Abstract
Starch production in all plants examined is altered by mutations of isoamylase-type starch-debranching enzymes (DBE), although how these proteins affect glucan polymer assembly is not understood. Various allelic mutations in the maize (Zea mays) gene sugary1 (su1), which codes for an isoamylase-type DBE, condition distinct kernel phenotypes. This study characterized the recessive mutations su1-Ref, su1-R4582::Mu1, and su1-st, regarding their molecular basis, chemical phenotypes, and effects on starch metabolizing enzymes. The su1-Ref allele results in two specific amino acid substitutions without affecting the Su1 mRNA level. The su1-R4582::Mu1 mutation is a null allele that abolishes transcript accumulation. The su1-st mutation results from insertion of a novel transposon-like sequence, designated Toad, which causes alternative pre-mRNA splicing. Three su1-st mutant transcripts are produced, one that is nonfunctional and two that code for modified SU1 polypeptides. The su1-st mutation is dominant to the null allele su1-R4582::Mu1, but recessive to su1-Ref, suggestive of complex effects involving quaternary structure of the SU1 enzyme. All three su1- alleles severely reduce or eliminate isoamylase-type DBE activity, although su1-st kernels accumulate less phytoglycogen and Suc than su1-Ref or su1-R4582::Mu1 mutants. The chain length distribution of residual amylopectin is significantly altered by su1-Ref and su1-R4582::Mu1, whereas su1-st has modest effects. These results, together with su1 allele-specific effects on other starch- metabolizing enzymes detected in zymograms, suggest that total DBE catalytic activity is the not the sole determinant of Su1 function and that specific interactions between SU1 and other components of the starch biosynthetic system are required.
Much of the metabolic activity in plants is directed toward production of starch from reduced carbon formed during photosynthesis. Starch that accumulates in leaves during the light phase of a diurnal cycle is degraded in the dark phase. Suc is transported from this source tissue to non-photosynthetic storage tissues, where it supplies Glc units for the synthesis of starch that serves as the carbon reserve for the subsequent generation. Starch biosynthesis is highly conserved throughout the plant kingdom, presumably owing to this central role in plant physiology and reproduction. Nevertheless, there is little understanding of the molecular mechanisms that determine how the precise structure of starch is achieved in either leaves or storage tissues.
Starch comprises two homopolymers of Glc, amylopectin, and amylose, and it is the ordered architectural structure of amylopectin, the most abundant component, that confers crystallinity to the starch granule (Imberty et al., 1991; Gallant et al., 1997). Amylopectin synthesis involves the combined actions of multiple isoforms of starch synthases (SS), which elongate linear chains via the introduction of α-(1→4) glycosidic bonds, and starch branching enzymes (BE), which catalyze the formation of α-(1→6) branch linkages (for reviews, see Smith, 1999; Kossman and Lloyd, 2000; Myers et al., 2000). The nonrandom placement of branch linkages and determination of chain length results in a complex, hierarchical structure that renders the molecule both crystalline and insoluble.
A broad body of genetic evidence in several plant species indicates that α-(1→6) glucan hydrolases, i.e. starch debranching enzymes (DBE), are necessary for the normal formation of crystalline starch granules. Two types of DBEs have been identified in plants, distinguished by their substrate specificities (Lee and Whelan, 1971; Doehlert and Knutson, 1991). Isoamylase-type DBEs cleave α(1→6) branch linkages in amylopectin and glycogen, but do not hydrolyze the chemically identical bonds in pullulan, an α(1→6)-linked maltotriose polymer. In contrast, pullulanase-type DBEs, also referred to as R-enzymes or limit-dextrinases (Manners, 1997), hydrolyze α(1→6) linkages of pullulan and to a lesser degree amylopectin, but have little or no activity toward glycogen. Sequence comparisons among plant and bacterial α(1→6) glucan hydrolases indicate that the pullulanase-type and isoamylase-type DBEs have been conserved separately in evolution (Beatty et al., 1999).
The first genetic evidence for DBE involvement in starch biosynthesis came from mutations of the maize (Zea mays) sugary1 (su1) gene, described a century ago in the scientific literature (Correns, 1901). These mutations result in the accumulation of soluble sugars and a water-soluble polysaccharide (WSP) termed phytoglycogen in the kernel (Morris and Morris, 1939). In su1 kernels, the reduction in amylopectin content approximately matches the abundance of phytoglycogen, suggesting a diversion from biosynthesis of the normal insoluble branched glucan to the water-soluble form. Similar to glycogen, phytoglycogen has approximately twice the frequency of branch linkages as amylopectin, a shorter average chain length, and a more uniform chain length distribution (Yun and Matheson, 1993). More recently, the accumulation of phytoglycogen also has been reported in mutants of rice, Arabidopsis, and Chlamydomonas (Mouille et al., 1996; Nakamura et al., 1996; Zeeman et al., 1998; Kubo et al., 1999). In each species, phytoglycogen accumulation correlates with a lack of DBE activity of the isoamylase type (Mouille et al., 1996; Rahman et al., 1998; Zeeman et al., 1998; Beatty et al., 1999; Kubo et al., 1999). In maize and Arabidopsis the mutation responsible for this specific phenotype has been shown directly to reside in a gene that codes for an isoamylase-type DBE (James et al., 1995; Zeeman et al., 1998), and indirect evidence indicates this is likely to be the case in Chlamydomonas and rice.
The first described su1 mutation (Correns, 1901) is now termed su1-Ref. As the allele name indicates, this mutation has served as the reference standard for most analyses of the phenotypic effects. Kernels homozygous for su1-Ref have a glassy, translucent, and shrunken appearance at maturity resulting from their altered carbohydrate composition. The total amount of phytoglycogen reported to be present in su1-Ref kernels has ranged from 25% to 35% of the total glucan (Creech, 1968; Shannon and Garwood, 1984). Several early reports indicated that su1-Ref kernels contained an altered BE activity, prompting speculation that phytoglycogen accumulation results from an alteration in the formation of branch linkages (Lavintman, 1966; Manners et al., 1968; Hodges et al., 1969; Tomalsky and Krisman, 1987). Previous analysis of su1-Ref mutant kernels also revealed the deficiency of a DBE, although that activity was of the pullulanase-type (Pan and Nelson, 1984). More recently, cloning of the Su1 gene revealed that it codes for a DBE of the isoamylase-type (James et al., 1995), and that su1-Ref kernels are deficient in both isoamylase- and pullulanase-type DBE isoforms (Beatty et al., 1999). This dual effect on both DBEs is also observed in endosperm tissue of rice su1 mutants (Nakamura et al., 1996). However, pullulanase-type DBE apparently is unaffected in Arabidopsis leaves or the unicellular algae Chlamydomonas when mutations affecting an isoamylase-type DBE are present (Zeeman et al., 1998; Dauvillee et al., 2000).
The complexity of the starch biosynthetic system, including the involvement of multiple isozymes of SS, BE, and DBE, so far has precluded biochemical reconstitution of an architecturally accurate synthetic system. Pleiotropic effects in most phytoglycogen-accumulating mutants also have complicated the analysis of the role of isoamylase-type DBEs in the starch biosynthetic process. In this study, we have begun to exploit the great deal of allelic diversity that is available for the maize su1 locus, toward the aim of establishing correlations between mutational effects on the SU1 polypeptide, specific enzymatic alterations, and defined changes in the chemical structures of glucan polymers. Three su1 alleles in near-isogenic lines were compared with the nonmutant allele Su1. Two of the mutant alleles, su1-Ref and the transposon-induced allele su1-R4582::Mu1 (James et al., 1995), condition severe glassy, shrunken, and translucent kernel phenotypes. The third, su1-starchy (su1-st), results in a relatively mild phenotype in which translucency is typically limited to the kernel crown (Dahlstrom and Lonnquist, 1964). This research examined the dominance relationships among these su1 alleles, the quantity of phytoglycogen and sugars that accumulate in the kernels, the chain length distributions of the polymers present, and the effects of each allele on the activities of DBEs and other starch metabolizing enzymes. The molecular nature of each mutation also was determined. In the course of this analysis, a novel transposon-like sequence, termed Toad, was identified at the su1-st locus. This element was shown to affect the pre-mRNA splicing of the exon in which it resides. The three su1- mutations had diverse effects on the compositions and structures of kernel glucans, as well as on the activities of other starch metabolizing enzymes. However, direct correlation was not established between the apparent level of isoamylase-type DBE activity and the observed chemical phenotype. Together, the findings suggest that the effects of each mutation varied according to the primary structure of the SU1 polypeptide, and may relate to the assembly-state of the SU1 holoenzyme.
RESULTS
Kernel Morphologies and Dominant/Recessive Relationships of the su1- Mutations
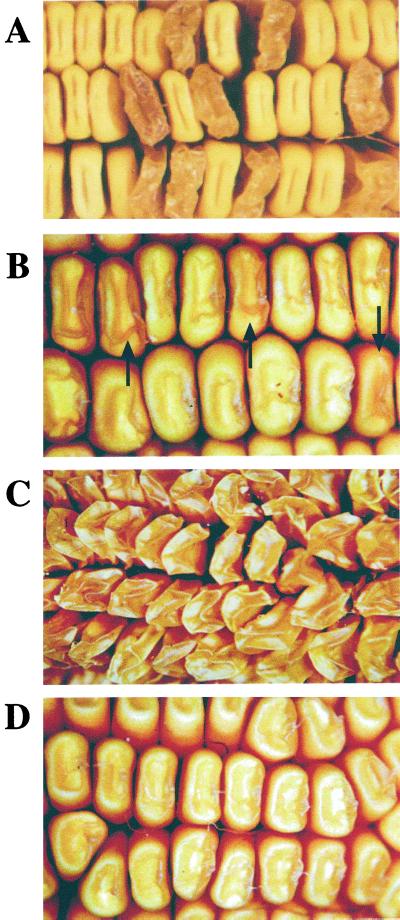
Self-pollinated ears from Su1/su1-Ref or Su1/su1-st heterozygous plants contain progeny kernels segregating at the ratio of 1:3 for the mutant and non-mutant phenotypes. The typical glassy, shrunken, and translucent phenotype of su1-Ref homozygous kernels is evident (Fig. 1A), and this is very similar to the kernel phenotype conditioned by su1-R4582::Mu1 (James et al., 1995). The su1-st kernel phenotype is relatively slight (Fig. 1B) compared with that conditioned by su1-Ref. These mutant kernels are plump and starchy with translucency and wrinkling only on the kernel crown. Also, the su1-st mutation varies considerably in expressivity with some kernels appearing near normal. Despite these distinct kernel phenotypes, su1-st is clearly identified as a mutation of the su1 locus because it fails to complement su1-Ref and by definitive molecular characterization (see following section).
Figure 1.
Kernel phenotypes of su1-mutants. A, The su1-Ref mutant phenotype. The ear shown was obtained by self-pollination of an su1-Ref/Su1 heterozygote. B, The su1-st mutant phenotype. The ear shown was obtained by self-pollination of an su1-st/Su1 heterozygote. Mutant kernels (examples indicated by the arrows) and wild-type kernels segregated at a frequency of approximately 1:3, respectively. C, Phenotype of su1-Ref/su1-st kernels. The ear shown is the result of a cross of an su1-st/su1-st plant as the male parent to an su1-Ref/su1-Ref plant as the female parent. D, Phenotype of su1-R4582::Mu1/su1-st kernels. The ear shown is the result of a cross of an su1-R4582::Mu1/su1-R4582::Mu1 plant as the male parent to an su1-st/su1-st plant as the female parent.
Previous genetic analyses of the dominant/recessive relationship between su1-st and wild-type (Su1) or su1-Ref alleles showed that the su1-st allele is recessive to both, although reciprocal crosses between su1-Ref and su1-st plants indicated subtle dosage effects of the su1-st allele (Dahlstrom and Lonnquist, 1964). This finding also was confirmed in this study, where progeny ears from reciprocal su1-Ref by su1-st crosses produced kernels that were uniformly shrunken, glassy, and translucent in appearance (Fig. 1C). Crosses also were made between su1-st plants and plants homozygous for su1-R4582::Mu1. The su1-R4582::Mu1 allele is predicted to be a null allele, based on the presence of a Mu1 transposable element in the first exon of the gene downstream of the translational start site (James et al., 1995). Progeny ears from the su1-st by su1-R4582::Mu1 cross were uniformly like su1-st homozygotes in appearance, indicating that the su1-st allele is dominant to su1-R4582::Mu1 (Fig. 1D). Given that su1-Ref and su1-R4582::Mu1 condition nearly identical kernel phenotypes, the different dominance relationships of these two alleles with su1-st most likely reflect the presence or absence of a SU1 polypeptide.
Steady-State Levels of Su1 mRNA in the Various su1- Mutants
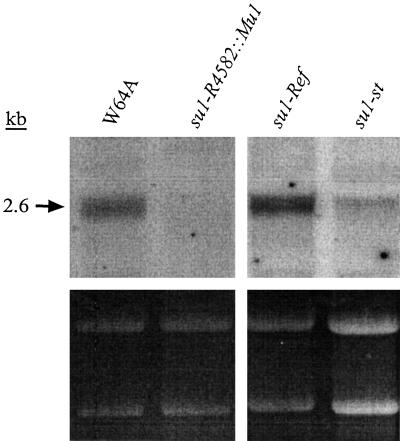
Gene expression of su1-Ref, su1-st, and su1- R4582::Mu1 relative to the nonmutant allele Su1 was characterized in kernels at a mid-developmental stage by RNA gel-blot analysis. Total RNA was isolated from kernels harvested 20 DAP and hybridized with an antisense RNA probe from the 3′ end of the Su1 cDNA. Kernels homozygous for su1-Ref produce transcripts of normal size that accumulate to wild-type levels (Fig. 2). In contrast, kernels homozygous for su1-R4582::Mu1 accumulated no detectable Su1 transcript. This supports the prediction that su1-R4582::Mu1 is a null allele. Homozygous su1-st kernels at this stage accumulated a relatively slight amount of apparently normal-sized transcript. Thus, the partial function of the su1 locus that is evident from the intermediate phenotype conditioned by su1-st (Fig. 1; Table I) is provided by a relatively small number of transcripts.
Figure 2.
RNA gel-blot analysis. Total RNA was extracted from endosperm of kernels harvested at 20 DAP, fractionated on a 1% (w/v) agarose-formaldehyde gel, and hybridized with Su1 cDNA antisense RNA. RNA concentration variations can be visualized by the ethidium bromide-stained rRNA, shown in the lower panel. The lanes shown were run simultaneously in the same gel.
Table I.
Carbohydrate analysis of nonmutant maize W64A kernels and sugary1 mutant kernels
| Year | DAP | Total Carbohydrate
|
Total mg/g | % Glucan Polymer
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Glc
|
Fru
|
Suc
|
WSPa
|
Starcha
|
WSP | Starch | |||||||||
| mg/gb | % | mg/gb | % | mg/gb | % | mg/gb | % | mg/gb | % | ||||||
| W64A | 2000 | 20 | 3.7 | 0.5 | 3.2 | 0.45 | 43 | 5.6 | 5.6 | 0.7 | 718 | 92.7 | 774 | 0.8 | 99.2 |
| W64A | 1999 | 30 | 2.4 | 0.3 | 2 | 0.2 | 31 | 3.6 | 15 | 1.8 | 820 | 94 | 870 | 2 | 98 |
| sul-ref | 2000 | 20 | 9.3 | 1.5 | 5.5 | 0.9 | 168 | 27.8 | 336 | 55.6 | 85 | 14.1 | 604 | 80 | 20 |
| sul-ref | 1999 | 30 | 20 | 3.8 | 4.4 | 0.8 | 109 | 20.5 | 274 | 52 | 121 | 22.8 | 528 | 70 | 30 |
| sul-4582 | 1998 | 25 | 15 | 2 | 5.5 | 0.8 | 110 | 16 | 465 | 66.5 | 103 | 15 | 699 | 80 | 20 |
| sul-st | 2000 | 20 | 9.1 | 1.2 | 7.6 | 1 | 104 | 13.6 | 198 | 26 | 444 | 58 | 763 | 31 | 69 |
| sul-st | 1998 | 20 | 12 | 1.9 | 9.3 | 1.5 | 123 | 19.7 | 242 | 38.7 | 240 | 38.1 | 626 | 50 | 50 |
| sul-st | 1997 | 30 | 3.6 | 0.5 | 2.2 | 0.3 | 30 | 4 | 52 | 6.8 | 683 | 88.5 | 771 | 7 | 93 |
The amount of WSP and starch were determined as glucose equivalents by measuring the amount of glucose produced following complete digestion with amyloglucosidase.
The results are expressed as milligram of carbohydrate by gram of dry wt.
The Molecular Nature of su1-Ref
Although the su1-Ref mutation has long been used for the production of traditional sweet corn varieties, the molecular basis for this mutation was not known. To characterize this allele, segments of su1-Ref kernel mRNA and genomic DNA were amplified by PCR, and their sequences were determined. All materials analyzed were obtained initially from the W64A genetic background. Sequence comparisons of su1-Ref to Su1 revealed three nucleotide changes that would result in amino acid substitutions, a C to G transversion at nucleotide (nt) 576, an A to G transition at nt 1,100, and a T to C transition at nt 1,819 (nucleotide and amino acid numbers refer to GenBank accession no. U18908). Translation would result in three amino acid changes relative to the wild-type polypeptide SU1, Phe to Leu at residue 163, Glu to Val at residue 338, and Trp to Arg at residue 578. This mutant polypeptide is designated SU1Ref. Similar analysis of commercial su1-Ref sweet corn varieties in the NK199 and Golden Cross Bantam backgrounds showed that only two of these nucleotide changes were present, specifically those resulting in the F163L and W578R substitutions. These two mutations, therefore, most likely were present in the progenitor su1-Ref allele that has been used in existing commercial sweet corn lines and introgressed into the inbred line W64A. The third mutation, E338V, most likely arose more recently and thus is not required for the phytoglycogen-accumulating phenotype conditioned by su1-Ref.
Sequence comparisons among α-(1→6) glucan hydrolases revealed numerous conserved sequence blocks, some of which are common to all members the α-amylase family (Jesperson et al., 1993) and some of which are specific to either the isoamylse-type or pullulanase-type DBE isoforms (Beatty et al., 1999). Two of the three substitutions in su1-Ref occur within conserved domains, specifically E338V in α-amylase region I, and W578R in the isomylase-specific domain IS-XII. The third mutation, F163L, lies just downstream of domain IS-VII but is not located in a conserved region of the enzyme. The only highly conserved residue affected in su1-Ref is W578, which is present in all isoamylases examined to date from plants and bacteria except that from Escherichia coli (Beatty et al., 1999). W578R, therefore, is likely to cause the defect in the SU1 isoamylase-type DBE in su1-Ref mutants.
Structure of the mRNA Transcripts from su1-st
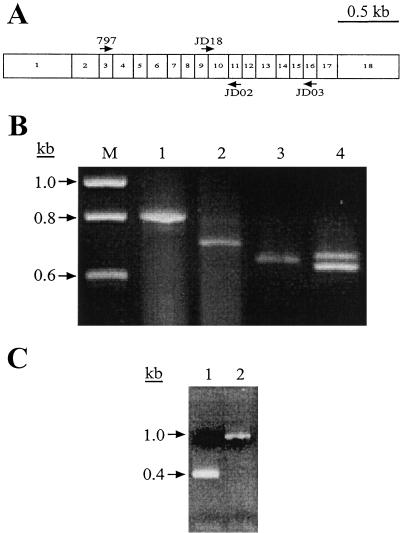
The molecular nature of the su1-st mutation also was characterized, again beginning with reverse transcriptase (RT)-PCR amplification of mRNA from homozygous mutant kernels. Primers 797 and JD02 from the Su1 cDNA sequence, which correspond to regions within exon 3 and exon 11 of the genomic sequence, respectively, were predicted to amplify a fragment of 800 bp (Beatty et al., 1997) (Fig. 3A). For Su1 and su1-Ref, a fragment of the predicted size was amplified (Fig. 3B, lane 1; and data not shown). The major RT-PCR product from su1-st kernels, however, was approximately 100 bp smaller than that obtained from wild-type kernels, although a minor product near the expected size also was seen (Fig. 3B, lane 2). Sequence analysis of the approximately 700-bp major product revealed that in this mutant mRNA, the splice donor site of exon 9 has been joined precisely to the acceptor site of exon 11, and that exon 10 is completely lacking. Loss of this 127-nucleotide exon would result in a shift in the open reading frame within exon 11 and the introduction of a premature translational stop codon. Therefore, this transcript, designated as transcript type I, is predicted to result in the translation of a truncated, nonfunctional polypeptide.
Figure 3.
PCR and RT-PCR analyses. A, Relative positions of oligonucleotide primers within the Su1 cDNA. Exons comprising the Su1 cDNA are represented by numbered boxes. Primers used for the detection of alternatively spliced transcripts are indicated by arrows above and below the boxes (depicting sense and antisense sequences, respectively). B, RT-PCR amplifications. DNA was reverse-transcribed from purified mRNA isolated from 20 DAP whole kernels homozygous for Su1 (lanes 1 and 3) and su1-st (lanes 2 and 4). Primer pair 797/JD02 was used for the amplifications in lanes 1 and 2, and primer pair JD03/JD18 was used for the amplifications in lanes 3 and 4. C, PCR amplification of genomic DNA from seedling tissue homozygous for Su1 (lane 1) and su1-st (lane 2) with primer pair JD19/JD20 to amplify the region of the gene containing the Toad element.
The preceding observation was difficult to reconcile with the fact that su1-st mutants display a kernel morphology and chemical phenotype quite distinct from those of the confirmed null mutation su1-R4582::Mu1 (Fig. 1; Table I). To search for minor transcripts in which exon 9 was correctly spliced to exon 10, a RT-PCR primer that bridges this splice donor site and acceptor site was synthesized. Kernel RNA was amplified by RT-PCR using this primer, JD18, and the downstream primer JD03 from within exon 16 (Fig. 3A). This resulted in two distinct su1-st mutant cDNA products, one slightly smaller than that obtained when Su1 kernel RNA was amplified with this primer pair, and the other slightly larger (Fig. 3B, lanes 3 and 4). These two transcripts are designated type II and type III, respectively. The observation of multiple su1-st transcripts, none of which is the size of the wild-type transcript, suggests that alternative splicing occurs in pre-mRNA transcripts from the su1-st locus.
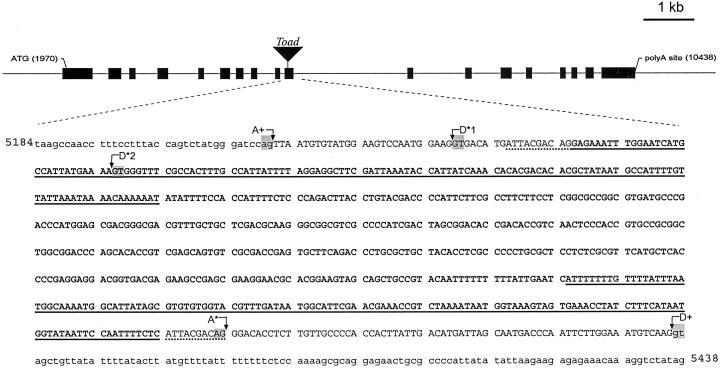
To further investigate the apparent deletion of exon 10 in transcript I, genomic DNA from a homozygous su1-st plant was amplified with oligonucleotide primers JD19 and JD20, derived from intron 9 and intron 10, respectively. This primer pair is designed to amplify a sequence of 370 bp from wild-type genomic DNA that includes all 127 bp of exon 10 flanked by portions of the adjacent introns. The predicted amplification product was obtained from genomic DNA from a Su1 plant, but a product of slightly greater than 1 kb was obtained from su1-st genomic DNA (Fig. 3C). Characterization of this mutant fragment revealed the insertion of a 638-bp sequence within exon 10, as well as duplication of 10 nt of the Su1 sequence flanking each end of the insertion (ATTACGACAG; Fig. 4). This 638-bp sequence is referred to as the “Toad ” genetic element.
Figure 4.
Nucleotide sequence analysis of the su1-st locus. The su1-st locus contains an insertion of the 638-bp Toad sequence (shown in bold) in exon 10. Intron sequence flanking exon 10 is designated by lowercase letters, and exon 10 sequence is designated by uppercase letters. Nucleotides underlined with a dotted line indicate the 10-bp target site duplication of the host sequence. Regions underlined with a solid line indicate 138-bp TIRs within the Toad element. The wild-type acceptor site for the end of intron 9 is designated A+. The intron formed for the creation of the type-II transcript is made by joining cryptic acceptor sites D*1 to A*. This results in a deletion of 18 nt of Su1 sequence from the mature transcript and complete removal of the Toad sequence. The intron formed for the type-III transcript is produced by the use of D*2 as the donor site and A* as the acceptor site, removing most of the Toad sequence as an intron, but leaving an insertion of 30 bp in the mature mRNA. The wild-type donor site for the start of intron 10 is indicated as D+. The highly conserved GT and AG dinucleotides found at the termini of most plant introns are highlighted at the indicated splice donor/acceptor sites. The nucleotides indicated correspond to the wild-type Su1 genomic sequence (GenBank accession no. AF030882).
Characterization of the su1-st genomic sequence enabled definition of the pre-mRNA splicing events that gave rise to the type-II and type-III transcripts. Nucleotide sequence of the shorter cDNA amplified by the JD18/JD03 primer pair (Fig. 3B), corresponding to the type-II transcript, revealed deletion of 18 nt within exon 10 (Fig. 4). This deletion extends from nucleotide 5,248 in the su1 genomic sequence through nucleotide 5,265 (GenBank accession no. AF030882). The latter nucleotide corresponds to the last residue of the direct repeat adjacent to both ends of the Toad element. This deletion maintains the open reading frame of the mRNA, resulting in a mutant polypeptide designated SU1st-II, that has a deletion of six amino acids (residues 440–445; GDMITT). The larger cDNA product, corresponding to the type-III transcript, contains an insertion of 30 nt relative to the nonmutant sequence. This insertion corresponds exactly to the first 30 nt of the inserted Toad element. In this instance the reading frame is again maintained with the insertion of 10 amino acids of the sequence EIWNHAIMLR between residues 446 and 447 of the nonmutant protein, resulting in SU1st-III. In the type-II transcript the entire Toad element contained in the genomic sequence is missing from the mRNA, and in the type-III transcript all but 30 nt of Toad have been removed. Thus it appears that the insertion or deletion mutations in the type-II and -III transcripts are generated through aberrant pre-mRNA splicing events induced by presence of the Toad element.
A Novel Transposon-Like Sequence within the su1-st Locus
The 638-bp insertion sequence found within exon 10 at the su1-st locus was subjected to a BLAST search of the public databases. This search failed to identify similar sequences within any genome, indicating that the 638-bp sequence is novel. Alignment of the nucleotide sequence of the element with its reverse complement identified terminal inverted repeat (TIR) regions of 138 bp, which are 86% identical. Class-II transposable elements in flowering plants, which include all non-retrotransposons, are characterized by the presence of TIRs that range from a few to more than 200 nucleotides (Finnegan, 1989). Also, insertion of a class-II transposable element typically results in the duplication of small region of host DNA at the insertion site, such that direct host repeat sequences flank the element (Federoff, 1989). Together, these transposon-like features of Toad are strong indicators that the 638-bp sequence is a transposable element (the sequence of Toad is available as GenBank accession no. AF317693). Open reading frames of 67, 70, and 94 codons are present within Toad, although none of the predicted polypeptides are significantly similar to sequences in the public database.
To investigate the approximate number of copies of the Toad elements, or homologous elements, within the maize genome, DNA from six inbred lines was subjected to high stringency gel-blot analysis with the Toad sequence as a hybridization probe. This analysis indicated that Toad is a low-copy element with no more than two to four copies in any of the genomes tested (data not shown). In DNA from a su1-st plant, two Toad sequences were observed, one corresponding to the element at the su1-st locus and the other corresponding to the single Toad sequence in the W64A genome. A detailed analysis of the Toad sequence revealed short blocks of similarity with other maize transposons, such as have been observed for many plant transposable elements (Nevers et al., 1986).
Glucan Structure Phenotypes
The phenotype conditioned by each su1- mutation with regard to the chemical properties and structures of the glucans present in developing kernels was determined. Aqueous kernel extracts were prepared from samples harvested at mid-developmental time points from several growing seasons, and separated by centrifugation into soluble and granular phases. The quantities of Suc, d-Glc, d-Fru, oligosaccharide, and glucan polymer in each phase were determined by specific analytical methods (Table I). The carbohydrate profile conditioned by either su1-R4582::Mu1 or su1-Ref is significantly altered relative to nonmutant kernels, whereas su1-st conditions an intermediate phenotype. The intermediate effects of the su1-st allele varied, however, among the samples, suggesting environmental factors may impact su1-st phenotypic expression.
Selected data from Table I highlight su1 allele-specific effects on carbohydrate composition. For example, approximately 4% of the total carbohydrate is in the form of Suc in both Su1 and su1-st kernels harvested 30 d after pollination (DAP) from the 1997 nursery, but higher Suc proportions are found in 20-DAP su1-st kernels from the 1998 and 2000 nurseries, approximately 14% and approximately 20%, respectively. Suc comprises 16% of the total carbohydrate in su1-R4582::Mu1 and approximately 20% to 28% of the total carbohydrate in su1-Ref kernels. The great majority of carbohydrate in nonmutant kernels at 20 or 30 DAP, approximately 94%, is in the form of starch. Granular starch is reduced to various levels in su1-st kernels, comprising approximately 88% of the 30 DAP sample from 1997, approximately 38% of the 20-DAP sample from 1998, and 58% of the 20-DAP sample from 2000. In contrast, only approximately 14% to 23% of the total carbohydrate in su1-Ref kernels and su1-R4582::Mu1 kernels is found in starch granules at comparable developmental stages. The relative severity of the phenotypes also is evident in the accumulation of WSP (i.e. phytoglycogen). This more highly-branched glucan accounts for approximately 52% to 66% of the total carbohydrate in the more phenotypically severe su1-R4582::Mu1 or su1-Ref kernels, whereas in su1-st kernels WSP comprised approximately 7% of the total in the 30 DAP sample and from 26 to approximately 39% in the 20 DAP samples.
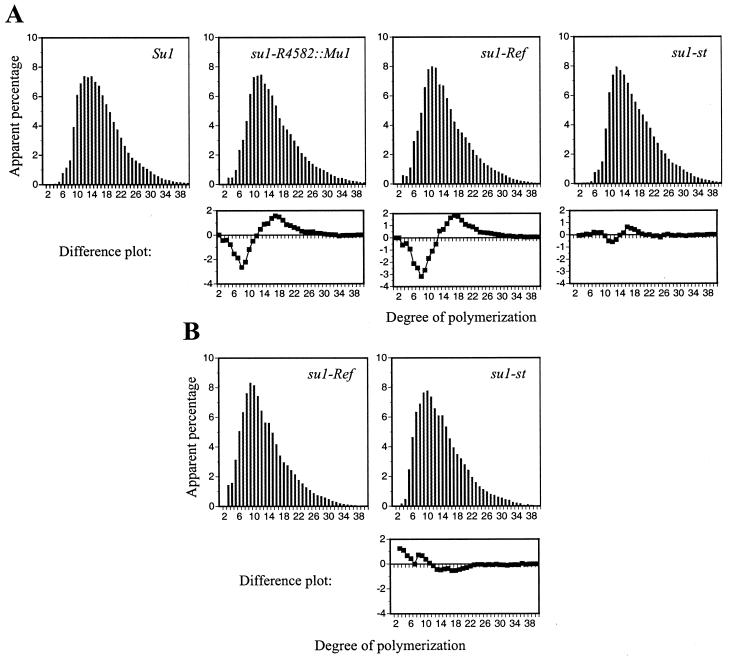
The structure of the amylopectin purified from granules of each mutant was analyzed by high performance anion exchange chromatography with pulsed amperometric detection (HPAEC-PAD) to determine if the su1- mutations resulted in altered chain length distributions. Analysis of starch from 30-DAP kernels revealed that the amylopectin component in su1-R4582::Mu1 and su1-Ref kernels is structurally different from wild-type amylopectin (Fig. 5A). The amylopectin-like glucans in su1-R4582::Mu1 and su1-Ref kernels are similar to each other, having more short chains, approximately 3 to 12 glucosyl units in length, and fewer intermediate-length chains, approximately 12 to 30 glucosyl units in length, relative to wild type. The su1-st chain length profile for amylopectin is intermediate between wild type and that of su1-R4582::Mu1 and su1-Ref with slightly more short chains and slightly fewer intermediate chains relative to wild type. Thus, the su1-st amylopectin-like glucan is structurally distinct from any of the others. A similar analysis of the WSP from su1-Ref and su1-st kernels also shows that these two soluble glucans have structural differences, as evidenced by a reduction in very short chains, less than six glucosyl units in length, in the su1-st WSP (Fig. 5B).
Figure 5.
HPAEC-PAD analysis of chain length distributions. A, Amylopectin from Su1, su1-R4582::Mu1, su1-Ref, and su1-st homozygous kernels harvested 30 DAP was debranched with Pseudomonas isoamylase. Chain length distributions were determined using HPAEC-PAD and normalized to the total peak area. Differences in chain lengths of su1-R4582::Mu1, su1-Ref, and su1-st amylopectin-like molecules relative to Su1 amylopectin are shown in difference plots below the respective mutant profiles. B, WSP from su1-Ref and su1-st kernels harvested 30 DAP was debranched and separated, as described for A. A difference plot comparing the WSP chain distributions is below the su1-st profile.
Enzyme Profile Phenotypes
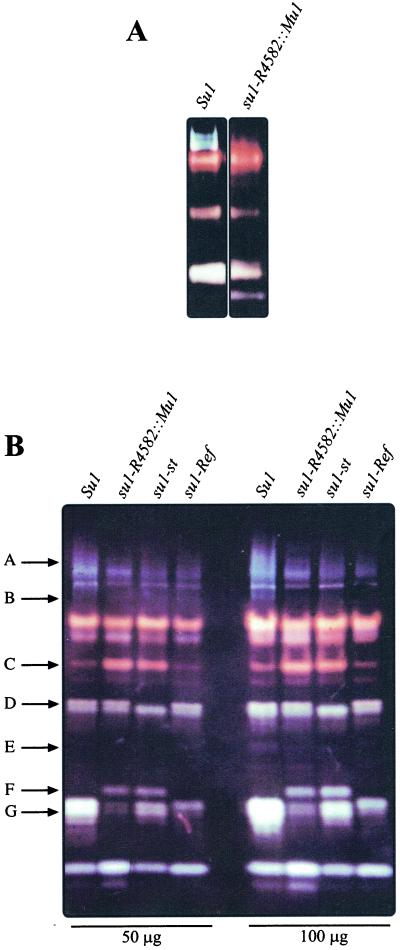
To examine the pleiotropic effects of su1 mutations on starch metabolizing enzyme activities, proteins from developing wild-type kernels and kernels homozygous for su1-Ref, su1-R4582::Mu1, and su1-st were separated by native PAGE and transferred to acrylamide gels containing solubilized starch. After incubation of the activity gels (i.e. zymograms) and staining with I2/KI, enzymes that modify starch structure are detected by alterations in the dark blue background color. Thus, DBE activities are visualized as light blue-staining bands, BE activities as red- or orange-staining bands, and amylolytic activities as regions where staining is reduced to white or very light color. The su1-R4582::Mu1 and Su1 extracts were compared initially on 5-cm gels (Fig. 6A), which showed that a series of light blue bands was completely missing in the mutant extract. These bands did not appear when red-pullulan, a pullulanase-type DBE-specific substrate, was included in the activity gels (data not shown). Considering that su1-R4582::Mu1 is a null mutation of a gene coding for an isoamylase-type DBE (see below), these data strongly suggest that the light blue bands represent the isoamylase activity coded for by su1.
Figure 6.
Native PAGE/activity gels (zymograms) of starch modifying activities. A, Twenty-five micrograms of total protein from Su1 and su1-R4582::Mu1 homozygous kernels harvested at 20 DAP was separated on a 5-cm native polyacrylamide gel, then electroblotted to a starch-containing gel. Starch modifying enzyme activities were visualized by staining with I2/KI. B, Fifty or 100 μg total protein from Su1, su1-R4582::Mu1, su1-st, and su1-Ref homozygous kernels harvested at 20 DAP was analyzed as described for A on 15-cm native PAGE/activity gels. Arrows A through G indicate starch modifying activities that exhibit allele-specific variation.
All three mutants also were analyzed on 15-cm activity gels, which reveal a larger number of starch metabolizing enzymes (Fig. 6B). In the nonmutant extract, the isoamylase activity was observed as a series of diffuse, light-blue bands at the top of the gel. This broad staining area labeled as arrow A is not detected in any of the three su1- mutants, and the light blue band labeled as arrow B also is missing in the mutants. Notably, the pattern in this region of the gel is the same in all three su1- mutants. Another light blue band was evident in the 15-cm gels, noted by arrow E. Duplicate gels including red-pullulan as the substrate showed an activity band of the same electrophorectic mobility, demonstrating that this signal indicates the pullulanase-type DBE (data not shown). This pullulanase-type enzyme activity was reduced in all three mutants relative to wild type, consistent with previous characterizations of DBEs in su1-Ref (Pan and Nelson, 1984; Beatty et al., 1999) and su1- mutants of rice (Kubo et al., 1999).
The su1- mutations also result in a wide variety of allele-specific changes in other starch modifying activities, as exhibited by altered staining intensity or protein mobility. An apparent BE activity (Fig. 3B, arrow C), as indicated by the red color of the band, is increased in staining intensity in su1-R4582::Mu1 and su1-st extracts compared with wild type, although this activity is not affected by su1-Ref. Also, a new apparent amylolytic activity, as suggested by the white color of the band, appears specifically in the su1-R4582::Mu1 and su1-st mutants (arrow F). An unknown starch modifying activity (arrow D) is not detected specifically in su1-st extracts. Furthermore, a doublet of apparent amylolytic activity (arrow G) is reduced in intensity in all three su1- mutant extracts, and the lower of these two bands is differentially affected by su1-Ref.
DISCUSSION
In this study, the direct molecular effects of three different su1- mutations on the isoamylase-type DBE specified by the gene were clearly determined. For su1-R4582::Mu1, the structure of the gene and lack of transcript indicate that the SU1 protein is completely missing in homozygous mutants. In contrast, both su1-Ref and su1-st produce transcripts that code for mutant SU1 polypeptides. Expression of su1-Ref appears to be normal at the level of mRNA accumulation, although the polypeptide abundance is reduced compared with wild type (Beatty et al., 1999). Thus, the SU1Ref mutant protein may be unstable relative to the wild-type SU1 protein. The fact that the kernel appearance and the chemical structure phenotype conditioned by su1-Ref closely resemble that caused by the null allele suggests that this mutant protein lacks enzymatic activity. SU1 isoamylase-type DBE activity was, in fact, shown to be severely reduced both in partially purified extracts (Beatty et al., 1999) and in the activity gels in this study. This loss of enzymatic activity is likely to result from the substitution in region IS-XII of the hydrophobic residue Trp, which is nearly universally conserved in all known isoamylase type enzymes by the charged residue Arg.
Despite their phenotypic similarities, differences do exist in the effects of su1-Ref and su1-R4582::Mu1. In particular, the former is dominant to su1-st with respect to kernel phenotype, whereas the latter is recessive. These differences may be explained by a complex effect of the su1-st allele on SU1 protein expression. The steady-state su1-st mRNA level is significantly reduced compared with wild type, and at least three different transcripts are produced from the mutant gene. Only two of these transcripts, type II and type III, are capable of producing full SU1 polypeptides, SU1st-II, and SU1st-III. At least one of these proteins must be present to explain the dominance of su1-st over the null allele su1-R4582::Mu1, although whether both forms accumulate cannot be discerned from these data. Furthermore, the mild kernel phenotype and intermediate chemical structure phenotype of homozygous su1-st kernels suggests that at least one of the SU1st-II and SU1st-III polypeptides possesses some enzymatic activity. This is likely to be a low level of activity, however, because the zymogram analysis showed the same result for su1-st and su1-R4582::Mu1. Alternatively, su1-st might not produce any active isoamylase-type DBE, in which case the partial function of this allele would have to be explained by a structural property of the polypeptide independent of starch hydrolytic activity.
The dominant/recessive relationships of the three mutant su1 alleles are likely to be explained by the multimeric nature of the SU1 holoenzyme. The rice isoamylase-type DBE exists as a high Mr homomultimer, estimated to comprise four to six monomers (Fujita et al., 1999), and the maize enzyme also migrates as a high Mr complex in gel filtration chromatography (Beatty et al., 1999). Because su1-R4582::Mu1 is a null allele, su1-st/su1-R4582::Mu1 heterozygous kernels are expected to assemble a relatively small amount of SU1 isoamylase in a complex comprising SU1st-II and/or SU1st-III, as would occur in homozygous su1-st kernels. Although such a complex would be reduced in abundance, it is likely to be enzymatically active and to provide some level of the DBE function needed for starch biosynthesis. The dominance of su1-Ref over su1-st most likely is explained by multimeric complexes containing SU1Ref, SU1st-II, and/or SU1st-III, which we propose are inactive despite the presence of some potentially functional subunits. According to this hypothesis, the abundance of the SU1st subunits must be low relative to SU1Ref, such that the occurrence of homomultimers lacking any SU1Ref subunits is very rare.
The potential production of two distinct mutant polypeptides from the su1-st gene is the consequence of the Toad element insertion within exon 10. The type-I transcript results from the skipping of exon 10 and the inserted Toad element during splicing. The splice donor and acceptor sites that border exon 10 are intact in the mutant, however, and the context of the sequence is such that they are not recognized by the splicing machinery. Plant introns normally are AU-rich, although the exons surrounding them are GC-rich (Wiebauer et al., 1988). Intron recognition is thought to occur in part because of the high A+U composition of a sequence, but also because of discrete borders that separate AU-rich from GC-rich regions (Luehrsen et al., 1994; Lorkovic et al., 2000). The Toad TIRs are AU-rich (69%), so that their insertion into the small exon 10 sequence may mask the AU-rich/GC-rich borders that exist in the nonmutant su1 gene. This would create a new, larger intron containing the exon 10 sequences. Additional examples of exon skipping and/or intron creation owing to AU-rich transposons are known. These include insertion of Ds2 within the Shrunken2 (Sh2) locus, creating sh2-m1 (Giroux et al., 1994), insertion of dSpm within the A2 locus, creating a2-m1 (Menssen et al., 1990), and insertion of dSpm at the Bronze1 locus, creating bz1-m13 (Kim et al., 1987).
The AU-rich composition of the Toad TIRs also is likely to activate cryptic splice sites within the Su1 sequence or the Toad element that result in formation of the type-II and type-III transcripts. Determination of 5′- and 3′-splice sites is mediated by downstream and upstream AU-rich tracts, respectively (Lou et al., 1993; McCullough et al., 1993). The Su1 sequence normally contains both the D*1 cryptic donor site and the A* cryptic acceptor site (see Fig. 4), although neither is recognized by the splice machinery. Repositioning of the A* sequence downstream of the Toad insertion, as part of the host site duplication, changes the AU-environment of the sequence and enlarges the physical distance between it and either cryptic donor site (D*1 or D*2). Thus, these sites are used by the splicing machinery only when Toad is present.
The zymogram profiles of enzyme activities clearly show that the su1 mutant alleles differ in their effects on specific BE activities and the activity of one or more amylolytic enzymes (Fig. 6). These observations suggest that the SU1Ref mutant polypeptide does have some functionality with respect to other enzymes, even if it lacks enzymatic function. One possible explanation is that either the SU1 or SU1Ref polypeptide can inhibit or alter the activities marked by arrows C and F in Figure 6, whether or not the proteins constitute an active isoamylase-type DBE. In contrast, su1-R4582::Mu1 causes complete absence of the SU1 protein, which may explain the changes in these activities. Similarly, the su1-st mutation most likely results in accumulation of only a small amount of SU1st-II and/or SU1st-III polypeptide, and thus is likely to condition the same pleiotropic effects as the null allele. The unique effect of su1-st on the loss of a starch modifying activity indicated by arrow D in Figure 6, which is distinct from that of Su1, su1-Ref, or su1-R4582::Mu1, is likely to arise from a structural property of SU1st-II and/or SU1st-III. This argument derives from the fact that this band does not vary depending on SU1 activity, because it is the same in the null mutant and the wild type. In summary, we propose that the overall function of the su1 gene depends on (a) the level of enzymatic activity, (b) the level of the polypeptide independent of enzyme activity, and (c) specific structural properties of the SU1 polypeptide again unrelated to enzyme activity.
The su1-Ref and su1-R4582::Mu1 mutations clearly affected the structure of amylopectin in starch granules with regard to the chain length distribution (Fig. 5). These data are consistent with the analysis of rice su1- mutants (Kubo et al., 1999), but differ from the analysis of Arabidopsis leaf starch in which an isoamylase mutation caused phytoglycogen accumulation without altering the structure of residual amylopectin (Zeeman et al., 1998). Both direct and indirect roles of DBE function in amylopectin biosynthesis have been proposed to account for the simultaneous occurrence of amylopectin and WSP in isoamylase-deficient mutants (Zeeman et al., 1998; Fujita et al., 1999; Myers et al., 2000). As far as can be determined from the activity gel profile, BEs are not altered significantly by su1-Ref, suggesting that changes in these enzymes are not responsible for the altered amylopectin structure. The change in chain length distribution is consistent with the hypothesis that the isoamylase-type DBE, and/or the pullulanase-type enzyme, directly modifies a precursor to amylopectin prior to its crystallization into the granule. One possibility is that the DBE(s) preferentially remove chains of a certain length, or chains positioned in some specific way within the newly branched region of the growing amylopectin molecule. An indirect role of the DBE, however, is not ruled out by these data. For example, the presence of phytoglycogen might alter the function of an SS or a BE, or altered supply of the substrates ADP-Glc or maltooligosaccharide might affect an SS activity such that it synthesizes chains of different lengths than normal.
The transcript characterization, the activity gel data, and the recessiveness of su1-st relative to su1-Ref, indicate that the level of isoamylase-type DBE activity is very low in su1-st mutants. The relatively mild kernel- and chemical structure phenotypes of su1-st homozygotes and su1-st/su1-R4582::Mu1 heterozygotes, therefore, support the notion that only a very small amount of SU1 isoamylase is sufficient for synthesis of significant quantities of normal or near normal amylopectin. In nonmutant endosperm there may be an excess of isoamylase-type DBE activity over what is needed to produce wild-type levels of amylopectin. A major reduction in this enzyme in su1-st mutants may still leave enough function to produce significant quantities of starch, as was seen in the 30 DAP kernels from the 1997-growing season. The variability in su1-st expressivity may be explained by environmental factors affecting the threshold level of isoamylase-type DBE activity required for normal starch biosynthesis.
This study has provided new information about the effects of mutations in the isoamylase-type DBE. The observation of numerous allele-specific effects on starch structure and the activities of other starch modifying enzymes implicates both enzymatic and nonenzymatic features of the SU1 polypeptide in starch biosynthesis. We expect that the pleiotropic effects of the su1- mutations are indicative of fundamental aspects of the starch biosynthetic mechanism.
MATERIALS AND METHODS
Plant Materials and Nomenclature
The nomenclature follows the standard maize (Zea mays) genetics format (Beavis et al., 1995). The mutant allele su1-st arose spontaneously in a population of Krug's yellow dent corn (Dahlstrom and Lonnquist, 1964) and was generously provided by Dr. Oliver Nelson (University of Wisconsin, Madison). All su1- mutant alleles were introgressed into the W64A inbred background by five generations of back crossing to the standard line.
RNA Gel-Blot Analyses
Total RNA was isolated from maize kernels harvested 20 DAP and subjected to gel-blot analysis as described previously (James et al., 1995; Gao et al., 1996). RNA gel blots were hybridized with a 387-bp antisense RNA probe from the 3′ end of the Su1 cDNA. The probe was synthesized as an in vitro run-off transcription product using the StripEZ T7 kit (catalog no. 1362, Ambion, Austin, TX). The transcription template was plasmid pSu1(BS), linearized at the unique BglII site within the Su1 cDNA. The RNA probe corresponds to nt 2,608 to nt 2,221 of the Su1 cDNA sequence (GenBank accession no. U18908) running in the antisense direction.
PCR, RT-PCR, and DNA Sequence Analysis
Sequencing of the su1-Ref coding sequence followed PCR and RT-PCR amplifications. PCR amplification of su1-Ref genomic DNA was used to obtain 5′-end sequence information with the primer pair JD01 (5′-GGC TCC CTC CCC TCC ACT TCC-3′) and 452 (5′- GGG ATC ATA CCA GCC ATT TGA-3′). Downstream sequence information for su1-Ref and su1-st was obtained following RT-PCR amplification of mRNA using the following primer pairs: 797 (5′-CTC AAA TGG CTG GTA TGA-3′) and JD02 (5′-CCA TTC CAC TCT GAC CAA ACG-3′); MB02A (5′-GGG GAA AAT CAT AAT CTT A-3′) and MB02 N (5′-TGG TAG ACG GTG ACA GCA-3′); JD03 (5′-ATT GTT CTT CCT TCT TAT CCC-3′) and MB03D (5′-TTG GTC AGA GTG GAA TGG-3′); JD18 (5′-ACC AGA GGA TGC AGT CTA TG-3′) and JD03. Primers JD19 (5′-TCC TTA TTT TGT CTT TTT AGG CAT TCT-3′) and JD20 (5′-GCA GAT GTT GGG AGG GTT AGT GTC T-3′) were used for PCR amplification of su1-st genomic DNA containing the Toad sequence. Template mRNA for RT-PCR was purified from total RNA using the PolyA+ Tract mRNA isolation system (catalog no. Z5300, Promega, Madison, WI). The HF One-step RT-PCR System (catalog no. 600164, Stratagene, La Jolla, CA) was used for the RT-PCR. Sequences were determined directly from the PCR products, except for the type-II and type-III transcripts produced from su1-st by the JD18/JD03 primer pair. In those instances the PCR products were first cloned using the pGEM-Teasy vector system (catalog no. A1360, Promega) and then sequenced from the recombinant plasmids. The amplification product from the JD19/JD20 primer pair, containing the Toad element and flanking su1 genomic sequences, also was cloned in the vector system, producing plasmid pJD19. Computational analyses were performed using the GCG Sequence Analysis Software Package (Genetics Computer Group, Madison, WI).
DNA Gel-Blot Analysis
Genomic DNAs were isolated from seedling leaves of a homozygous su1-st plant in the W64A background and inbred lines W64A, Oh43, Mo17, B73, and F1 hybrids B77/B79 and Q66/Q67 according to published procedure (Dellaporta et al., 1983), digested with BamHI and EcoRI, electrophoresed, and transferred to nylon membranes. The blots were hybridized at 65°C in a buffer of 5× SSC, 5× Denhardt's solution, 0.1% (w/v) SDS, and 10 μg mL−1 sonicated, denatured salmon sperm DNA. The hybridization probe was the 0.5-kb EcoRI/PvuII fragment from pJD19 (containing the central portion of the Toad element and one TIR), labeled with 32P using the RTS Radprime Kit (catalog no. 10387–017, Life Technologies/Gibco-BRL, Cleveland). Blots were washed one time in 2× SSC, 0.1% (w/v) SDS at 65°C for 15 min and once in 0.2× SSC, 0.1% (w/v) SDS at 65°C for 15 min.
Carbohydrate Extraction and Quantification
Immature kernels were harvested 20 to 30 DAP and stored at −80°C. Insoluble starch and soluble carbohydrates were extracted from five whole kernels as follows. After removal of the pericarp and embryo by dissection, endosperm tissue was ground in water in a chilled mortar and pestle, and the volume was brought to 4 mL. Water was removed from a 1-mL portion of the homogenate by drying in an oven at 70°C, and the kernel dry weight was determined from the remaining material. The remaining 3-mL portion of the homogenate was centrifuged at 10,000g for 5 min. The pellet was washed in cold water and again centrifuged at 10,000g for 5 min. The supernatant fractions from both centrifugations, containing soluble carbohydrates, were pooled and incubated in a boiling water bath for 10 min. The pellet fraction, consisting mostly of insoluble starch granules, was dissolved in 100% (v/v) dimethyl sulfoxide (DMSO) and incubated in a boiling water bath for 30 min.
Total glucan oligomer or polymer content was measured in terms of milligram carbohydrate/gram dry weight following complete digestion of the insoluble starch fraction and the soluble carbohydrate fraction with amyloglucosidase. These assays used a commercial Glc assay kit (catalog no. 207748, Boehringer Mannheim, Basel) according to the manufacturer's protocol. The quantities of Suc, d-Fru, and d-Glc were measured in terms of milligram carbohydrate/gram dry weight by specific enzymatic assays (catalog no. 716260, Boehringer Mannheim).
Starch Structural Analysis
Approximately 20 mg Glc equivalents of solubilized granular starch in 100% (v/v) DMSO was precipitated by overnight incubation in 4 volumes absolute ethanol at −20°C. During this step, we presume that lipids have been removed from the carbohydrate fraction. After centrifugation at 2,000g for 5 min, the pellet was dissolved in 2 mL of 10 mm NaOH. Amylopectin and amylose were separated by gel permeation chromatography on Sepharose CL-2B columns (1.8-m height; 1.8-cm diameter). Samples were eluted with 10 mm NaOH at a rate of 30 mL/h. Aliquots of each 3-mL fraction (100 μL) were mixed with 400 μL of I2/KI (0.67% [w/v] I2 and 3.33% [w/v] KI) solution, and maximal absorbance was determined by measuring the spectrum from 400 to 700 nm. Fractions containing amylopectin or amylose were pooled, dialyzed against water at 4°C, lyophilized, and stored at room temperature.
Characterization of the chain length distribution by HPAEC-PAD chromatography was performed as previously described (Fontaine et al., 1993). Briefly, the lyophilized amylopectin obtained from the gel permeation column was dissolved in 90% (v/v) DMSO, quantified, diluted to 1 mg mL−1, and digested to completion with 0.2 units mL−1 Pseudomonas sp isoamylase (catalog no. E-ISAMY, Megazyme International, Bray, County Wicklow, Ireland) during an overnight incubation at 45°C. Samples (50 μL) were injected into the Dionex HPAEC-PAD system for characterization of the chain length distribution. The area under each peak of chain length 3 to 40 Glc units was integrated and the apparent frequency of each chain length was calculated as the percentage of total peak area.
Activity Gel Analysis of Starch Modifying Enzymes
Frozen kernels (5 g) were ground to a fine powder in liquid nitrogen with a mortar and pestle, and the tissue was suspended in 5 mL of buffer containing 50 mm sodium acetate, pH 6, 20 mm dithiothreitol. All of the lysates were centrifuged at 50,000g for 1 h at 4°C, which removes phytoglycogen from the su1- mutant extracts. The supernatant was stored at −80°C. Total proteins (50 or 100 μg) were separated on a native polyacrylamide gel (16 cm × 20 cm × 0.15 cm). The resolving gel contained 7% (w/v) acrylamide (29:1 acrylamide-bisacrylamide was used throughout) and 375 mm Tris-HCl, pH 8.8. The stacking gel contained 4% (w/v) acrylamide and 63 mm Tris-HCl, pH 6.8. Electrophoresis was conducted at 4°C, 25 V cm−1 for 4 h using a Protean II cell (Bio-Rad Laboratories, Hercules, CA) in an electrode buffer of 25 mm Tris, 192 mm Gly, pH 8.8, and 2 mm dithiothreitol. At the end of the run, the gel was electroblotted to a polacrylamide gel of the same size containing 7% (w/v) acrylamide, 0.3% (w/v) potato starch (Sigma, St. Louis), and 375 mm Tris-HCl, pH 8.8. The transfer was performed overnight at 20 V in the electrode buffer at room temperature. Starch metabolic activities were observed by staining the gel with I2/KI solution, and the gel was photographed immediately. The 5-cm activity gels were identical except that the Mini-Protean II cell (Bio-Rad Laboratories) was used, 25 μg of the sample was applied, and electrophoresis was for 45 min.
ACKNOWLEDGMENTS
We thank Oliver Nelson for providing the original su1-st material, Katie Dilks for technical assistance in carbohydrate analysis, and Paul Scott for assistance with HPAEC-PAD analysis. Nucleotide sequencing was performed at the Iowa State University Nucleic Acid Sequencing and Synthesis Facility.
Footnotes
This work was supported by the U.S. Department of Agriculture (grant no. 99–35304–8642 to M.G.J. and A.M.M.), by a U.S. Department of Agriculture National Needs Fellowship in Plant Biotechnology (grant no. 98–38420–5838 to J.R.D.), and by the National Science Foundation (grant no. DIR–9113593 to the Iowa State University Signal Transduction Training Group). This is a journal paper (no. J–19209) of the Iowa Agriculture and Home Economics Experiment Station (Ames; project no. 3593).
LITERATURE CITED
- Beatty MK, Myers AM, James MG. Genomic nucleotide sequence of a full-length wild type allele of the maize sugary1 (su1) gene (accession no. AF030882) (PGR 97-187) Plant Physiol. 1997;115:1731. [Google Scholar]
- Beatty MK, Rahman A, Cao H, Woodman W, Lee M, Myers AM, James MG. Purification and molecular genetic characterization of ZPU1, a pullulanase-type starch debranching enzyme from maize. Plant Physiol. 1999;119:255–266. doi: 10.1104/pp.119.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beavis W, Berlyn M, Burr B, Chandler V, Coe E, Fauron C, Nelson O, Polacco M, Rodermel S, Sachs M. A standard for maize genetics nomenclature. Maize Genet Coop Newsl. 1995;69:182–184. [Google Scholar]
- Correns C. Bastarde zwischen maisrassen, mit besonder Berucksichtung der Xenien. Bibliotheca Bot. 1901;53:1–161. [Google Scholar]
- Creech RG. Carbohydrate synthesis in maize. Adv Agron. 1968;20:275–322. [Google Scholar]
- Dahlstrom DE, Lonnquist JH. A new allele at the sugary-1 locus in maize. J Hered. 1964;55:242–246. [Google Scholar]
- Dauvillee D, Mestre V, Colleoni C, Slomianny M-C, Mouille G, Delrue B, d'Hulst C, Bliard C, Nuzillard J-M, Ball S. The debranching enzyme complex missing in glycogen accumulating mutants of Chlamydomonas reinhardtiidisplays an isoamylase-type specificity. Plant Sci. 2000;157:145–156. doi: 10.1016/s0168-9452(00)00256-9. [DOI] [PubMed] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. A plant version of DNA minipreparation: version II. Plant Mol Biol Rep. 1983;1:19–21. [Google Scholar]
- Doehlert DC, Knutson CA. Two classes of starch debranching enzymes from developing maize kernels. J Plant Physiol. 1991;138:566–572. [Google Scholar]
- Federoff N. Maize transposable elements. In: Berg D, Howe M, editors. Mobile DNA. Washington, DC: American Society of Microbiology; 1989. pp. 375–411. [Google Scholar]
- Finnegan DJ. Eukaryotic transposable elements and genome evolution. Trends Genet. 1989;5:103–107. doi: 10.1016/0168-9525(89)90039-5. [DOI] [PubMed] [Google Scholar]
- Fontaine T, Hulst CD, Maddelein M-L, Routier F, Pepin TM, Decq A, Wieruszeski J-M, Delrue B, Van den Koornhuyse N, Bossu J-P. Toward an understanding of the biogenesis of the starch granule. J Biol Chem. 1993;268:16223–16230. [PubMed] [Google Scholar]
- Fujita N, Kubo A, Francisco PB, Nakakita M, Harada K, Minaka N, Nakamura Y. Purification, characterization, and cDNA structure of isoamylase from developing endosperm of rice. Planta. 1999;208:283–293. doi: 10.1007/s004250050560. [DOI] [PubMed] [Google Scholar]
- Gallant DJ, Bouchet B, Baldwin PM. Microscopy of starch: evidence of a new level of granule organization. Carbohydr Polym. 1997;32:177–191. [Google Scholar]
- Gao M, Fisher DK, Kim K-N, Shannon JC, Guiltinan MJ. Evolutionary conservation and expression patterns of maize starch branching enzyme I and IIb genes suggests isoform specialization. Plant Mol Biol. 1996;30:1223–1232. doi: 10.1007/BF00019554. [DOI] [PubMed] [Google Scholar]
- Giroux MJ, Clancy M, Baier J, Ingham L, McCarty D, Hannah LC. De novo synthesis of an intron by the maize transposable element dissociation. Proc Natl Acad Sci USA. 1994;91:12150–12154. doi: 10.1073/pnas.91.25.12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges HF, Creech RG, Loerch JD. Biosynthesis of phytoglycogen in maize endosperm: the branching enzyme. Biochim Biophys Acta. 1969;185:70–79. doi: 10.1016/0005-2744(69)90283-6. [DOI] [PubMed] [Google Scholar]
- Imberty A, Buleon A, Tran V, Perez S. Recent advances in knowledge of starch structure. Starch/Staerke. 1991;43:375–384. [Google Scholar]
- James MG, Robertson DS, Myers AM. Characterization of the maize gene sugary1, a determinant of starch composition in kernels. Plant Cell. 1995;7:417–429. doi: 10.1105/tpc.7.4.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jesperson HM, MacGregor EA, Henrissat B, Sierks MR, Svensson B. Starch- and glycogen-debranching and branching enzymes: prediction of structural features of the catalytic (β/α)8-barrel domain and evolutionary relationship to other amylolytic enzymes. J Protein Chem. 1993;12:791–805. doi: 10.1007/BF01024938. [DOI] [PubMed] [Google Scholar]
- Kim HY, Schiefelbein JW, Raboy V, Furtek DB, Nelson OE., Jr RNA splicing permits expression of a maize gene with a defective suppressor-mutator transposable element insertion in an exon. Proc Natl Acad Sci USA. 1987;84:5863–5867. doi: 10.1073/pnas.84.16.5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kossman J, Lloyd J. Understanding and influencing starch biochemistry. Crit Rev Biochem Mol Biol. 2000;35:141–196. [PubMed] [Google Scholar]
- Kubo A, Fujita N, Harada K, Matsuda T, Satoh H, Nakamura Y. The starch-debranching enzymes isoamylase and pullulanase are both involved in amylopectin biosynthesis in rice endosperm. Plant Physiol. 1999;121:399–409. doi: 10.1104/pp.121.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavintman N. The formation of branched glucans in sweet corn. Arch Biochem Biophys. 1966;116:1–8. doi: 10.1016/0003-9861(66)90004-x. [DOI] [PubMed] [Google Scholar]
- Lee EYC, Whelan WJ. Glycogen and starch debranching enyzmes. In: Boyer P, editor. The Enzymes. Vol. 5. New York: Academic Press; 1971. pp. 191–234. [Google Scholar]
- Lorkovic ZJ, Kirk DAW, Lambermon MHL, Filipowicz W. Pre-mRNA splicing in higher plants. Trends Plant Sci. 2000;5:160–167. doi: 10.1016/s1360-1385(00)01595-8. [DOI] [PubMed] [Google Scholar]
- Lou H, McCullough AJ, Schuler MA. 3′-Splice site selection in dicot plant nuclei is position dependent. Mol Cell Biol. 1993;13:4485–4493. doi: 10.1128/mcb.13.8.4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luehrsen KR, Taha S, Walbot V. Nuclear pre-RNA processing in higher plants. Prog Nucleic Acids Res Mol Biol. 1994;47:149–193. doi: 10.1016/s0079-6603(08)60252-4. [DOI] [PubMed] [Google Scholar]
- Manners D. Observations on the specificity and nomenclature of starch debranching enzymes. J Appl Glycosci. 1997;44:83–85. [Google Scholar]
- McCullough AJ, Lou H, Schuler MA. Factors affecting authentic 5′-splice site selection in plant nuclei. Mol Cell Biol. 1993;13:1323–1331. doi: 10.1128/mcb.13.3.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menssen A, Hohmann S, Martin W, Schnable PS, Peterson PA, Saedler H, Gierl A. The En/Spm transposable element of Zea mayscontains splice sites at the termini generating a novel intron from a dSpm element in the A2 gene. EMBO J. 1990;9:3051–7. doi: 10.1002/j.1460-2075.1990.tb07501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris DZ, Morris CT. Glycogen in the seed of Zea mays. J Biol Chem. 1939;130:535–544. [Google Scholar]
- Mouille G, Maddelein M-L, Libessart N, Talaga P, Decq A, Delrue B, Ball S. Preamylopectin processing: a mandatory step for starch biosynthesis in plants. Plant Cell. 1996;8:1353–1366. doi: 10.1105/tpc.8.8.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers AM, Morell MK, James MG, Ball SG. Recent progress toward understanding the amylopectin crystal. Plant Physiol. 2000;122:989–997. doi: 10.1104/pp.122.4.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Umemoto T, Takahata Y, Komae K, Amano E, Satoh H. Changes in structure of starch and enzyme activities affected by sugary mutations in developing rice endosperm: possible role of starch debranching enzyme (R-enzyme) in amylopectin biosynthesis. Physiol Plant. 1996;97:491–498. [Google Scholar]
- Nevers P, Shepherd NS, Saedler H. Plant transposable elements. Adv Bot Res. 1986;12:103–203. [Google Scholar]
- Pan D, Nelson OE. A debranching enzyme deficiency in endosperms of the sugary-1mutants of maize. Plant Physiol. 1984;74:324–328. doi: 10.1104/pp.74.2.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman A, Wong K-S, Jane J-L, Myers AM, James MG. Characterization of SU1 isoamylase, a determinant of storage starch structure in maize. Plant Physiol. 1998;117:425–435. doi: 10.1104/pp.117.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon JC, Garwood DL. Genetics and physiology of starch development. In: Whistler RL, Bemiller JN, Paschall EF, editors. Starch: Chemistry and Technology. Orlando, FL: Academic Press; 1984. pp. 25–86. [Google Scholar]
- Smith AM. Making starch. Curr Opin Plant Biol. 1999;2:223–229. doi: 10.1016/S1369-5266(99)80039-9. [DOI] [PubMed] [Google Scholar]
- Tomalsky DS, Krisman CR. The degree of branching in (α1, 4)-(α1, 6)-linked glucopolysaccharides is dependent on intrinsic properties of the branching enzymes. Eur J Biochem. 1987;168:393–397. doi: 10.1111/j.1432-1033.1987.tb13432.x. [DOI] [PubMed] [Google Scholar]
- Wiebauer K, Herrero JJ, Filipowicz W. Nuclear pre-mRNA processing in plants: distinct modes of 3′-splice-site selection in plants and animals. Mol Cell Biol. 1988;8:2042–51. doi: 10.1128/mcb.8.5.2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun S, Matheson NK. Structures of the amylopectins of waxy, normal, amylose-extender, and wx:ae genotypes and of the phytoglycogen of maize. Carbohydr Res. 1993;243:307–321. doi: 10.1016/0008-6215(93)87035-q. [DOI] [PubMed] [Google Scholar]
- Zeeman SC, Umemoto T, Lue W-L, Au-Yeung P, Martin C, Smith AM, Chen J. A mutant of Arabidopsislacking a chloroplastic isoamylase accumulates both starch and phytoglycogen. Plant Cell. 1998;10:1699–1711. doi: 10.1105/tpc.10.10.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]