Abstract
Noonan syndrome (NS) is characterized by distinctive craniofacial appearance, short stature, and congenital heart disease. Approximately 80% of individuals with NS harbor mutations in genes whose products are involved in the RAS/mitogen-activating protein kinase (MAPK) pathway. However, the underlying genetic causes in nearly 20% of individuals with NS phenotype remain unexplained. Here, we report four de novo RRAS2 variants in three individuals with NS. RRAS2 is a member of the RAS subfamily and is ubiquitously expressed. Three variants, c.70_78dup (p.Gly24_Gly26dup), c.216A>T (p.Gln72His), and c.215A>T (p.Gln72Leu), have been found in cancers; our functional analyses showed that these three changes induced elevated association of RAF1 and that they activated ERK1/2 and ELK1. Notably, prominent activation of ERK1/2 and ELK1 by p.Gln72Leu associates with the severe phenotype of the individual harboring this change. To examine variant pathogenicity in vivo, we generated zebrafish models. Larvae overexpressing c.70_78dup (p.Gly24_Gly26dup) or c.216A>T (p.Gln72His) variants, but not wild-type RRAS2 RNAs, showed craniofacial defects and macrocephaly. The same dose injection of mRNA encoding c.215A>T (p.Gln72Leu) caused severe developmental impairments and low dose overexpression of this variant induced craniofacial defects. In contrast, the RRAS2 c.224T>G (p.Phe75Cys) change, located on the same allele with p.Gln72His in an individual with NS, resulted in no aberrant in vitro or in vivo phenotypes by itself. Together, our findings suggest that activating RRAS2 mutations can cause NS and expand the involvement of RRAS2 proto-oncogene to rare germline disorders.
Keywords: Noonan syndrome, RRAS2, RASopathies, exome sequencing, macrocephaly, zebrafish, functional profiling, RAS/MAPK
Main Text
Noonan syndrome (NS [MIM: 163950]) is an autosomal-dominant or -recessive disorder characterized by distinctive craniofacial features, short stature, and congenital heart disease.1, 2 NS is one of the developmental syndromes caused by mutations in molecules involved in the RAS/MAPK (RAF/MEK/ERK) signaling pathway, termed collectively as the RASopathies.3 The RAS/MAPK pathway regulates cell proliferation, differentiation, survival, and apoptosis.4 Previous studies of genes associated with NS and related disorders, including Costello syndrome (CS [MIM: 218040]) and cardio-facio-cutaneous syndrome (CFCS [MIM: 115150]),5, 6, 7, 8, 9 have unified these clinically overlapping disorders as RASopathies in which dysregulation of the RAS/MAPK pathway is a common molecular basis. To date, mutations in genes encoding many components of the RAS/MAPK pathway, including PTPN11 (MIM: 176876), KRAS (MIM: 190070), SOS1 (MIM: 182530), RAF1 (MIM: 164760), SHOC2 (MIM: 602775), CBL (MIM: 165360), BRAF (MIM: 164757), NRAS (MIM: 164790), RRAS (MIM: 165090), have been reported as causes of NS or NS-like syndromes.1
The emergence of whole-exome sequencing (WES) has accelerated the discovery of RASopathy genes. We identified RIT1 (MIM: 609591) mutations in individuals with NS using WES.10 Subsequently, mutations in A2ML1 (MIM: 610627), RASA2 (MIM: 601589), SOS2 (MIM: 601247), LZTR1 (MIM: 600574),1 PPP1CB (MIM: 600590),11 and MRAS (MIM: 608435)12 have been reported as causes of NS or NS-like syndromes, although the contribution of each locus to the pathogenicity of NS is modest. Such discoveries suggest that the remaining ∼20% of individuals with NS that remain undiagnosed molecularly will likely harbor rare, possibly private, mutations in a host of hitherto uncharacterized genes. As such, we have undertaken an investigational paradigm in which we pair WES on individuals with NS-related disorders with in vitro and in vivo studies of candidate pathogenic variants. In the present study, we report the identification of three individuals with de novo RRAS2 (MIM: 600098) variants. Our genetic and functional data support a causal role for de novo dominant alleles as drivers of NS-like pathology in humans.
First, we performed WES on 27 individuals with clinically diagnosed or suspected NS or NS-related disorders without known RASopathy mutations (details in Supplemental Material and Methods). This study was approved by the Ethics Committee of the Tohoku University School of Medicine. Written informed consent was obtained from all subjects involved in the study or from their parents. We filtered the identified variants by minor allele frequency (MAF) in population databases such as 1000 Genomes, dbSNP, ExAC Browser, and Human Genetic Variation Database (absent or <0.01) and by functional prediction (missense, nonsense, indel, or splicing). We prioritized genes whose products are members of the RAS/MAPK pathway. We found a RRAS2 (GenBank: NM_012250.6; c.70_78dup) variant in individual NS462 and two RRAS2 variants (c.216A>T and c.224T>G) in individual NS833 (Table 1). Segregation analysis in the parents revealed that all three variants arose de novo. Parentage in each family was confirmed by the thousands of variants in common between child and parent in the trio exomes. RRAS2, also called TC21, encodes a member of the RAS GTPase superfamily and is expressed ubiquitously, with the highest levels in the heart, placenta, and skeletal muscle.13 RRAS2 is an evolutionarily conserved protein and exhibits 55% amino acid identity to RAS proteins.14
Table 1.
Clinical Features in the Scoring System of Noonan Syndrome Proposed by van der Burgt15 in Individuals with RRAS2 Mutations
| Individual | NS462 | NS833 | HU1 |
|---|---|---|---|
| Sex | female | female | male |
| Age at evaluation | 6 years | 4 years | 3 years |
| Initial diagnosis | NS or CFC | NS | undiagnosed |
| RRAS2 variant | c.70_78dup (p.Gly24_Gly26dup) | c.[216A>T;224T>G], p.[(Gln72His);(Phe75Cys)] | c.215A>T (p.Gln72Leu) |
| Facial dysmorphology | typical | typical | suggestive |
| Cardiac feature | pulmonic stenosis | − | dilated cardiomyopathy |
| Short stature (SD) | − (+0.2) | + (−2.4) | + (−5.9) |
| Pectus abnormalities | − | pectus excavatum | pectus excavatum |
| Family history | simplex | simplex | simplex |
| Intellectual disability (ID), cryptorchidism, and lymphatic dysplasia | − | mild ID | severe ID, cryptorchidism |
Subsequently, we sequenced all coding exons of RRAS2 in 191 molecularly undiagnosed individuals suspected of having NS or NS-related disorders, but we did not identify any additional rare (i.e., <0.01 MAF in population databases) nonsynonymous variants. In parallel, we performed WES on samples from an undiagnosed individual HU1 with severe failure to thrive and his healthy parents. We identified a de novo RRAS2 c.215A>T variant in HU1 and confirmed the segregation of the variant by Sanger sequencing. The c.215A>T (p.Gln72Leu) variant has been found in epithelial ovarian tumors,13 juvenile myelomonocytic leukemia,15 and other cancers (reported in COSMIC: COSG1128). Importantly, the codon Gln72 corresponds to codon Gln61 in KRAS, HRAS, and NRAS, a hotspot for mutations in several cancers (Figures 1A–1C).16 This variant has also been reported to induce activation of ERK17 and to elevate neoplastic transformation.13, 17 Individual HU1 was 3 years old at the time of last examination and was the third child of non-consanguineous parents. He had short stature, pectus excavatum, intellectual disability, and cryptorchidism (Supplemental Note; Tables 1 and S1). His facial appearance was not typical for NS at age 3 years, however, retrospective inspection of the images taken in his infancy revealed that he had a facial gestalt of NS. Therefore, he fulfilled the criteria for NS proposed by van der Burgt.18 In total, we identified rare RRAS2 variants in 3 of 219 individuals suspected of having NS or NS-related disorders. These data suggest that, similar to other recently discovered NS-associated genes, RRAS2 mutations likely account for a small fraction of NS.
Figure 1.
RRAS2 Mutations Identified in Individuals with Noonan Syndrome
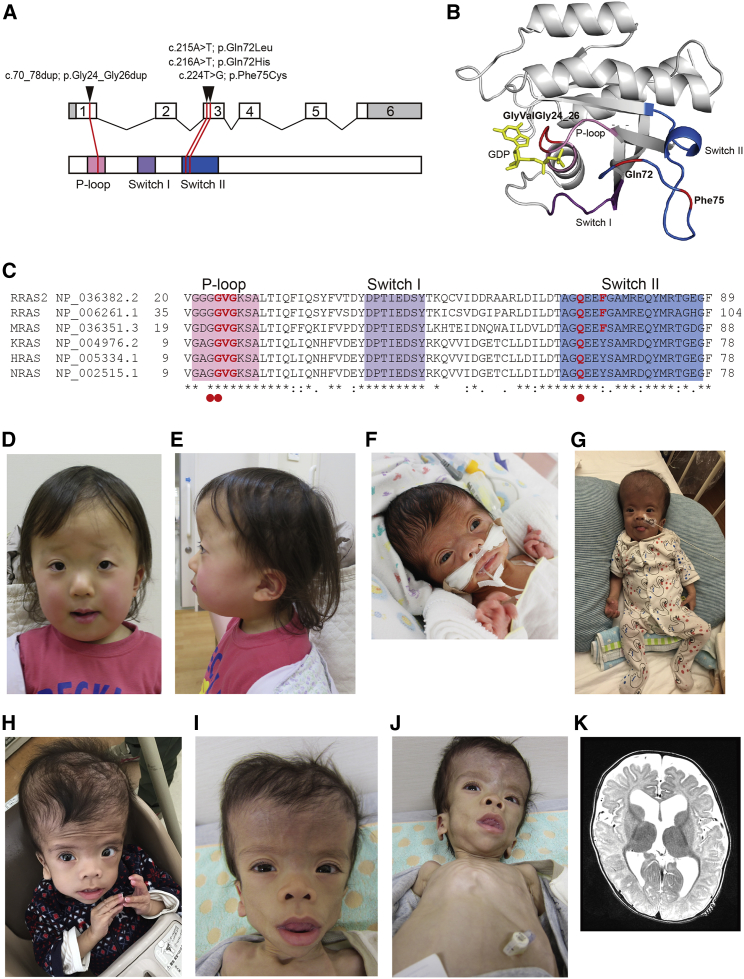
(A) Exon-intron structure of RRAS2 (upper) and functional domains of RRAS proteins (lower). RRAS2 variants in individuals with Noonan syndrome (NS) were located in phosphate-binding loop or switch II region.
(B) Crystal structure of human RRAS2. Phosphate-binding loop, switch I, switch II, GDP, and residues mutated in individuals with Noonan syndrome (NS) are highlighted in pink, purple, blue, yellow, and red, respectively.
(C) Partial amino acid sequence alignment of human RRAS2, RRAS, MRAS, KRAS, HRAS, and NRAS. High conservation of residues of Gly24-Gly26 and Gln72 through all paralogs are shown. Conservation of residues at Phe75 was limited in the RRAS subfamily. Red circles indicate hotspot residues mutated in various cancers in KRAS, HRAS, and NRAS.
(D and E) Photos of individual NS833. She had the definitive facial appearance of NS at 4 years of age.
(F–J) Photos of individual HU1 at (F) neonatal period, (G) 7 months, (H) 22 months, and (I and J) 3 years of age.
(K) Axial slice of brain magnetic resonance image of HU1 taken at 6 months of age shows ventriculomegaly and widened subarachnoid spaces in the frontal and temporal lobes.
Individual NS462, who harbored RRAS2 c.70_78dup, was 6 years old at her last clinical examination; she is the second child of healthy non-consanguineous parents. She presented with macrocephaly, pulmonic stenosis, hemangiomas, mild myopia, and left hearing impairment (Tables 1 and S1). Microarray analysis did not reveal gross duplication or deletion. Her initial diagnosis was CFCS or NS. RRAS2 c.70_78dup, which encodes p.Gly24_Gly26dup, maps to a phosphate-binding loop (P loop) in a domain that is conserved across the RAS family (Figures 1A–1C). This duplication has already been identified in a human uterine leiomyosarcoma cell line, SK-UT-l,19 and it is known to activate ERK20 and its transforming activity.19 We also sequenced DNA from the individual’s hair and fingernails and confirmed the presence of RRAS2 c.70_78dup, suggesting that the variation was not a somatic mutation in peripheral blood (data not shown).
Individual NS833 was 4 years old at the time of last examination, and she is the first child of healthy non-consanguineous parents. She presented with macrocephaly with enlarged ventricles, short stature (−2.4 SD), pectus excavatum, skin manifestations, and mild intellectual disability (Figures 1D and 1E; Tables 1 and S1). She was diagnosed with NS at 2 years 4 months. WES detected two RRAS2 variants, c.216A>T and c.224T>G. Sanger sequencing confirmed the presence of c.216A>T and c.224T>G in the same allele in NS833 (Figure S1). The c.216A>T (p.Gln72His) variant has been found in endometrioid carcinoma (reported in COSMIC: COSG1128) and results in an amino acid change at the same residue as p.Gln72Leu, which activates ERK. In contrast, c.224T>G (p.Phe75Cys) has not been reported thus far. In this context, we presumed that aberrant activation of the RAS/MAPK pathway underlies NS of the individuals with RRAS2 variants.
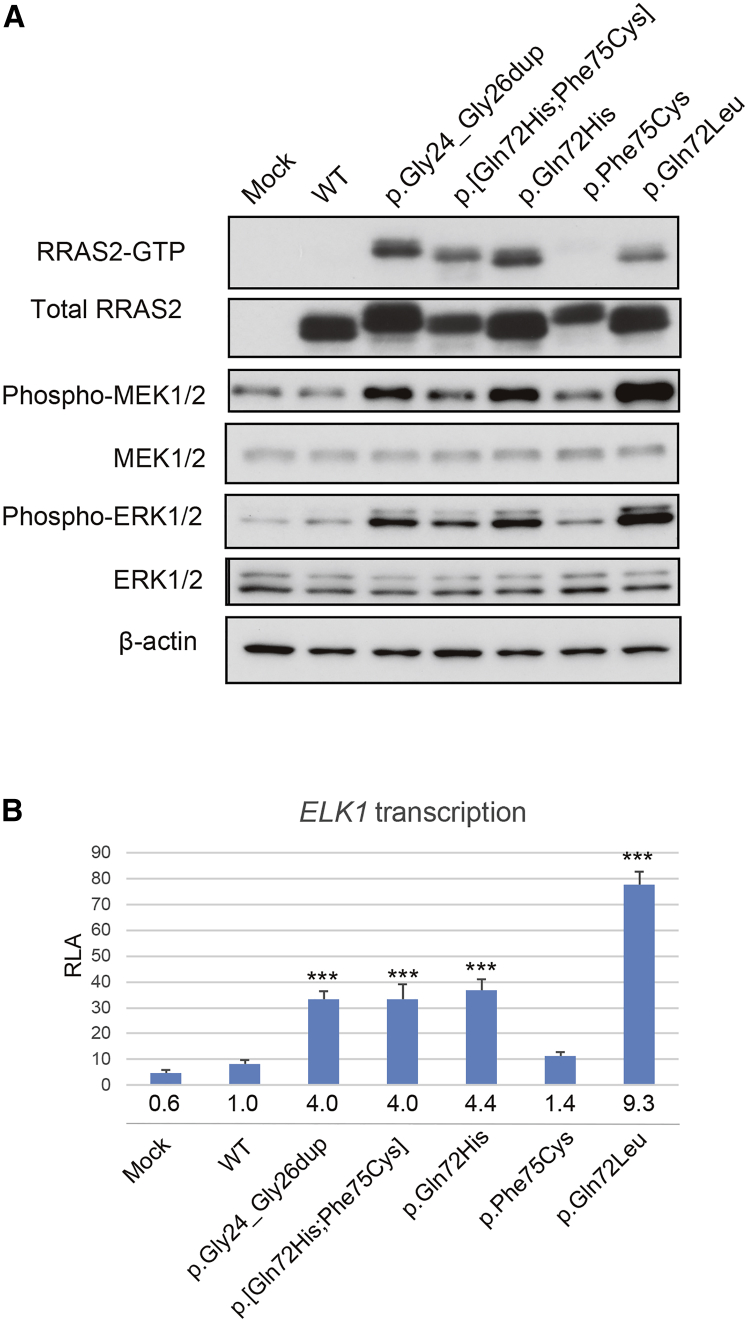
To test the pathogenicity of the discovered RRAS2 mutations, we performed pull-down assays with the Raf-Ras-binding domain (RBD, residues 1–149) of RAF1.21 We transfected a RRAS2 wild-type (WT) or mutant cDNAs in HEK293 cells and pulled-down activated RAS. The RRAS2 levels in cells expressing p.[Gln72His;Phe75Cys] and p.Phe75Cys were decreased compared to WT (consistent results across three independent experiments). Furthermore, compared to WT RRAS2 we observed increased binding of activated RAS for each of RRAS2 p.Gly24_Gly26dup, p.[Gln72His;Phe75Cys], p.Gln72His, and p.Gln72Leu but not p.Phe75Cys alone, suggesting that the first four alleles are pathogenic but that the fifth is benign (Figure 2A). Next, we conducted immunoblotting of the cell lysates expressing WT or mutant RRAS2 using anti-phospho-MEK1/2 and phospho-ERK1/2 antibodies. We observed increased levels of phospho-MEK1/2 and phospho-ERK1/2 in the cells expressing RRAS2 p.Gly24_Gly26dup, p.[Gln72His;Phe75Cys], p.Gln72His, and p.Gln72Leu. These results suggest that p.Gly24_Gly26dup, p.[Gln72His;Phe75Cys], p.Gln72His, and p.Gln72Leu mutants had increased affinity toward RAF and activate MEK/ERK.
Figure 2.
Functional Assays of RRAS2 Variants
(A) Representative immunoblots of three independent experiments. HEK293 cells transfected with WT or mutant RRAS2 constructs or empty vector (Mock) were used for pull-down assays and immunoblotting. RRAS2-guanosine triphosphate (GTP) which was pulled down using RAF1-RBD agarose, total RRAS2, phospho-MEK1/2, total MEK1/2, phospho-ERK1/2, total ERK1/2, and β-actin as a loading control were shown. WT, wild-type.
(B) Stimulation of ELK transcription by RRAS2 mutants. ELK-GAL4 and GAL4-luciferase trans-reporter vectors were transiently co-transfected with RRAS2 constructs into unstimulated HEK293 cells. Relative luciferase activity (RLA) was normalized to the activity of a co-transfected control vector (phRLnull-luc) expressing Renilla reniformis luciferase. Folds under each bar were calculated as a multiple of WT. Data are presented as mean ± SD; n = 3 per group. WT, wild-type. ∗∗∗p < 0.001 compared with WT.
Our pull-down assays and immunoblotting data suggested downstream activation of the RAS/MAPK pathway. To confirm these findings, we performed reporter assays using ELK-1, a transcriptional factor downstream of RAS/MAPK, whose transactivation in cells expressing RASopathy-gene mutations is known to increase.5 We transfected HEK293 cells with WT or mutant RRAS2 expression constructs, a pFR-luc trans-reporter vector, a pFA2-ELK1 vector, and a phRLnull-luc vector and measured their relative luciferase activity (RLA). Consistent with our earlier data, we observed a significant increase in RLA in cells transfected with RRAS2 p.Gly24_Gly26dup, p.[Gln72His;Phe75Cys], p.Gln72His, and p.Gln72Leu but not in cells transfected with p.Phe75Cys alone (Figure 2B). These results suggest that RRAS2 p.Gly24_Gly26dup, p.[Gln72His;Phe75Cys], p.Gln72His, and p.Gln72Leu as well as other RASopathy mutations stimulate the signaling pathway leading to ELK1 activation.
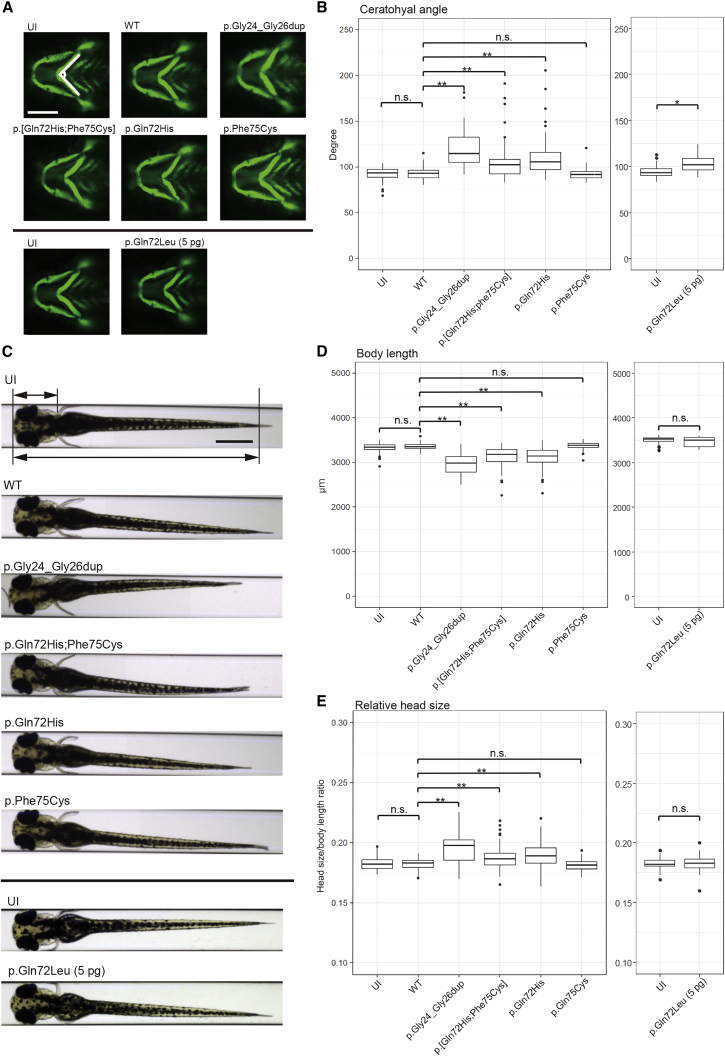
Our in vitro data suggest that aberrant activation of RAS/MAPK are likely drivers of the pathology seen in our NS-affected case subjects. To test this model in vivo, we turned to zebrafish, a useful model for testing defective RAS signaling,22, 23, 24 whose genome also contains a rras2 ortholog (96% identical; 98% similar for human versus zebrafish protein). We took advantage of the fact that quantitative measurements of the craniofacial skeleton demarcated by a transgene expressing GFP under the col1a1 promoter Tg(−1.4col1a1:egfp) are a useful method for assessing the effect of alleles that impact craniofacial development.25 Given the in vitro data, we asked whether expression of each of the human RRAS2 mutant mRNAs might recapitulate aspects of pathology observed in affected individuals. To test this possibility, we injected WT mRNA of human RRAS2 or mRNA encoding each of the variants (or combination thereof) into transgenic embryos. We noticed that injection of 25 pg of p.Gln72Leu mRNA was lethal, but injections of the same dose of WT or the other mutant mRNAs represented the highest non-lethal dose. Therefore, we started with embryos which were injected with 25 pg of WT or mutant mRNA except for p.Gln72Leu. At 11 h post fertilization (hpf), we observed significant elongation of the yolk in RRAS2 p.Gly24_Gly26dup, p.[Gln72His;Phe75Cys], and p.Gln72His embryos compared to WT (Figure S2); differences between p.Phe75Cys and WT were not significant. We then raised embryos to 3 days post fertilization (dpf) and measured the ceratohyal angle, a mandibular structure which reflects the width and bluntness of the head.26 The ceratohyal angle was significantly increased in larval batches injected with mRNA encoding p.Gly24_Gly26dup, p.[Gln72His;Phe75Cys], and p.Gln72His (Figures 3A and 3B) but was indistinguishable from WT upon injection of p.Phe75Cys mRNA. These results were consistent with our in vitro data and prior genetic observations in tumors, all of which point toward the mutants driving hyperactivation of MAPK signaling.
Figure 3.
Morphology of Zebrafish Larvae Injected with Wild-Type (WT) or Mutant RRAS2 mRNA at 3 Days Post Fertilization (dpf)
(A) Tg(−1.4col1a1:egfp) transgenic embryos, in which cartilage cells are marked by EGFP, were injected at the 1- to 4-cell stage with RNA encoding RRAS2-WT or variants identified in affected individuals. The angle of the ceratohyal cartilage was measured at 3 dpf. Representative images of an uninjected control (UI) and RRAS2-variant mRNA injected larvae are depicted. Scale bar: 200 μm.
(B) Quantification of the ceratohyal angle. Left: we injected 25 pg mRNA encoding each indicated RRAS2 condition. n = 61–80 embryos per batch. n.s., not significant; ∗∗p < 0.001 by Dunnett’s test. Right: 5 pg of p.Gln72Leu endoding mRNA was injected. n = 39–56 embryos per batch. ∗p < 0.01 by Student’s t test. The thick line in the box represents median value; the bottom and top lines of the box represent first and third quartiles, respectively; the whiskers extend from the hinge to the lowest or highest value that is within 1.5-fold of interquartile range from the hinge; the filled circles are outliers.
(C) The body (lower arrow) and head (upper arrow) length were measured at 3 dpf. Representative images of an uninjected control (UI) and RRAS2-mRNA injected larvae are shown. Scale bar: 500 μm.
(D and E) Quantification of body length (D) and relative head size, which was the value of the head length divided by the value of the body length (E). Left: we injected 25 pg of each RRAS2 mRNA. n = 61–82 embryos per batch. n.s., not significant; ∗∗p < 0.001 by Dunnett’s test. Right: 5 pg of p.Gln72Leu encoding RNA was injected. n = 36–53 embryos per batch. n.s., not significant by Student’s t test. The thick line in the box represents median value; the bottom and top lines of the box represent first and third quartiles, respectively; the whiskers extend from the hinge to the lowest or highest value that is within 1.5-fold of interquartile range from the hinge; the filled circles are outliers.
As a second test, we also measured body length and relative head length (head length divided by body length) at 3 dpf. Expression of RRAS2 p.Gly24_Gly26dup, p.[Gln72His;Phe75Cys], and p.Gln72His led to significant decreases in body length compared to WT RRAS2 (Figures 3C and 3D). Relative head length in RRAS2 p.Gly24_Gly26dup, p.[Gln72His;Phe75Cys], and p.Gln72His larvae were also greater than in WT larvae (Figure 3E). Finally, we also observed an increased incidence of pericardial effusion in RRAS2 p.Gly24_Gly26dup, p.[Gln72His;Phe75Cys], and p.Gln72His larvae (Figure S3) that might underscore a structural heart defect.
To test the p.Gln72Leu variant, we established the highest tolerated dose of mRNA and injected it into embryos to observe developmental defects in live larvae. The embryos injected with 5 pg of p.Gln72Leu mRNA were viable and showed significantly increased ceratohyal angles compared to the uninjected controls (Figure 3B, right), although the body length and relative head size of p.Gln72Leu-injected larvae were not different from those of uninjected controls (right panels of Figures 3D and 3E). Together, these data suggest that expression of RRAS2 encoding p.Gly24_Gly26dup, p.[Gln72His;Phe75Cys], p.Gln72His, and p.Gln72Leu induced craniofacial patterning defects in zebrafish larvae that correspond to symptoms relevant to our affected individuals and further support our in vitro studies.
We describe de novo RRAS2 mutations identified in three individuals showing a NS phenotype. All three individuals with RRAS2 mutations had macrocephaly and typical or suggestive facial appearance of NS. Regarding the other features, they had clinical manifestations sufficient to fulfill the diagnostic criteria by van der Burgt, though they were not common among all individuals. HU1, who had more severe phenotype such as failure to thrive and severe developmental delay, harbored a p.Gln72Leu variant. This variant elicited a potent effect in our in vitro and in vivo assays, suggesting a possible genotype-phenotype correlation. However, the identification of a larger RRAS2 allelic series will be required to enable establishment of such correlations.
Individual NS833 has two de novo variants, p.Gln72His and p.Phe75Cys, in cis. p.Gln72His has been found in endometrioid carcinoma and is likely pathogenic because of its homologous position to the hotspot Gln61 in KRAS, NRAS, and HRAS.16 Our functional data agree with that prediction. In contrast, the second variant is likely benign. Multiple in silico analyses predicted that p.Phe75Cys exerts an adverse effect (Table S2), which is only partially consistent with our functional analyses wherein we observe decreased protein levels compared to WT. It is formally possible that this variant is detrimental to protein function but not in a fashion that would give rise to NS-associated pathologies, especially since the allele is absent from population databases.
Our zebrafish model expressing RRAS2 mutations showed elongated shapes of developing embryos, reduced body length, macrocephaly, and craniofacial defects. Injection of the same dose of p.Gln72Leu mRNA as those of the other variants induced embryonic death, suggesting stronger impact of this mutation. We observed significant differences in ceratohyal angle in larvae with lower dose injection of p.Gln72Leu mRNA, but not in body length and relative head size. Increases of ceratohyal angle may be a sensitive marker in zebrafish models of RASopathies. Zebrafish models expressing BRAF, HRAS (MIM: 190020), and PTPN11 mutations were approved for functional studies by ClinGen’s RASopathy Expert Panel.27 Notably, craniofacial defects, including wide heads and/or hypoplasia of the ventral side of the head, have been observed in zebrafish models expressing PTPN11,28 HRAS,29 NRAS,26 RIT1,10 and zebrafish a2ml130 mutations. Furthermore, elongation of developing embryos at 10–12 hpf has been reported in zebrafish models of RASopathies expressing BRAF, MAP2K1 (MIM: 176872), MAP2K2 (MIM: 601263),31 RIT1,10 and NRAS.26 In addition, reduced body length has been seen in zebrafish models expressing PTPN1128 and HRAS29 mutations. Such similarities among RASopathy models suggest common underlying mechanisms led by the dysregulation of the RAS/MAPK pathway.
In summary, we identified four de novo RRAS2 variants in three individuals with NS, three of which are likely drivers of pathology through the hyperactivation of the RAS/MAPK pathway. Together with an accompanying study showing RRAS2 mutations in individuals with NS or NS-like phenotype,32 these findings broaden our understanding of roles of RRAS2 in human development, expanding the mutational landscape of NS and related disorders. Our work also highlights how, given the rarity of the remaining genes for this group of disorders, the combination of genetic, in vitro, and in vivo studies might be necessary to establish the identity of causal loci.
Declaration of Interests
N.K. is a paid consultant and holds founder stock in Rescindo Therapeutics. The other authors declare no competing interests.
Acknowledgments
The authors thank the individuals who participated in this study and their families. We also thank Drs. Kenji Kurosawa, Seiji Mizuno, Hiroshi Kawame, Tsutomu Ogata, Yoichi Matsubara, and other physicians for sending samples of patients, and Ikumi Umeki, Yoko Tateda, Kumi Kato, Miyuki Tsuda, Mami Kikuchi, Makiko Nakagawa, and Kiyotaka Kuroda for their technical assistance. We also acknowledge the technical support of the Biomedical Research Core of Tohoku University Graduate School of Medicine. This work was supported by US National Institutes of Health grant MH106826 to E.E.D., AMED under grants to Y.A. (JP18ek0109241 and JP18ek0109278) and N.M. (JP18ek0109280, JP18dm0107090, JP18ek0109301, JP18ek0109348, and JP18kk020501), and Japan Society for the Promotion of Science (JSPS) KAKENHI Grant Numbers JP16K15522 to Y.A., JP17H01539 to N.M., JP17H06994 to A.F., and JP15KK0293 to T.N.
Published: May 23, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.04.014.
Contributor Information
Tetsuya Niihori, Email: tniihori@med.tohoku.ac.jp.
Nicholas Katsanis, Email: katsanis@cellbio.duke.edu.
Web Resources
1000 Genomes, http://www.internationalgenome.org/
COSMIC, https://cancer.sanger.ac.uk/cosmic/gene/analysis?ln=RRAS2
ExAC Browser, http://exac.broadinstitute.org/
Human Genetic Variation Database (HGVD), http://www.genome.med.kyoto-u.ac.jp/SnpDB/
MutationTaster, http://www.mutationtaster.org/
OMIM, http://www.omim.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
PROVEAN, http://provean.jcvi.org
R statistical software, https://www.r-project.org/
Supplemental Data
References
- 1.Aoki Y., Niihori T., Inoue S., Matsubara Y. Recent advances in RASopathies. J. Hum. Genet. 2016;61:33–39. doi: 10.1038/jhg.2015.114. [DOI] [PubMed] [Google Scholar]; Aoki, Y., Niihori, T., Inoue, S., and Matsubara, Y. (2016). Recent advances in RASopathies. J. Hum. Genet. 61, 33-39. [DOI] [PubMed]
- 2.Johnston J.J., van der Smagt J.J., Rosenfeld J.A., Pagnamenta A.T., Alswaid A., Baker E.H., Blair E., Borck G., Brinkmann J., Craigen W., Members of the Undiagnosed Diseases Network Autosomal recessive Noonan syndrome associated with biallelic LZTR1 variants. Genet. Med. 2018;20:1175–1185. doi: 10.1038/gim.2017.249. [DOI] [PMC free article] [PubMed] [Google Scholar]; Johnston, J.J., van der Smagt, J.J., Rosenfeld, J.A., Pagnamenta, A.T., Alswaid, A., Baker, E.H., Blair, E., Borck, G., Brinkmann, J., Craigen, W., et al.; Members of the Undiagnosed Diseases Network (2018). Autosomal recessive Noonan syndrome associated with biallelic LZTR1 variants. Genet. Med. 20, 1175-1185. [DOI] [PMC free article] [PubMed]
- 3.Tidyman W.E., Rauen K.A. The RASopathies: developmental syndromes of Ras/MAPK pathway dysregulation. Curr. Opin. Genet. Dev. 2009;19:230–236. doi: 10.1016/j.gde.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tidyman, W.E., and Rauen, K.A. (2009). The RASopathies: developmental syndromes of Ras/MAPK pathway dysregulation. Curr. Opin. Genet. Dev. 19, 230-236. [DOI] [PMC free article] [PubMed]
- 4.Aoki Y., Niihori T., Narumi Y., Kure S., Matsubara Y. The RAS/MAPK syndromes: novel roles of the RAS pathway in human genetic disorders. Hum. Mutat. 2008;29:992–1006. doi: 10.1002/humu.20748. [DOI] [PubMed] [Google Scholar]; Aoki, Y., Niihori, T., Narumi, Y., Kure, S., and Matsubara, Y. (2008). The RAS/MAPK syndromes: novel roles of the RAS pathway in human genetic disorders. Hum. Mutat. 29, 992-1006. [DOI] [PubMed]
- 5.Aoki Y., Niihori T., Kawame H., Kurosawa K., Ohashi H., Tanaka Y., Filocamo M., Kato K., Suzuki Y., Kure S., Matsubara Y. Germline mutations in HRAS proto-oncogene cause Costello syndrome. Nat. Genet. 2005;37:1038–1040. doi: 10.1038/ng1641. [DOI] [PubMed] [Google Scholar]; Aoki, Y., Niihori, T., Kawame, H., Kurosawa, K., Ohashi, H., Tanaka, Y., Filocamo, M., Kato, K., Suzuki, Y., Kure, S., and Matsubara, Y. (2005). Germline mutations in HRAS proto-oncogene cause Costello syndrome. Nat. Genet. 37, 1038-1040. [DOI] [PubMed]
- 6.Niihori T., Aoki Y., Narumi Y., Neri G., Cavé H., Verloes A., Okamoto N., Hennekam R.C.M., Gillessen-Kaesbach G., Wieczorek D. Germline KRAS and BRAF mutations in cardio-facio-cutaneous syndrome. Nat. Genet. 2006;38:294–296. doi: 10.1038/ng1749. [DOI] [PubMed] [Google Scholar]; Niihori, T., Aoki, Y., Narumi, Y., Neri, G., Cave, H., Verloes, A., Okamoto, N., Hennekam, R.C.M., Gillessen-Kaesbach, G., Wieczorek, D., et al. (2006). Germline KRAS and BRAF mutations in cardio-facio-cutaneous syndrome. Nat. Genet. 38, 294-296. [DOI] [PubMed]
- 7.Schubbert S., Zenker M., Rowe S.L., Böll S., Klein C., Bollag G., van der Burgt I., Musante L., Kalscheuer V., Wehner L.E. Germline KRAS mutations cause Noonan syndrome. Nat. Genet. 2006;38:331–336. doi: 10.1038/ng1748. [DOI] [PubMed] [Google Scholar]; Schubbert, S., Zenker, M., Rowe, S.L., Boll, S., Klein, C., Bollag, G., van der Burgt, I., Musante, L., Kalscheuer, V., Wehner, L.E., et al. (2006). Germline KRAS mutations cause Noonan syndrome. Nat. Genet. 38, 331-336. [DOI] [PubMed]
- 8.Rodriguez-Viciana P., Tetsu O., Tidyman W.E., Estep A.L., Conger B.A., Cruz M.S., McCormick F., Rauen K.A. Germline mutations in genes within the MAPK pathway cause cardio-facio-cutaneous syndrome. Science. 2006;311:1287–1290. doi: 10.1126/science.1124642. [DOI] [PubMed] [Google Scholar]; Rodriguez-Viciana, P., Tetsu, O., Tidyman, W.E., Estep, A.L., Conger, B.A., Cruz, M.S., McCormick, F., and Rauen, K.A. (2006). Germline mutations in genes within the MAPK pathway cause cardio-facio-cutaneous syndrome. Science 311, 1287-1290. [DOI] [PubMed]
- 9.Tartaglia M., Mehler E.L., Goldberg R., Zampino G., Brunner H.G., Kremer H., van der Burgt I., Crosby A.H., Ion A., Jeffery S. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat. Genet. 2001;29:465–468. doi: 10.1038/ng772. [DOI] [PubMed] [Google Scholar]; Tartaglia, M., Mehler, E.L., Goldberg, R., Zampino, G., Brunner, H.G., Kremer, H., van der Burgt, I., Crosby, A.H., Ion, A., Jeffery, S., et al. (2001). Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat. Genet. 29, 465-468. [DOI] [PubMed]
- 10.Aoki Y., Niihori T., Banjo T., Okamoto N., Mizuno S., Kurosawa K., Ogata T., Takada F., Yano M., Ando T. Gain-of-function mutations in RIT1 cause Noonan syndrome, a RAS/MAPK pathway syndrome. Am. J. Hum. Genet. 2013;93:173–180. doi: 10.1016/j.ajhg.2013.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]; Aoki, Y., Niihori, T., Banjo, T., Okamoto, N., Mizuno, S., Kurosawa, K., Ogata, T., Takada, F., Yano, M., Ando, T., et al. (2013). Gain-of-function mutations in RIT1 cause Noonan syndrome, a RAS/MAPK pathway syndrome. Am. J. Hum. Genet. 93, 173-180. [DOI] [PMC free article] [PubMed]
- 11.Gripp K.W., Aldinger K.A., Bennett J.T., Baker L., Tusi J., Powell-Hamilton N., Stabley D., Sol-Church K., Timms A.E., Dobyns W.B. A novel rasopathy caused by recurrent de novo missense mutations in PPP1CB closely resembles Noonan syndrome with loose anagen hair. Am. J. Med. Genet. A. 2016;170:2237–2247. doi: 10.1002/ajmg.a.37781. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gripp, K.W., Aldinger, K.A., Bennett, J.T., Baker, L., Tusi, J., Powell-Hamilton, N., Stabley, D., Sol-Church, K., Timms, A.E., and Dobyns, W.B. (2016). A novel rasopathy caused by recurrent de novo missense mutations in PPP1CB closely resembles Noonan syndrome with loose anagen hair. Am. J. Med. Genet. A. 170, 2237-2247. [DOI] [PMC free article] [PubMed]
- 12.Higgins E.M., Bos J.M., Mason-Suares H., Tester D.J., Ackerman J.P., MacRae C.A., Sol-Church K., Gripp K.W., Urrutia R., Ackerman M.J. Elucidation of MRAS-mediated Noonan syndrome with cardiac hypertrophy. JCI Insight. 2017;2:e91225. doi: 10.1172/jci.insight.91225. [DOI] [PMC free article] [PubMed] [Google Scholar]; Higgins, E.M., Bos, J.M., Mason-Suares, H., Tester, D.J., Ackerman, J.P., MacRae, C.A., Sol-Church, K., Gripp, K.W., Urrutia, R., and Ackerman, M.J. (2017). Elucidation of MRAS-mediated Noonan syndrome with cardiac hypertrophy. JCI Insight 2, e91225. [DOI] [PMC free article] [PubMed]
- 13.Chan A.M., Miki T., Meyers K.A., Aaronson S.A. A human oncogene of the RAS superfamily unmasked by expression cDNA cloning. Proc. Natl. Acad. Sci. USA. 1994;91:7558–7562. doi: 10.1073/pnas.91.16.7558. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chan, A.M., Miki, T., Meyers, K.A., and Aaronson, S.A. (1994). A human oncogene of the RAS superfamily unmasked by expression cDNA cloning. Proc. Natl. Acad. Sci. USA 91, 7558-7562. [DOI] [PMC free article] [PubMed]
- 14.Graham S.M., Vojtek A.B., Huff S.Y., Cox A.D., Clark G.J., Cooper J.A., Der C.J. TC21 causes transformation by Raf-independent signaling pathways. Mol. Cell. Biol. 1996;16:6132–6140. doi: 10.1128/mcb.16.11.6132. [DOI] [PMC free article] [PubMed] [Google Scholar]; Graham, S.M., Vojtek, A.B., Huff, S.Y., Cox, A.D., Clark, G.J., Cooper, J.A., and Der, C.J. (1996). TC21 causes transformation by Raf-independent signaling pathways. Mol. Cell. Biol. 16, 6132-6140. [DOI] [PMC free article] [PubMed]
- 15.Stieglitz E., Taylor-Weiner A.N., Chang T.Y., Gelston L.C., Wang Y.D., Mazor T., Esquivel E., Yu A., Seepo S., Olsen S. The genomic landscape of juvenile myelomonocytic leukemia. Nat. Genet. 2015;47:1326–1333. doi: 10.1038/ng.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]; Stieglitz, E., Taylor-Weiner, A.N., Chang, T.Y., Gelston, L.C., Wang, Y.D., Mazor, T., Esquivel, E., Yu, A., Seepo, S., Olsen, S., et al. (2015). The genomic landscape of juvenile myelomonocytic leukemia. Nat. Genet. 47, 1326-1333. [DOI] [PMC free article] [PubMed]
- 16.Prior I.A., Lewis P.D., Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012;72:2457–2467. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]; Prior, I.A., Lewis, P.D., and Mattos, C. (2012). A comprehensive survey of Ras mutations in cancer. Cancer Res. 72, 2457-2467. [DOI] [PMC free article] [PubMed]
- 17.Graham S.M., Cox A.D., Drivas G., Rush M.G., D’Eustachio P., Der C.J. Aberrant function of the Ras-related protein TC21/R-Ras2 triggers malignant transformation. Mol. Cell. Biol. 1994;14:4108–4115. doi: 10.1128/mcb.14.6.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]; Graham, S.M., Cox, A.D., Drivas, G., Rush, M.G., D’Eustachio, P., and Der, C.J. (1994). Aberrant function of the Ras-related protein TC21/R-Ras2 triggers malignant transformation. Mol. Cell. Biol. 14, 4108-4115. [DOI] [PMC free article] [PubMed]
- 18.van der Burgt I. Noonan syndrome. Orphanet J. Rare Dis. 2007;2:4. doi: 10.1186/1750-1172-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]; van der Burgt, I. (2007). Noonan syndrome. Orphanet J. Rare Dis. 2, 4. [DOI] [PMC free article] [PubMed]
- 19.Huang Y., Saez R., Chao L., Santos E., Aaronson S.A., Chan A.M. A novel insertional mutation in the TC21 gene activates its transforming activity in a human leiomyosarcoma cell line. Oncogene. 1995;11:1255–1260. [PubMed] [Google Scholar]; Huang, Y., Saez, R., Chao, L., Santos, E., Aaronson, S.A., and Chan, A.M. (1995). A novel insertional mutation in the TC21 gene activates its transforming activity in a human leiomyosarcoma cell line. Oncogene 11, 1255-1260. [PubMed]
- 20.Rong R., He Q., Liu Y., Sheikh M.S., Huang Y. TC21 mediates transformation and cell survival via activation of phosphatidylinositol 3-kinase/Akt and NF-kappaB signaling pathway. Oncogene. 2002;21:1062–1070. doi: 10.1038/sj.onc.1205154. [DOI] [PubMed] [Google Scholar]; Rong, R., He, Q., Liu, Y., Sheikh, M.S., and Huang, Y. (2002). TC21 mediates transformation and cell survival via activation of phosphatidylinositol 3-kinase/Akt and NF-kappaB signaling pathway. Oncogene 21, 1062-1070. [DOI] [PubMed]
- 21.Inoue S., Moriya M., Watanabe Y., Miyagawa-Tomita S., Niihori T., Oba D., Ono M., Kure S., Ogura T., Matsubara Y., Aoki Y. New BRAF knockin mice provide a pathogenetic mechanism of developmental defects and a therapeutic approach in cardio-facio-cutaneous syndrome. Hum. Mol. Genet. 2014;23:6553–6566. doi: 10.1093/hmg/ddu376. [DOI] [PubMed] [Google Scholar]; Inoue, S., Moriya, M., Watanabe, Y., Miyagawa-Tomita, S., Niihori, T., Oba, D., Ono, M., Kure, S., Ogura, T., Matsubara, Y., and Aoki, Y. (2014). New BRAF knockin mice provide a pathogenetic mechanism of developmental defects and a therapeutic approach in cardio-facio-cutaneous syndrome. Hum. Mol. Genet. 23, 6553-6566. [DOI] [PubMed]
- 22.Tsai I.C., McKnight K., McKinstry S.U., Maynard A.T., Tan P.L., Golzio C., White C.T., Price D.J., Davis E.E., Amrine-Madsen H., Katsanis N. Small molecule inhibition of RAS/MAPK signaling ameliorates developmental pathologies of Kabuki Syndrome. Sci. Rep. 2018;8:10779. doi: 10.1038/s41598-018-28709-y. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tsai, I.C., McKnight, K., McKinstry, S.U., Maynard, A.T., Tan, P.L., Golzio, C., White, C.T., Price, D.J., Davis, E.E., Amrine-Madsen, H., and Katsanis, N. (2018). Small molecule inhibition of RAS/MAPK signaling ameliorates developmental pathologies of Kabuki Syndrome. Sci. Rep. 8, 10779. [DOI] [PMC free article] [PubMed]
- 23.Bögershausen N., Tsai I.C., Pohl E., Kiper P.O., Beleggia F., Percin E.F., Keupp K., Matchan A., Milz E., Alanay Y. RAP1-mediated MEK/ERK pathway defects in Kabuki syndrome. J. Clin. Invest. 2015;125:3585–3599. doi: 10.1172/JCI80102. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bogershausen, N., Tsai, I.C., Pohl, E., Kiper, P.O., Beleggia, F., Percin, E.F., Keupp, K., Matchan, A., Milz, E., Alanay, Y., et al. (2015). RAP1-mediated MEK/ERK pathway defects in Kabuki syndrome. J. Clin. Invest. 125, 3585-3599. [DOI] [PMC free article] [PubMed]
- 24.Magini P., Pippucci T., Tsai I.C., Coppola S., Stellacci E., Bartoletti-Stella A., Turchetti D., Graziano C., Cenacchi G., Neri I. A mutation in PAK3 with a dual molecular effect deregulates the RAS/MAPK pathway and drives an X-linked syndromic phenotype. Hum. Mol. Genet. 2014;23:3607–3617. doi: 10.1093/hmg/ddu070. [DOI] [PubMed] [Google Scholar]; Magini, P., Pippucci, T., Tsai, I.C., Coppola, S., Stellacci, E., Bartoletti-Stella, A., Turchetti, D., Graziano, C., Cenacchi, G., Neri, I., et al. (2014). A mutation in PAK3 with a dual molecular effect deregulates the RAS/MAPK pathway and drives an X-linked syndromic phenotype. Hum. Mol. Genet. 23, 3607-3617. [DOI] [PubMed]
- 25.Hutson M.R., Keyte A.L., Hernández-Morales M., Gibbs E., Kupchinsky Z.A., Argyridis I., Erwin K.N., Pegram K., Kneifel M., Rosenberg P.B. Temperature-activated ion channels in neural crest cells confer maternal fever-associated birth defects. Sci. Signal. 2017;10:10. doi: 10.1126/scisignal.aal4055. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hutson, M.R., Keyte, A.L., Hernandez-Morales, M., Gibbs, E., Kupchinsky, Z.A., Argyridis, I., Erwin, K.N., Pegram, K., Kneifel, M., Rosenberg, P.B., et al. (2017). Temperature-activated ion channels in neural crest cells confer maternal fever-associated birth defects. Sci. Signal. 10, 10. [DOI] [PMC free article] [PubMed]
- 26.Runtuwene V., van Eekelen M., Overvoorde J., Rehmann H., Yntema H.G., Nillesen W.M., van Haeringen A., van der Burgt I., Burgering B., den Hertog J. Noonan syndrome gain-of-function mutations in NRAS cause zebrafish gastrulation defects. Dis. Model. Mech. 2011;4:393–399. doi: 10.1242/dmm.007112. [DOI] [PMC free article] [PubMed] [Google Scholar]; Runtuwene, V., van Eekelen, M., Overvoorde, J., Rehmann, H., Yntema, H.G., Nillesen, W.M., van Haeringen, A., van der Burgt, I., Burgering, B., and den Hertog, J. (2011). Noonan syndrome gain-of-function mutations in NRAS cause zebrafish gastrulation defects. Dis. Model. Mech. 4, 393-399. [DOI] [PMC free article] [PubMed]
- 27.Gelb B.D., Cavé H., Dillon M.W., Gripp K.W., Lee J.A., Mason-Suares H., Rauen K.A., Williams B., Zenker M., Vincent L.M., ClinGen RASopathy Working Group ClinGen’s RASopathy Expert Panel consensus methods for variant interpretation. Genet. Med. 2018;20:1334–1345. doi: 10.1038/gim.2018.3. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gelb, B.D., Cave, H., Dillon, M.W., Gripp, K.W., Lee, J.A., Mason-Suares, H., Rauen, K.A., Williams, B., Zenker, M., and Vincent, L.M.; ClinGen RASopathy Working Group (2018). ClinGen’s RASopathy Expert Panel consensus methods for variant interpretation. Genet. Med. 20, 1334-1345. [DOI] [PMC free article] [PubMed]
- 28.Jopling C., van Geemen D., den Hertog J. Shp2 knockdown and Noonan/LEOPARD mutant Shp2-induced gastrulation defects. PLoS Genet. 2007;3:e225. doi: 10.1371/journal.pgen.0030225. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jopling, C., van Geemen, D., and den Hertog, J. (2007). Shp2 knockdown and Noonan/LEOPARD mutant Shp2-induced gastrulation defects. PLoS Genet. 3, e225. [DOI] [PMC free article] [PubMed]
- 29.Santoriello C., Deflorian G., Pezzimenti F., Kawakami K., Lanfrancone L., d’Adda di Fagagna F., Mione M. Expression of H-RASV12 in a zebrafish model of Costello syndrome causes cellular senescence in adult proliferating cells. Dis. Model. Mech. 2009;2:56–67. doi: 10.1242/dmm.001016. [DOI] [PMC free article] [PubMed] [Google Scholar]; Santoriello, C., Deflorian, G., Pezzimenti, F., Kawakami, K., Lanfrancone, L., d’Adda di Fagagna, F., and Mione, M. (2009). Expression of H-RASV12 in a zebrafish model of Costello syndrome causes cellular senescence in adult proliferating cells. Dis. Model. Mech. 2, 56-67. [DOI] [PMC free article] [PubMed]
- 30.Vissers L.E., Bonetti M., Paardekooper Overman J., Nillesen W.M., Frints S.G., de Ligt J., Zampino G., Justino A., Machado J.C., Schepens M. Heterozygous germline mutations in A2ML1 are associated with a disorder clinically related to Noonan syndrome. Eur. J. Hum. Genet. 2015;23:317–324. doi: 10.1038/ejhg.2014.115. [DOI] [PMC free article] [PubMed] [Google Scholar]; Vissers, L.E., Bonetti, M., Paardekooper Overman, J., Nillesen, W.M., Frints, S.G., de Ligt, J., Zampino, G., Justino, A., Machado, J.C., Schepens, M., et al. (2015). Heterozygous germline mutations in A2ML1 are associated with a disorder clinically related to Noonan syndrome. Eur. J. Hum. Genet. 23, 317-324. [DOI] [PMC free article] [PubMed]
- 31.Anastasaki C., Estep A.L., Marais R., Rauen K.A., Patton E.E. Kinase-activating and kinase-impaired cardio-facio-cutaneous syndrome alleles have activity during zebrafish development and are sensitive to small molecule inhibitors. Hum. Mol. Genet. 2009;18:2543–2554. doi: 10.1093/hmg/ddp186. [DOI] [PMC free article] [PubMed] [Google Scholar]; Anastasaki, C., Estep, A.L., Marais, R., Rauen, K.A., and Patton, E.E. (2009). Kinase-activating and kinase-impaired cardio-facio-cutaneous syndrome alleles have activity during zebrafish development and are sensitive to small molecule inhibitors. Hum. Mol. Genet. 18, 2543-2554. [DOI] [PMC free article] [PubMed]
- 32.Capri Y., Flex E., Krumbach O.H.F., Carpentieri G., Ceccetti S., Lißewski C., Adariani S.R., Schanze D., Brinkmann J., Piard J. Activating mutations of RRAS2 are a rare cause of Noonan syndrome. Am. J. Hum. Genet. 2019;104:1223–1232. doi: 10.1016/j.ajhg.2019.04.013. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]; Capri, Y., Flex, E., Krumbach, O.H.F., Carpentieri, G., Ceccetti, S., Lißewski, C., Adariani, S.R., Schanze, D., Brinkmann, J., Piard, J., et al. (2019). Activating mutations of RRAS2 are a rare cause of Noonan syndrome. Am. J. Hum. Genet. 104, this issue. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.