Abstract
Aberrant signaling through pathways controlling cell response to extracellular stimuli constitutes a central theme in disorders affecting development. Signaling through RAS and the MAPK cascade controls a variety of cell decisions in response to cytokines, hormones, and growth factors, and its upregulation causes Noonan syndrome (NS), a developmental disorder whose major features include a distinctive facies, a wide spectrum of cardiac defects, short stature, variable cognitive impairment, and predisposition to malignancies. NS is genetically heterogeneous, and mutations in more than ten genes have been reported to underlie this disorder. Despite the large number of genes implicated, about 10%–20% of affected individuals with a clinical diagnosis of NS do not have mutations in known RASopathy-associated genes, indicating that additional unidentified genes contribute to the disease, when mutated. By using a mixed strategy of functional candidacy and exome sequencing, we identify RRAS2 as a gene implicated in NS in six unrelated subjects/families. We show that the NS-causing RRAS2 variants affect highly conserved residues localized around the nucleotide binding pocket of the GTPase and are predicted to variably affect diverse aspects of RRAS2 biochemical behavior, including nucleotide binding, GTP hydrolysis, and interaction with effectors. Additionally, all pathogenic variants increase activation of the MAPK cascade and variably impact cell morphology and cytoskeletal rearrangement. Finally, we provide a characterization of the clinical phenotype associated with RRAS2 mutations.
Keywords: Noonan syndrome, RRAS2, RASopathies, RAS, MAPK
Main Text
Noonan syndrome (NS [MIM: PS163950]) is one of the most common monogenic disorders affecting development and growth.1 The phenotype of NS comprises a distinctive facies (e.g., hypertelorism, downslanting palpebral fissures, ptosis, and low-set/posteriorly rotated ears), cardiac abnormalities (a wide spectrum of congenital heart defects and cardiomyopathy), postnatally reduced growth, skeletal defects (chest and spine), cryptorchidism, bleeding diathesis, as well as variable neurocognitive impairment and predisposition to malignancies,1, 2 most commonly juvenile myelomonocytic leukemia (JMML [MIM: 607785]).3 NS is generally transmitted as an autosomal-dominant trait and is genetically heterogeneous. So far, pathogenic variants in more than ten genes have been reported as causative events underlying this disorder.4 While mutations in PTPN11 (MIM: 176876), SOS1 (MIM: 182530), RAF1 (MIM: 164760), and RIT1 (MIM: 609591) have been documented to occur most frequently,5, 6, 7, 8, 9, 10, 11 a smaller proportion of cases has been ascribed to mutations in other functionally related genes, including NRAS (MIM: 164790), KRAS (MIM: 190070), BRAF (MIM: 164757), MAP2K1 (MIM: 176872), SOS2 (MIM: 601247), LZTR1 (MIM: 600574), MRAS (MIM: 608435), and RASA2 (MIM: 601589).12, 13, 14, 15, 16, 17, 18, 19, 20 Although the causal link between mutations in a subset of these genes and the disorder still remains to be confirmed,4 the accumulated molecular evidence strongly supports the view that NS is caused by upregulated intracellular traffic through the RAS-mitogen-activated protein kinase (MAPK) signaling pathway.21, 22 Other disorders clinically related to NS (e.g., cardio-facio-cutaneous syndrome [MIM: PS115150], Costello syndrome [MIM: 218040], neurofibromatosis type 1 [MIM: 162200], Legius syndrome [MIM: 611431], Mazzanti syndrome [MIM: 607721], and Noonan syndrome with multiple lentigines [MIM: PS151100]) are also caused by mutations in genes encoding key proteins of the RAS-MAPK signaling backbone or upstream regulators (i.e., CBL, HRAS, KRAS, NF1, SPRED1, SHOC2, BRAF, MAP2K1, and MAP2K2).21, 22 In all these related conditions, termed RASopathies, increased signaling through RAS and the MAPK cascade can result from upregulated activity of RAS proteins, enhanced function of upstream signal transducers (e.g., proteins positively controlling RAS function) or downstream RAS effectors, as well as from the inefficient signaling switch-off by feedback mechanisms (e.g., neurofibromin and CBL loss of function). More recently, the use of whole-exome sequencing (WES) has allowed the discovery of RASopathy-associated genes encoding signal transducers or modulators that do not belong to the canonical RAS-MAPK pathway, but when functionally perturbed, are predicted to impact RAS signaling by still poorly characterized circuits.20, 23, 24, 25, 26, 27, 28, 29
A remarkable finding of the molecular genetics of NS and other RASopathies is the occurrence of conserved themes in the mechanism of disease. This applies in particular to mutations affecting genes encoding the various members of the RAS superfamily of GTPases that have been implicated in these disorders, including KRAS, HRAS, NRAS, RRAS, MRAS, and CDC42.11, 12, 13, 14, 20, 23, 24, 25, 26, 30 Missense mutations in these genes affect a small number of highly conserved amino acid residues that lead to overactivation of these proteins by decreasing/impairing their GTPase activity in response to GTPase-activating proteins (GAPs), increasing guanine nucleotide exchange factor (GEF)-independent GDP release, altering binding properties to effectors, or a combination of these mechanisms.31 Notably, while these germline mutations may affect the same residues that are generally mutated in cancer, multiple lines of evidence indicate that RASopathy-causing changes are generally less activating than their respective cancer-associated somatic lesions.21
Despite the large number of genes implicated in NS and related phenotypes, about 10%–20% of affected individuals with a convincing clinical diagnosis of NS do not have mutations in currently known RASopathy-associated genes, indicating that other unidentified genes contribute to this disorder. Through the use of complementary approaches based on “functional candidacy” (parallel sequencing of selected gene panels containing functionally related candidate genes) or WES, we identified RRAS2 (MIM: 600098; GenBank: NM_012250.5) as a gene implicated in NS. We provide structural, biochemical, and functional data to support the causal link between RRAS2 mutations and NS, outline the mechanisms by which mutations perturb RRAS2 function, and characterize the clinical phenotype associated with these gene lesions.
Subjects from six unrelated families were included in the study. Clinical data and DNA samples were collected from the participating families after written informed consent was obtained. DNA samples were stored and used under research projects approved by the Review Boards of the participating institutions. Because of a suspected RASopathy, subjects 1, 2, 3-III-1, and 5 were referred for diagnostic genetic testing by sequencing of an “extended” panel of RASopathy-associated genes designed to include a set of candidate disease genes selected in the frame of the NSEuroNet Consortium, while subjects 4 and 6 were analyzed by WES (Supplemental Subjects and Methods). In five cases, the RRAS2 variant (c.68G>T [p.Gly23Val], c.65_73dup [p.Gly22_Gly24dup], c.70_78dup [p.Gly24_Gly26dup], c.208G>A [p.Ala70Thr], c.215A>T [p.Gln72Leu]) arose de novo (i.e., it was not identified in parental blood DNA samples). In family 3, mutation scan in one affected family member (3-III-1) identified the heterozygous c.208G>A missense change, and subsequent co-segregation analysis confirmed the occurrence of the variant in three similarly affected relatives. All variants were validated by Sanger sequencing. In all cases, no other candidate variant was identified, further supporting the clinical relevance of this finding. In subject 4, the RRAS2 variant was detected in both amniocyte and peripheral blood DNA, at 44% and 46% of reads, respectively, indicating the heterozygous mutation was present in the germline of the subject. The clinical data of the affected subjects from the six families are shown in Table 1, facial features of four affected individuals as well as the pedigree of family 3 are presented in Figure 1, and a detailed clinical history is provided in the Supplemental Note. Taken together, the identified RRAS2 variants included three different nucleotide substitutions predicting missense changes of highly conserved amino acid residues (Gly23, Ala70, and Gln72) among RRAS2 orthologs and paralogs (Figure S1). Alterations to the corresponding positions in other GTPases of the RAS superfamily have already been reported to cause RASopathies or to contribute to oncogenesis (Table S1). In the remaining cases, we identified two small in-frame duplications (p.Gly22_Gly24dup, p.Gly24_Gly26dup) affecting the well-established mutational hotspot of RAS proteins (Figure 2A). Of note, p.Gly22_Gly24dup had previously been reported as somatic event in an uterine leiomyosarcoma specimen,32 and other similar, but not identical, small in-frame duplications affecting these residues have also been reported in association with different cancers in the Catalogue of Somatic Mutations in Cancer (COSMIC database). The two small in-frame duplications and c.68G>T (p.Gly23Val) and c.215A>T (p.Gln72Leu) substitutions were absent from general population databases, while the c.208G>A (p.Ala70Thr) change had previously been reported in two subjects in gnomAD (heterozygous state, frequency < 0.00001) (Table S2). Multiple in silico prediction algorithms uniformly rated these changes as deleterious/pathogenic (Table S2).
Table 1.
Clinical Features and Genotype of Individuals with RRAS2 Variants
| Subject 1 | Subject 2 |
Family 3 |
Subject 4 | Subject 5 | Subject 6 | ||||
|---|---|---|---|---|---|---|---|---|---|
| 3-II-1 | 3-II-2 | 3-III-1 | 3-III-2 | ||||||
| Origin | Algerian | Sri Lanka | German | Indian | Serbian | South American/ Ashkenazi | |||
| Gender | M | M | F | F | F | M | M | F | M |
| Age at last visit | 7 y 11 m | 12 y 2 m | 32 y | 40 y | 7 y 1 m | 1 y 7 m | 2 weeks | 8 y 10 m | 22 m (last measurement 18 m) |
| RRAS2 variant | c.65_73dup (p.Gly22_Gly24dup) | c.68G>T (p.Gly23Val) | c.208G>A (p.Ala70Thr) | c.208G>A (p.Ala70Thr) | c.208G>A (p.Ala70Thr) | c.208G>A (p.Ala70Thr) | c.215A>T (p.Gln72Leu) | c.208G>A (p.Ala70Thr) | c.70_78dup (p.Gly24_Gly26dup) |
| Inheritance | de novo | de novo | presumed paternal | presumed paternal | maternal | maternal | de novo | de novo | de novo |
| Prenatal features | NE, PH | PH | NA | NA | NA | N | NE, fetal ventriculo-megaly and cardiac abnormalities | NE | PH, LGA |
| Birth measurements: weight, length, OFC (weeks GA) | 3,730 g, 50.5 cm, 37 cm (35) | 3,180 g, 46.5 cm, 35 cm (35) | NA | 3,740 g, 51 cm, 36 cm | 3,110 g, 48 cm, 36 cm (39) | 2,440 g, 48 cm, 32 cm (35) | 2,400 g (33) | NA | 3,600 g, 51 cm, 38 cm (35) |
| Feeding difficulties | PF | PF, TF | NA | NA | PF | N | NA | N | N |
| Height at last examination | 125.5 cm (+0.3 SD) | 139.5 (−1.5 SD) 85 cm (−3.3 SD)a | 160 cm (−1.3 SD) | 170 cm (+0.3 SD) | 108 cm (−3.0 SD) | 78 cm (−1.8 SD) | NA | 122 cm (−2.1 SD) | 84.5 cm (+0.5 SD) |
| Weight | 27.5 kg (+0.5 SD) | 32.5 kg (−1.4 SD) | NA | 59 kg (+0.1 SD) | 18.6 kg (−1.8 SD) | 11 kg (−0.4 SD) | NA | 22 kg (−1.9 SD) | 12.5 kg (+0.7 SD) |
| OFC | 54 cm (+1.2 SD) | 57 cm (+2.5 SD) | 52.5 cm (−2.2 SD) | 55.5 cm (+0.2 SD) | 52 cm (+0.4 SD) | 49 cm (+0.2 SD) | NA | 52.5 cm (+0.2 SD) | 54.5 cm (+5.0 SD) |
| Cryptorchidism | N | N | NA | NA | NA | N | hypoplastic scrotum | NA | N |
| Congenital heart defect | SVAoS | VSD | VSD | N | N | N | TOF | AVSD, multiple VSDs | N |
| Lymphatic anomalies | N | N | N | N | N | N | N | N | N |
| Facial anomalies | typical NS | typical NS | suggestive NS | very mild in adulthood | typical NS | typical NS | multiple anomalies | suggestive NS | typical NS |
| Development | N | mild MD, mild LD | N | N | mild MD, mild LD | N | NA | N | mild global delay |
| Neurology | N | Chiari malformation | N | N | N | N | non-obstructive hydrocephalus | N | mild ventriculomegaly, hypotonia |
| Skeletal | N | N | N | N | N | N | 11 rib pairs, proximally placed thumb, spinal canal stenosis | pectus excavatum | N |
| Hematology & oncology | N | lymphopenia | N | N | N | N | thrombocytopenia | N | N |
| Skin and hair | glabellar heamangioma | N | N | N | atopic dermatitis, | N | N | N | glabellar hemangioma |
| Ocular | N | strabismus | N | strabismus | hyperopia, bilateral ptosis | N | NA | N | strabismic amblyopia, esotropia |
| Other malformations/anomalies | N | GH deficiency, GH treatment from age 4 y | unilateral duplex kidney | N | multiple allergies, bronchitis | N | labyrinth dysplasia, anteriorly placed anus | minor hippocampal malformation on brain MRI | N |
Abbreviations: AVSD, atrioventricular septal defect; F, female; GA, gestational age; GD, global delay; GH, growth hormone; LD, learning difficulties; LGA, large for gestational age; M, male; m, months; MD, motor delay; N, none/normal; NA, not applicable/not available; NE, nuchal edema; OFC, occipitofrontal head circumference; PF, poor feeding reported; PH, polyhydramnios; SVAoS, supravalvular aortic stenosis; TF, tube feeding (>4 weeks); TOF, Tetralogy of Fallot; y, years.
Before onset of growth hormone treatment at age 3 y 6 m.
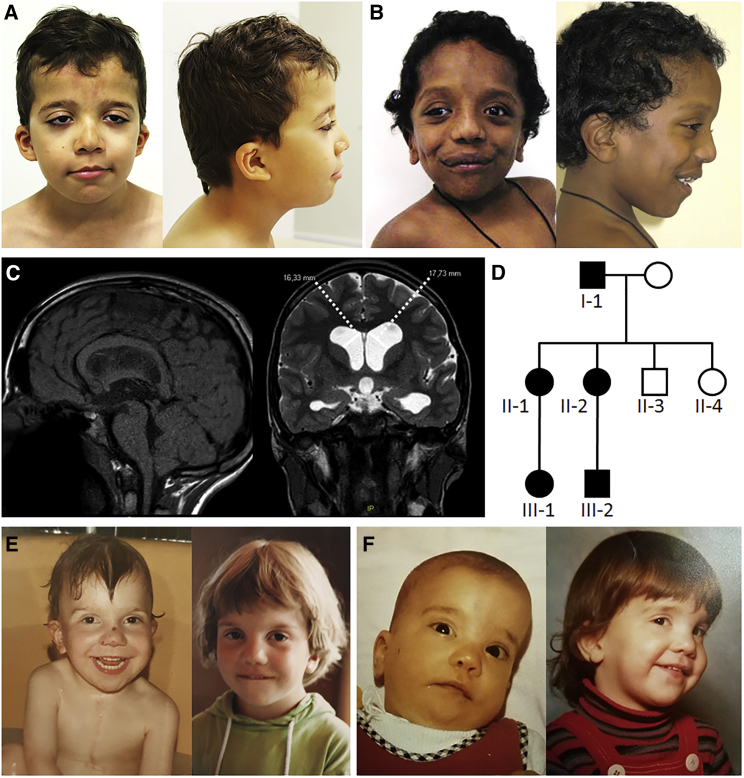
Figure 1.
Clinical Features of Individuals with Heterozygous Noonan Syndrome-Causing RRAS2 Variants
(A) Clinical appearance of subject 1 at 7 years and 11 months. Note the distinctive NS features, including bitemporal narrowing, downslanting palpebral fissures, ptosis, low-set ears, and low posterior hairline.
(B) Facial features of subject 2 at 2 years and 6 months. Facial features overlap those characterizing subject 1, even though a “coarse” face is also observed.
(C) Subject 2 brain MRI at 11 years and 9 months showing Chiari type 1 malformation and bilateral ventricular dilatation.
(D) Pedigree of family 3.
(E) Clinical appearance of subject 3-II-1 at the age of 11 months and 4.5 years.
(F) Facial features of subject 3-II-2 at 9 months and 5 years. The NS facial gestalt of subjects 3-II-1 and 3-II-2 became less obvious in adulthood.
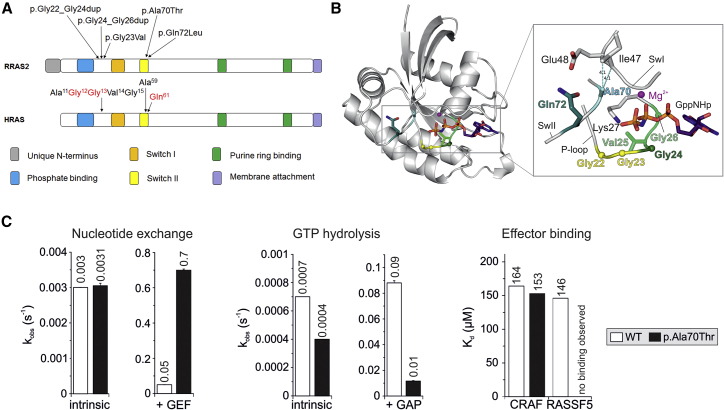
Figure 2.
RRAS2 Structure and Location and Functional Impact of Noonan Syndrome-Causing Variants
(A) Schematic representation of RRAS2 and HRAS proteins. Conserved motifs critical for tight guanine nucleotide binding and hydrolysis, and position of the disease-causing RRAS2 variants are illustrated together with the homologous residues of HRAS. The three residues representing the mutational hotspots of oncogenic HRAS mutations are shown in red.
(B) Structural modeling of RRAS2 variants. A structural model of the active GTP-bound RRAS2 protein highlights the relative position of the disease-causing missense or insertion mutations. All RRAS2 mutations affect residues that are located in the nucleotide binding active site region, which contains integral elements involved in GDP/GTP binding, GTP hydrolysis, and interactions with regulators (GEFs and GAPs) and effectors.
(C) Biochemical assessment of RRAS2p.Ala70Thr. RRAS2WT and RRAS2p.Ala70Thr proteins were biochemically characterized regarding their nucleotide exchange (left), GTP hydrolysis (middle), and effector binding (right) properties. The nucleotide exchange reaction was measured in the absence (intrinsic) and in the presence of the catalytic RASGEF domain of mouse RASGRF1, while the catalytic activity of the GTPase was assessed in the absence (intrinsic) and in the presence of the p120 RASGAP GAP domain. The RAS-binding and RAS association domains of CRAF and RASSF5 were used to evaluate the binding behavior of the RRAS2p.Ala70Thr mutant to RAS effectors. Overall, the data indicate that the p.Ala70Thr change leads to an accumulation of the protein in its GTP-bound active state, resulting to an increased signaling activity. The missense change, however, is predicted to differentially impact on the diverse downstream signaling pathways.
RRAS2 (RAS related 2, also known as TC21, teratocarcinoma 21) is a member of the RAS superfamily of GTPases, originally described in 1990.33 The protein shares the same four conserved functional domains with HRAS, KRAS, and NRAS, and about 55% amino acid sequence homology with HRAS (Figure 2A), which reaches 80% when considering the region between residues 5 to 120 (i.e., excluding the hypervariable tail at the C terminus).34, 35 RRAS2 controls multiple cellular processes, including proliferation, survival, and migration, and its functional dysregulation has been documented to contribute to oncogenesis.34, 36, 37 Indeed, a number of oncogenic RRAS2 variants have been reported, including the p.Gly23Val, p.Ala70Thr, and p.Gln72Leu changes, in a variety of solid tumors (Table S1). More recently, the p.Gln72Leu change in RRAS2 has been identified in subjects with isolated JMML,38 which represents the archetypal somatic RASopathy. Notably, germline mutations in other RAS genes affecting analogous codons to those observed in the present cases have also been identified (Table S1), including the missense mutation p.Gln87Leu in RRAS (homologous to p.Gln72Leu in RRAS2), previously reported in individuals having features reminiscent of NS.23
In order to decipher the consequences of the observed amino acid changes and the small in-frame duplications on the molecular structure of RRAS2, we performed structural modeling. A closer view into the active site of RRAS2 structure in its active form (Figure 2B, left) revealed that the identified RRAS2 mutations affect residues localized around the nucleotide binding pocket of the GTPase. The corresponding amino acids, including Gly22-Gly26, Ala70, and Gln72, do not only play a critical role in GDP/GTP exchange and GTP hydrolysis but also are involved in stabilization of the switch regions (Figure 2B, right), which are the binding sites for both RRAS2 regulators (GEFs and GAPs) and effectors.39 Specifically, the amino acid stretch encompassing Gly22 to Gly26 constitutes part of the phosphate-binding loop (P loop; residues Gly21 to Ser28) that is responsible for binding to the phosphate groups of either GTP or GDP. These residues play a critical role in nucleotide binding and hydrolysis by contacting both the β-γ phosphates of GTP (shown as GppNHp, a non-hydrolyzable GTP analog in Figure 2B) and residues 67 to 69 of the switch II region (SwII; Asp68 to Arg84). Val25 stabilizes the P loop by contacting Val92, Ser94, and Ser100. The Gly22-to-Gly24 and Gly24-to-Gly26 duplications were predicted to destabilize the P loop and result in increased nucleotide exchange and decreased GTP hydrolysis reactions. Differently, Ala70 and Gln72 are located in the switch II region of the GTPase and are directly involved in Mg2+ coordination and GTP hydrolysis reaction. Additionally, Ala70 and Gln72 stabilize the switch I region (SwI; Phe39-Ser50) by contacting Ile47 and Glu48, respectively. Based on these considerations, the NS-associated amino acid changes were expected to affect various aspects of RRAS2 biochemical behavior, including a faster nucleotide exchange, an impaired GTP hydrolysis, and a decrease in GEF, GAP, and effector interactions. Subsequent biochemical analysis of RRAS2p.Ala70Thr clearly confirmed these structural predictions, as assessment of the intrinsic and stimulated nucleotide exchange demonstrated a significantly increased response of the RRAS2p.Ala70Thr protein to GEF as compared to wild-type RRAS2 (Figure 2C). In contrast, the GTP hydrolysis reactions of the mutant were reduced compared to the wild-type protein. Particularly, the GAP-stimulated GTPase activity of RRAS2p.Ala70Thr was significantly decreased (9-fold) (Figure 2C). Finally, the binding properties to two RRAS2 effectors, RAF1 (CRAF) and RASSF5, were assessed. While the affinity of the interaction with CRAF was comparable to that of wild-type RRAS2, binding to RASSF5 was abolished (Figure 2C). This suggests the p.Ala70Thr change leads to a structural rearrangement of RRAS2 switch II, which is a key binding site for RASSF5 but not for CRAF. Overall, these data support that the p.Ala70Thr change leads to an accumulation of RRAS2 in its GTP-bound active state, which predicts an increase in signaling activity. The impaired binding to RASSF5, however, suggest a possible differential impact of the missense change on downstream signaling pathways.
RRAS2 shares downstream effectors with the other members of the RAS subfamily;35 however, little information exists about the function of this protein in cellular processes and development. Similarly, scant data exist on the specific role of this protein in intracellular signaling as well as on the extent of functional overlap with the other RAS proteins implicated in RASopathies. To explore the consequences of NS-associated RRAS2 mutations on the intracellular signaling pathways affected in NS, the signaling flows through the MAPK and phosphatidylinostiol-3 kinase (PI3K)-AKT cascades were evaluated using transient expression in HEK293T cells. Expression of all mutants resulted in variably enhanced ERK phosphorylation compared to cells overexpressing the wild-type protein (Figure 3A). Notably, RRAS2p.Ala70Thr and RRAS2p.Gln72Leu were observed to constitutively promote increased ERK phosphorylation, while only a slight increase was observed basally in cells expressing the RRAS2p.Gly22_Gly24dup and RRAS2p.Gly23Val mutants. However, this slight increase substantially strengthened after stimulation with EGF. This activating role of p.Gly22_Gly24dup is in line with previous evidence supporting the gain-of-function role of short insertional mutations in the P loop of other members of the RAS family.40 Based on previous data indicating that upregulated RRAS2 promotes tumorigenesis in a PI3K-dependent manner,41 the impact of NS-associated mutants on PI3K-AKT signaling was also assessed. No significant difference in the extent of AKT phosphorylation was documented, indicating a specific functional link between RRAS2 and the MAPK signaling cascade, at least in the present experimental conditions. In line with these findings, Rras2 KO mice showed a downmodulation of Erk activation and unaltered levels of phosphorylated Akt.42
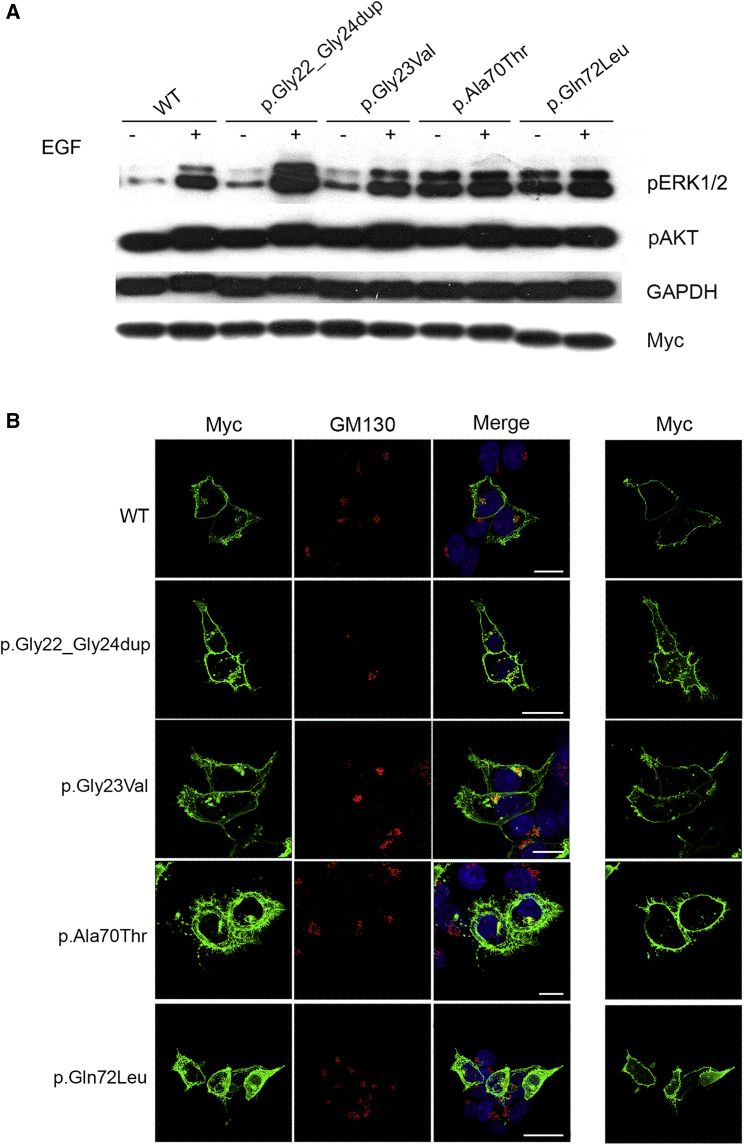
Figure 3.
Biochemical and Functional Characterization of Noonan Syndrome-Causing RRAS2 Variants
(A) ERK and AKT phosphorylation assays. HEK293T cells were transfected with the indicated Myc-tagged RRAS2 constructs. Following starvation (18 h) and EGF stimulation (30 ng/mL for 15 min), ERK and AKT phosphorylation levels were evaluated using a mouse monoclonal anti-phospho-p44/42 ERK (Thr202/Tyr204) antibody and a rabbit polyclonal anti-phospho-AKT (Ser473) antibody, respectively. To assess myc-RRAS2 protein levels, 20 μg of total lysates were immunoblotted with a mouse monoclonal anti-Myc antibody. Membranes were re-probed with mouse monoclonal anti-GAPDH antibody for protein normalization. Representative blot of three performed experiments are shown.
(B) RRAS2 subcellular localization showed by confocal laser scanning microscopy (CLSM) observations (left). Assays were performed on HEK293T cells starved overnight and stained with an anti-Myc mouse monoclonal antibody, followed by goat anti-mouse Alexa Fluor-488 (green), and an anti-GM130 (Golgi marker) rabbit polyclonal antibody, followed by goat anti-rabbit Alexa Fluor-594 (red). Nuclei are visualized by DAPI staining (blue). Co-localization areas were detected in yellow. CLSM observation were also performed at the basal level of cells to show the distinctive pattern of adhesion-like structures and cytoskeletal rearrangement in cells expressing the RRAS2 mutants (right). In all panels, bars correspond to 21 μm.
RAS proteins interact with multiple signaling platforms, which allow these proteins to differentially control multiple signaling pathways.43 Such complex behavior is attained by their dynamic interaction with the plasma membrane and other intracellular membranes (i.e., endosomes, endoplasmic reticulum, and Golgi). To explore any perturbing effect of mutations on the subcellular localization and distribution of RRAS2, including possible preferential targeting to specific intracellular domains, confocal laser scanning microscopy analysis was performed in HEK293T cells transiently expressing Myc-tagged RRAS2 constructs under starved condition. Similarly to the wild-type protein, a fraction of all RRAS2 mutant proteins co-localized with GM130, indicating their targeting to the Golgi apparatus, and the remainder were largely found at the plasma membrane (Figure 3B, left), indicating that mutations do not cause any overt subcellular redistribution of the GTPase. Notably, transient expression of all mutants was found to variably impact cell morphology and cytoskeletal rearrangement, with all mutant proteins promoting spreading and adhesion (Figure 3B, right). Taken together, these experimental data suggest that NS-associated RRAS2 mutations variably upregulate MAPK signaling and are likely to affect cellular processes depending on cytoskeleton rearrangement similar to observations of RASopathy-causing KRAS mutants.44
Our findings establish RRAS2 germline mutations as a cause of NS. Although previous screening of a cohort of 116 subjects with a clinical diagnosis of NS without a genetic explanation did not identify germline pathogenic RRAS2 variants,45 the present collaborative effort allowed to identify six unrelated affected individuals. Of the case subjects reported here, two individuals carrying de novo germline NS-causing RRAS2 variants (subjects 1 and 2) were identified among 1,220 samples addressed to Robert Debré Hospital, Paris, for diagnostic testing for NS, between February 2016 and September 2018. Within the same period, 181 of these subjects were found to carry a PTPN11 mutation. At the University Hospital of Magdeburg, screening of a multigene panel including RRAS2 in a cohort of 280 subjects with a tentative diagnosis of NS and negative results for mutations in previously known genes yielded two RRAS2 mutation-positive cases. Finally, no putative RRAS2 mutation was identified among 150 case subjects with a clinical diagnosis of NS from Ospedale Pediatrico Bambino Gesù, Rome. Overall, these findings indicate that RRAS2 mutations are rare events in NS.
The phenotypes associated with the two RRAS2 mutation hotspots were found to fit well within the clinical spectrum of NS even though they appeared variable in terms of severity. While individuals 1, 2, 5, and 6 had features fitting typical NS, the phenotype in some affected members of family 3 was relatively mild. On the other hand, subject 4 showed a complex and particularly severe phenotype with multiple congenital anomalies and neonatal lethality. Of note, prenatal features (nuchal edema, polyhydramnios, and/or cardiomyopathy) were reported in five of six subjects, and none showed pulmonary valve stenosis or hypertrophic cardiomyopathy. While the small size of the studied cohort does not allow us to outline specific genotype-phenotype correlations, we hypothesize that such variable expressivity likely reflects the differential strength of individual variants to perturb RRAS2 function and intracellular signaling. Consistent with the collected functional data, p.Gln72Leu (analogous to p.Gln61Leu in HRAS, NRAS, and KRAS) is a strong activating mutation and has not been observed to occur as a germline event in HRAS, KRAS, or NRAS. Similar differences in the biological and phenotypic consequences have previously been reported for HRAS, NRAS, and KRAS,12, 13, 14, 30, 31, 46, 47, 48, 49, 50, 51, 52, 53 including the positions corresponding to the presently identified RRAS2 mutations. The genotype-phenotype correlations in HRAS are illustrative and correlate well with the present findings: while p.Ala59Thr has been associated with Costello syndrome and p.Gly12Val has been reported with severe expression of Costello syndrome,46 p.Gln61Leu and other changes at this codon have only been reported as somatic events in cancer (Table S1).
A noticeable finding of this study is the observation of a diverse impact of the p.Ala70Thr on RRAS2 binding to CRAF/RAF1 and RASSF5. These data suggest the possibility that multiple signaling pathways downstream of RRAS2 may contribute to dysregulation of cellular processes (e.g., cell proliferation). As expected, a variable hyperactivation of the MAPK pathway resulting from the hyperactive state of the GTPase and unaltered binding to CRAF was observed for the NS-causing RRAS2Ala70Thr protein. Remarkably, impaired binding of this mutant to RASSF5, a known tumor suppressor protein negatively modulating YAP1 levels through activation of the Hippo pathway, was also observed. YAP1 is a transcriptional cofactor promoting cell proliferation, which undergoes RASSF5-mediated phosphorylation and degradation.54 The impaired binding of RRAS2 to RASSF5 raises the possibility that a less effective Hippo-mediated control of YAP1 levels may contribute to disease pathogenesis in NS.
Among RAS GTPases, RRAS2 exhibits the highest amino acid identity to HRAS, KRAS, and NRAS.35 Somatic mutations in RRAS2 have been established to contribute to oncogenesis, even though in a substantially restricted tumor type and less frequently compared to HRAS, KRAS, and NRAS. Consistently, it was originally demonstrated that RRAS2 proteins containing amino acid substitutions analogous to those with oncogenic role in HRAS, KRAS, and NRAS have transforming properties comparable to the strong transforming activity of RAS oncoproteins and similarly promote constitutive activation of the MAPK cascade.55 Our findings, which are in line with the data presented in an accompanying report by Niihori et al. published in this issue,56 further extend these observations by demonstrating the clinical relevance of a narrow spectrum of germline pathogenic variants in RRAS2 as the event underlying a small fraction of NS cases via upregulation of MAPK signaling. Further studies are required to more accurately define the precise mechanisms and circuits linking upregulated RRAS2 function and RAS-MAPK signaling dysregulation.
Declaration of Interests
K.G.M. declares no additional conflicts of interest beyond her employment affiliation. L.M.V. is a former employee of GeneDx. All the other authors declare no competing interests.
Acknowledgments
The authors thank the subjects and their families for participating in this study. This work was supported by the ERN-ITHACA networking (A.V. and M.T.) and grants from E-Rare (NSEuroNet, ERARE15-pp-063) to H.C., M.R.A. (01GM1602B), M.T., and M.Z. (01GM1602A), AIRC (IG21614) and Ministero della Salute (Ricerca Corrente 2017, 2018) to M.T., German Federal Ministry of Education and Research - BMBF (German Network for RASopathy Research “GeNeRARe”) to M.R.A. (01GM1519D and 01GM1902C) and M.Z. (01GM1519A), and the German Research Foundation through the Collaborative Research Center 974 (SFB 974) “Communication and Systems Relevance during Liver Injury and Regeneration” to M.R.A and grant number ZE 524-10/1 to M.Z.
Published: May 23, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.04.013.
Contributor Information
Marco Tartaglia, Email: marco.tartaglia@opbg.net.
Martin Zenker, Email: martin.zenker@med.ovgu.de.
Accession Numbers
The accession numbers for the five RRAS2 variants reported in this paper are ClinVar: SCV000902249–SCV000902253.
Web Resources
Exome Aggregation Consortium (ExAC) Browser, http://exac.broadinstitute.org/
GenBank, https://www.ncbi.nlm.nih.gov/genbank
GeneMatcher, https://genematcher.org
MutationAssessor, http://mutationassessor.org/r3/
MutationTaster, http://mutationtaster.org
MutPred2, http://mutpred.mutdb.org/
OMIM, http://www.omim.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
PROVEAN, http://provean.jcvi.org/index.php
Supplemental Data
References
- 1.Roberts A.E., Allanson J.E., Tartaglia M., Gelb B.D. Noonan syndrome. Lancet. 2013;381:333–342. doi: 10.1016/S0140-6736(12)61023-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tartaglia M., Gelb B.D., Zenker M. Noonan syndrome and clinically related disorders. Best Pract. Res. Clin. Endocrinol. Metab. 2011;25:161–179. doi: 10.1016/j.beem.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strullu M., Caye A., Lachenaud J., Cassinat B., Gazal S., Fenneteau O., Pouvreau N., Pereira S., Baumann C., Contet A. Juvenile myelomonocytic leukaemia and Noonan syndrome. J. Med. Genet. 2014;51:689–697. doi: 10.1136/jmedgenet-2014-102611. [DOI] [PubMed] [Google Scholar]
- 4.Grant A.R., Cushman B.J., Cavé H., Dillon M.W., Gelb B.D., Gripp K.W., Lee J.A., Mason-Suares H., Rauen K.A., Tartaglia M. Assessing the gene-disease association of 19 genes with the RASopathies using the ClinGen gene curation framework. Hum. Mutat. 2018;39:1485–1493. doi: 10.1002/humu.23624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tartaglia M., Mehler E.L., Goldberg R., Zampino G., Brunner H.G., Kremer H., van der Burgt I., Crosby A.H., Ion A., Jeffery S. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat. Genet. 2001;29:465–468. doi: 10.1038/ng772. [DOI] [PubMed] [Google Scholar]
- 6.Tartaglia M., Pennacchio L.A., Zhao C., Yadav K.K., Fodale V., Sarkozy A., Pandit B., Oishi K., Martinelli S., Schackwitz W. Gain-of-function SOS1 mutations cause a distinctive form of Noonan syndrome. Nat. Genet. 2007;39:75–79. doi: 10.1038/ng1939. [DOI] [PubMed] [Google Scholar]
- 7.Roberts A.E., Araki T., Swanson K.D., Montgomery K.T., Schiripo T.A., Joshi V.A., Li L., Yassin Y., Tamburino A.M., Neel B.G., Kucherlapati R.S. Germline gain-of-function mutations in SOS1 cause Noonan syndrome. Nat. Genet. 2007;39:70–74. doi: 10.1038/ng1926. [DOI] [PubMed] [Google Scholar]
- 8.Zenker M., Horn D., Wieczorek D., Allanson J., Pauli S., van der Burgt I., Doerr H.-G., Gaspar H., Hofbeck M., Gillessen-Kaesbach G. SOS1 is the second most common Noonan gene but plays no major role in cardio-facio-cutaneous syndrome. J. Med. Genet. 2007;44:651–656. doi: 10.1136/jmg.2007.051276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pandit B., Sarkozy A., Pennacchio L.A., Carta C., Oishi K., Martinelli S., Pogna E.A., Schackwitz W., Ustaszewska A., Landstrom A. Gain-of-function RAF1 mutations cause Noonan and LEOPARD syndromes with hypertrophic cardiomyopathy. Nat. Genet. 2007;39:1007–1012. doi: 10.1038/ng2073. [DOI] [PubMed] [Google Scholar]
- 10.Razzaque M.A., Nishizawa T., Komoike Y., Yagi H., Furutani M., Amo R., Kamisago M., Momma K., Katayama H., Nakagawa M. Germline gain-of-function mutations in RAF1 cause Noonan syndrome. Nat. Genet. 2007;39:1013–1017. doi: 10.1038/ng2078. [DOI] [PubMed] [Google Scholar]
- 11.Kouz K., Lissewski C., Spranger S., Mitter D., Riess A., Lopez-Gonzalez V., Lüttgen S., Aydin H., von Deimling F., Evers C. Genotype and phenotype in patients with Noonan syndrome and a RIT1 mutation. Genet. Med. 2016;18:1226–1234. doi: 10.1038/gim.2016.32. [DOI] [PubMed] [Google Scholar]
- 12.Cirstea I.C., Kutsche K., Dvorsky R., Gremer L., Carta C., Horn D., Roberts A.E., Lepri F., Merbitz-Zahradnik T., König R. A restricted spectrum of NRAS mutations causes Noonan syndrome. Nat. Genet. 2010;42:27–29. doi: 10.1038/ng.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schubbert S., Zenker M., Rowe S.L., Böll S., Klein C., Bollag G., van der Burgt I., Musante L., Kalscheuer V., Wehner L.-E. Germline KRAS mutations cause Noonan syndrome. Nat. Genet. 2006;38:331–336. doi: 10.1038/ng1748. [DOI] [PubMed] [Google Scholar]
- 14.Zenker M., Lehmann K., Schulz A.L., Barth H., Hansmann D., Koenig R., Korinthenberg R., Kreiss-Nachtsheim M., Meinecke P., Morlot S. Expansion of the genotypic and phenotypic spectrum in patients with KRAS germline mutations. J. Med. Genet. 2007;44:131–135. doi: 10.1136/jmg.2006.046300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarkozy A., Carta C., Moretti S., Zampino G., Digilio M.C., Pantaleoni F., Scioletti A.P., Esposito G., Cordeddu V., Lepri F. Germline BRAF mutations in Noonan, LEOPARD, and cardiofaciocutaneous syndromes: molecular diversity and associated phenotypic spectrum. Hum. Mutat. 2009;30:695–702. doi: 10.1002/humu.20955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nava C., Hanna N., Michot C., Pereira S., Pouvreau N., Niihori T., Aoki Y., Matsubara Y., Arveiler B., Lacombe D. Cardio-facio-cutaneous and Noonan syndromes due to mutations in the RAS/MAPK signalling pathway: genotype-phenotype relationships and overlap with Costello syndrome. J. Med. Genet. 2007;44:763–771. doi: 10.1136/jmg.2007.050450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamamoto G.L., Aguena M., Gos M., Hung C., Pilch J., Fahiminiya S., Abramowicz A., Cristian I., Buscarilli M., Naslavsky M.S. Rare variants in SOS2 and LZTR1 are associated with Noonan syndrome. J. Med. Genet. 2015;52:413–421. doi: 10.1136/jmedgenet-2015-103018. [DOI] [PubMed] [Google Scholar]
- 18.Cordeddu V., Yin J.C., Gunnarsson C., Virtanen C., Drunat S., Lepri F., De Luca A., Rossi C., Ciolfi A., Pugh T.J. Activating Mutations Affecting the Dbl Homology Domain of SOS2 Cause Noonan Syndrome. Hum. Mutat. 2015;36:1080–1087. doi: 10.1002/humu.22834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen P.-C., Yin J., Yu H.-W., Yuan T., Fernandez M., Yung C.K., Trinh Q.M., Peltekova V.D., Reid J.G., Tworog-Dube E. Next-generation sequencing identifies rare variants associated with Noonan syndrome. Proc. Natl. Acad. Sci. USA. 2014;111:11473–11478. doi: 10.1073/pnas.1324128111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Higgins E.M., Bos J.M., Mason-Suares H., Tester D.J., Ackerman J.P., MacRae C.A., Sol-Church K., Gripp K.W., Urrutia R., Ackerman M.J. Elucidation of MRAS-mediated Noonan syndrome with cardiac hypertrophy. JCI Insight. 2017;2:e91225. doi: 10.1172/jci.insight.91225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tartaglia M., Gelb B.D. Disorders of dysregulated signal traffic through the RAS-MAPK pathway: phenotypic spectrum and molecular mechanisms. Ann. N Y Acad. Sci. 2010;1214:99–121. doi: 10.1111/j.1749-6632.2010.05790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rauen K.A. The RASopathies. Annu. Rev. Genomics Hum. Genet. 2013;14:355–369. doi: 10.1146/annurev-genom-091212-153523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flex E., Jaiswal M., Pantaleoni F., Martinelli S., Strullu M., Fansa E.K., Caye A., De Luca A., Lepri F., Dvorsky R. Activating mutations in RRAS underlie a phenotype within the RASopathy spectrum and contribute to leukaemogenesis. Hum. Mol. Genet. 2014;23:4315–4327. doi: 10.1093/hmg/ddu148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gripp K.W., Aldinger K.A., Bennett J.T., Baker L., Tusi J., Powell-Hamilton N., Stabley D., Sol-Church K., Timms A.E., Dobyns W.B. A novel rasopathy caused by recurrent de novo missense mutations in PPP1CB closely resembles Noonan syndrome with loose anagen hair. Am. J. Med. Genet. A. 2016;170:2237–2247. doi: 10.1002/ajmg.a.37781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinelli S., Krumbach O.H.F., Pantaleoni F., Coppola S., Amin E., Pannone L., Nouri K., Farina L., Dvorsky R., Lepri F., University of Washington Center for Mendelian Genomics Functional Dysregulation of CDC42 Causes Diverse Developmental Phenotypes. Am. J. Hum. Genet. 2018;102:309–320. doi: 10.1016/j.ajhg.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Young L.C., Hartig N., Boned Del Río I., Sari S., Ringham-Terry B., Wainwright J.R., Jones G.G., McCormick F., Rodriguez-Viciana P. SHOC2-MRAS-PP1 complex positively regulates RAF activity and contributes to Noonan syndrome pathogenesis. Proc. Natl. Acad. Sci. USA. 2018;115:E10576–E10585. doi: 10.1073/pnas.1720352115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Motta M., Fidan M., Bellacchio E., Pantaleoni F., Schneider-Heieck K., Coppola S., Borck G., Salviati L., Zenker M., Cirstea I.C., Tartaglia M. Dominant Noonan syndrome-causing LZTR1 mutations specifically affect the kelch domain substrate-recognition surface and enhance RAS-MAPK signaling. Hum. Mol. Genet. 2018;28:1007–1022. doi: 10.1093/hmg/ddy412. [DOI] [PubMed] [Google Scholar]
- 28.Bigenzahn J.W., Collu G.M., Kartnig F., Pieraks M., Vladimer G.I., Heinz L.X., Sedlyarov V., Schischlik F., Fauster A., Rebsamen M. LZTR1 is a regulator of RAS ubiquitination and signaling. Science. 2018;362:1171–1177. doi: 10.1126/science.aap8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steklov M., Pandolfi S., Baietti M.F., Batiuk A., Carai P., Najm P., Zhang M., Jang H., Renzi F., Cai Y. Mutations in LZTR1 drive human disease by dysregulating RAS ubiquitination. Science. 2018;362:1177–1182. doi: 10.1126/science.aap7607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aoki Y., Niihori T., Kawame H., Kurosawa K., Ohashi H., Tanaka Y., Filocamo M., Kato K., Suzuki Y., Kure S., Matsubara Y. Germline mutations in HRAS proto-oncogene cause Costello syndrome. Nat. Genet. 2005;37:1038–1040. doi: 10.1038/ng1641. [DOI] [PubMed] [Google Scholar]
- 31.Gremer L., Gilsbach B., Ahmadian M.R., Wittinghofer A. Fluoride complexes of oncogenic Ras mutants to study the Ras-RasGap interaction. Biol. Chem. 2008;389:1163–1171. doi: 10.1515/BC.2008.132. [DOI] [PubMed] [Google Scholar]
- 32.Huang Y., Saez R., Chao L., Santos E., Aaronson S.A., Chan A.M. A novel insertional mutation in the TC21 gene activates its transforming activity in a human leiomyosarcoma cell line. Oncogene. 1995;11:1255–1260. [PubMed] [Google Scholar]
- 33.Drivas G.T., Shih A., Coutavas E., Rush M.G., D’Eustachio P. Characterization of four novel ras-like genes expressed in a human teratocarcinoma cell line. Mol. Cell. Biol. 1990;10:1793–1798. doi: 10.1128/mcb.10.4.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bos J.L. Ras-like GTPases. Biochim. Biophys. Acta. 1997;1333:M19–M31. doi: 10.1016/s0304-419x(97)00015-2. [DOI] [PubMed] [Google Scholar]
- 35.Nakhaei-Rad S., Haghighi F., Nouri P., Rezaei Adariani S., Lissy J., Kazemein Jasemi N.S., Dvorsky R., Ahmadian M.R. Structural fingerprints, interactions, and signaling networks of RAS family proteins beyond RAS isoforms. Crit. Rev. Biochem. Mol. Biol. 2018;53:130–156. doi: 10.1080/10409238.2018.1431605. [DOI] [PubMed] [Google Scholar]
- 36.Graham S.M., Oldham S.M., Martin C.B., Drugan J.K., Zohn I.E., Campbell S., Der C.J. TC21 and Ras share indistinguishable transforming and differentiating activities. Oncogene. 1999;18:2107–2116. doi: 10.1038/sj.onc.1202517. [DOI] [PubMed] [Google Scholar]
- 37.Erdogan M., Pozzi A., Bhowmick N., Moses H.L., Zent R. Signaling pathways regulating TC21-induced tumorigenesis. J. Biol. Chem. 2007;282:27713–27720. doi: 10.1074/jbc.M703037200. [DOI] [PubMed] [Google Scholar]
- 38.Stieglitz E., Taylor-Weiner A.N., Chang T.Y., Gelston L.C., Wang Y.-D., Mazor T., Esquivel E., Yu A., Seepo S., Olsen S. The genomic landscape of juvenile myelomonocytic leukemia. Nat. Genet. 2015;47:1326–1333. doi: 10.1038/ng.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vetter I.R., Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 40.Klockow B., Ahmadian M.R., Block C., Wittinghofer A. Oncogenic insertional mutations in the P-loop of Ras are overactive in MAP kinase signaling. Oncogene. 2000;19:5367–5376. doi: 10.1038/sj.onc.1203909. [DOI] [PubMed] [Google Scholar]
- 41.Larive R.M., Moriggi G., Menacho-Márquez M., Cañamero M., de Álava E., Alarcón B., Dosil M., Bustelo X.R. Contribution of the R-Ras2 GTP-binding protein to primary breast tumorigenesis and late-stage metastatic disease. Nat. Commun. 2014;5:3881. doi: 10.1038/ncomms4881. [DOI] [PubMed] [Google Scholar]
- 42.Larive R.M., Abad A., Cardaba C.M., Hernández T., Cañamero M., de Álava E., Santos E., Alarcón B., Bustelo X.R. The Ras-like protein R-Ras2/TC21 is important for proper mammary gland development. Mol. Biol. Cell. 2012;23:2373–2387. doi: 10.1091/mbc.E12-01-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hancock J.F. Ras proteins: different signals from different locations. Nat. Rev. Mol. Cell Biol. 2003;4:373–384. doi: 10.1038/nrm1105. [DOI] [PubMed] [Google Scholar]
- 44.Gremer L., Merbitz-Zahradnik T., Dvorsky R., Cirstea I.C., Kratz C.P., Zenker M., Wittinghofer A., Ahmadian M.R. Germline KRAS mutations cause aberrant biochemical and physical properties leading to developmental disorders. Hum. Mutat. 2011;32:33–43. doi: 10.1002/humu.21377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ceremsak J.J., Yu A., Esquivel E., Lissewski C., Zenker M., Loh M.L., Stieglitz E. Germline RRAS2 mutations are not associated with Noonan syndrome. J. Med. Genet. 2016;53:728. doi: 10.1136/jmedgenet-2016-103889. [DOI] [PubMed] [Google Scholar]
- 46.Quélin C., Loget P., Rozel C., D’Hervé D., Fradin M., Demurger F., Odent S., Pasquier L., Cavé H., Marcorelles P. Fetal costello syndrome with neuromuscular spindles excess and p.Gly12Val HRAS mutation. Eur. J. Med. Genet. 2017;60:395–398. doi: 10.1016/j.ejmg.2017.03.014. [DOI] [PubMed] [Google Scholar]
- 47.Carta C., Pantaleoni F., Bocchinfuso G., Stella L., Vasta I., Sarkozy A., Digilio C., Palleschi A., Pizzuti A., Grammatico P. Germline missense mutations affecting KRAS Isoform B are associated with a severe Noonan syndrome phenotype. Am. J. Hum. Genet. 2006;79:129–135. doi: 10.1086/504394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gripp K.W., Hopkins E., Sol-Church K., Stabley D.L., Axelrad M.E., Doyle D., Dobyns W.B., Hudson C., Johnson J., Tenconi R. Phenotypic analysis of individuals with Costello syndrome due to HRAS p.G13C. Am. J. Med. Genet. A. 2011;155A:706–716. doi: 10.1002/ajmg.a.33884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gripp K.W., Sol-Church K., Smpokou P., Graham G.E., Stevenson D.A., Hanson H., Viskochil D.H., Baker L.C., Russo B., Gardner N. An attenuated phenotype of Costello syndrome in three unrelated individuals with a HRAS c.179G>A (p.Gly60Asp) mutation correlates with uncommon functional consequences. Am. J. Med. Genet. A. 2015;167A:2085–2097. doi: 10.1002/ajmg.a.37128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bertola D., Buscarilli M., Stabley D.L., Baker L., Doyle D., Bartholomew D.W., Sol-Church K., Gripp K.W. Phenotypic spectrum of Costello syndrome individuals harboring the rare HRAS mutation p.Gly13Asp. Am. J. Med. Genet. A. 2017;173:1309–1318. doi: 10.1002/ajmg.a.38178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pantaleoni F., Lev D., Cirstea I.C., Motta M., Lepri F.R., Bottero L., Cecchetti S., Linger I., Paolacci S., Flex E. Aberrant HRAS transcript processing underlies a distinctive phenotype within the RASopathy clinical spectrum. Hum. Mutat. 2017;38:798–804. doi: 10.1002/humu.23224. [DOI] [PubMed] [Google Scholar]
- 52.Altmüller F., Lissewski C., Bertola D., Flex E., Stark Z., Spranger S., Baynam G., Buscarilli M., Dyack S., Gillis J. Genotype and phenotype spectrum of NRAS germline variants. Eur. J. Hum. Genet. 2017;25:823–831. doi: 10.1038/ejhg.2017.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stark Z., Gillessen-Kaesbach G., Ryan M.M., Cirstea I.C., Gremer L., Ahmadian M.R., Savarirayan R., Zenker M. Two novel germline KRAS mutations: expanding the molecular and clinical phenotype. Clin. Genet. 2012;81:590–594. doi: 10.1111/j.1399-0004.2011.01754.x. [DOI] [PubMed] [Google Scholar]
- 54.Nussinov R., Zhang M., Tsai C.J., Liao T.J., Fushman D., Jang H. Autoinhibition in Ras effectors Raf, PI3Kα, and RASSF5: a comprehensive review underscoring the challenges in pharmacological intervention. Biophys. Rev. 2018;10:1263–1282. doi: 10.1007/s12551-018-0461-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Graham S.M., Cox A.D., Drivas G., Rush M.G., D’Eustachio P., Der C.J. Aberrant function of the Ras-related protein TC21/R-Ras2 triggers malignant transformation. Mol. Cell. Biol. 1994;14:4108–4115. doi: 10.1128/mcb.14.6.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Niihori T., Nagai K., Fujita A., Ohashi H., Okamoto N., Okada S., Harada A., Kihara H., Arbogast T., Funayama R. Germline activating RRAS2 mutations cause Noonan syndrome. Am. J. Hum. Genet. 2019;104:1233–1240. doi: 10.1016/j.ajhg.2019.04.014. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.