Abstract
Background
Neonatal screening for Pompe disease is complicated by difficulties in predicting symptom onset in patients with the common c.-32-13T>G (IVS1) variant/null (i.e. fully deleterious) acid α-glucosidase (GAA) genotype. This splicing variant occurs in 90% of Caucasian late onset patients, and is associated with a broad range of symptom onset.
Methods
We analyzed a cohort of 143 compound heterozygous and 10 homozygous IVS1 patients, and we assessed ages at symptom onset, the presence of cis-acting single nucleotide variants (SNVs), and performed splicing analysis and enzyme activity assays.
Findings
In compound heterozygous IVS1 patients, the synonymous variant c.510C>T was uniquely present on the IVS1 allele in 9/33 (27%) patients with childhood onset, but was absent from 110 patients with onset in adulthood. GAA enzyme activity was lower in fibroblasts from patients who contained c.510C>T than it was in patients without c.510C>T. By reducing the extent of leaky wild-type splicing, c.510C>T modulated aberrant splicing caused by the IVS1 variant. The deleterious effect of c.510C>T was also found in muscle cells, the main target cells in Pompe disease. In homozygous IVS1 patients, the c.510C>T variant was absent in 4/4 (100%) asymptomatic individuals and present in 3/6 (50%) symptomatic patients. In cells from homozygous IVS1 patients, c.510C>T caused reduced leaky wild-type splicing.
Interpretation
c.510C>T is a genetic modifier in compound heterozygous and homozygous IVS1 patients. This finding is important for neonatal screening programs for Pompe disease.
Fund
This work was funded by grants from Sophia Children's Hospital Foundation (SSWO, grant S17–32) and Metakids (2016–063).
Keywords: Modifying factor, Pompe disease, Lysosomal storage disease, Pre-mRNA splicing, c.510C>T
Research in context.
Evidence before this study
Many reports from our own group as well as from many colleagues have been published on the clinical heterogeneity of patients with late onset Pompe disease that carry the common c.-32-13T>G (IVS1) variant in the acid α-glucosidase (GAA) gene. Onset of symptoms can vary from the age of 0 to >60 years, even for patients with identical GAA disease-associated variants. This has resulted in the hypothesis that modifying factors for Pompe disease exist.
Added value of this study
Our findings present the first genetic modifier identified in Pompe disease that can explain why certain compound heterozygous IVS1 patients develop symptoms early in life and why certain homozygous IVS1 patients develop symptoms at all. This is important for newborn screening programs, for genetic counseling, and for deciding when to start with enzyme replacement therapy treatment.
Implications of all the available evidence
We identified a variant that so far has often not been reported in standard DNA diagnostics. However, it is a simple DNA test that can be included in standard diagnostics. This is important to ensure a timely start of treatment before irreversible muscle damage has occurred. It will also provide important information for patients and their parents that participate in newborn screening programs, because at present the diagnosis of late onset Pompe disease poses much uncertainty on how and when this diagnosis will impact their lives. A more accurate prediction contributes to better genetic counseling.
Alt-text: Unlabelled Box
1. Introduction
Pompe disease, a metabolic myopathy caused by lysosomal glycogen accumulation, results in progressive vacuolization of muscle cells. Patients with the most severe classic infantile form present with symptoms shortly after birth and, if left untreated, die from cardiorespiratory insufficiency within the first year of life. Patients with onset in childhood or adulthood become wheelchair and/or ventilator dependent at some point in their life [1]. Pompe disease is a monogenic autosomal recessive disorder caused by disease-associated variants in the acid α-glucosidase (GAA) gene. Over 400 disease-associated variants are known and described in the Pompe mutation database (www.pompecenter.nl). Whereas classic infantile patients carry two fully deleterious GAA variants, those with onset in childhood or adulthood usually carry one fully deleterious variant in combination with a milder variant. In the Caucasian population, 90% of patients with onset at childhood or adulthood carry the c.-32-13T>G (IVS1) variant on one allele in combination with a deleterious variant on the second allele (Suppl. Table S1) [[2], [3], [4], [5], [6], [7], [8], [9], [10], [11], [12], [13]]. However, symptom onset in these patients varies broadly, ranging from <1 to 62 years of age. This suggests the possibility that unidentified modifying factors are able to modulate age at symptom onset [5,6,[14], [15], [16], [17], [18]]. This is also indicated by the relatively low frequency of patients with the homozygous IVS1 genotype, suggesting that many remain asymptomatic [11,17,19,20]. The genetics of Pompe disease is explained in more detail in the supplementary text.
Since 2006, enzyme replacement therapy (ERT) has been available for Pompe disease. ERT in these patients improves survival and motor function, normalizes left ventricular cardiac hypertrophy, and stabilizes respiratory capacity [21]. To enable fast diagnosis and early treatment, newborn screening (NBS) programs have been introduced in a number of countries in Asia, North America and Europe [22]. Such programs have shown that most patients have a genotype consistent with the late onset form of Pompe disease, and that this complicates the prediction of disease onset and severity, and also the timing for starting ERT treatment.
As these findings have increased the need to identify factors that can modulate the onset of Pompe disease, we aimed to identify a putative modifier in patients with the common IVS1 variant. Genomic DNA analysis of 153 patients with Pompe disease that carry the IVS1 variant was performed using IVS1 allele-specific PCR. This resulted in the identification of the c.510C>T variant to be exclusively present on the IVS1 allele in a subset of patients. This variant was only identified in compound heterozygous IVS1 patients with childhood onset of symptoms. It was also identified in 3 out of 6 symptomatic patients that were homozygous for IVS1. RT-(q)PCR analysis showed that the variant negatively influences pre-mRNA splicing in the context of the IVS1 splicing variant, resulting in lowered expression of residual full-length GAA mRNA and GAA enzymatic activity. These findings are important for the estimation of age at symptom onset and for deciding when to start ERT treatment. Reports on the presence of c.510C>T should be included in the genetic diagnostic analysis of Pompe disease.
2. Materials & methods
2.1. Patients
Patients enrolled in this study were all diagnosed with Pompe disease on the basis of GAA enzyme deficiency and the presence of two disease-associated variants in the GAA gene, at least one of which was c.-32-13T>G. One hundred thirty-one compound heterozygous patients enrolled in this study had a confirmed fully deleterious variant on the second allele that did not produce any functional GAA enzymatic activity, as determined on the basis of clinical information (see www.pompecenter.nl). Nine patients contained a deleterious GAA variant on the second allele based on in silico prediction, while for three patients the disease-associated variant on the second allele was not identified. There was no preferential enrichment of patients based on the disease-associated variant on the 2nd allele in any of the groups (childhood onset, adult onset, presence or absence of c.510C>T) analyzed. RT-PCR analysis was restricted where possible to patients that had a nonsense or frameshift variant on the second allele. We were unable to avoid this for the analysis of one patient (patient 16) which had an in-frame deletion of exon 18. Patients' age at symptom onset was determined by experienced clinicians on the basis of the description of the first symptoms. Guidelines for determining symptom onset were used as described in van der Ploeg et al., 2017 [23]. Briefly, symptomatic patients had skeletal muscle and/or respiratory involvement. In patients at Erasmus MC, age at symptom onset ranged from 0 to 67 years. Childhood onset patients are patients younger than 18 years of age at onset of disease who do not have hypertrophic cardiomyopathy. Adult onset patients are patients that are 18 years of age or older at onset of disease. Biopsies from skin obtained using the punching method as described [24]. Open muscle biopsies were taken as described previously [19]. Patients analyzed in more detail in this study are listed in Supplementary Tables S2 and S3. All patients provided informed consent. Analysis of the patients analyzed in this study was approved by the medical ethics committee of the participating institutes.
2.2. Allele-specific PCR and sequencing
DNA was isolated from either fibroblasts, leukocytes or myoblasts. Allele-specific primers were designed based on the sequence surrounding the c.-32-13T>G variant with either T or G as the last nucleotide of the primer and the requirements stated in the guidelines for each PCR kit (see below). Two different PCR reactions were performed to obtain allele specific results upstream and downstream of the c.-32-13T>G variant. Amplification of a region of 3 kilobases (kb) upstream of IVS1 required an optimized protocol using the Advantage GC2 PCR kit (Clontech). Amplification 1 kb downstream of IVS1 was performed using a standard PCR protocol. Sanger sequencing of the amplified products was performed using a ABI3730XL DNA Analyzer and was analyzed for the presence of single nucleotide variants (SNVs) with ApE software.
2.3. RT-PCR
RT-PCR and RT-qPCR measurements were conducted as previously described [25]. In brief, cells were harvested using the standard protocol of the RNeasy® kit (Qiagen) which included the DNase treatment (Qiagen). cDNA was generated using the iScript™ cDNA synthesis kit (Bio-Rad) with 500 ng input per reaction. RT-PCR was performed using the Advantage GC 2 kit (Clontech). RT-qPCR analysis was performed using iTaq™ universal SYBR® Green Supermix (Bio-Rad) with a CFX96 RT-system (Bio-Rad). RT-qPCR data were normalized relative to beta-actin expression. PCR primers are listed in Supplementary Table S4.
2.4. GAA enzymatic activity assay
The enzymatic activity of GAA in fibroblasts was determined as described using a one-step protocol with 4-methylumbelliferone-α-d-glucopyranoside (4-MU) (Sigma) as a substrate [16]. Protein lysates were incubated for one hour, and fluorescence was measured at 365/448 nm with a Varioskan™ system (Thermo-Fisher). Protein concentration of the lysates was determined using the BCA protein assay (Pierce) performed as instructed in the protocol. Samples of Fig. 2C were measured at the diagnostic department of the Erasmus MC using a standardized protocol. All other samples were measured on research basis and were normalized using individual 14 as an internal control.
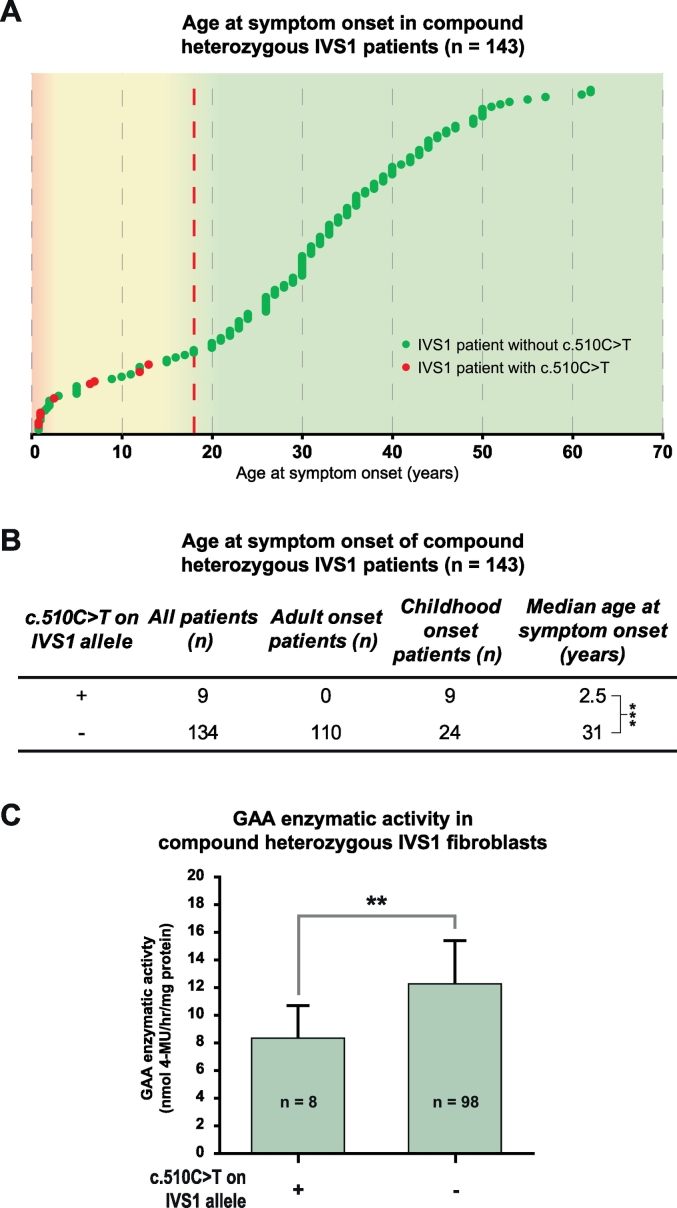
Fig. 2.
Association of c.510C>T with age at symptom onset and GAA enzymatic activity in compound heterozygous IVS1 patients. (A) Distribution of ages at symptom onset in compound heterozygous IVS1 patients with c.510C>T (red symbols) and without c.510C>T (green symbols). Each dot in the graph represents one patient. The dashed red line indicates the cut-off age of 18 years between patients with childhood onset and adult onset of symptoms. (B) Median age at symptom onset in compound heterozygous IVS1 patients with and without c.510C>T. *** p < 0.001. (C) GAA enzymatic activity in fibroblasts from compound heterozygous IVS1 patients with and without c.510C>T. ** p < 0.01.
2.5. Minigene
The minigenes were generated by site directed mutagenesis of a pcDNA3.1 construct containing the genomic region of GAA exon 1 to 3 (GRCh38/hg38, chr17:80,101,086-80,105,998), according to the manufacturer's protocol (QuikChange® II Site-Directed Mutagenesis, Agilent, primers Suppl. Table S4) [29]. Minigenes were transfected in HEK293T cells using Lipofectamine 2000 (Life Technologies) according to the manufacturer's protocol (per sample in 12-well format: 4 μl Lipofectamine and 1.6 μg plasmid DNA). Two days after transfection, RNA was isolated using the RNeasy® mini kit (Qiagen), and 800 ng of total RNA was reverse transcribed with iScript™ (Bio-Rad). RT-(q)PCRs were performed as described above. RT-qPCR data were normalized relative to Neomycin expression, which is constitutively expressed from the pcDNA3.1 vector, to normalize for transfection efficiency.
2.6. Statistics
Analyses were performed using the R statistical package, version 3.4.3. To correct for the non-random distribution of the data and familial connections, a Generalized Estimating Equation (geepack package in R) was used to analyze the differences in age at symptom onset between the patient groups with or without the c.510C>T variant present on the IVS1 allele. All other statistical analyses were performed using the standard t-test (random distribution) or the Wilcoxon test (non-random distribution). All biological data is shown using the average ± SD of three biological replicates.
3. Results
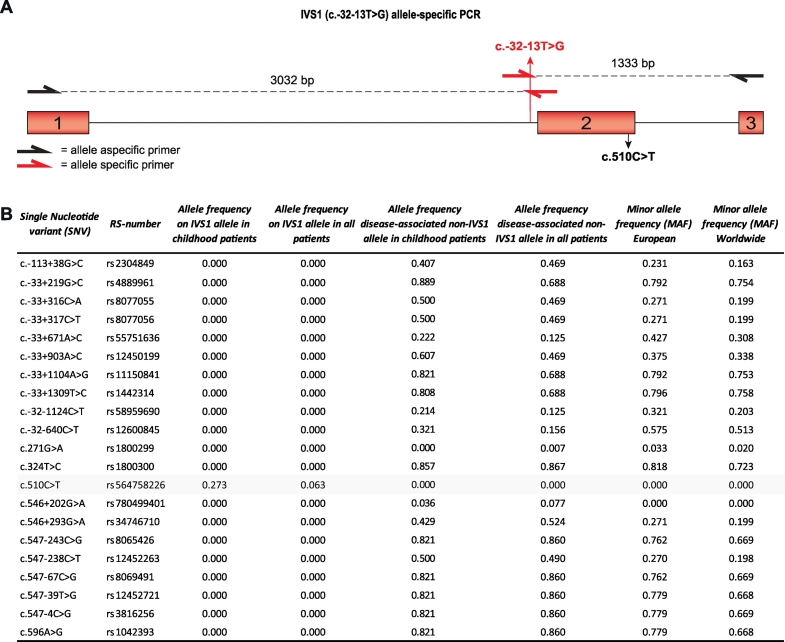
The IVS1 variant is located at −13 nucleotides (nt) of GAA exon 2 in the polypyrimidine (pY) tract (Fig. 1A). GAA exon 2 contains the translation initiation codon, is relatively large (546 nt), and is preceded by an intron of 2664 nt. We hypothesized that cis-acting variants might be present near the IVS1 variant that modulate the activity of the IVS1 allele. To test this, we used long-range allele-specific PCR and Sanger sequencing of a 4.4 kb region comprising exon 1–3. In a cohort of 143 compound heterozygous IVS1 patients that contained a deleterious variant on the second allele, we identified 21 SNVs (Fig. 1B). Interestingly, all but one were located on non-IVS1 alleles, except for the synonymous variant c.510C>T, which was found in a subset of patients exclusively on the IVS1 allele. The c.510C>T variant was found in 9 patients, all of whom had symptom onset in childhood (see Table S2 for patient details). While c.510C>T occurred in combination with the IVS1 variant in 9/33 (27%) of the population of IVS1 patients with childhood onset, it occurred in 0/110 (0%) of patients with symptom onset in adulthood (Fig. 1B). The c.271G>A variant (GAA2; pD91N), which has previously been reported to lower the affinity of GAA for glycogen [26], was absent from the IVS1 allele, and occurred in only 1 adult patient on the non-IVS1 allele, indicating that its frequency is low and that it cannot account for phenotypic differences within our cohort. Fig. 2A shows the ages at symptom onset in compound heterozygous IVS1 patients with and without c.510C>T. Median age at symptom onset was 2.5 years in patients with c.510C>T, against 31 for patients without c.510C>T (p ≤.001) (Fig. 2B). Similar results were found for age at diagnosis (5.4 years in those with c.510C>T versus 37 in those without) (Suppl. Fig. S1A and B). To illustrate that the disease-associated variant on the second allele did not play a role in phenotypic heterogeneity, we analyzed 57 patients that all shared the same IVS1/c.525del GAA genotype. These patients also varied widely in age at symptom onset: two patients that contained c.510C>T had symptom onset at 1 and 6.5 years, while 55 patients without c.510C>T had symptom onset ranging from 0.5–61 years (Suppl. Fig. S1C). The GAA enzymatic activity in fibroblasts from compound heterozygous patients with c.510C>T was lower than in patients without c.510C>T (Fig. 2C). We conclude that c.510C>T is linked to the IVS1 variant and, when present, is associated with early onset of symptoms and low GAA enzymatic activity.
Fig. 1.
Allele frequencies of SNVs around GAA IVS1. (A) Cartoon of the genomic region of GAA spanning exon 1–3. Primers used for allele-specific amplification of the IVS1 allele are highlighted in red (Supplementary Table S4). Exonic regions are highlighted as red boxes, and introns are indicated by lines. (B) List of 21 SNVs in GAA exons 1–3 with RS-numbers and allele frequencies (MAF was obtained from gnomAD: http://gnomad.broadinstitute.org/).
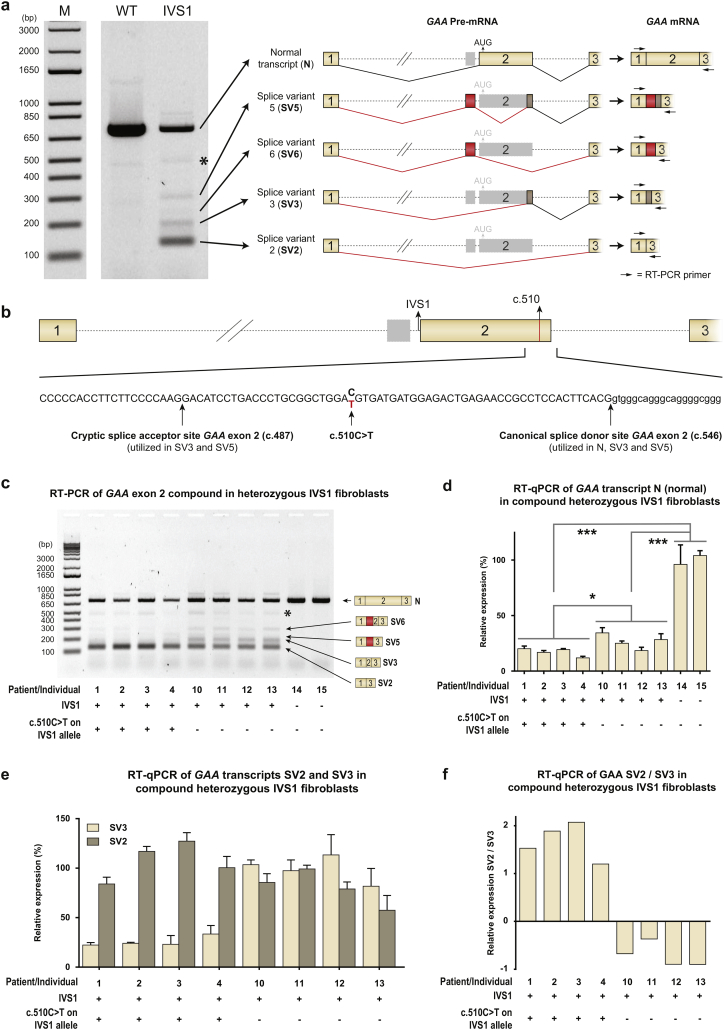
The IVS1 variant affects pre-mRNA splicing and induces full or partial skipping of GAA exon 2 (products splice variant (SV) 2 and SV3 in Fig. 3A, respectively) [15,25,27,28] and utilization of a pseudoexon in intron 1 (products SV5 and SV6) [29]. It also allows a low level (10–15%) of leaky wild-type splicing (product N). The c.510C>T variant is located close to the cryptic splice site in exon 2 and the canonical splice donor of exon 2 (Fig. 3B), suggesting that it might affect splicing. To test this, we performed flanking exon RT-PCR analysis of primary fibroblasts obtained from patients with and without c.510C>T (Fig. 3C). Exon 2 splicing was altered in cells that contained c.510C>T: expression of N and SV3 were lowered relative to cells that lacked c.510C>T. To quantify these results, we performed RT-qPCR analysis using primers that specifically amplified splice isoforms. Relative to cells that lacked c.510C>T, this showed the following: decreased expression of normal splicing (product N) (Fig. 3D) and cryptic splicing (product SV3)(Fig. 3E); unchanged expression of fully skipped product (SV2) (Fig. 3E); and increased SV2/SV3 ratios (Fig. 3F) in cells that contained c.510C>T (see Suppl. Tables S2 and S3 for patient details). Importantly, the decreased expression of N resulted in a significant decrease of GAA enzymatic activity in these patients (Suppl. Fig. S2). To establish that these changes were caused solely by c.510C>T rather than by other genetic variations, we used a minigene system in which the genomic region of GAA exons 1–3 was cloned in an expression vector (Suppl. Fig. S3A) [29]. After transfection into HEK293T cells, splicing was analyzed by PCR. In the minigene system, the IVS1 variant reproduced the aberrant splicing seen endogenously in fibroblasts [29]. Flanking exon RT-PCR analysis showed that, when combined with the IVS1 variant, the c.510C>T variant changed the splicing pattern of exon 2 (Suppl. Fig. S3B). RT-qPCR analysis indicated that c.510C>T reduced expression of N (Suppl. Fig. S3C) and SV3 (Suppl. Fig. S3D), and increased expression of SV2 (Suppl. Fig. S3D) and SV2/SV3 ratios (Suppl. Fig. S3E). In the minigene lacking IVS1, c.510C>T did not change expression of N (Suppl. Fig. S3C). We conclude that, by lowering the extent of leaky wild-type splicing, c.510C>T modulates the aberrant splicing caused by IVS1, and that c.510C>T in the absence of IVS1 does not affect wild-type splicing.
Fig. 3.
c.510C>T worsens splicing outcome in compound heterozygous IVS1 patients. (A) Left: RT-PCR analysis of GAA exons 1–3 of fibroblasts from a healthy control (WT) and a compound heterozygous IVS1 patient (IVS1). The second allele in this patient did not produce mRNA. M indicates the DNA size marker in base pairs (bp). Right: Cartoons of the major splice products. Numbered boxes represent canonical exons. Unnumbered boxes represent facultative exons or parts of exons: a pseudoexon in intron 1 (in red) or the C-terminal part of exon 2, which is derived from utilization of the cryptic splice site at c.487 (in brown). Dotted lines represent introns. Continuous lines represent splicing events. *: structural variant (see Suppl. Fig. S4). (B) Position of c.510C>T in GAA exon 2. Positions of the cryptic and canonical splice sites of exon 2 are also indicated. Numbered boxes represent canonical exons. The unnumbered gray box represent the pseudoexon in intron 1. Dotted lines represent introns. (C) RT-PCR analysis of GAA exon 1 to 3 in fibroblasts from compound heterozygous IVS1 patients with and without c.510C>T. Splice products N, SV6, SV5, SV3 and SV2 are indicated on the right. *: structural variant (see Suppl. Fig. S4). The patients or individuals analyzed are indicated and listed in Supplementary Tables S2 and S3. (D) As (C), but now analyzed using RT-qPCR for the wildtype splice product N. (E) As (C), but now analyzed using RT-qPCR for the aberrant splice products SV2 and SV3. (F) Ratio of SV2/SV3. In healthy control cells, expression of SV2 and SV3 was too low to allow quantification. Data in d-e represent means ± SD (n= 3 biological replicates). * = p < 0.05, *** = p < 0.001.
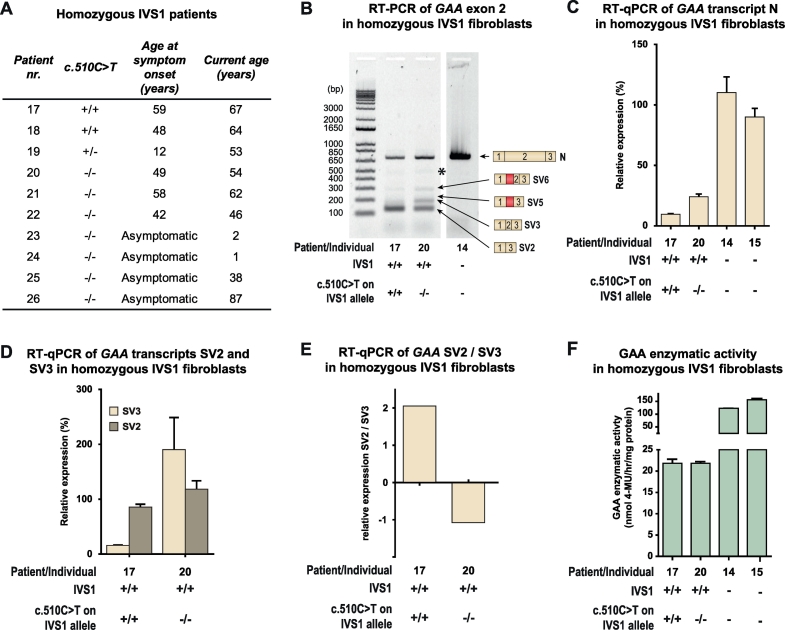
The enzymatic activity of GAA in compound heterozygous patients is derived from the IVS1 allele via leaky wild-type splicing, which produces 10–15% of normal mRNA transcript and full-length GAA protein. Homozygous IVS1 patients should have twice the amount of this activity, reaching 20–30%, which, using the assay conditions employed here, would be above the disease threshold. Consistent with this fact, only few homozygous IVS1 patients have been reported in the literature. In our cohort 10 unrelated individuals homozygous for IVS1 were present (Fig. 4A): six patients developed symptoms at a median age of 48.5 years (range 12–59 years); five patients (18–22) have been described previously [19]; two individuals currently aged 38 and 87 years are asymptomatic (25 and 26); and two individuals currently aged 1 and 2 year (23 and 24) were diagnosed from NBS programs and are asymptomatic (Fig. 4A). Sanger sequencing revealed that 3/6 (50%) symptomatic homozygous IVS1 patients carried c.510C>T: patients 17 and 18 carried c.510C>T at homozygous state, and patient 19 carried it at heterozygous state (Fig. 4A). None of the asymptomatic patients carried c.510C>T. In fibroblasts from homozygous IVS1 patients, c.510C>T caused reduced normal splicing (product N) and an increased SV2/SV3 ratio, as it did in fibroblasts from compound heterozygous IVS1 patients (Fig. 4B–E). The GAA enzyme activity in fibroblasts from the homozygous IVS1 patient (patient 17 in Fig. 4F) who carried c.510C>T was in the patient range (16 nmol/h/mg protein), which was consistent with the low expression of product N and the development of symptoms. Patient 20, who was homozygous for IVS1 but did not carry c.510C>T, also had GAA enzyme activity in the patient range (17 nmol/h/mg protein), which was consistent with the development of symptoms. It is likely that another putative modifying factor plays a role in this patient. These findings indicate that c.510C>T accelerates the development of symptoms by worsening splicing outcome in homozygous IVS1 patients in the same way as in compound heterozygous IVS1 patients.
Fig. 4.
c.510C>T is associated with symptom onset and worsening of splicing outcome in homozygous IVS1 patients. (A) Genotypes and symptom onset in 10 homozygous IVS1 patients. (B) RT-PCR analysis of GAA exon 1 to 3 in fibroblasts from homozygous IVS1 patients with and without c.510C>T. Products N, SV6, SV5, SV3 and SV2 are indicated on the right. *: structural variant (see Suppl. Fig. S4). (C) Quantification using RT-qPCR of normal splicing (product N) in fibroblasts from two homozygous IVS1 patients, one carrying the c.510C>T variant homozygously, and one without c.510C>T. For comparison, expression in fibroblasts from a healthy control is shown (individual 14). (D) As (C), but now of aberrant splice products SV2 and SV3. In fibroblasts from healthy controls, SV2 and SV3 expression was undetectable and could not therefore be quantified. (E) Ratio of SV2 and SV3. (F) Enzymatic activity of GAA. Data in C-D and F represent means ± SD (n= 3 biological replicates).
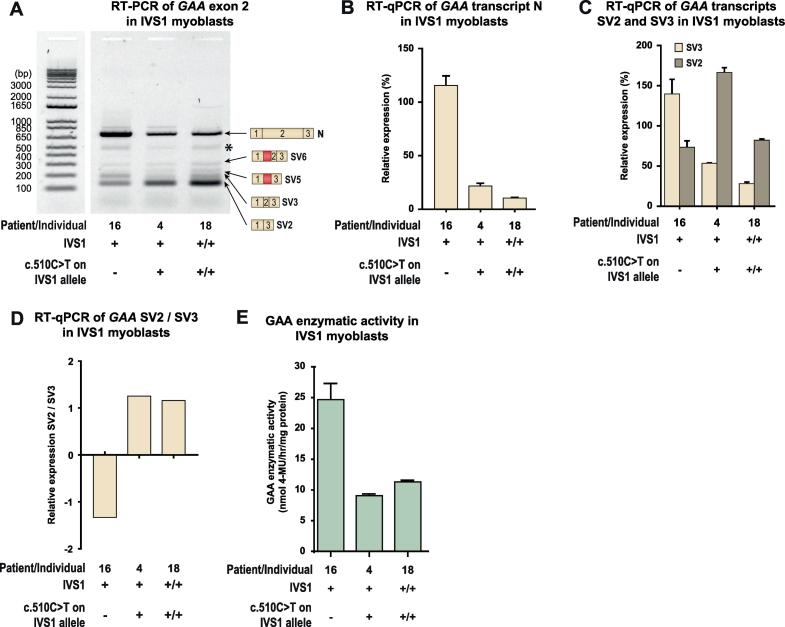
Pre-mRNA splicing can be cell-type dependent. Because skeletal muscle is the tissue most affected in Pompe disease, we examined the effect of c.510C>T on splicing in muscle cells. Myoblasts were available from three patients (Fig. 5A). The first was patient 16, who was compound heterozygous for IVS1 and lacked c.510C>T, but whose second allele contained the c.2481 + 102_2646 + 31del (delex18) variant. As this is an in-frame deletion of exon 18 that produces normal levels of a truncated mRNA transcript (translated into an inactive GAA protein), it produces high levels of product N, which obscured analysis of product N from the IVS1 allele (Fig. 5B). The second patient was patient 4, who was compound heterozygous for IVS1 and c.510C>T, in which the second allele carried the c.1548G>A nonsense variant leading to nonsense-mediated decay of mRNA produced from this allele [25]. The third, patient 18, was homozygous for IVS1 and c.510C>T. RT-PCR analysis in these three patients showed that the presence of c.510C>T was associated with the characteristic splicing pattern observed in fibroblasts, in which expression of SV3 is reduced (Fig. 5A). RT-qPCR analysis confirmed that SV3 expression was reduced and that the ratio of SV2/SV3 was increased in cells that contained c.510C>T (Fig. 5C and D). In myoblasts, the c.510C>T variant was associated with lower GAA enzyme activity in the compound heterozygous IVS1 patients (compare patient 16 and 4), while the activity in the homozygous IVS1 patient (patient 18) was well within the patient range (Fig. 5E). We noted that the expression levels of exon 2 splicing products N, SV2, and SV3 were higher in patient 4 compared to patient 16, although their GAA enzyme activities were similar. We speculate that this might indicate differences in mRNA turnover, but further experiments are required to address this. We conclude that the deleterious effect that c.510C>T has on the aberrant splicing caused by IVS1 in myoblasts is similar to its effect in fibroblasts.
Fig. 5.
c.510C>T worsens splicing outcome in skeletal muscle cells from compound heterozygous and homozygous IVS1 patients. (A) Flanking exon RT-PCR of GAA exon 2 in myoblasts. Patient 16: compound heterozygous for IVS1, c.510C>T absent; Patient 4: compound heterozygous for IVS1, c.510C>T present on IVS1 allele; Patient 18: homozygous for IVS1, homozygous for c.510C>T. Splice products N, SV6, SV5, SV3 and SV2 are indicated. *: structural variant (see Suppl. Fig. S4). Splice product N (normally spliced product) is highly expressed in patient 16 due to the presence of the c.2481+102_2646+31del variant on the second allele (which allows normal expression of GAA product N). (B) Quantification of splice product N using RT-qPCR. (C) Quantification of splice products SV2 and SV3. (D) Ratio of SV2 and SV3. (E) GAA enzymatic activity. Data in C-D and F represent means ± SD (n= 3 biological replicates).
4. Discussion
The most frequent disease-associated variant in the Caucasian population is the IVS1 variant, which occurs in a large majority (90%) of childhood and adult onset patients [[2], [3], [4], [5], [6], [7], [8], [9], [10], [11], [12], [13]]. Previous work has identified 12 haplotypes in Caucasian IVS1 patients based on 17 single nucleotide polymorphisms that were distributed over the entire GAA gene [16]. Here we zoomed in on a 4.4 kb region around the IVS1 variant and showed that the IVS1 allele comes in two flavors: linked to either c.510C or c.510 T. We used allele-specific PCR, which we find to be a useful approach to providing more detailed information on haplotypes. The c.510C>T variant is synonymous, and we noted that its presence is often not included in official DNA diagnostic reports, something that warrants manual examination and special attention in future reports. The fact that c.510C>T has an allele frequency of 0.27 in the childhood-onset IVS1 patients in our cohort indicates that it is a relatively frequent variant within this population. Symptoms developed in childhood in all 9/9 (100%) carriers of c.510C>T. This is relevant to regular diagnostics and to newborn screening programs, which are urgently needed for predicting symptom onset and for deciding when ERT treatment should be started in newly diagnosed late onset Pompe disease (LOPD) patients [22]. Results from NBS programs indicate that a new group of asymptomatic LOPD patients is coming into being, and that among patients and their parents there is anxiety and uncertainty regarding the timing of symptom onset and when treatment with ERT will be necessary [31].
The exceptionally broad spectrum of symptom onset within compound heterozygous IVS1 patients observed in many studies has reinforced the suspicion that there are modifying factors for Pompe disease [[2], [3], [4], [5], [6], [7], [8], [9], [10], [11], [12], [13]]. In theory, residual activity from the second allele could explain clinical heterogeneity in compound heterozygous IVS1 patients. This would potentially explain a later onset of symptoms. However, it has been reported that in Pompe disease, the second allele is a null allele in the far majority of cases [9,11,32]. This was confirmed in our cohort, in which >90% of patient- including those in the group with adult onset of symptoms- contained a fully deleterious variant on the second allele. In addition, we showed a broad range of age at symptom onset in a group of 57 patients with an identical GAA genotype (IVS1; c.525del) (Suppl. Fig. S1C). Therefore, the second allele in compound heterozygous IVS1 patients cannot explain the differences in age at symptom onset observed in our cohort.
The median age at symptom onset for compound heterozygous patients carrying c.510C>T was 2.5 years. However, we noted that patients that contained c.510C>T showed some heterogeneity in symptom onset that ranged from <1 to 14 years. Furthermore, not all childhood-onset patients carried the c.510C>T variant. This suggests that there are likely additional modifying factors. For example, other SNVs might be present in cis with the GAA gene that modulate some aspect of GAA expression or GAA protein function such as folding. We have analyzed a 4.4 kB region of genomic DNA around IVS1 using allele-specific PCR, but it will be difficult with this approach to extend the analysis beyond this region due to the limitations of the length of the PCR product that can be obtained. It will be interesting to investigate this idea in the future. Although previous work has reported on the angiotensin-converting enzyme (ACE) I/D polymorphism and alpha actinin 3 (ACTN3) p.R577X polymorphism to be involved in disease onset [33], no such involvement of the ACE I/D polymorphism was found in symptom onset in the Dutch cohort of 131 patients or in a multicenter cohort from the late-onset treatment study (LOTS) with 88 patients [34,35]. This does indeed suggest that there are additional modifying factors.
We found that c.510C>T worsens the outcome of aberrant splicing caused by the IVS1 variant. The IVS1 variant weakens the splice acceptor of exon 2 to reduce the level of normal splicing (N) to 10–15%, and to induce full skipping of exon 2 (SV2) or partial skipping (SV3) by utilizing a cryptic splice acceptor in exon 2 [15,25,27,28]. We recently found that the IVS1 variant induces utilization of a pseudoexon in intron 1 (SV5 and SV6) [29]. Antisense oligonucleotides (AONs) that prevented inclusion of the pseudoexon were able to restore normal exon 2 splicing almost completely in the context of IVS1 [29,30]. The c.510C>T variant did not affect utilization of the pseudoexon (data not shown). Instead, it reduced leaky wild-type splicing (N) and utilization of the cryptic splice site of exon 2 (SV3) (Fig. 3C–F, Fig. 4B–E, Suppl. Fig. S3B–D, Fig. 5C and D). These results indicate that exon 2 splicing can be modulated by sequences in exon 2 itself, as well as in the intronic pseudoexon. This is consistent with findings that showed that AONs directed at exon 2 sequences were also able to elevate normal splicing [36]. These findings highlight the importance of diagnostic screening and the potential for development of new treatment strategies for variants involved in splicing such as c.510C>T [37].
Although few symptomatic homozygous IVS1 patients have been reported in the literature [11,17,19,20], a large number of compound heterozygous IVS1 patients have been reported in Europe, with many more in countries in North and South America, Australia and New Zealand [[2], [3], [4], [5], [6], [7], [8], [9], [10], [11], [12], [13],23]. This suggests that many homozygous IVS1 patients remain asymptomatic, but that a subset develops symptoms [11,19]. Due to NBS programs, the group of identified asymptomatic individuals who are homozygous for IVS1 is increasing, bringing challenges regarding the prediction of symptom onset and when to start with ERT treatment that are similar to those described above for the compound heterozygous IVS1 patients. In our cohort, 3/6 (50%) of symptomatic homozygous IVS1 patients carried c.510C>T, and when c.510C>T was present, 3/3 (100%) of homozygous IVS1 patients developed symptoms. This indicates not only that c.510C>T has a strong predictive value for symptom onset in homozygous IVS1 patients, but also that there are probably additional modifying factors for homozygous IVS1 patients. We also observed that the effects of c.510C>T on exon 2 splicing in patient-derived fibroblasts were similar as in myoblasts obtained from muscle biopsies. This indicates that c.510C>T modulates splicing via shared molecular targets, which is in line with the similar effects of the IVS1 variant on exon 2 splicing in fibroblasts and induced pluripotent stem cell-derived skeletal muscle cells [29].
In conclusion, we report the identification of c.510C>T as a genetic modifier of disease onset in patients with Pompe disease that are compound heterozygous or homozygous for the common IVS1 GAA variant. We recommend the following interpretation of newborn screening results and regular diagnostics: compound heterozygous IVS1 patients who carry c.510C>T have the prognosis to develop symptoms at childhood. Compound heterozygous patients without c.510C>T may develop symptoms at any age. Homozygous IVS1 patients who carry c.510C>T, either at heterozygous or at homozygous state, have the prognosis to develop symptoms at any age, while homozygous IVS1 patients without c.510C>T may remain asymptomatic or may develop symptoms at any age. Future work should determine whether there are additional genetic modifiers in cis or in trans with GAA, and, if so, how these may modulate the clinical course of Pompe disease.
Funding source statement
This work was funded by grants from Sophia Children's Hospital Foundation (SSWO, grant S17–32) and Metakids (2016–063). These funding sources did not have any further role in the writing of the manuscript or the decision to submit it for publication. None of the authors have been paid by a pharmaceutical company or other agency to write this article. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Acknowledgments
Acknowledgements
We thank the members of the Molecular stem cell biology group for the critical discussions. We thank Dimitris Rizopoulos for the help with the statistical analysis of the data and David Alexander for critical review of the manuscript. We dedicate this work in memory of our colleague Dr. Elisabetta Pasquini, who recently passed away.
Authors contributions
Study concept and design: Atze Bergsma, Stijn in ‘t Groen, Jeroen van den Dorpel, Hannerieke van den Hout, Nadine van der Beek, Ans van der Ploeg, Pim Pijnappel.
Acquisition of data: Atze Bergsma, Stijn in ‘t Groen, Jeroen van den Dorpel, Hannerieke van den Hout, Nadine van der Beek, Ans van der Ploeg, Pim Pijnappel.
Analysis and interpretation of data: All authors.
Drafting the manuscript: Atze Bergsma, Stijn in ‘t Groen, Jeroen van den Dorpel, Ans van der Ploeg, Pim Pijnappel.
Critical revision of the manuscript: All authors.
Statistical analysis: Atze Bergsma, Stijn in ‘t Groen, Pim Pijnappel.
Obtained funding: Atze Bergsma, Ans van der Ploeg, Pim Pijnappel.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.03.048.
Appendix A. Supplementary data
Supplementary material
References
- 1.van der Ploeg A.T., Reuser A.J. Pompe's disease. Lancet. 2008;372(9646):1342–1353. doi: 10.1016/S0140-6736(08)61555-X. [DOI] [PubMed] [Google Scholar]
- 2.Angelini C., Semplicini C., Tonin P., Filosto M., Pegoraro E., Soraru G. Progress in enzyme replacement therapy in glycogen storage disease type II. Ther Adv Neurol Disord. 2009;2(3):143–153. doi: 10.1177/1756285609103324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Figueroa-Bonaparte S., Llauger J., Segovia S., Belmonte I., Pedrosa I., Montiel E. Quantitative muscle MRI to follow up late onset Pompe patients: a prospective study. Sci Rep. 2018;8(1):10898. doi: 10.1038/s41598-018-29170-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuperus E., Kruijshaar M.E., Wens S.C.A., de Vries J.M., Favejee M.M., van der Meijden J.C. Long-term benefit of enzyme replacement therapy in Pompe disease: a 5-year prospective study. Neurology. 2017;89(23):2365–2373. doi: 10.1212/WNL.0000000000004711. [DOI] [PubMed] [Google Scholar]
- 5.Loscher W.N., Huemer M., Stulnig T.M., Simschitz P., Iglseder S., Eggers C. Pompe disease in Austria: clinical, genetic and epidemiological aspects. J Neurol. 2018;265(1):159–164. doi: 10.1007/s00415-017-8686-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Montalvo A.L., Bembi B., Donnarumma M., Filocamo M., Parenti G., Rossi M. Mutation profile of the GAA gene in 40 Italian patients with late onset glycogen storage disease type II. Hum Mutat. 2006;27(10):999–1006. doi: 10.1002/humu.20374. [DOI] [PubMed] [Google Scholar]
- 7.Mori M., Haskell G., Kazi Z., Zhu X., DeArmey S.M., Goldstein J.L. Sensitivity of whole exome sequencing in detecting infantile- and late-onset Pompe disease. Mol Genet Metab. 2017;122(4):189–197. doi: 10.1016/j.ymgme.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Papadimas G.K., Terzis G., Methenitis S., Spengos K., Papadopoulos C., Vassilopoulou S. Body composition analysis in late-onset Pompe disease. Mol Genet Metab. 2011;102(1):41–43. doi: 10.1016/j.ymgme.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 9.Regnery C., Kornblum C., Hanisch F., Vielhaber S., Strigl-Pill N., Grunert B. 36 months observational clinical study of 38 adult Pompe disease patients under alglucosidase alfa enzyme replacement therapy. J Inherit Metab Dis. 2012;35(5):837–845. doi: 10.1007/s10545-012-9451-8. [DOI] [PubMed] [Google Scholar]
- 10.Scheidegger O., Leupold D., Sauter R., Findling O., Rosler K.M., Hundsberger T. 36-months follow-up assessment after cessation and resuming of enzyme replacement therapy in late onset Pompe disease: data from the Swiss Pompe registry. J Neurol. 2018;265(12):2783–2788. doi: 10.1007/s00415-018-9065-7. [DOI] [PubMed] [Google Scholar]
- 11.Semplicini C., Letard P., De Antonio M., Taouagh N., Perniconi B., Bouhour F. Late-onset Pompe disease in France: molecular features and epidemiology from a nationwide study. J Inherit Metab Dis. 2018;41(6):937–946. doi: 10.1007/s10545-018-0243-7. [DOI] [PubMed] [Google Scholar]
- 12.van der Meijden J.C., Kruijshaar M.E., Harlaar L., Rizopoulos D., van der Beek N., van der Ploeg A.T. Long-term follow-up of 17 patients with childhood Pompe disease treated with enzyme replacement therapy. J Inherit Metab Dis. 2018;41(6):1205–1214. doi: 10.1007/s10545-018-0166-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Witkowski G., Konopko M., Rola R., Lugowska A., Ryglewicz D., Sienkiewicz-Jarosz H. Enzymatic replacement therapy in patients with late-onset Pompe disease - 6-year follow up. Neurol Neurochir Pol. 2018;52(4):465–469. doi: 10.1016/j.pjnns.2018.05.002. [DOI] [PubMed] [Google Scholar]
- 14.Herzog A., Hartung R., Reuser A.J., Hermanns P., Runz H., Karabul N. A cross-sectional single-Centre study on the spectrum of Pompe disease, German patients: molecular analysis of the GAA gene, manifestation and genotype-phenotype correlations. Orphanet J Rare Dis. 2012;7:35. doi: 10.1186/1750-1172-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huie M.L., Chen A.S., Brooks S.S., Grix A., Hirschhorn R. A de novo 13 nt deletion, a newly identified C647W missense mutation and a deletion of exon 18 in infantile onset glycogen storage disease type II (GSDII) Hum Mol Genet. 1994;3(7):1081–1087. doi: 10.1093/hmg/3.7.1081. [DOI] [PubMed] [Google Scholar]
- 16.Kroos M.A., Pomponio R.J., Hagemans M.L., Keulemans J.L., Phipps M., DeRiso M. Broad spectrum of Pompe disease in patients with the same c.-32-13T->G haplotype. Neurology. 2007;68(2):110–115. doi: 10.1212/01.wnl.0000252798.25690.76. [DOI] [PubMed] [Google Scholar]
- 17.Laforet P., Laloui K., Granger B., Hamroun D., Taouagh N., Hogrel J.Y., The French Pompe registry Baseline characteristics of a cohort of 126 patients with adult Pompe disease. Rev Neurol (Paris) 2013;169(8–9):595–602. doi: 10.1016/j.neurol.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 18.Muller-Felber W., Horvath R., Gempel K., Podskarbi T., Shin Y., Pongratz D. Late onset Pompe disease: clinical and neurophysiological spectrum of 38 patients including long-term follow-up in 18 patients. Neuromuscul Disord. 2007;17(9–10):698–706. doi: 10.1016/j.nmd.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 19.Musumeci O., Thieme A., Claeys K.G., Wenninger S., Kley R.A., Kuhn M. Homozygosity for the common GAA gene splice site mutation c.-32-13T>G in Pompe disease is associated with the classical adult phenotypical spectrum. Neuromuscul Disord. 2015;25(9):719–724. doi: 10.1016/j.nmd.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 20.Sharma M.C., Schultze C., von Moers A., Stoltenburg-Didinger G., Shin Y.S., Podskarbi T. Delayed or late-onset type II glycogenosis with globular inclusions. Acta Neuropathol. 2005;110(2):151–157. doi: 10.1007/s00401-005-1026-4. [DOI] [PubMed] [Google Scholar]
- 21.Schoser B., Stewart A., Kanters S., Hamed A., Jansen J., Chan K. Survival and long-term outcomes in late-onset Pompe disease following alglucosidase alfa treatment: a systematic review and meta-analysis. J Neurol. 2017;264(4):621–630. doi: 10.1007/s00415-016-8219-8. [DOI] [PubMed] [Google Scholar]
- 22.Bodamer O.A., Scott C.R., Giugliani R. Pompe disease newborn screening working G. Newborn Screen Pompe Dis Pediatr. 2017;140(Suppl. 1):S4–S13. doi: 10.1542/peds.2016-0280C. [DOI] [PubMed] [Google Scholar]
- 23.van der Ploeg A.T., Kruijshaar M.E., Toscano A., Laforet P., Angelini C., Lachmann R.H. European consensus for starting and stopping enzyme replacement therapy in adult patients with Pompe disease: a 10-year experience. Eur J Neurol. 2017;24(6):768–e31. doi: 10.1111/ene.13285. [DOI] [PubMed] [Google Scholar]
- 24.Zuber T.J. Punch biopsy of the skin. Am Fam Physician. 2002;65(6) (1155-8, 61-2, 64) [PubMed] [Google Scholar]
- 25.Bergsma A.J., Kroos M., Hoogeveen-Westerveld M., Halley D., van der Ploeg A.T., Pijnappel W.W. Identification and characterization of aberrant GAA pre-mRNA splicing in pompe disease using a generic approach. Hum Mutat. 2015;36(1):57–68. doi: 10.1002/humu.22705. [DOI] [PubMed] [Google Scholar]
- 26.van Diggelen O.P., Oemardien L.F., van der Beek N.A., Kroos M.A., Wind H.K., Voznyi Y.V. Enzyme analysis for Pompe disease in leukocytes; superior results with natural substrate compared with artificial substrates. J Inherit Metab Dis. 2009;32(3):416–423. doi: 10.1007/s10545-009-1082-3. [DOI] [PubMed] [Google Scholar]
- 27.Boerkoel C.F., Exelbert R., Nicastri C., Nichols R.C., Miller F.W., Plotz P.H. Leaky splicing mutation in the acid maltase gene is associated with delayed onset of glycogenosis type II. Am J Hum Genet. 1995;56(4):887–897. [PMC free article] [PubMed] [Google Scholar]
- 28.Dardis A., Zanin I., Zampieri S., Stuani C., Pianta A., Romanello M. Functional characterization of the common c.-32-13T>G mutation of GAA gene: identification of potential therapeutic agents. Nucleic Acids Res. 2014;42(2):1291–1302. doi: 10.1093/nar/gkt987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Wal E., Bergsma A.J., van Gestel T.J.M., SLM In 't Groen, Zaehres H., Arauzo-Bravo M.J. GAA deficiency in Pompe disease is alleviated by exon inclusion in iPSC-derived skeletal muscle cells. Mol Ther Nucleic Acids. 2017;7:101–115. doi: 10.1016/j.omtn.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van der Wal E., Bergsma A.J., Pijnenburg J.M., van der Ploeg A.T., Pijnappel W. Antisense oligonucleotides promote exon inclusion and correct the common c.-32-13T>G GAA splicing variant in Pompe disease. Mol Ther Nucleic Acids. 2017;7:90–100. doi: 10.1016/j.omtn.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pruniski B., Lisi E., Ali N. Newborn screening for Pompe disease: impact on families. J Inherit Metab Dis. 2018;41(6):1189–1203. doi: 10.1007/s10545-018-0159-2. [DOI] [PubMed] [Google Scholar]
- 32.Wens S.C., van Gelder C.M., Kruijshaar M.E., de Vries J.M., van der Beek N.A., Reuser A.J. Phenotypical variation within 22 families with Pompe disease. Orphanet J Rare Dis. 2013;8:182. doi: 10.1186/1750-1172-8-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Filippi P., Saeidi K., Ravaglia S., Dardis A., Angelini C., Mongini T. Genotype-phenotype correlation in Pompe disease, a step forward. Orphanet J Rare Dis. 2014;9:102. doi: 10.1186/s13023-014-0102-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baek R.C., Palmer R., Pomponio R.J., Lu Y., Ma X., McVie-Wylie A.J. The influence of a polymorphism in the gene encoding angiotensin converting enzyme (ACE) on treatment outcomes in late-onset Pompe patients receiving alglucosidase alfa. Mol Genet Metab Rep. 2016;8:48–50. doi: 10.1016/j.ymgmr.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuperus E., van der Meijden J.C., SLM In 't Groen, Kroos M.A., Hoogeveen-Westerveld M., Rizopoulos D. The ACE I/D polymorphism does not explain heterogeneity of natural course and response to enzyme replacement therapy in Pompe disease. PLoS One. 2018;13(12):e0208854. doi: 10.1371/journal.pone.0208854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goina E., Peruzzo P., Bembi B., Dardis A., Buratti E. Glycogen reduction in myotubes of late-onset Pompe disease patients using antisense technology. Mol Ther. 2017;25(9):2117–2128. doi: 10.1016/j.ymthe.2017.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bergsma A.J., van der Wal E., Broeders M., van der Ploeg A.T., Pim Pijnappel W.W.M. Alternative splicing in genetic diseases: improved diagnosis and novel treatment options. Int Rev Cell Mol Biol. 2018;335:85–141. doi: 10.1016/bs.ircmb.2017.07.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material