have a 2.1-kb region showing 78% identity, flanked by divergent 5′ and 3′ regions. Sbe2a has high expression levels in vegetative tissues and moderate expression in developing kernels, whereas the expression of Sbe2b is restricted to very high levels in kernels during development. These expression patterns suggest that the combinations of isoforms, which interact to form the transitory starch formed in leaf chloroplasts, are different than the combinations responsible for the reserve starch formed in amyloplasts of kernel endosperm. It is interesting that while sequences closely related to the maize SBEII class are found in many plant species, the SBEIIa isoform may be specific to monocots. T-Blast analysis using the N-terminal divergent domain of SBEIIa reveals significant similarity only to genes from rice and wheat, whereas more conserved sequences common to all SBEII proteins detect orthologues in a large number of plant species, including Arabidopsis (M Guiltinan, unpublished data). The lack of a clear SBEIIa homolog in any of the dicot species studied to date may indicate that this isoform evolved after the monocot-dicot divergence and raises questions as to its functional significance in monocots.
The genomic DNA and/or cDNA sequences encoding each of the three SBE isoforms have been cloned and sequenced (Baba et al., 1991; Fisher et al., 1993, 1995; Gao et al., 1997; Kim et al., 1998, 1999), however, mutant phenotypes resulting in the loss of enzyme activity have been identified for only SBEIIb (Vineyard and Bear, 1952; Fisher et al., 1993; Stinard et al., 1993). Compared with wild-type controls, this mutant, ae, accumulates 20% less endosperm starch, which has a reduced amount of branching and therefore longer branch chains (Garwood et al., 1976). Our understanding of how these three SBE isoforms interact with each other and other enzymes in vivo to determine starch structure would be greatly improved by the discovery of mutants lacking SBEIIa activity.
Reverse genetic strategies have been used to successfully isolate mutants in several plant species (for review, see Maes et al., 1999). In maize, the transposon family Mutator shows exceptionally high rates of forward mutation and thus has been useful in mutagenesis studies (for review, see Chandler and Hardeman, 1992). Reverse genetic strategies using Mutator transposons have identified mutants of previously characterized genes such as An1 (Benson et al., 1995) and Zag1 (Mena et al., 1996). The goal of this work was to identify and characterize Mutator insertional mutants for Sbe2a to gain an understanding of the role of SBEIIa in starch synthesis.
RESULTS
Identification of sbe2a::Mu Mutants
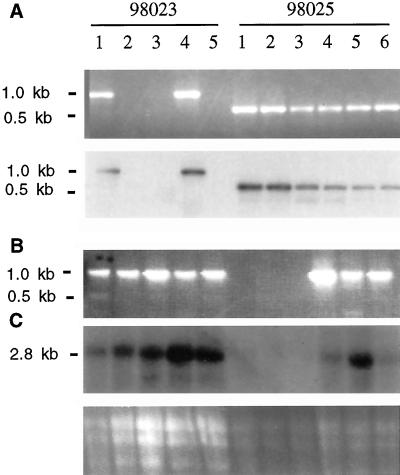
To investigate the in vivo role of SBEIIa in maize, a reverse genetic analysis was conducted using the Mu transposable element. The Trait Utility System for Corn screening system at Pioneer Hi-bred International identified 11 unrelated F1 plants as potentially containing a Mu insertion within the gene encoding SBEIIa; this mutation is designated sbe2a::Mu. F2 progeny from these 11 plants were screened for the presence of sbe2a::Mu by amplification using Sbe2a and MuTIR specific primers, followed by hybridization of products with the full length cDNA of Sbe2a. Two of these F2 lines (98,023 and 98,025) included plants in which a Mu transposon is located within the gene that codes for SBEIIa. The mutation in the 98,025 population is designated sbe2a::Mu (Fig. 1A).
Figure 1.
Identification of sbe::Mu mutants and expression of Sbe2a in individuals from two F2 populations (98023-1 through 5 and 98025-1 through 6). A, Detection of sbe2a::Mu alleles in two independent F2 populations via PCR using primers 2A2 and MuTIR. Gel electrophoresis of amplification products (top) and hybridization of amplification products to Sbe2a cDNA clone (bottom). B, Detection of wild-type Sbe2a alleles via PCR using primers 2A1 and 2A2. Presence of Mutator insertion interferes with PCR resulting in no band in samples from homozygous plants. Combined with part A identifies homozygous wild-type individuals (98023-2, 3, 5), heterozygotes (98023-1, 4, and 98025-4, 5, 6), and homozygous mutants (98025-1, 2, 3). C, Northern blot using seedling tissue hybridized with Sbe2a cDNA clone (top). Gel electrophoresis of RNA showing sample loading (bottom).
Cloning of the 5′-Genomic Fragment
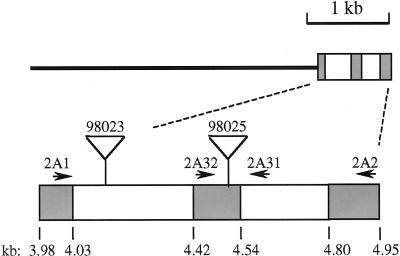
The genomic fragment from the inbred line B73 containing the 5′-untranscribed region, promoter, and 5′ portion of the Sbe2a gene was isolated, cloned, and sequenced (GenBank accession no. AF228486) to aid in the detection of transposition events at the insertion site identified in the sbe2a::Mu mutants and to aid in the interpretation of Sbe2a expression and function. Of 3 × 105 pfu screened, only one unique isolate hybridized to the 5′-gene-specific Sbe2a cDNA fragment. This isolate contains 4 kb of sequence 5′ to the cDNA sequence reported by Gao et al. (1997), in addition to two exons corresponding to 1 to 173 bp of the cDNA clone and two introns (Fig. 2).
Figure 2.
Structure of the genomic sequence from the isolated clone (0–4.71 kb) and PCR products (4.71–5.08 kb). The thin line indicates the region 5′ of the known cDNA sequence. Shaded boxes indicate exons and open boxes indicate introns. Lower diagram includes Mutator insertion sites for lines 98023 (approximately position 4.135) and 98025 (between 4.507/4.508) as determined via size and sequence of PCR products. Diagram also indicates positions of Sbe2a-specific primers (2A1, 2A32, 2A31, and 2A2). Corresponding locations from the Sbe2a cDNA sequence are noted (cDNA bp).
Comparison of DNA sequence data from amplification products using Sbe2a and Mu-specific primers to the sequence of the genomic Sbe2a fragment indicates the position of the Mu elements within the first intron in line 98,023, and within the second exon in line 98,025 (Fig. 2). The insertion site within the exon is between codons 43 and 44 relative to the Sbe2a cDNA sequence (Gao et al., 1997).
Verification of the Lack of SBEIIa in sbe2a::Mu Mutants
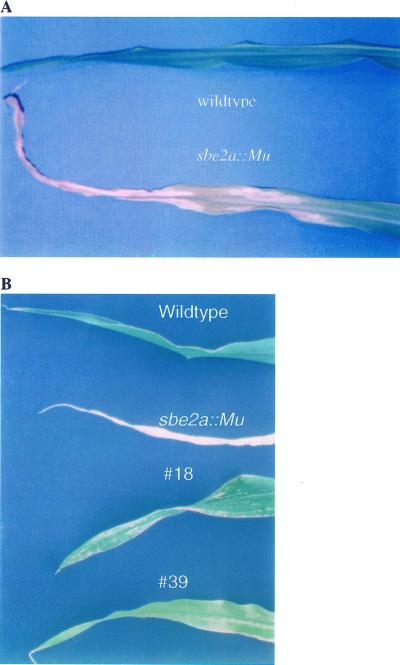
To confirm the successful disruption of Sbe2a expression resulting from the Mu insertion, homozygous mutants were identified (Fig. 1B), and the expression of Sbe2a in their seedling tissue was investigated using northern hybridization (Fig. 1C). Homozygous mutants of line 98,025 (plants 1, 2, and 3) lack expression of Sbe2a, whereas heterozygous mutants (plants 4, 5, and 6) show moderate levels of expression. The homozygous sbe2a::Mu mutants of line 98,025 (plants 1, 2, and 3) had a striking phenotype resembling early senescence. After approximately 10 d after emergence (DAE), leaves at the third node from the youngest visible leaves developed necrotic zones on the leaf margins. As leaves developed these zones expanded into large, necrotic zones on the margins of the leaves, fading into chlorotic zones closer to the mid-vein of the leaves (Fig. 3). At sexual maturity, these three plants failed to produce ears and showed a dramatic loss of vigor likely due to necrosis and chlorosis of the majority of its leaf tissue. Heterozygous and homozygous wild-type siblings showed no signs of early senescence and displayed normal development of ears. Self-pollinated heterozygotes produce kernels that do not segregate in appearance.
Figure 3.
Leaf phenotypes of sbe2a::Mu mutant and their controls. A, Leaf phenotypes of sbe2a::Mu mutant and wild-type controls from line 98025. Leaves at node 5 from line 98025 were photographed from a homozygous sbe2a::Mu mutant and its full-sibling wild-type control at 30 DAE. B, Leaf phenotypes of plants with the reduced senescence phenotype and sbe2a::Mu mutant and wild-type controls. Leaves at node 7 were photographed from a homozygous sbe2a::Mu mutant (sbe2a::Mu) and its full-sibling wild-type control (Wildtype) alone with the two plants displaying reduced senescence (#18 and #39) at 50 DAE.
Homozygous mutants from full siblings of line 98,023 show levels of Sbe2a expression comparable with wild type (data not shown). The ability of these mutants to express Sbe2a is not unusual, for removal of transposons that lie within introns during splicing has been previously observed (Chandler and Hardeman, 1992). Mutants from this line were not studied further.
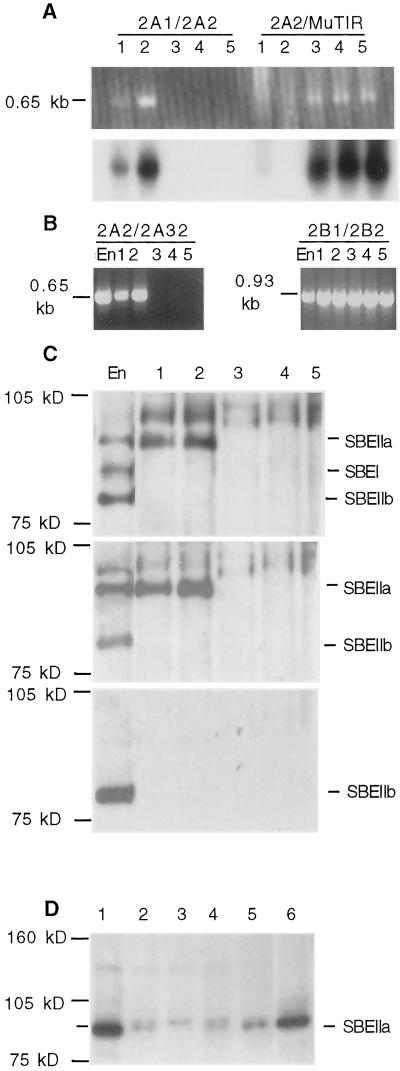
Western analysis of protein extracts from mature, dry kernels using the SBEIIa antibody revealed that whereas SBEIIa is detectable in W64A and ae-ref extracts, it is not detectable in sbe2a::Mu extracts (Fig. 4D). Additionally, SBEIIa was detected in protein extracts from an independent mutant, which is wild type for Sbe2a and contains a Mu insertion at a separate locus. Leaf protein extract from a sbe2a::Mu/Sbe2a heterozygote also shows a very strong band of the expected mobility for SBEIIa (approximately 90 kD). Note that the amount of SBEIIa in dried kernels is significantly reduced as compared with fresh kernels or leaf tissue. A cross-reacting protein of slightly higher Mr was visible in the extracts from dry kernels of all genotypes. This is consistent with previous observations using this antibody (see “Materials and Methods”).
Figure 4.
Molecular analysis of sbe2a::Mu mutants. A through C, Selection of homozygous sbe2a::Mu mutants (lanes 3–5) and wild-type controls (lanes 1 and 2) and the expression patterns of SBEs as compared with inbred line W64A 20 DAP endosperm (En). D, Western analysis of mutant and control genotypes in kernels. A, Identification of sbe2a::Mu mutants and their full-sibling wild-type controls from a BC1F2 population. PCR was used to detect the wild-type allele (primers 2A1/2A2) and the mutant allele (primers 2A2/MuTIR) using DNA extracted from seedlings. Gel electrophoresis of PCR products (top) and hybridization to Sbe2a cDNA clone (bottom). B, RT-PCR analysis of homozygous sbe2a::Mu mutants and their full-sibling wild-type controls to detect transcripts in 30 DAE leaf tissue using primers 2A2 and 2A32 and Sbe2b primers 2B1 and 2B2. C, Western analysis of homozygous sbe2a::Mu mutants and their full-sibling wild-type controls to detect the presence of SBEI, SBEIIa, and SBEIIb in 30 DAE leaf tissue using the non-specific SBEI antibody (top). The high Mr proteins cross-reacting with this antibody are unknown. Identity of SBEIIa and SBEIIb were verified using antibodies to SBEIIa (middle) and SBEIIb (bottom). D, Western analysis of homozygous sbe2a::Mu mutants and controls to detect the presence of SBEIIa in dry kernel tissue using the SBEIIa antibody. Endosperm protein extracts are from fresh sweet corn (lane 1) and from dry kernels of an unrelated Mu mutant wild type for Sbe2a (lane 2), sbe2a:Mu(lane 3), ae-ref (lane 4), and W64A wild type (lane 5). Lane 6 contains protein extract from leaves of an sbe2a::Mu/Sbe2a heterozygote.
Characterization of sbe2a::Mu Mutants
To determine the effects of a lack of SBEIIa on the structure of transitory starch, homozygous mutant plants along with homozygous wild-type controls were selected from a BC1F2 family produced from the self pollination of plant 98133-2, which is heterozygous for sbe2a::Mu. From this segregating population, three homozygous mutant plants and two homozygous wild-type controls were selected using PCR with Sbe2a and MuTIR specific primers, followed by hybridization to the full length Sbe2a cDNA (Fig. 4A). The sequences of these amplification products indicate that the Mu transposon was within the second exon in the same position as identified earlier (data not shown).
RNA was extracted from leaves 30 DAE and analyzed using reverse transcriptase (RT)-PCR to investigate the expression of Sbe2a in homozygous sbe2a::Mu mutants (Fig. 4B). The sbe2a::Mu mutants do not express Sbe2a at detectable levels, compared with easily detectable levels in wild-type controls. RNA from both sbe2a::Mu mutants and their wild-type controls can be used to amplify products via RT-PCR using Sbe2b primers. Western analysis was conducted with sbe2a::Mu mutants to confirm the absence of SBEIIa (Fig. 4C). Extractions from 30 DAE leaves show undetectable levels of SBEIIa in sbe2a::Mu mutants and high levels of SBEIIa present in wild-type controls. Levels of SBEI and SBEIIb were undetectable in both wild type and sbe2a::Mu leaves, whereas both proteins are easily detectable in the W64 endosperm control. The ability to detect Sbe2b transcripts whereas SBEIIb cannot be detected may be due to the higher sensitivity of RT-PCR versus western analysis or may be due to the Sbe2b primers binding to the sbe2a::Mu allele downstream of the Mu insertion.
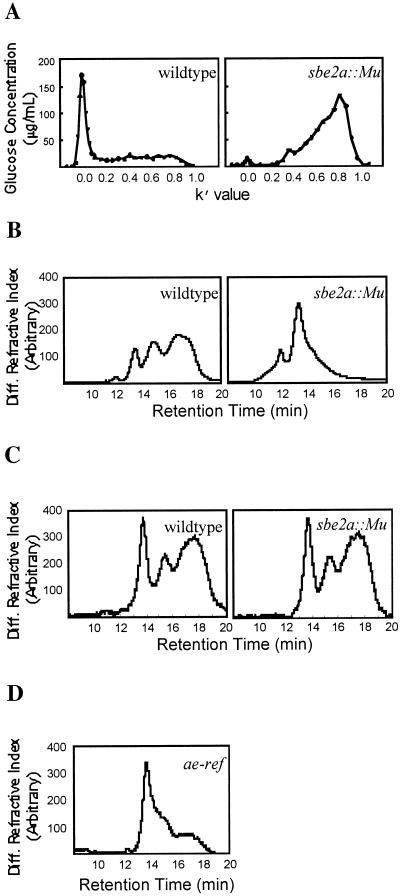
The structure of leaf starch produced by the sbe2a::Mu mutants and wild-type controls was determined using size exclusion chromatography (SEC) of whole starch (Fig. 5A) and high-performance size exclusion chromatography (HPSEC) of debranched starch (Fig. 5B). SEC of leaf starch from sbe2a::Mu mutants shows a much greater proportion of low Mr material than material eluting at the void volume compared with wild-type controls (Fig. 5A). The broad response observed with sbe2a::Mu mutant starch with a peak at k′ of 0.8 corresponds to either amylose or low Mr amylopectin. If this material were amylose, the increased amount of material observed in this region would be consistent with a lower overall degree of branching. HPSEC analysis of the debranched wild-type leaf starch (Fig. 5B) shows three major peaks, in general agreement with previously published analyses of wild-type maize endosperm starch (Klucinec and Thompson, 1998). The first peak results from the long chains of debranched amylose, and the second and third peaks represent the long and short chains of debranched amylopectin, respectively (Klucinec and Thompson, 1998). When debranched wild type and sbe2a::Mu leaf starch are compared (Fig. 5B), the peak corresponding to the short chains of debranched amylopectin is much diminished for the sbe2a::Mu leaf starch.
Figure 5.
Analysis of starch samples from sbe2a::Mu 30 DAE leaves and endosperm. A, SEC of whole starches from leaves of homozygous sbe2a::Mu mutants (right) and their full-sibling wild-type controls (left). Mutants and wild-type controls were selected from a BC1F2 population. B, HPSEC of debranched starches from leaves of homozygous sbe2a::Mu mutant (right) and their full-sibling wild-type controls (left). Mutant and wild-type controls were selected from a BC1F2 population. C, HPSEC of debranched endosperm starches from kernels of homozygous sbe2a::Mu mutant (right) and W64A wild-type controls (left). The mutant was selected from a BC3F3 population. D, HPSEC of debranched starch from endosperm of homozygous amylose extender mutant (ae-ref).
The structure of endosperm starch from mature, dry kernels of a sbe2a::Mu mutant isolated from a BC3F3 population was determined using HPSEC of debranched starch (Fig. 5C). HPSEC of debranched endosperm starch produced indistinguishable chromatograms for the sbe2a::Mu mutant and the wild-type control. For comparison, the structure of endosperm starch from ae-ref was determined (Fig. 5D). The peak corresponding to the short chains of debranched amylopectin is much diminished in comparison with the sbe2a::Mu and the wild-type endosperm starches.
Correlation of Phenotype and Genotype
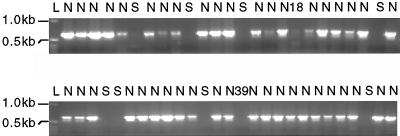
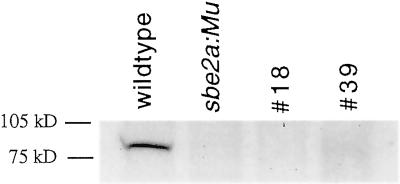
A correlation study was conducted to determine whether the early senescence phenotype displayed by sbe2a::Mu mutants was due to the lack of SBEIIa or due to mutations at a linked locus (Fig. 6). Of the 52 BC1F2 plants scored, 43 plants exhibited normal phenotypes and contained at least one wild-type Sbe2a allele. Eight plants exhibited early senescence and lacked wild-type Sbe2a alleles. The two remaining plants, numbers 18 and 39, were scored as senescent at 10 to 32 DAE, and likewise scored as lacking wild-type Sbe2a alleles. However, when left to mature to 50 DAE, the developing leaves on these plants had a much reduced level of senescence compared with full sibling senescent mutants, as senescence was restricted to small sectors throughout the leaves instead of extensive necrosis (Fig. 3). Total protein extractions from leaves taken from these two plants at 55 DAE show undetectable levels of SBEIIa compared to wild-type control (Fig. 7), suggesting that the reduced phenotype is not due to excision events at the Sbe2a locus. Plant number 39 failed to produce an ear, however plant number 18 was successfully self-fertilized. Preliminary data show that the kernels appear similar to wild-type kernels and the resulting progeny appear to segregate for levels of senescence in a non-Mendelian manner. This segregation pattern indicates that plant number 18 had a reduced expressivity of the mutant phenotype and that the degree of senescence is likely to be influenced by multiple factors. This would suggest that the senescence phenotype is either due to the sbe2a::Mu mutation or to a mutation in a locus within 5.36 map units of Sbe2a based upon the perfect cosegregation of Sbe2a and early senescence in a population of 52 plants.
Figure 6.
Segregation of senescence phenotype and the wild-type Sbe2a allele in a BC1F2 population. The wild-type Sbe2a allele was amplified from DNA extracted from each of 52 plants at 32 DAE using primers 2A2 and 2A32. The molecular mass of two of the bands within the 1-kb ladder (L) are shown to the left. The phenotype observed for each plant is indicated above each lane as normal (N) or senescent (S). Plants 18 and 39 displayed senescence at 10 to 32 DAE, and reduced senescence after 50 DAE.
Figure 7.
Western detection of SBEIIa in plants with the reduced senescence phenotype and sbe2a::Mu mutant and wild-type controls. Total protein was extracted and blotted using leaves at node 9 from homozygous sbe2a::Mu mutant and its full-sibling wild-type control along with the two plants displaying reduced senescence (nos. 18 and 39) at 55 DAE. The western blot was probed using the SBEIIa specific antibody.
DISCUSSION
Analyses of the leaf starch in the sbe2a::Mu mutants isolated in this study, together with previous knowledge, indicate a similar in vivo role for SBEIIa in leaf chloroplasts to that performed by SBEIIb in kernel amyloplasts. Both SBEIIa and SBEIIb have similar in vitro properties and high homology of their cDNA sequences, but differ in their expression pattern (Guan and Preiss, 1993; Takeda et al., 1993; Gao et al., 1996), which is consistent with the lack of similarity between the promoter sequences of Sbe2a and Sbe2b found in this study.
This relationship between SBEIIa and SBEIIb is reflected in the similarity of the starches found in the leaves of sbe2a::Mu mutants and the mature kernels of ae-ref mutants in that both exhibit a much lower proportion of the short chains of amylopectin than the wild-type controls. One may conclude that starch from sbe2a::Mu mutant leaves has little or no normal amylopectin. In this respect, the effects of the sbe2a::Mu mutation on leaf starch are even more pronounced than the effects of the ae-ref mutation on kernel starch (Fig. 5, B and D). The chromatogram in Figure 5D is in excellent agreement with similar analyses of commercial ae-type maize starches (Klucinec and Thompson, 1998).
The lack of SBEIIa has been found to have no effect on the branching of endosperm starch (Fig. 5C). This result may be due to the ability of SBEIIb to compensate for any lack of SBEIIa activity. One must bear in mind the possibility that the relationship among branch points might be altered without a difference in the chain length distribution (Thompson, 2000). We have not investigated this possibility. Alternatively, SBEIIa may simply not play an important role in determining endosperm starch structure. Analysis of fine structure of endosperm starch using multiple mutants will shed light on these questions.
In this study, SBEIIa was present at detectable levels in leaves whereas SBEI was not detected. Dang and Boyer (1988), however, attributed starch branching activity in leaves to both SBEIIa and SBEI based upon their elution profile as compared with that from endosperm tissue. Gao et al. (1997) also conclude that SBEI is present in leaves due to the presence of RNA transcripts from leaves that hybridize to the Sbe1 cDNA. The inability to detect SBEI in leaves in this study suggests that the enzyme with SBEI-like activity detected by Dang and Boyer is likely to be due to an as-yet-unidentified SBE. Alternatively, since the SBEI antibody was raised against a polypeptide sequence from the amino end of SBEI, it is also possible that the SBEI-like activity in leaves was due to a truncated form of SBEI that is missing the amino end sequence needed for detection using our SBEI antibody. The elution profile from leaves (Dang and Boyer, 1988) does not conflict with these possible explanations since the elution of the protein showing SBEI-like activity does differ from that of SBEI from endosperm. Detailed studies using isoform specific antibodies and measuring SBE activity of SBE mutants are needed to clarify the identity of the protein responsible for the SBEI-like activity found in leaves.
An early senescence phenotype was found to be tightly linked to Sbe2a, although two sbe2a::Mu mutant plants had a reduced senescence phenotype compared with the other eight sbe2a::Mu mutants observed. If the early senescence is conferred by sbe2a::Mu, the reduced senescence phenotype may be considered as having a reduced expressivity likely to be in part due to differences in genetic background from those sbe2a::Mu mutants with extreme senescence. In fact, progeny of one of these two mutant plants vary in the degree of senescence, suggesting a variable level of expressivity. This observation, however, does not eliminate the possibility that the early senescence phenotype is not conferred by sbe2a::Mu, but by a locus tightly linked to Sbe2a. It is clear that further work is needed to firmly establish the relationship between Sbe2a and the early senescence phenotype.
The inability of 15 of 16 of the sbe2a::Mu mutants observed in this study to produce ears is likely to be a direct consequence of the severe reduction in photo-assimilate due to the early senescence phenotype. Alternatively, it is possible that the lack of ears may reflect a more direct role of SBEIIa in the early development of the female inflorescence. The discovery of an sbe2a::Mu mutant plant in the population used for the correlation study with reduced senescence that successfully produced an ear will allow the opportunity to explore these questions and to investigate the role SBEIIa plays in starch biosynthesis in the kernel.
The identification of sbe2a::Mu mutants and the isolation of the genomic 5′ region of Sbe2a opens new opportunities to explore leaf starch biosynthesis and the interaction of SBEIIa with other enzymes involved in starch synthesis. These studies in turn may lead to the development of starch with a broader spectrum of branching structure than is currently available via unmodified ae mutants.
MATERIALS AND METHODS
Identification of Putative sbe2a::Mu
Plant populations of maize (Zea mays) segregating for multiple Mutator (Mu) insertions were screened for the presence of a Mu transposon within Sbe2a using a Mu-TIR specific primer (MuTIR9242: 5′-AGAGAAGCCAACGCCA(AT) CGCCTC(CT) ATTTCGTC-3′) and several Sbe2a specific primers (2A1: 5′-GTGCTCTTCAGGAGGAAGGACGCTT-3′, 2A2: 5′-ACCCCGAAACCCTTCCAACATTGGGT-3′, 2A31: 5′-GTGCCCTAACCAACAAT-3′, 2A32: 5′-TGCGCTGGTGCTCCTGGAAAGGTACT-3′). Amplification was conducted by personnel at Pioneer Hi-bred International in a 50-μL reaction containing 10% (w/v) Suc, 10 mm MgCl2, 10% (v/v) dimethyl sulfoxide (DMSO), 0.25 mm dNTP, 1 μm MuTIR primer, 1 μm target primer, 1× HotTub buffer (Amersham, Buckinghamshire, UK), 0.036 units HotTub DNA Polymerase (Amersham), and 50 ng of DNA. Reactions were denatured at 94°C for 2 min and then run for 35 cycles at 94°C for 2 min, 62°C for 1 min, 72°C for 2 min, with a final extension of 2 min at 72°C. Of the total 50 μL of amplification products, 20 μL were separated and visualized on 2% (w/v) agarose gels. Out of approximately 40,000 plants screened, 11 were identified as potentially containing Mu insertions within Sbe2a. F2 seed from these 11 families were analyzed further to identify Mu insertional mutants for Sbe2a (sbe2a::Mu).
Cloning of the 5′-Genomic Fragment
A genomic fragment containing the 5′ end of Sbe2a was isolated from an EMBL-3 genomic library prepared from maize inbred B73 seedlings (Clonetech, Palo Alto, CA) as described by Sambrook et al. (1989). Approximately 3 × 105 pfu were transferred to nylon membranes (Hybond N+, Amersham) following manufacturer's instructions. These filters were probed with a gene-specific fragment of Sbe2a cDNA containing the first 380 bp (Gao et al., 1997) labeled by random hexamer labeling (Boehringer Mannheim, Indianapolis) using the following hybridization conditions. Filters were prehybridized at 65°C for 2 h in 0.5 m Na2HPO4 (pH 7.2), 7% (w/v) SDS, and 40 μg/mL denatured salmon sperm DNA (Church and Gilbert, 1984). Labeled probe was added to prehybridization solution and incubated at 65°C for 12 h. Filters were washed for 15 min at 65°C, twice in 5% SEN (5% [w/v] SDS, 1 mm EDTA, 0.04 m Na2HPO4, pH 7.2) and once in 1% SEN (1% [w/v] SDS, 1 mm EDTA, 0.04 m Na2HPO4, pH 7.2). One unique plaque strongly hybridized to the probe and was purified through four rounds of screening. The phage DNA was isolated as described by Chisholm (1988). PstI fragments hybridizing to the gene-specific Sbe2a probe were subcloned into pBluescript SK− (Stratagene, La Jolla, CA) using standard techniques (Sambrook et al., 1989). Plasmids were then isolated using a Wizardprep kit (Promega, Madison, WI) and sequenced at the DNA Sequencing Facility at the Pennsylvania State University. Sequence data from both strands were compared and compiled using software from DNASTAR, Inc. (Madison, WI).
Plant Material
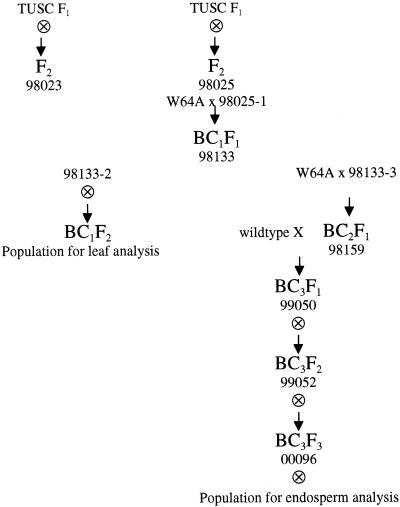
Plants were field grown in the summer at the Pennsylvania State University Horticultural Research Farm (Rock Springs, PA). Plants were grown in the fall and spring under standard greenhouse conditions with supplemental lighting on a 14-h-d/10-h-night cycle. Plants determined to contain Mu insertions within Sbe2a were backcrossed to W64A create a population for leaf phenotype analysis (Fig. 8). Plants containing Mu insertions within Sbe2a were also crossed to wild-type controls and self-pollinated through several generations to generate kernels for endosperm analysis (Fig. 8).
Figure 8.
Pedigree of populations used in this study. Independent F1 plants were identified by Pioneer personnel (Trait Utility System for Corn F1). Self progeny of these plants were crossed to inbred line W64A to create BC1F2 and BC3F3 populations for analysis. Plant 98025-1 was identified as homozygous for sbe2a::Mu (see “Results”). Plants 98133-2, 98133-3, 98159, 99050, and 99052 were determined to be heterozygous for sbe2a::Mu via PCR genotyping (data not shown). Plant 00096 was similarly identified via PCR to be homozygous for sbe2a:Mu (data not shown).
Genotyping
Genomic DNA was extracted from leaf tissue at approximately 10 DAE following the microprep procedure described by Dellaporta (1994). The resulting pellet was dissolved in 200 μL of Tris-EDTA, of which 1 μL was used for amplification via PCR. Each 25-μL reaction contained 2.5 units of Taq DNA polymerase (Promega), 1× Promega buffer, 2.5 mm MgCl2, 0.4 μm each primer, and approximately 50 ng of target DNA. To detect sbe2a::Mu alleles, the Mu-TIR primer was used in combination with one of two Sbe2a specific primers (2A1 or 2A2). To detect Sbe2a alleles lacking a Mutator transposon at a specific area, two Sbe2a specific primers flanking the area in question were used in combination (e.g. 2A1 and 2A2). After heating the reactions to 80°C, 1 mm total dNTP was added to each reaction before resuming the following conditions in a thermal cycler (ERICOMP, San Diego): one cycle of 5 min at 94°C, 40 cycles of 45 s at 94°C, 1 min at 60°C, 2 min at 72°C, with a final extension of 5 min at 72°C. Amplification products were separated on 2% (w/v) agarose gels using standard procedures (Sambrook et al., 1989).
Amplification products were probed with Sbe2a and sequenced to verify the initial identification of sbe2a::Mu mutants. Following gel electrophoresis, amplification products were transferred to nylon membrane and probed with the full-length Sbe2a cDNA clone (Gao et al., 1997) as described above with the following modifications. Filters were hybridized for 3 h, washed once in 5% SEN and twice in 1% SEN for 15 min and exposed to film for approximately 2 h. Amplification products to be sequenced were excised from the agarose gel and purified using a filter unit (Millipore, Bedford, MA) prior to being sequenced.
RNA Expression Analysis
Total RNA was isolated from 30 DAE leaf tissue as described by DeVries et al. (1988). Total RNA was fractionated, transferred to Hybond-N+ as described above, and probed following the same methods as in genomic Southern hybridizations with RNase-free solutions. RT-PCR was performed using a Superscript pre-amplification system (Life Technologies/Gibco-BRL, Cleveland) followed by PCR amplification as described by the manufacturer using gene specific primers to amplify Sbe2a transcripts (2A2 and 2A32) and Sbe2b transcripts (2B1: 5′-TCGCTGCGTTGTCCTCTCTA-3′ and 2B2: 5′-GCTCTCCCTTTTACTGTTGT-3′). Amplification products were separated on 2% (w/v) agarose gels, excised, purified, and sequenced as described above.
Protein and Starch Granule Extraction from Leaves and Fresh Sweet Corn Endosperm
Total protein and starch were extracted from one 4-g sample of 30 DAE leaves of individuals from a BC1F2 population (Fig. 8), 60 DAE leaves of a sbe2a::Mu/Sbe2a heterozygote from a BC3F3 population, and from fresh sweet corn purchased at a local grocery store. Leaf tissues from BC1F2 individuals were harvested at the end of the 14-h-light cycle, flash frozen with liquid nitrogen, and stored at −70°C, whereas leaf tissue of the sbe2a::Mu/Sbe2a heterozygote was harvested, frozen, and immediately freeze dried. Whole sweet corn kernels were dissected from ears and frozen in liquid nitrogen.
Tissue samples from BC1F2 individuals were then ground in liquid nitrogen with a mortar and pestle, and 1 mL of protein extraction buffer containing 100 mm sodium citrate, pH 7.0, 5 mm dithioerythritol (Dang and Boyer, 1988) and protease inhibitors (Complete, Boehringer Mannheim) per gram of tissue was added. The homogenates were centrifuged at 4°C for 10 min at 6,000g. Pellets were resuspended in 10 mL of extraction buffer and homogenized using a tissue homogenizer (Polytron PT10/35, Kinematica, Switzerland). Homogenates were centrifuged as above. The combined supernatants served as a crude extract of total proteins. Starch granules contained in the pellet were resuspended in cold water, filtered successively through 400 and 200 μm mesh and centrifuged as described above. The resulting pellet was washed four times with acetone and dried overnight at 40°C.
Tissue samples from the BC3F3 sbe2a::Mu/Sbe2a heterozygotes and the fresh sweet corn were then ground in liquid nitrogen with a mortar and pestle, and 4 mL of protein extraction buffer containing 100 mm sodium citrate, pH 7.0, 5 mm dithioerythritol (Dang and Boyer, 1988) was added. The homogenates were centrifuged at 4°C for 10 min at 6,000g and the supernatant used for western analysis.
Protein and Starch Granule Extraction from Mature Dry Kernels
Twenty mature dry kernels were harvested from self-pollinated ears of wild-type, sbe2a::Mu mutant and amylose extender mutant (ae-ref) respectively and placed in a cold mortar. After adding liquid nitrogen, kernels were ground into a fine powder and transferred into 15-mL centrifugation tubes. In each tube, 3 mL of protein extraction buffer containing 100 mm sodium citrate, pH 7.0, 5 mm dithioerythritol (Dang and Boyer, 1988) was added and placed on ice for 10 min before being centrifuged at 4°C for 10 min at 6,000g. The supernatants were stored at −80 and used for western blotting.
Endosperm starch from dried kernels was obtained by the method of Boyer and Liu (1985) with modifications. For the W64A wild-type and ae-ref mutant ears, 20 kernels were randomly selected and soaked in 50 mL of 0.02 m, pH 6.5 sodium acetate, 0.01 m mercuric chloride solution. For the sbe2a::Mu mutant ear six kernels were randomly selected and soaked in 20 mL of the same solution. After 24 h, the embryo was removed and the same volume of fresh solution was added for an additional 24 h. Endosperm and pericarps were homogenized for 10 min in a mortar and pestle. The free starch granules were washed through a 105-μm mesh. The fibrous materials not passing through the mesh were returned to the mortar and ground five more times. Starch granules passing through the mesh were further purified by extracting 10 times with a mixture of 0.05 m sodium chloride and toluene. The starch was then washed with deionized water three times, with 95% (v/v) ethanol one time, and with acetone one time before drying at room temperature.
Western Analysis
Total protein concentrations from extracts were estimated using a protein assay (Bio-Rad Laboratories, Hercules, CA) based on the Bradford method (Stoscheck, 1990). From these crude extracts of total proteins, 10 μg were fractionated on a 7.5% (w/v) Tris-HCl poly-acrylamide gel at 90 V for 4 h using a Mini Protean apparatus (Bio-Rad Laboratories) following manufacturer's instructions. Proteins were then blotted onto Immobulin PVDF membrane (Millipore, Bedford, MA) using a HEP-1 semi-dry electroblotting device (Owl Scientific, Cambridge, MA) and Towbin's buffer (Towbin et al., 1979). Immunodetection of SBEI, SBEIIa, and SBEIIb was performed using an enhanced chemiluminescence western-blotting detection kit (Amersham). Antibodies were generated against peptides from unique regions of SBEI (amino acids 502–520: DYLKNKDDSEWSMGEIAHT), SBEIIa (amino acids 77–91: EASSGVEAEERPELS), and SBEIIb (amino acids 143–161: RRIRSDIDEHEGGLTAFSR) by injection of multiple antigen peptides in Freund's adjuvant into New Zealand white rabbits (Research Genetics, Huntsville, AL). The antibody to SBEIIb is isoform specific, detecting a protein of 80 kD. However the antibody to SBEIIa binds to SBEIIa (90 kD) and as of yet unidentified proteins; the antibody to SBEI binds to SBEI, SBEIIa, and SBEIIb (84, 90, and 80 kD, respectively) and other as of yet unidentified proteins (Martha James, personal communication).
Starch Analysis
Starch granules were solubilized in 90% (v/v) DMSO by heating in a boiling water bath for 3 h with periodic vortexing. Insoluble materials consisting mainly of small particles of cellular debris were pelleted by microcentrifugation for 5 min at full speed. The starch was precipitated from the supernatant by adding ethanol to 80% (v/v), and pelleted by centrifuging for 5 min at 2,500g. The starch was washed once with 80% (v/v) ethanol and three times with acetone and dried overnight at 50°C.
Size exclusion chromatography of dispersed starch was performed using Sepharose CL-2B (Supelco, Bellefonte, PA) and 0.01 m NaOH (pH 12) as the mobile phase following the method of Klucinec and Thompson (1998). One-hundred 5-mL fractions were collected. The total carbohydrate content of the fractions was determined as described by DuBois et al. (1956).
HPSEC of debranched starches was conducted using the method of Klucinec and Thompson (1998). Briefly, between 2.5 and 5 mg of starch was mixed with a sufficient quantity of 90% (v/v) DMSO to result in a 5% (w/v) starch solution. The mixtures were heated in a boiling water bath for 10 min to disperse the starch. Samples were diluted 1:8 in 50°C 0.05 n sodium acetate buffer, pH 3.75. After cooling to 37°C, 0.5 units of isoamylase (Megazyme USA, Bozeman, MT) were added; these samples were held at 37°C with constant agitation for 24 h. The digested samples were diluted 1:10 with DMSO, heated in a boiling water bath for 10 min, and centrifuged at 6,000g for 10 min. A 50-μL sample was injected into the chromatograph.
Correlation Study
A population of 52 BC1F2 plants (Fig. 8) were scored from 10 DAE, when the mutant phenotype was first apparent, until 32 DAE to ensure mutant phenotypes were not due to temporary environmental factors. At 32 DAE, DNA was extracted from each plant and used to score each plant as described above using primers 2A1 and 2A2. Western protein analysis for plants numbers 18 and 39 was conducted as described above using 55 DAE leaf tissue. Retention times were converted to capacity factors (k′) based on the retention time of waxy starch (k′ = 0, corresponding to void volume) and Glc (k′ = 1, corresponding to salt volume).
ACKNOWLEDGMENTS
The authors wish to thank Dr. Kyung-Nam Kim, Shuqing Zhao, and Jennifer Tomscha for helpful advice.
Footnotes
This work was supported by the U.S. Department of Energy, Bioscience Program (grant no. DE–FG02–96ER20234 to M.J.G., J.C.S., and D.B.T.) and by the Pennsylvania State Agricultural Experiment Station (grant no. CRIS 3303).
LITERATURE CITED
- Baba T, Kimura K, Mizuno K, Etoh H, Ishida Y, Shida O, Arai Y. Sequence conservation of the catalytic regions of amylolytic enzymes in maize branching enzyme-I. Biochem Biophys Res Commun. 1991;181:87–94. doi: 10.1016/s0006-291x(05)81385-3. [DOI] [PubMed] [Google Scholar]
- Benson RJ, Johal GS, Crane VC, Tossberg JT, Schnable PS, Meeley RB, Briggs SP. Cloning and characterization of the maize An1 gene. Plant Cell. 1995;7:75–84. doi: 10.1105/tpc.7.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer CD, Liu K-C. The interaction of endosperm genotype and genetic background: I. Differences in chromatographic profiles of starches from nonmutant and mutant endosperms. Staerke. 1985;37:73–79. [Google Scholar]
- Boyer CD, Preiss J. Multiple forms of (1,4)-α-d-glucan, (1,4)-α-d-glucan-6-glycosyl transferase from developing Zea mays L. kernels. Carbohydr Res. 1978;61:321–334. [Google Scholar]
- Chandler VL, Hardeman KJ. The Mu elements of Zea mays. Adv Genet. 1992;30:77–122. doi: 10.1016/s0065-2660(08)60319-3. [DOI] [PubMed] [Google Scholar]
- Chisholm D. A convenient moderate-scale procedure for obtaining DNA from bacteriophage λ. Biotechniques. 1988;7:255–257. [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang PL, Boyer CD. Maize leaf and kernel starch synthases and starch branching enzymes. Pytochemistry. 1988;27:1255–1259. [Google Scholar]
- Dellaporta S. Plant DNA miniprep and microprep: versions 2.1–2.3. In: Freeling M, Walbot V, editors. The Maize Handbook. New York: Springer-Verlag; 1994. pp. 522–525. [Google Scholar]
- DeVries S, Hoge H, Bisseling T. Isolation of total and polysomal RNA from plant tissues. Plant Mol Biol Manual. 1988;B6:1–5. [Google Scholar]
- DuBois M, Gilles KA, Hamilton JK, Rebers PA, Smith F. Colorimetric method for determination of sugars and related substances. Anal Chem. 1956;28:350–356. [Google Scholar]
- Fisher DK, Boyer CD, Hannah LC. Starch branching enzyme II from maize endosperm. Plant Physiol. 1993;102:1045–1046. doi: 10.1104/pp.102.3.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher DK, Kim KN, Gao M, Boyer CD, Guiltinan MJ. A cDNA encoding starch branching enzyme I from maize endosperm. Plant Physiol. 1995;108:1313–1314. doi: 10.1104/pp.108.3.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Fisher DK, Kim KN, Shannon JC, Guiltinan MJ. Evolutionary conservation and expression patterns of maize starch branching enzyme I and IIb genes suggests isoform specialization. Plant Mol Biol. 1996;30:1223–1232. doi: 10.1007/BF00019554. [DOI] [PubMed] [Google Scholar]
- Gao M, Fisher DK, Kim KN, Shannon JC, Guiltinan MJ. Independent genetic control of maize starch-branching enzymes IIa and IIb: isolation and characterization of a Sbe2a cDNA. Plant Physiol. 1997;114:69–78. doi: 10.1104/pp.114.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garwood DL, Shannon JC, Creech RG. Starches of endosperms possessing different alleles at the amylose-extender locus in Zea mays L. Cereal Chem. 1976;53:355–364. [Google Scholar]
- Guan HP, Preiss J. Differentiation of the properties of the branching isozymes from maize (Zea mays) Plant Physiol. 1993;102:1269–1273. doi: 10.1104/pp.102.4.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KN, Fisher DK, Gao M, Guiltinan MJ. Genomic organization and promoter activity of the maize Starch branching enzyme I gene. Gene. 1998;216:233–243. doi: 10.1016/s0378-1119(98)00339-4. [DOI] [PubMed] [Google Scholar]
- Kim KN, Gao M, Fisher DK, Guiltinan MJ. Molecular cloning and characterization of the Amylose-Extender gene encoding starch branching enzyme IIB in maize. Plant Mol Biol. 1999;38:945–956. doi: 10.1023/a:1006057609995. [DOI] [PubMed] [Google Scholar]
- Klucinec JD, Thompson DB. Fractionation of high-amylose maize starches by differential alcohol precipitation and chromatography of the fractions. Cereal Chem. 1998;75:887–896. [Google Scholar]
- Maes T, De Keukeleire P, Gerats T. Plant tagnology. Trends Plant Sci. 1999;4:90–96. doi: 10.1016/s1360-1385(99)01375-8. [DOI] [PubMed] [Google Scholar]
- Mena M, Ambrose BA, Meeley RB, Briggs SP, Yanofsky MF, Schmidt RJ. Diversification of C-function activity in maize flower development. Science. 1996;274:1537–1540. doi: 10.1126/science.274.5292.1537. [DOI] [PubMed] [Google Scholar]
- Myers AM, Morell MK, James MG, Ball SG. Recent progress toward understanding biosynthesis of the amylopectin crystal. Plant Physiol. 2000;122:989–997. doi: 10.1104/pp.122.4.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Smith AM, Denyer K, Martin C. The synthesis of the starch granule. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:67–87. doi: 10.1146/annurev.arplant.48.1.67. [DOI] [PubMed] [Google Scholar]
- Stinard PS, Robertson DS, Schnable PS. Genetic isolation, cloning and analysis of Mutator- induced, dominant antimorph of the maize amylose-extender1 locus. Plant Cell. 1993;5:1555–1566. doi: 10.1105/tpc.5.11.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoscheck CM. Quantitation of protein. Methods Enzymol. 1990;182:50–69. doi: 10.1016/0076-6879(90)82008-p. [DOI] [PubMed] [Google Scholar]
- Takeda Y, Guan HP, Preiss J. Branching of amylose by the branching isoenzymes of maize endosperm. Carbohydr Res. 1993;240:253–263. [Google Scholar]
- Takeda Y, Shitaozono T, Hizukuri S. Molecular structure of corn starch. Staerke. 1988;40:51–54. [Google Scholar]
- Thompson DB. On the non-random nature of amylopectin branching. Carbohydr Polym. 2000;43:223–239. [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vineyard ML, Bear RP. Amylose content. Maize Genet Coop Newsl. 1952;26:5. [Google Scholar]