Abstract
Background
There is a lack of information as to which molecular processes, present at diagnosis, favor tumour escape from standard-of-care treatments in cervical cancer (CC). RAIDs consortium (www.raids-fp7.eu), conducted a prospectively monitored trial, [BioRAIDs (NCT02428842)] with the objectives to generate high quality samples and molecular assessments to stratify patient populations and to identify molecular patterns associated with poor outcome.
Methods
Between 2013 and 2017, RAIDs collected a prospective CC sample and clinical dataset involving 419 participant patients from 18 centers in seven EU countries. Next Generation Sequencing has so far been carried out on a total of 182 samples from 377 evaluable (48%) patients, allowing to define dominant genetic alterations. Reverse phase protein expression arrays (RPPA) was applied to group patients into clusters. Activation of key genetic pathways and protein expression signatures were tested for associations with outcome.
Findings
At a median follow up (FU) of 22 months, progression-free survival rates of this FIGO stage IB1-IV population, treated predominantly (87%) by chemoradiation, were65•4% [CI95%: 60•2-71.1]. Dominant oncogenic alterations were seen in PIK3CA (40%), while dominant suppressor gene alterations were seen in KMT2D (15%) and KMT2C (16%). Cumulative frequency of loss-of-function (LOF) mutations in any epigenetic modulator gene alteration was 47% and it was associated with PIK3CA gene alterations in 32%. Patients with tumours harboring alterations in both pathways had a significantly poorer PFS. A new finding was the detection of a high frequency of gains of TLR4 gene amplifications (10%), as well as amplifications, mutations, and non-frame-shift deletions of Androgen receptor (AR) gene in 7% of patients. Finally, RPPA protein expression analysis defined three expression clusters.
Interpretation
Our data suggests that patient population may be stratified into four different treatment strategies based on molecular markers at the outset.
Fund
European Union's Seventh Program grant agreement No 304810.
Keywords: Bioraids study, Prospective database, Cervical cancers, Whole exome sequencing, Reverse phase protein array, Epigenetics pathways, PI3KCA, Patient stratification
Research in context.
Evidence before this study
The Cancer Genome Atlas Research Network (TCGA) published the molecular characterizations of its cervical cancer (CC) dataset, providing a wealth of information on tumour subtypes. However, clinical annotation of TCGA samples does not include complete data on tumour stage and treatment outcome. Therefore, it has not been possible so far to associate molecular data with response to treatment efficacy.
Added value of this study
BioRAIDS is the first prospectively collected extensive database from Central and Western EU countries. The aim of the European BioRAIDS study was to characterize newly diagnosed cervical cancer from a biological and a clinical view, in order to identify new pre-treatment prognostic factors and to provide a scientific rationale for novel treatment combinations. FIGO stage at inclusion and the revised staging (FIGO 2018) are compared with treatment categories and outcome. Close to 90% of BioRAIDs patients received chemoradiation, as compared to surgery, which was the primary treatment in the TCGA dataset.
Implications of all the available evidence
We identified genetic alterations in PIK3CA and in epigenetic modulators that were associated with a poor prognosis. Our data suggest that new treatment combinations may improve outcome in subsets of cervical cancer patients.
Alt-text: Unlabelled Box
1. Background
Despite effective prophylactic vaccines, cervical cancer remains the second most commonly diagnosed cancer in women. It accounts for more than a quarter of a million deaths worldwide each year [1] with the highest impact in countries and regions with suboptimal access to health care. The etiology of >90% of cervical cancers is the persistent infection with high-risk human papillomavirus types (HPV) which inactivate normal cellular controls via the viral proteins E6 and E7, progressively leading to genomic instability and accumulation of molecular alterations. While prognosis is related to FIGO stage and the presence or absence of lymph node metastases [2], the role of specific molecular patterns that resist DNA repair interference (platinum compounds and chemoradiation) is poorly documented. The Cancer Genome Atlas Research Network [3] has published molecular characterizations of its cervical cancer (CC) dataset, but correlations with outcome based on specific treatments are awaited. While some patients can be treated with surgery up to stage IB2 and IIA, the gold standard of stages IB2-III cervical cancer treatment is chemoradiation. Two thirds of the “core” TCGA samples analyzed previously originated from patients in which surgery was reportedly the primary treatment.
For stages ≥ IB2 at presentation, 30–40% of patients relapse within 2–3 years [4]. While improved therapeutic strategies over the last decade using IMRT (intensity modulated radiotherapy) to the pelvis and “preventive” para-aortic lymph node irradiation in patients with high risk disease and positive pelvic nodes, do make an impact on local control and outcome, there remains a high variability in treatment strategies. Image guided brachytherapy in a recently published multicenter cohort study (RetroEMBRACE) demonstrated excellent 3-year local control rates of 93% and 79% for patients with FIGO stage IIB and IIIB disease, respectively. However, the 5-year actuarial disease-specific OS was 65% [5].
A recent report identified a subset of virally associated tumours that presented a molecular profile distinct from that of typical HPV+ tumours and exhibited poor treatment response, indicating molecular and clinical similarities with HPV- tumours [6]. Patient stratification based on molecular patterns associated with poor outcome is a first step towards targeted strategies. In the current manuscript, we report on the first prospectively collected extensive database from Central and Western EU countries with a focus on correlating molecular patterns with tumour pathophysiology, response to treatment and outcome.
2. Methods
2.1. Patient recruitment, samples collection and treatment
The EU funded RAIDs Network (Rational Molecular Assessment and Innovative Drug Selection, www.raids-fp7.eu) collected a prospective CC sample and dataset BioRAIDs: NCT02428842 (n = 419).
The clinical protocol together with tumour sampling procedures and quality control of samples and treatments in 18 European centers (seven European countries) have been previously published. [7,8]. A signed informed consent for the participation in the study protocol was a prerogative, prior to inclusion and sampling. Quality assurance and source data verification was performed according to a risk adapted approach and a pre-defined essential study data. Frozen and formalin-fixed, paraffin-embedded tumour samples, blood as well as frozen sera specimen were transferred to common repositories for centralized processing, pathology review, and quality assessments at Erasmus University, Rotterdam, prior to DNA sequencing, analysis and storage at SeqOmics Ltd., Hungary. Ethical review was conducted according to national requirements and in accordance with the Declaration of Helsinki [9,10]. All tumour and serum samples and clinical data were registered in a common database using a unique barcode system assuring data privacy. A radiotherapy dummy run was performed before patient recruitment to homogenize contouring among centers (Rivin del Campo et al., Radioth Oncol 2017, Supplementary material). Treatment decisions were based on guidelines defined in the clinical protocol as detailed in Ngo et al., 2015 [7] and in Supplementary material. Since FIGO staging at the time of patient accrual did not take into account lymph node staging, for the purpose of the present analysis, the patient population is shown both as a function of initial FIGO staging as well as the new FIGO 2018 staging, upstaging early stage patients with lymph node positivity to stage IIIC and down staging patients with previous stage IVA to stage IIIC2.
2.2. HPV typing
All samples included in this study were analyzed for HPV type, using the SPF10 primer set and INNO-LiPA HPV genotyping extra line probe assay (Fujirebio Europe, Gent, Belgium) according to the manufacturers protocol. For DNA isolation, one to five 10 μm tissue sections were cut depending on the size of the tumour biopsy. DNA was isolated using the automated Tissue Preparation System (Siemens Healthcare Diagnostics, NY, USA).
2.3. WES & targeted sequencing and bioinformatics analyses
Paired-end whole exome sequencing of 87 samples and paired-end targeted gene panel sequencing (608 genes) of 96 paired tumour/normal samples was performed at SeqOmics (Hungary) on a HiSeq2500 platform to date. The sequencing was performed to reach an average depth of coverage of ~150× for WES and ~730× for targeted sequencing. The data were further processed by the Institut Curie bioinformatics pipeline. Somatic alterations (point mutations, insertions/deletions and copy number changes) were identified from the aligned sequences of matched-samples using dedicated tools (Supplementary methods). Biologically relevant variants were selected regarding their functional impact per gene category (oncogene, tumour suppressor gene, or uncharacterized) and compared with patients' outcome. Detailed information on genomic and bioinformatics analyses is available in the Supplementary Methods section.
The future integration of additional BioRAIDS data such as the ongoing longitudinal ctDNA analysis, of transcriptomics, methylome and TME characteristics will decipher the pathway complexity in more detail and hopefully assist innovative treatment orientations.
2.4. Reverse phase protein arrays (RPPA)
154 baseline samples, 103 post-treatment samples were processed. Arrays were labeled with 194 specific antibodies except for the negative control(s) where primary antibodies were omitted, using an Autostainer Plus (AGILENT). All primary antibodies used in RPPA have been previously tested by Western Blotting to assess their specificity for the protein of interest. Detailed information is available in the Supplementary Methods section [11,12].
2.5. Biostatistical analysis
Quantitative data were described as median with range. Qualitative data were presented as numbers and proportions leaving out missing data. Survival curves were constructed according to Kaplan-Meier and were compared using the log-rank test. Overall Survival (OS) and Progression-Free Survival (PFS) were calculated from the date of inclusion to the date of event, (disease related death for OS and relapse/or disease related death for PFS). Patients without event at the date of last follow-up were censored at this time. Univariate and multivariate Cox proportional hazard models were performed to determine prognostic factors associated with relapse or death. All analyses were performed using R version 3·4·4 software.
3. Findings
3.1. Patient population eligible for analysis
Four hundred and nineteen patients with histologically proven cervical carcinoma cases were prospectively recruited between 2013 and 2016 in 18 centers from seven European countries [7,8]. Fourty two patients were not eligible for analysis due to: inclusion error [26], death before start of treatment [1], investigator/patient decision [6], second cancer [4], lost to follow up [3], or missing data [2]. At a median follow up time of 22 months [min = 1·1 – max = 41·2], PFS data is available for 377 patients.
3.2. Staging procedures
As defined by protocol, the FIGO staging based on clinical examination was confirmed by magnetic resonance imaging (MRI) in all 377 patients (100%). Median tumour size by MRI was 48 mm [range: 9–167]. Tumour size ≥4 cm (≥stage IIA2) was present in 248/377 (74·5%) patients. Additional imaging was by CT scan in 230 patients (61%), positron electron tomography (PET) scan in 185 patients (49%). Lymphadenectomy for lymph node staging purposes was performed in a total of 130 (34%) patients, depending on center policy. According to the FIGO classification in use at the time of trial conduct 291 (77%), of the evaluable patients were considered stage I-II and 86 (23%) stage III-IV. If patients with positive lymph nodes are being reclassified according to the new FIGO 2018 classification [13], into IIIC1 (pelvic node positive) and IIIC2 (aortic node positive), 137 (36%) patients are to be considered stages I-II and 240 (64%) stages III-IV. PFS according to FIGO stages are illustrated in Supplementary Figs 1 & 2. Pretreatment PET-scan imaging was carried out in 13/56 (23%) of surgical patients, in 138/263 (52%) of chemoradiation patients and in 34/58 (74%) neoadjuvant chemotherapy (NACT) patients. Positive lymph nodes (by pathology or by imaging) were diagnosed in a total of 216/377 (57·3%) patients.
3.3. Treatment allocation
Primary treatment allocation as a function of restaging according to the FIGO 2018 classification is shown in Table 1. First line surgical management was the preferred management in early stage (IB1 and IB2) as well as in pelvic lymph node positive disease (IIIC1), whereas chemoradiation was the treatment of choice in higher stages (stages II and III). Only 15% (n = 56) of the population were allocated to first line surgery, while the majority of the patients (n = 263; 70%) were allocated to the chemoradiation (CR) group. The patients treated with NACT had stage III-IV advanced or metastatic disease, representing 15% (n = 58) of the population. In the chemoradiation group, 21 patients could not receive concomitant chemotherapy and 34 patients received less than the planned four to five cycles of chemotherapy for reasons of tolerance (see Supplementary data).
Table 1.
Treatment allocation as a function of FIGO staging.
| FIGO 2014 |
FIGO 2018 |
Primary treatment allocation |
|||||
|---|---|---|---|---|---|---|---|
| Stage | Number | Stage | Number | (%) | Surgery | Chemo radiation | NACT |
| IB1 | 34 | IB1 | 25 | 7% | 18 | 7 | 0 |
| IB2 | 62 | IB2 | 25 | 7% | 10 | 9 | 6 |
| IIA | 28 | IIA | 14 | 4% | 0 | 14 | 0 |
| IIB | 167 | IIB | 73 | 19% | 2 | 67 | 4 |
| IIIA | 11 | IIIA | 2 | 1% | 0 | 1 | 1 |
| IIIB | 42 | IIIB | 18 | 5% | 0 | 17 | 1 |
| IIIC1 | 147 | 39% | 23 | 108 | 16 | ||
| IIIC2 | 40 | 11% | 1 | 28 | 11 | ||
| IVA | 18 | IVA | 18 | 5% | 1 | 11 | 6 |
| IVB | 15 | IVB | 15 | 4% | 1 | 1 | 13 |
| TOTAL | 377 | 377 | 100% | 56 | 263 | 58 | |
3.4. First line and follow on treatments received in the first 6 months
Surgery: While surgical resection was the first intention-to-treat in a limited population (n = 56), a surgical resection in the first 6 months period was deemed necessary in a total of 136 (36% of all) patients (see Supplementary data). Lymphadenectomy was carried out in 202 (53%) of all patients, as a staging procedure (34%), or for treatment (19%) purposes. (Additional information in Supplementary).
Radiotherapy: Treatment consisted in external beam radiation therapy (EBRT) with concomitant platinum based chemotherapy in 295 (78%) of the population as per protocol and previous publications [7,14]; in EBRT alone (n = 34) or in brachytherapy alone (n = 2). A total of 331/377 patients (87%) received radiotherapy either as primary or as follow on treatment after surgery or NACT. Following pelvic irradiation, an additional boost was administered to the tumour by EBRT (n = 66) and/or by brachytherapy (n = 284). Twenty-two patients received no additional external radiotherapy boost. Forty-three received no brachytherapy. (Additional information in Supplementary).
Neoadjuvant Chemotherapy: A combination of Taxol and Carboplatin was the first treatment in 55 patients (Fig. 1). Additional loco-regional treatment was administered in 44/55 (80%) patients. Second line chemotherapy was administered as needed in advanced/refractory disease.
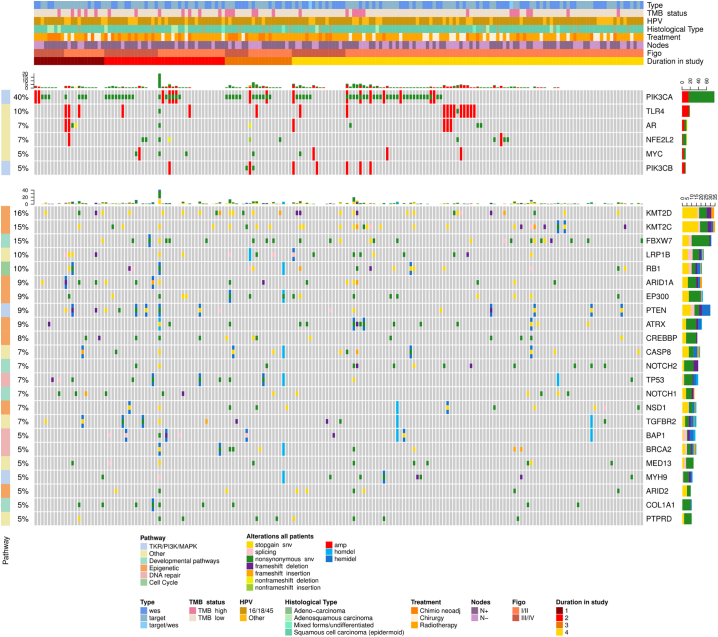
Fig. 1.
Oncoprints are presented, showing the most frequently altered (higher than 5% of the cohort) oncogenes and tumour suppressor genes, combining both small and large-scale variations together with clinical information (WES and target panel combined), using the R package Complex Heatmap (v1.17.1). Each column represents a patient. Key oncogene and tumour suppressor gene alterations are listed by name (right column), by frequency in the population (left column), by alteration type (colour code) by each clinical data detail for each individual patient (heading). Headings define the type of mutational analysis (full exome or targeted 607 gene panel), the tumour mutational burden (high/low; cut-off = 8mutations/Mb), the HPV clade (7, 9, other), the histological type (squamous, adenocarcinoma, other), first treatment received (surgery, chemoradiation, NACT), node status (positive/negative), FIGO stage (2014) not taking into account lymph node involvement and regrouped into I-II versus III-IV. Quantile PFS ordered by time interval from start to date of progression: 1. PD prior to 6 Mo, 2. 6–12 Mo, 3. 12–18 Mo, 4. DFS at last follow up.
3.5. Survival based on clinical characteristics (Table 2)
Table 2.
PFS according to clinical and tumour characteristics.
|
Univariate |
Multivariate |
|||||
|---|---|---|---|---|---|---|
| HR | IC95%(HR) | p-value | HR | IC95%(HR) | p-value | |
| Age | 0·47 | |||||
| <50 | 1 | |||||
| ≥50 | 1·14 | [0·79; 1·65] | ||||
| Tobacco consumption | 0·62 | |||||
| Never | 1 | |||||
| Yes. current or past (>1PA) | 0·91 | [0·63; 1·31] | ||||
| BMI | 0·01 | 0·03 | ||||
| <25 | 1 | 1 | ||||
| ≥25 | 0·63 | [0·43; 0·91] | 0·66 | [0·45; 0·96] | ||
| ECOG | 0·002 | 0·04 | ||||
| 0 | 1 | 1 | ||||
| 1–2 | 2·10 | [1·32; 3·28] | 1·70 | [1·04; 2·77] | ||
| Histological type | 0·73 | |||||
| Squamous cell carcinoma | 1 | |||||
| Adenocarcinoma | 1·11 | [0·65; 1·88] | ||||
| Adenosquamous. clear cell. mixed + undifferentiated | 0·75 | [0·30; 1·84] | ||||
| Hemoglobin (g/dl) | 0·06 | 0·34 | ||||
| ≤10 | 1 | 1 | ||||
| >10 | 0·61 | [0·37; 1·01] | 0·76 | [0·44; 1·32] | ||
| FIGO 2014 | <0·001 | |||||
| I | 1 | |||||
| II | 1·87 | [1·09; 3·22] | ||||
| III | 4·90 | [2·66; 9·02] | ||||
| IV | 3·64 | [1·83; 7·21] | ||||
| FIGO 2018 (integrates lymph nodes status under IIIC) | <0·001 | 0·004 | ||||
| I | 1 | 1 | ||||
| II | 2·14 | [0·86; 5·35] | 1·83 | [0·72; 4·63] | ||
| III | 4·04 | [1·76; 9·29] | 3·25 | [1·40; 7·57] | ||
| IV | 5·71 | [2·23; 14·60] | 3·46 | [1·28; 9·31] | ||
| HPV type (based on hybridisation test) | 0·17 | 0·56 | ||||
| Clade 9 (HPV 16.31.33.35.52.58) | 1 | 1 | ||||
| Clade 7 (HPV 18.39.45.59.68) | 1·47 | [0·96;2·24] | 1·32 | [0·85; 2·05] | ||
| Others or Negatives | 1·38 | [0·76; 2·52] | 1·30 | [0·69; 2·44] | ||
Median overall survival is presently not reached; survival rates at 24 months are 85·2% [CI95%: 80·7–89·8], progression-free survival rates are 65·4% [CI95%: 60·2–71·1]. As expected, a highly significant correlation with outcome was seen for both FIGO 2014 and FIGO 2018 stages (Tables 1 and 2 and Supplementary Figs 1 & 2). We chose to integrate the new FIGO 2018 staging in the multivariate Cox regression model and the results of multivariate analysis identified FIGO 2018, ECOG performance index and BMI as correlated with PFS. Histological type was not related to outcome.
3.6. The genetic landscape of treatment-naive cervical cancer
In our cohort of 182 patients [out of the 377 evaluable BioRAIDS patients (48%)] analyzed, using next generation sequencing (NGS) all novel and previously confirmed significantly altered genes reported by The Cancer Genome Atlas [3] were detected in the RAIDs dataset; the most frequent alterations are represented in Fig. 1. PIK3CA mutations (Supplementary Fig 3) and/or gene amplifications were the most frequently diagnosed oncogenic alteration, present in 40% of BioRAIDs patients. The most frequently diagnosed suppressor gene alterations were loss-of-function (LOF) mutations in KMT2A-D (Lysine methyl transferase) genes leading to defective histone H3K4 methylation. The cumulative frequency of tumours harboring any suppressor gene alterations in the epigenetic pathway (involving KMT2C, KMT2D, EP300, ARID1A, ARID2, ATRX, CREBBP, KMT2A, KDM5C) was 45% of which 32% also had alterations in PI3KCA. Relevant to the viral etiology and possible influencing immune function, a high level of TLR4 (toll like receptor-4) amplifications (10%), not previously reported to our knowledge, was detected in the BioRAIDs population. We confirmed the presence of CD274/PD-L1 and PDCD1LG2/PD-L2 amplifications at a low frequency [5/182 patients, data not shown]. Another new finding was the presence of amplifications, mutations, and non-frame-shift deletions of AR (Androgen receptor) in 7% of patients, presently of unknown significance. The presence of an APOBEC signature could only be tested in samples with full exome sequencing and the numbers are too small to test for significant correlations with PIK3CA hotspot mutation.
RPPA protein expression analysis [11] of 154 samples defined three expression clusters (Supplementary Figs 4 & 5). Significant features of RAIDs cluster 1 corresponded to an “EMT” signature, including high phospho-YAP (p = 3·11e-06) and phospho-MET (p = 4·74e-03), high MET (p = 2·42e-11), high NOTCH (p = 4·14e-17) and low E-CADHERIN (p = 2·41e-06). Significant features of cluster 2 included high NBS1 (p = 5·85e-11), MRE11 (p = 6·09e-11), FANCD2 (p = 1·69e-09), phosphorylated FANCD2 (p = 6·37e-07), and HSP90alpha (p = 4·31e-06), as well as low levels of phosphorylated AKT (p = 5.48e-18), EGFR (p = 6.39e-11), HER3 (p = 1.96e-09), HER2 (p = 4.79e-09) and p70 S6K (p = 1.01e-05) compared to the two other clusters. Significant features of cluster 3 included phosphorylated forms of NFkB (p = 8.29e-09), p70 S6 kinase (p = 2·82e-04), AKT (p = 4·36e-04), p38 MAPK (p = 7·32e-03), ERK1/2 (p = 1·32e-02), and EGFR (p = 2·67e-02).
3.7. Molecular signatures predictive of outcome following standard therapy
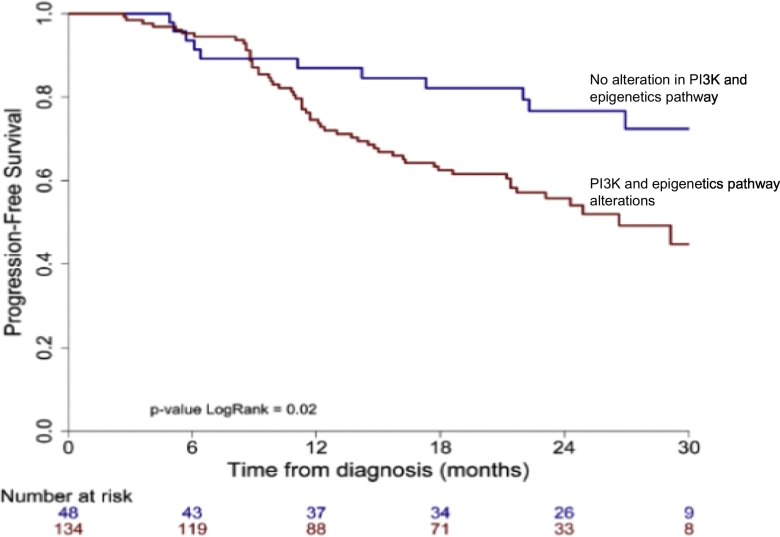
When patients were clustered in quantiles of PFS (ONCOPRINT: Fig. 1), those patients who remained progression free at the time of analysis (PFS, yellow line), had visibly less alterations in genes involved in Tyrosine Kinase receptor/PI3K pathway and in suppressor functions related to the following epigenetic enzyme modifications (KMT2C, KMT2D, KMT2A, KDM5C, EP300, CREBBP, ARID1A, ARID2, ATRX). When these pathway alterations were pooled in a “metagene”, Kaplan Meyer progression free survival estimates (Fig. 2) confirmed a poorer PFS (p = 0·02) for patients with tumours harboring at least one molecular alteration in this metagene as opposed to patients with no such alteration. While the medium number of variants increased slightly with FIGO stage this was not statistically significant, since the confidence intervals are very large (Supplementary Fig. 6).
Fig. 2.
Progression free survival according to the presence/absence of alterations in a METAGENE composed of any of the following alterations in the PI3K pathway and/or in Enzymes involved in Epigenetic Signaling. The list of genes included in this analysis was predominantly (but not exclusively) oncogenes from the Tyrosine Kinase receptor/PI3K pathway: PI3KCA and PI3KCB mutations as well as PTEN in association with the following (predominantly suppressor) genes with loss of function alterations in Epigenetic enzymes: KMT2C, KMT2D, KMT2A, KDM5C, EP300, CREBBP, ARID1A, ARID2, ATRX.
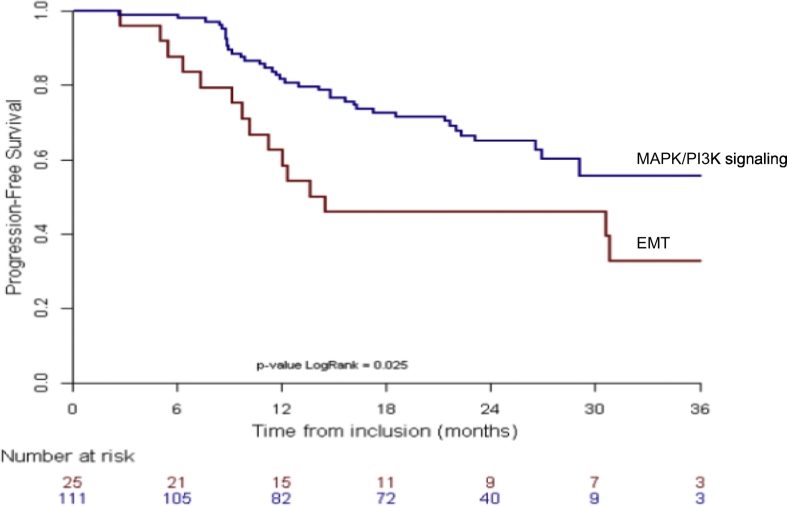
Among the 154 tumours analyzed using RPPA, 136 patients were evaluable for outcome. While there was no difference in overall survival, our data showed a significant poorer PFS for the cluster “EMT” as compared to the other two clusters combined (Fig. 3) (p = 0·03). There was no correlation between WES analysis and RPPA data, likely due to the limited antibody targets used in RPPA as compared to a full exome analysis.
Fig. 3.
Progression free survival by RPPA analysis from the total evaluable population with available RPPA data (n = 136). Comparison of outcome of cluster “EMT” (Red) versus pooled gene expression clusters of DNA damage and MAPK/PI3K signaling (Blue), suggests a poorer PFS for the patients whose tumours showed EMT associated gene expression. Gene expression of epigenetically active enzymes was not assessed. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.8. Interpretation
Patient stratification according to outcome categories based on genomic variants, needs a prospectively accrued, sufficiently large cohort of tumour samples together with supervised standard treatments. Chemoradiation was the first line of treatment in >2/3 of patients and chemoradiation or radiation was administered in 87% of all patients if first line treatments were pooled with Supplementary treatments received during the first six months. This is in stark contrast with the predominant surgical treatment (70%) reported in other series [3,15]. In case of a pelvic lymph node detection prior to treatment, most centers perform primary chemoradiation to avoid the morbidity of adding radical surgery to radiotherapy. In the BioRAIDS population, only 36% of patients had a surgical tumour resection, either as a first approach in small tumours (15%) or as a secondary resection (16%) in case of an incomplete or doubtful response to chemoradiation. Some positive nodes are detected only during surgery leading to additional radiotherapy or chemoradiation. Reclassifying BioRAIDS patients according to the new FIGO 2018 stage resulted in well differentiated PFS curves (Supplementary data).
Integrative bioinformatics analysis suggests 4 privileged treatments for tumours associated with resistance to present treatments directed to DNA repair interference.
1: PIK3CA activating mutations and amplifications, present in 40% of tumours, were frequently associated with LOF in epigenetic regulator genes. The cumulative presence of both (in 34% of the population), but not the PIK3 pathway alone as suggested before [16] was associated with a significantly poorer PFS. PI3K activity contributes to diverse functional roles in cellular metabolism, immune function and cell motility in cancer [17] and is associated with treatment resistance. In a patient group with PIK3 pathway mutations we previously reported a negative impact on patient outcome by the addition of Cetuximab, an EGFR inhibitor [14] to chemoradiation. Recent evidence suggests that PIK3CA genetic alterations may induce centrosome amplification and increase tolerance to spontaneous genome doubling, thereby contributing to irreversible genomic changes in cancer [18]. While much effort has been devoted to targeting PIK3CA, tolerance to specific PI3K isoforms has been so far a rate limiting step. It was recently suggested that toxicities may be contained through intermittent dosing and nanoparticle delivery [17]. Many selective inhibitors for each PI3K isoform are in clinical trials and activity was shown associated with PI3K specific mutations [19].
2: Epigenetic LOF alterations were seen in 47% BioRAIDs tumours assessed so far. While epigenetic alterations (involving DNA methylation and covalent histone modifications) are increasingly reported in solid tumours, this is to our knowledge the first report emphasizing not only a frequent loss-of-function (LOF) mutations in KMT2A-D genes, leading to defective histone H3K4 methylation in cervical cancers, but also linking these to outcome. KMT2A-D have already been reported in many other solid tumours (reviewed by [20]) and have been associated with increased micro satellite instability (MSI) in colorectal carcinoma. Of interest to oncologists is that pan histone deacetylase inhibitors (HDACi), such as Vorinostat (Zolinza®), which affects the alterations regulation of histone and non-histone proteins by modifying their post-translational acetylation, is likely to be relevant for cervical cancer treatment. [21] [22]. In preclinical models, Banerjee et al. [23] showed that Vorinostat significantly reduced E6 and E7 activity, abrogated viral DNA amplification and inhibited host DNA replication. >20 epigenome targeting drugs are in clinical development across cancer types, they have a broad spectrum of epigenetic activities. Vorinostat (Zolinza®), initially marketed for the treatment of cutaneous T-cell lymphoma, and more recently Romidepsin (Istodax®) are approved drugs.
3: Immune checkpoint inhibition (ICI) has significantly changed cancer therapy showing impressive durable responses across cancer types. There is only limited data on ICI treatment of PDL-1 positive cervical cancer to date; a 17% partial response rate was documented in recurrent disease [24]. The viral etiology of CC will affect the host immune response and the phenotype of the tumour microenvironment (TME). The presence of double stranded genomic or viral DNA in the cytosol has been recently shown to lead to activation of the cGAS-STING cytosolic DNA sensing pathway and the down-stream non-canonical NFkB inflammatory signaling pathway [25]. Collectively, HPV viruses also cause genetic amplifications, some of which may specifically impact inflammation and the immune response [26]: TLR4 (toll like receptor 4), was amplified in 10% of our series [not previously reported to our knowledge], PDL-1 and PDL-2 amplifications were present at a low frequency (3%). Loss of function of CASP8 (caspase 8), has been linked to apoptosis, necrosis and inflammation and together with amplification of TLR4 genes might serve as biomarker for an “inflamed” TME. Furthermore, the poor prognostic RPPA Cluster 1 was characterized by biomarkers of EMT and associated with upregulated PD-L1 and B7-H4. These features have been controlled by immunohistochemistry in a parallel project defining the tumour microenvironment (ongoing) with excellent correlation. EMT biomarkers may allow the identification of patients that could potentially benefit from ICI in the future. Additional data on gene methylation or miRNA which had not been planned in the present project upfront may shed light on the mechanism of enhanced gene expression. Moreover, recent preclinical data further suggested that HDAC inhibition by Vorinostat may potentiate effects of ICI through upregulation of PD-L1 and HLA-DR on tumour cells, [27] suggesting a potential synergy by combining ICI and HDACi in this patient population.
4: LOF mutations linked to deficient DNA repair were seen in ATRX (9%), BAP1 (5%), BRCA2 (5%) while the FANCB oncogene was mutated or amplified in 3% of tumours. RPPA also identified differences in expression and activation of DNA repair proteins among the three clusters, suggesting that some RPPA clusters might be more sensitive to PARP inhibitor than others. Treatment downstaging, through the use of a PARPi, rather than chemoradiation, may be envisioned in the context of a clinical trial. Compared to the omnipresent p53 and RB alterations in high grade serous ovarian cancer, the respective mutation frequencies 7% and 10% in CC were rare, consistent with the predominant HPV effects on the viral oncoproteins E7 and E6 to establish the permissive milieu by destabilizing the pRB and p53 family proteins [23] rather than by mutation.
3.9. Future prospects for clinical decision making
While routine molecular diagnostic testing has yet to be introduced to guide personalized cervical cancer treatment, the BioRAIDs dataset defined activated genetic pathways and expression signatures that are associated with poor outcome based on the present standard therapies. IMRT and prophylactic irradiation to para-aortic lymph nodes in high risk patients has been shown to mitigate risk and improve outcome, yet, despite occasional abscopal effects, they cannot effectively target micro or macro metastases outside of the radiation field. Higher rates of abscopal effect in many tumour sites may be reached through the integration of radiation or chemoradiation with immunotherapy and combination trials (NICOL, PRIMMO) are ongoing.
Key biomarkers for CC diagnostics appear dominated by alterations in the PI3K pathway, in genes coding for epigenetic remodeling enzymes, for DNA repair and for inflammatory pathways. Pathway specific druggability, using distinct compounds to define the genomic correlates for drug specific sensitivity or resistance in cell lines and tumours as well as drug synergies and synthetic lethality need to be tested in future preclinical and clinical studies. For the purpose of translational studies, the continuous pretreatment collection of frozen tissues in advanced stage CC appears mandatory.
Acknowledgments
Acknowledgments
We thank Charlotte Lecerf, Claire Morel, Choumouss Kamoun, Sébastien Armanet, Marie-Emmanuelle Legrier, Sinette Ngoumou Mabiala, Corine M Beaufort, Kirsten Ruigrok-Ritstier, Coralie Errera, Dora Latinovics, Judit Cseklye, Peter Bihari, Anika Hävemeier, Isabel Brito, Bérengère Ouine, Aurélie Cartier, for their precious help in the conduct of the RAIDs project.
Funding
This project has received founding from the European Union's Seventh Program for research, technological development and demonstration under grant agreement No 304810.
Contributors
-
•
Suzy Scholl and Maud Kamal coordinated the project and wrote the manuscript.
-
•
Marina Popovic, Anne de la Rochefordiere, Aliosa Mandic, Nina Samet, Marius Craina, Sanne Samuel, Madalin Margan, Henry Zijlmans, Peter Hillemanns, Sorin Dema, Akis Dema, Goran Malenkovic, Branislav Djuran, Anne Floquet, Delphine Garbay, Frédéric Guyon, Pierre Emmanuel Colombo, Michel Fabbro8, Christine Kerr8, Charlotte Ngo, Fabrice Lecuru, Charles Coutant, Frédéric Marchal, Nathalie Mesgouez-Nebout, Virginie Fourchotte, Jean Guillaume Feron, MD, Philippe Morice, Pauline Wimberger, Jean-Marc Classe, Coraline Dubot, Noreen Gleeson and Gemma Kenter included patients.
-
•
Katarina Koprivsek coordinated imaging.
-
•
Elodie Girard, Pierre Gestraud, Nicolas Servant, Philippe Hupe and Balazs Balint performed bioinfomatics' analyses.
-
•
Sylvain Dureau and Alexia Savignoni performed the biostatistical analyses
-
•
Heiko von der Leyen coordinated the clinical trial in Eastern European countries
-
•
Mathieu Minsat, Eleonor Rivin del Campo, Anne de la Rochefordiere and Eric Deutsch coordinated radiotherapy procedures and included patients.
-
•
Attila Kereszt and Istvan Nagy performed sequencing analyses.
-
•
Els (Petronella) Bern provided scientific feedback and participated in the manuscript design.
-
•
Ekaterina Jordanova performed HPV typing.
-
•
Nicolas de Saint-Jorre was in charge of the eCRF.
-
•
Leanne de Koning performed RPPA analyses and was involved in the manuscript preparation.
-
•
Pierre Fumoleau, Roman Rouzier participated to manuscription preparation.
Declaration of interests
All authors declared: NO conflict of interest.
Data sharing
The study protocol with the statistical analysis plan and informed consent form are publicly available [7]. Frozen tumour material collected for the study, including raw and processed samples are stored in a public biobank (https://www.ibbl.lu), IBBL (Integrated BioBank of Luxembourg), an autonomous not-for-profit institute dedicated to supporting biomedical research for the benefit of patients. Samples can be made available immediately beginning “with publication” of the present data on a collaborative basis, aiming to complete RAIDS sample analysis and data integration. Sample access will need a review of the analytical plan by the board of the RAIDS consortium and comply with the Sample Access Governance of IBBL which can be accessed following a signed data access agreement. Bioinformatics data, including a data dictionary defining each field in the set, is stored at Institut Curie. Access criteria for bioinformatics data will be after approval of a proposal by a board including RAIDS investigators. De-identified participant clinical data is stored at Institut Curie (26 rue d’Ulm 75005 Paris France).
Main contact is suzy.scholl@curie.fr and maud.kamal@curie.fr.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.03.069.
Appendix A. Supplementary data
Supplementary material 1
Supplementary material 2
References
- 1.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015 Mar;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Marth C., Landoni F., Mahner S., McCormack M., Gonzalez-Martin A., Colombo N. Cervical cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol Off J Eur Soc Med Oncol. 2018 Oct 1;29(Supplement_4):iv262. doi: 10.1093/annonc/mdy160. [DOI] [PubMed] [Google Scholar]
- 3.Cancer genome atlas research network, Albert Einstein College of Medicine, analytical biological services, Barretos Cancer hospital, Baylor College of Medicine, Beckman Research Institute of City of Hope, et al. integrated genomic and molecular characterization of cervical cancerNature. 2017 16;543(7645):378–384. doi: 10.1038/nature21386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DiSilvestro P.A., Ali S., Craighead P.S., Lucci J.A., Lee Y.-C., Cohn D.E. Phase III randomized trial of weekly cisplatin and irradiation versus cisplatin and tirapazamine and irradiation in stages IB2, IIA, IIB, IIIB, and IVA cervical carcinoma limited to the pelvis: a gynecologic oncology group study. J Clin Oncol Off J Am Soc Clin Oncol. 2014 Feb 10;32(5):458–464. doi: 10.1200/JCO.2013.51.4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sturdza A., Pötter R., Fokdal L.U., Haie-Meder C., Tan L.T., Mazeron R. Image guided brachytherapy in locally advanced cervical cancer: improved pelvic control and survival in RetroEMBRACE, a multicenter cohort study. Radiother Oncol J Eur Soc Ther Radiol Oncol. 2016;120(3):428–433. doi: 10.1016/j.radonc.2016.03.011. [DOI] [PubMed] [Google Scholar]
- 6.Gleber-Netto F.O., Rao X., Guo T., Xi Y., Gao M., Shen L. Variations in HPV function are associated with survival in squamous cell carcinoma. JCI Insight. 2019 Jan 10;4(1) doi: 10.1172/jci.insight.124762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ngo C., Samuels S., Bagrintseva K., Slocker A., Hupé P., Kenter G. From prospective biobanking to precision medicine: BIO-RAIDs - an EU study protocol in cervical cancer. BMC Cancer. 2015 Nov 4;15:842. doi: 10.1186/s12885-015-1801-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Samuels S., Balint B., von der Leyen H., Hupé P., de Koning L., Kamoun C. Precision medicine in cancer: challenges and recommendations from an EU-funded cervical cancer biobanking study. Br J Cancer. 2016 Dec 6;115(12):1575–1583. doi: 10.1038/bjc.2016.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.World Medical Association World medical association declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA. 2013 Nov 27;310(20):2191–2194. doi: 10.1001/jama.2013.281053. [DOI] [PubMed] [Google Scholar]
- 10.Emanuel E.J. Reconsidering the declaration of Helsinki. Lancet Lond Engl. 2013;381(9877):1532–1533. doi: 10.1016/s0140-6736(13)60970-8. May 4. [DOI] [PubMed] [Google Scholar]
- 11.Akbani R., Becker K.-F., Carragher N., Goldstein T., de Koning L., Korf U. Realizing the promise of reverse phase protein arrays for clinical, translational, and basic research: a workshop report: the RPPA (reverse phase protein Array) society. Mol Cell Proteomics MCP. 2014 Jul;13(7):1625–1643. doi: 10.1074/mcp.O113.034918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Troncale S., Barbet A., Coulibaly L., Henry E., He B., Barillot E. NormaCurve: a SuperCurve-based method that simultaneously quantifies and normalizes reverse phase protein array data. PloS One. 2012;7(6):e38686. doi: 10.1371/journal.pone.0038686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhatla N., Aoki D., Sharma D.N., Sankaranarayanan R. Cancer of the cervix uteri. Int J Gynaecol Obstet Off Organ Int Fed Gynaecol Obstet. 2018 Oct;143(Suppl. 2):22–36. doi: 10.1002/ijgo.12611. [DOI] [PubMed] [Google Scholar]
- 14.de la Rochefordiere A., Kamal M., Floquet A., Thomas L., Petrow P., Petit T. PIK3CA pathway mutations predictive of poor response following standard Radiochemotherapy ± Cetuximab in cervical Cancer patients. Clin Cancer Res Off J Am Assoc Cancer Res. 2015 Jun 1;21(11):2530–2537. doi: 10.1158/1078-0432.CCR-14-2368. [DOI] [PubMed] [Google Scholar]
- 15.Ojesina A.I., Lichtenstein L., Freeman S.S., Pedamallu C.S., Imaz-Rosshandler I., Pugh T.J. Landscape of genomic alterations in cervical carcinomas. Nature. 2014 Feb 20;506(7488):371–375. doi: 10.1038/nature12881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bahrami A., Hasanzadeh M., Hassanian S.M., ShahidSales S., Ghayour-Mobarhan M., Ferns G.A. The potential value of the PI3K/Akt/mTOR signaling pathway for assessing prognosis in cervical cancer and as a target for therapy. J Cell Biochem. 2017;118(12):4163–4169. doi: 10.1002/jcb.26118. [DOI] [PubMed] [Google Scholar]
- 17.Fruman D.A., Chiu H., Hopkins B.D., Bagrodia S., Cantley L.C., Abraham R.T. The PI3K pathway in human disease. Cell. 2017 Aug 10;170(4):605–635. doi: 10.1016/j.cell.2017.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berenjeno I.M., Piñeiro R., Castillo S.D., Pearce W., McGranahan N., Dewhurst S.M. Oncogenic PIK3CA induces centrosome amplification and tolerance to genome doubling. Nat Commun. 2017. 1773;8(1):24. doi: 10.1038/s41467-017-02002-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Juric D., de Bono J.S., LoRusso P.M., Nemunaitis J., Heath E.I., Kwak E.L. A first-in-human, phase I, dose-escalation study of TAK-117, a selective PI3Kα isoform inhibitor, in patients with advanced solid malignancies. Clin Cancer Res Off J Am Assoc Cancer Res. 2017 Sep 1;23(17):5015–5023. doi: 10.1158/1078-0432.CCR-16-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roy D.M., Walsh L.A., Chan T.A. Driver mutations of cancer epigenomes. Protein Cell. 2014 Apr;5(4):265–296. doi: 10.1007/s13238-014-0031-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marks P.A. Histone deacetylase inhibitors: a chemical genetics approach to understanding cellular functions. Biochim Biophys Acta. 2010 Dec;1799(10−12):717–725. doi: 10.1016/j.bbagrm.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Minucci S., Pelicci P.G. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006 Jan;6(1):38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 23.Banerjee N.S., Moore D.W., Broker T.R., Vorinostat Chow L.T. A pan-HDAC inhibitor, abrogates productive HPV-18 DNA amplification. Proc Natl Acad Sci U S A. 2018;115(47):E11138–E11147. doi: 10.1073/pnas.1801156115. 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frenel J.-S., Le Tourneau C., O'Neil B., Ott P.A., Piha-Paul S.A., Gomez-Roca C. Safety and efficacy of pembrolizumab in advanced, programmed death ligand 1-positive cervical Cancer: results from the phase Ib KEYNOTE-028 trial. J Clin Oncol Off J Am Soc Clin Oncol. 2017 Dec 20;35(36):4035–4041. doi: 10.1200/JCO.2017.74.5471. [DOI] [PubMed] [Google Scholar]
- 25.Bakhoum S.F., Ngo B., Laughney A.M., Cavallo J.-A., Murphy C.J., Ly P. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature. 2018 25;553(7689):467–472. doi: 10.1038/nature25432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang X., Chen G.T., Wang Y.Q., Xian S., Zhang L., Zhu S.M. TLR4 promotes the expression of HIF-1α by triggering reactive oxygen species in cervical cancer cells in vitro-implications for therapeutic intervention. Mol Med Rep. 2018 Feb;17(2):2229–2238. doi: 10.3892/mmr.2017.8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Terranova-Barberio M., Thomas S., Ali N., Pawlowska N., Park J., Krings G. HDAC inhibition potentiates immunotherapy in triple negative breast cancer. Oncotarget. 2017 Dec 26;8(69):114156–114172. doi: 10.18632/oncotarget.23169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
Supplementary material 2