Abstract
Background
Circulating tumor DNA (ctDNA) isolated from plasma contains genetic mutations that can be representative of those found in primary tumor tissue DNA. These samples can provide insights into tumoral heterogeneity in patients with advanced gastric cancer (AGC). Although trastuzumab has been shown to be effective in first-line therapy for patients with metastatic gastric cancer with overexpression of human epidermal growth factor receptor 2 (HER2), the mechanism of AGC resistance is incompletely understood.
Methods
In this prospective study, we used targeted capture sequencing to analyze 173 serial ctDNA samples from 39 AGC patients. We analyzed cancer cell fractions with PyClone to understand the clonal population structure in cancer, and monitored serial samples during therapy. Serial monitoring of ctDNA using the molecular tumor burden index (mTBI), identified progressive disease before imaging results (mean: 18 weeks).
Findings
We reconstructed the clonal structure of ctDNA during anti-HER2 treatment, and identified 32 expanding mutations potentially related to trastuzumab resistance. Multiple pathways activating in the same patients revealed heterogeneity in trastuzumab resistance mechanisms in AGC. In patients who received chemotherapy, mTBI was validated for the prediction of progressive disease, with a sensitivity of 94% (15/16). A higher mTBI (≥1%) in pretreatment ctDNA was also a risk factor for progression-free survival.
Conclusions
Analysis of ctDNA clones based on sequencing is a promising approach to clinical management, and may lead to improved therapeutic strategies for AGC patients.
Fund
This work was supported by grants from the National International Cooperation Grant (to J.X.; Project No. 2014DFB33160).
Keywords: Advanced gastric cancer, Circulating tumor DNA (ctDNA), Trastuzumab, Resistance mechanism, Monitoring
Research in context.
Evidence before this study
Trastuzumab has previously been shown to be an effective first-line therapy for patients with metastatic gastric cancer with overexpression of human epidermal growth factor receptor 2 (HER2); however, the mechanism of resistance and how resistant clones evolve in advanced gastric cancer (AGC) is incompletely understood. Circulating tumor DNA (ctDNA) sequencing, a method of liquid biopsy, provides a potential tool for real-time monitoring of the tumor during treatment. However, tumor heterogeneity limits the value of the ctDNA detecting method, which is based on a single gene or a few mutation positions, in predicting treatment outcomes or clinical prognoses.
Added value of this study
In this study, based on targeted capture sequencing and temporal evolution analyzing, ctDNA clonal mutations derived from tumor tissue were used in monitoring tumor burden, and 32 expanding mutations potentially related to trastuzumab resistance were identified. Moreover, in this study, we also confirmed that, in patients who received chemotherapy, mTBI was validated for the prediction of progressive disease with a sensitivity of 94% (15/16). A higher mTBI (≥1%) of the clonal structure in pretreatment ctDNA was also a risk factor for progression-free survival.
Implications of all the available evidence
This study provides a broad, clinically applicable method of using ctDNA to predict disease progression, and highlights the potential prognostic utility of mTBI for the identification of high-risk groups. It demonstrates for the first time that the co-occurrence of multiple resistant mutations in high-heterogeneity AGC, and suggests the possibility of combination of trastuzumab with other targeted agents.
Alt-text: Unlabelled Box
1. Introduction
Imaging methods combined with serum biomarkers have been commonly used to evaluate the efficacy of therapy for solid tumors [1]. Serum-based protein biomarkers, such as carcinoembryonic antigen (CEA), carbohydrate antigen 125 (CA125), and carbohydrate antigen 19-9 (CA19-9), have limitations with regard to sensitivity and specificity [[2], [3], [4], [5]]. Imaging has limitations in measuring bone, pleural, and peritoneal disease [6]. In addition, radiation exposure limits the frequency of using computerized tomography (CT) scans [7]. There is an urgent need for a highly sensitive, standardized and validated blood-based assay that may be used for the accurate detection of early relapse and/or progression of disease.
Non-invasive circulating tumor DNA (ctDNA) sequencing in plasma provides a potential tool for real-time monitoring of tumor load [[8], [9], [10]], and may be used in concert with imaging modalities to improve accuracy [11] as well as to improve the early detection of non-measurable metastases and occult lesions [12]. Molecular characteristics in ctDNA can provide clues for the temporal evolution of tumor-resistant clones [13]. Although many studies have described the use of ctDNA in late-stage tumors, the detection of a single gene or a few mutation positions to predict treatment outcome or clinical prognosis has limited value because of tumor heterogeneity among patients [[14], [15], [16], [17], [18]]. These methods have been unable to detect mutations that occur in additional regions along with tumor evolution, which may cause secondary resistance. The current clinical application of whole-exome sequencing is limited by its high cost [19]. In terms of calculating the ctDNA fraction, methodology based on mean variant allele frequency (VAF) of all detected mutations allows the use of ctDNA for molecular monitoring in patients, but could underestimate the ctDNA fraction in cell-free DNA (cfDNA) because of intra-tumor heterogeneity [20]. Currently, no widely applicable and accurate ctDNA-based method has been developed to monitor ctDNA and to explain resistance mechanisms in highly heterogeneous solid tumors such as advanced gastric cancer (AGC).
Studies on tumor evolution have provided evidence that clonal mutations (truncal mutations, occurring early and central on the phylogenetic trees) exist in all or almost all tumor cells [21,22]. Compared with subclonal mutations, these clonal mutations present a higher cancer cell fraction (CCF) and VAF in ctDNA when they are released into peripheral blood, compared with subclonal mutations (Fig. S1). With the development of a pan-cancer panel, targeted capture sequencing may cost-effectively detect gene regions to identify cancer-associated mutations in ctDNA for clonal structure reconstruction and the exploration of potential drug resistance mechanisms during treatment [23]. The mean VAF of clonal mutations, defined as molecular tumor burden index (mTBI), could help standardize the applicability and accuracy of ctDNA in monitoring tumor burden in highly heterogeneous gastric carcinoma [24]. Serial ctDNA sequencing may reveal drug-resistance mutations during treatment [21].
In this study, we validated ctDNA clonal mutations derived from tumor tissue, and determined the mTBI of serial ctDNA samples in patients with AGC who were undergoing anti-human epidermal receptor 2 (HER2) treatment. We demonstrated that the mTBI could be used as a sensitive biomarker to predict disease progression compared to standard imaging modalities in AGC patients, and that targeted capture identified potential trastuzumab-resistant clones during disease progression. We further validated the feasibility of using mTBI in AGC patients receiving chemotherapy, and showed that mTBI can serve as a predictive marker of treatment outcome.
2. Materials and methods
2.1. Patients and samples
Patients with AGC were drawn from the Fifth Medical Center, General Hospital of PLA, Beijing, China. The cohorts included 21 AGC patients receiving chemotherapy plus trastuzumab, and 18 AGC patients receiving chemotherapy alone. Patients all signed a written consent form prior to the study. Study protocols were approved by the respective institutional review boards. According to the Response Evaluation Criteria in Solid Tumors (RECIST), version 1.1, we evaluated the clinical response of all patients every 6–8 weeks.
Serial peripheral blood (10 mL) and matched formalin-fixed, paraffin-embedded (FFPE) tissues were obtained (details in Supplementary Materials and Methods).
2.2. Pan-cancer panel sequencing
DNA was isolated from all normal cell samples, serial peripheral blood, and matched tissues samples using commercial kits (Qiagen, Hilden, Germany). The KAPA Library Preparation Kit (Kapa Biosystems, Wilmington, MA, USA) was next used to prepare indexed Illumina NGS libraries. A custom SeqCap EZ Library (Roche NimbleGen, Madison, WI, USA) was used to perform target enrichment according to the manufacturer's protocol. Coordinates of selected regions and genes of each version are listed in Table S1.
2.3. Identification of somatic mutations in tumor tissue and ctDNA
Somatic mutations were detected in circulating and tissue DNA. Non-synonymous mutations annotated by ANNOVAR were used in clonal structure reconstruction (details in Supplementary Materials and Methods).
2.4. Clone structure and mTBI analysis
PyClone was employed to analyze the clonal structure, based on a Bayesian clustering method. An independent input was used to analyze the clonal structure in ctDNA and the tissue for ctDNA at baseline and matched tissue samples, respectively. For serial ctDNA, multiple inputs of each sample were used to analyze serial clonal population. Cancer cell fraction was calculated with the mean of predicted cellular frequencies. The cluster with the highest mean VAF was identified as the clonal cluster, and mutations in this cluster were clonal mutations. Meanwhile, other clusters and mutations were considered subclonal. In each ctDNA sample, mTBI was analyzed using the mean VAF of clonal mutations. The ΔmTBI was calculated based on the mTBI of the first ctDNA sample.
2.5. Pathway analysis of expanding clones
SIFT and PolyPhen2 were used for predicting the functional impact of an amino acid substitution caused by mutations. WebGestalt carried out pathway enrichment analysis to investigate the distribution of genes affected by somatic mutations and CNVs within the KEGG database [25].
2.6. Statistical analysis
The linear association between CNV and ΔmTBI was tested with Pearson correlation analysis. Multivariate Cox proportional hazards analysis (enter method) was performed considering the clinical characteristics and mTBI at baseline. Kaplan-Meier survival plots were generated for mTBI at baseline using log-rank tests. All statistical analyses were performed with SPSS (v.21.0; STATA, College Station, TX, USA) or GraphPad Prism (v. 6.0; GraphPad Software, La Jolla, CA, USA) software. Statistical significance was defined as a two-sided P-value of <0.05.
3. Results
3.1. Patient characteristics and mutation detection
Patients with AGC with HER2 overexpression, who received chemotherapy plus trastuzumab at the Affiliated Hospital, Academy of Military Medical Sciences, Beijing, China, were enrolled from July 2013 to January 2017. HER2-positive status was defined as immunohistochemistry (IHC) 3+ or IHC 2+/fluorescence in situ hybridization (FISH)+. Tumor tissues and serial plasma samples were collected from twenty-one AGC patients (P01–P21) receiving chemotherapy plus trastuzumab (Table 1). Seventeen patients were confirmed by FISH. Eight patients (38%) were IHC 2+ and nine patients (43%) were IHC 3+. A mean of five (2–9) plasma samples were available for mutation detection. Targeted capture sequencing revealed a mean effective depth of coverage of 464× in tissues and 1673× in plasma samples (Table S2). A total of 121 and 146 functional mutations were identified in 14 paired tissue and plasma samples, respectively, with a detection rate of 100% (Table S2). All pretreatment plasma samples presented at least one tumor-confirmed single nucleotide variant (SNV) or insertion-deletion (InDel). In addition, 31 and 36 copy number variations (CNVs) were detected in paired tissue and plasma samples, respectively. Most frequently, CNVs occurred in the ERBB2, CDK12, TOP2A, CCNE1, MET, and RARA genes. The CNV positive predictive values of ERBB2 obtained by sequencing, according to FISH results from 12 patients (6 patients with IHC 2+ and 6 patients with IHC 3+), were 58.33% (7/12 patients) and 66.67% (8/12 patients) in tissue and plasma, respectively. Four paired samples (P03, P13, P15, and P19) were negative, and another paired sample (P07) was plasma-positive but tissue-negative. The remaining two patients (P10 and P20) had immunohistochemistry (IHC) scores of 3+ and positive sequencing in both tissue and plasma samples (Fig. S2). These results suggest that intra-tumor heterogeneity influences CNV analysis in both tissue biopsy and plasma.
Table 1.
Clinical characteristics of patients with AGC.
| Characteristic | Patients (n = 21) |
|---|---|
| Age (years) | |
| Median (range) | 58 (35–61) |
| Sex, no. (%) | |
| Male | 17 (81) |
| Female | 4 (19) |
| ECOG performance status | |
| Median (range) | 1 (0–2) |
| Stage, no. (%) | |
| IIIA | 2 (10) |
| IIIC | 2 (10) |
| IV | 17 (80) |
| Tumor differentiation, no. (%) | |
| Well/Moderate | 8 (38) |
| Poor | 13 (62) |
| Lauren type, no. (%) | |
| Diffuse | 4 (19) |
| Intestinal | 7 (33) |
| Mixed | 10 (48) |
| Gastroesophageal junction involvement, no. (%) | |
| Yes | 10 (48) |
| No | 11 (52) |
| Liver involvement, no. (%) | |
| Yes | 15 (71) |
| No | 6 (28) |
| Lung involvement, no. (%) | |
| Yes | 10 (48) |
| No | 11 (52) |
| Prior chemotherapy regimens, no. | |
| Median (range) | 0 (0–3) |
Abbreviations: ECOG, Eastern Cooperative Oncology Group
3.2. Consistency of clonal mutation between tissue and ctDNA
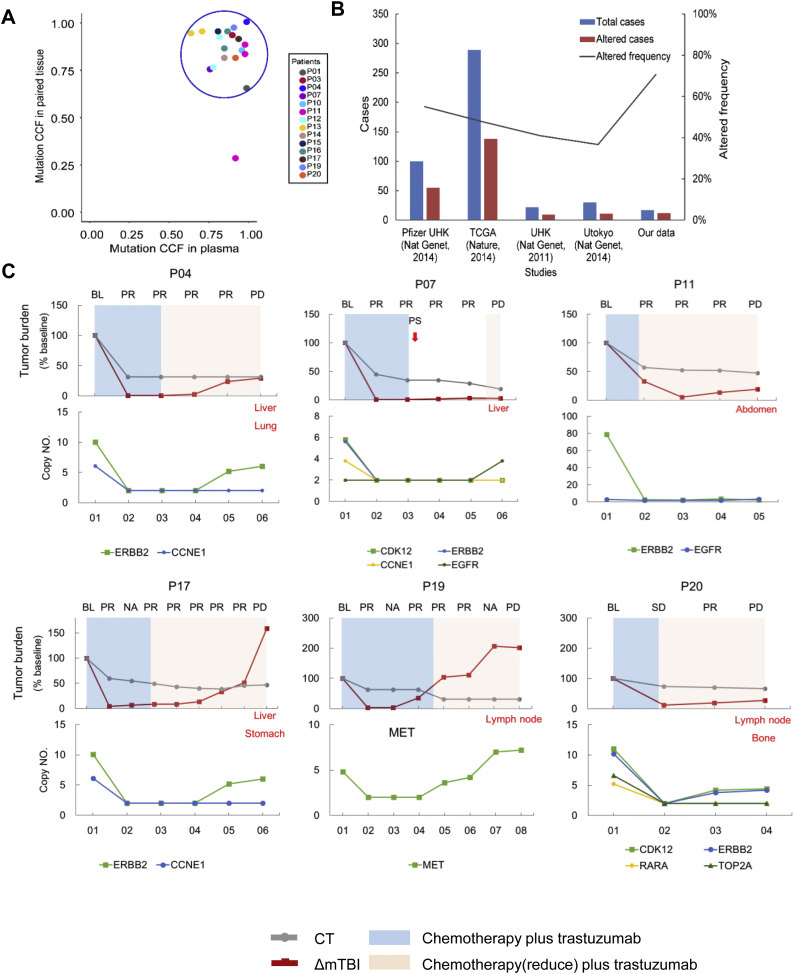
We investigated whether clonal mutations in plasma samples were derived from the matched tumor that presented the highest CCF. Mutations in 14 paired samples were clustered separately, using a Bayesian algorithm with PyClone [26]. An average cluster number of 10 (2−21) was obtained in pretreatment ctDNA. CCF of each mutation was predicted by PyClone. Clusters with the highest average VAF were identified as clonal. The clonal cluster with the highest CCF contained 1–3 mutations. Of the total of 20 clonal mutations identified in ctDNA, 19 mutations were identified in tissue, with median CCF values of 89% (95% CI, 81%–93%) in ctDNA and 88% (95% CI, 81%–94%) in tissues (Fig. 1A). Only one mutation in P11 (PAX5 p.V132I) presented as a clonal mutation, with mutated TP53 and PTPRD in ctDNA but not in tissue (CCF in plasma = 91%, CCF in tissue = 28%). Further validation of this clonal mutation in ctDNA during the disease progression of P11 showed that PAX5 p.V132I was still clustered with mutated TP53 and PTPRD, with the highest CCF. This likely reflects what we already know about sampling bias of selected tissue specimens, which confounds resolution of the clonal status of mutations, and illustrates the problems of using tissue alone as the gold standard [27].
Fig. 1.
Clonal mutations detected in paired samples and used to monitor serial ctDNA from patients who received chemotherapy plus trastuzumab. (A) Relationship between clonal mutations in baseline ctDNA and matched tissues. (B) Patient counts and fractions of detected TP53 mutation in different studies. (C) The mTBI of ctDNA before treatment and during progressive disease. Top: Tumor burden changed based on baseline status. The gray line indicates CT imaging results. The red line indicates ΔmTBI results. Progressive metastases are marked in red font at PD. Bottom: serial changes of CNV detected in ctDNA. Patient P07 received palliative surgery with continuous evaluation of PR. BL, baseline; PR, partial response; PD, progressive disease; NA, no available CT result; PS, palliative surgery; mTBI, molecular tumor burden index; CT, computed tomography. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. ctDNA monitoring with clonal mutations
Having demonstrated that clonal mutations with the highest CCF in ctDNA were derived from a tissue truncal clone, we next determined whether the mean VAF of a clonal mutation, defined as mTBI, could predict disease progression in AGC patients. Previous studies have reported that a single gene could be used to monitor tumor load [15]. We analyzed TP53, which mutated with the highest recurrence (55%) in gastric cancer [[28], [29], [30], [31]], and obtained a patient coverage of 70.6% (12/17, Fig. 1B) in pretreatment ctDNA.
We continuously monitored mTBI in plasma samples collected approximately every 2–4 treatment cycles in 21 patients (Table S4). Clinical progression of disease (PD) was evaluated in 15 of these patients, and ΔmTBI was calculated based on mTBI of the first ctDNA sample (12/15 was baseline). We found that ΔmTBI increased before (6 patients) or at (9 patients) PD. This approach provided 3–48 weeks (mean of 18 weeks, Fig. 1C and Fig. S3) lead time in the detection of PD compared to RECIST (Response Evaluation Criteria in Solid Tumors, version 1.1) during conventional follow-up with CT. In addition, the average CNV detected in serial ctDNA samples was significantly associated with ΔmTBI (Pearson r = 0.41, P = .002, Fig. S4). For P16, both CT and ΔmTBI increased at cycle ten of anti-HER2 therapy, eventually leading to palliative surgery. In five other non-PD patients, mTBI showed slight fluctuations, with a maximum increase of 0.6% (Fig. S5).
3.4. Expanding clones and resistance to trastuzumab
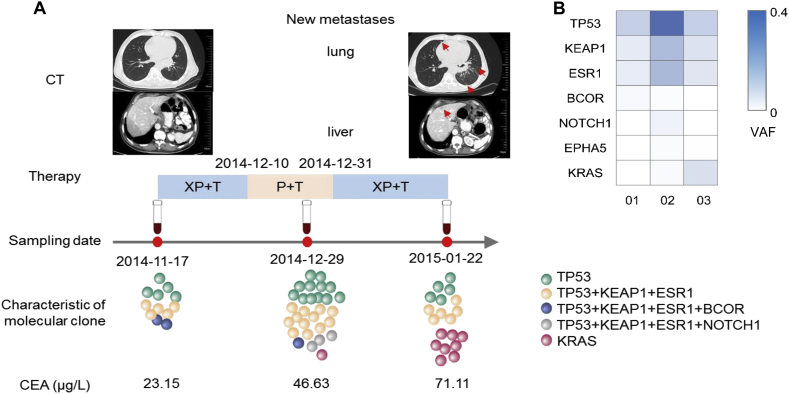
Subclones in ctDNA were revealed to change dynamically while monitoring the therapeutic response in serial ctDNA samples. The clonal evolution of P19, who underwent three cycles of chemotherapy (capecitabine/cis‑platinum) plus trastuzumab, showed a new subclone with a KRAS Q61R mutation that emerged in ctDNA at the seventh week of treatment. This subclone expanded at the ninth week, when CT imaging showed new metastases in lung and liver (Fig. 2).
Fig. 2.
Changes in ctDNA, imaging, and serum biomarker during progressive disease in patient P19. (A) CT: Imaging shows new metastasis in lung and liver. Therapy: treatment strategies and procedures. Sampling date: ctDNA sampled at baseline, the second treatment cycle, and when disease progressed. Characteristics of molecular clones: The diagram illustrates clone structure and selection in serial ctDNA. CEA: The levels of CEA increased during treatment. (B) Mutations identified in serial ctDNA. CEA, carcinoembryonic antigen; XP, capecitabine (xeloda)/cis‑platinum; T, trastuzumab; P, cis‑platinum; VAF, variated allele frequency.
To assess whether expanding clones were related to resistance to trastuzumab, mutations at PD and baseline of each patient were compared. Out of 112 mutations at baseline, 59 (52.68%) were detected after the treatment. Meanwhile, 86 secondary mutations emerged at PD. Mutations detected at PD with absolute increased VAF values of ≥2% or with a CCF increase of ≥2-fold compared with that of baseline samples were considered high-confidence expanding clones. Genes with increases in CNV in ERBB2 were also identified as expanding clones. Analysis of all 10 patients with paired PD and baseline ctDNA samples revealed 32 expanding mutations that were identified with high confidence in nine patients, including a KRAS CNV in a patient with a short benefit time (18 weeks, Table S5). The key pathways most frequently altered by these expanding mutations were MAPK (7 mutant genes, P = 5.09e-03) and RAS (8 mutant genes, P = 6.11e-03).
Next, we grouped all 32 mutations into three levels based on the reported clinical or biological evidence, as follows. Level 1, published clinical or biological evidence supports the variation associated with the resistance to trastuzumab; Level 2, published clinical or biological evidence supports the activated gene (2a) or pathway (2b) associated with the resistance to trastuzumab, and the variation was predicted to functional by both SIFT and PloyPhen-2 [32,33]; Level 3, expanding clones harbor the variation, but no published clinical or biological evidence supports its association with trastuzumab resistance (Fig. 3A). Level 1 included only CNVs of EGFR and MET [34,35]. Ninety-one percent of the mutations had no clear clinical or biological evidence supporting their relationship with trastuzumab resistance, although 35% of the mutations occurred in resistant genes or pathways (Fig. 3B). Interestingly, we found that multiple pathways were simultaneously activated in the same patients. Clone temporal evolution of serial ctDNA samples during the treatment of P07 showed a decline in the abundance of TP53 p.H193R, which was a potential functional mutation in the p53 pathway (level 2b, Clone 3, Fig. 3C). The abundance of this clone increased as disease progressed. Meanwhile, a new EGFR CNV clone emerged in ctDNA (Level 1, Clone 5, Fig. 3C), which was reported to be related to trastuzumab resistance by increasing signaling from HER2/EGFR complexes. During the disease progression of P14, the abundance of clonal KRAS CNV (Level 2b, Clone 1, Fig. 3D) and subclonal IGF1R p.P1257S (Level 2a, Clone 8, Fig. 3D) mutation increased in serial ctDNA samples. These observations may reflect heterogeneity in mechanisms of drug resistance in AGC.
Fig. 3.
Expanding mutations and trastuzumab resistance mechanism in AGC. (A) Levels of evidence of expanding mutations related to resistance mechanism, based on clinical or biological evidence reported and on predicted function. (B) Distribution of levels of expanding mutations. (C) Clonal temporal evolution in ctDNA of patient P07 during treatment. TP53 mutation (Clone 3) and EGFR CNV (Clone 5) expanded at progressive disease. (D) Clonal temporal evolution in ctDNA of patient P14 during treatment. KRAS CNV (Clone 1) and IGF1R mutations (Clone 8) expanded at progressive disease.
3.5. Validation of mTBI for the prediction of progressive disease
In addition to the chemotherapy plus trastuzumab cohort, we also validated mTBI sensitivity for the prediction of progressive disease in a group of patients that received chemotherapy alone. Eighteen patients, including two patients with baseline blood only, were enrolled in the chemotherapy group (Table S6). We analyzed 72 serial samples of ctDNA from 16 patients, without knowledge of their clinical results according to RECIST 1.1. The median mTBI was 1% (ranging from 0 to 65.89%). A cutoff value of 0.6% increased the amount of mTBI, which was previously mentioned to be the maximum increase in non-PD patients, and was therefore selected to improve specificity. In 15 of 16 patients, mTBI increased to exceed the cutoff before or at clinical PD, demonstrating 94% sensitivity (Fig. 4A). Analysis of mTBI detected disease progress earlier than did CT in three patients, with an average lead time of 9 weeks. The only false-negative patient (PC05), with peritoneal metastasis, received four cycles of treatment with oxaliplatin/capecitabine and progressed due to malignant pleural effusion.
Fig. 4.
Validation of mTBI in relation to progressive disease and treatment outcome. (A) Increasing mTBI predicts progressive disease during the period (x-axis) in 16 patients receiving chemotherapy only. Red triangles indicate an increase in mTBI exceeding the cutoff (≥0.6%). Gray triangles indicate an increase in mTBI below the cutoff (≥0.6%), or no increase. Purple vertical lines indicate clinical progressive disease.
(B) Chemo validation group. Kaplan-Meier analysis of progression-free survival in patients with high pretreatment mTBI (≥1%) compared with patients with low pretreatment mTBI (<1%) in AGC. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.6. Pretreatment mTBI as a prognostic factor in AGC
We next analyzed whether the mTBI of pretreatment ctDNA could be used as an indicator to evaluate clinically relevant risk groups in AGC. Baseline ctDNA samples were collected from 31 patients who had received chemotherapy plus trastuzumab, or chemotherapy alone, and had been followed up for 14 to 173 weeks (median = 67 weeks). Progression-free survival (PFS) was shorter in patients with high (≥1%) mTBI (median PFS, 21 weeks; 95% CI, 18–74 weeks) than patients with lower (<1%) mTBI (median PFS, 49 weeks; 95% CI, 13.5–31.5 weeks; HR, 13.75; 95% CI, 2.69–70.24; P = .002; Fig. 4B; Table 2). Analysis in both subgroups of chemotherapy plus trastuzumab and chemotherapy alone, also showed pretreatment mTBI as a prognostic factor (chemotherapy plus trastuzumab subgroup, P = .002; chemotherapy subgroup, P = .066; Fig. S6). Other thresholds of pretreatment of mTBI were evaluated, and were found to be associated with worse outcomes (pretreatment mTBI of 0.8%, P = .026; pretreatment mTBI of 2%, P = .027; Fig. S7). HER2 positivity was significantly associated with PFS, as determined by COX multivariate analysis (P = .037). HER2-positive patients had a significant longer PFS than did HER2-negative patients. Greater age was another clinical characteristic that was significantly associated with OS (P = .021). However, pretreatment mTBI and HER2 status had no correlation with overall survival (OS).
Table 2.
Multivariate Cox proportional hazards analysis.
| Variable | PFS |
OS |
||
|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |
| Age (≥65 years versus <65 years) | 0.79(0.28–2.25) | 0.658 | 0.16(0.03–0.76) | 0.021* |
| Gastroesophageal junction involvement (yes versus no) | 0.52(0.15–1.73) | 0.283 | 1.39(0.34–5.69) | 0.647 |
| Differentiation (poor versus well/moderate) | 0.45(0.12–1.66) | 0.231 | 1.69(0.29–9.7) | 0.556 |
| Lauren (diffuse versus intestinal/mixed) | 0.91(0.21–3.98) | 0.9 | 4.49(0.74–27.31) | 0.103 |
| HER2 status (positive versus negative) | 0.26(0.07–0.92) | 0.037* | 0.58(0.09–3.64) | 0.561 |
| Liver or lung involvement (yes versus no) | 0.72(0.2–2.57) | 0.617 | 2.34(0.45–12.07) | 0.311 |
| Baseline mTBI (≥1% versus <1%) | 13.75(2.69–70.24) | 0.002* | 8.19(0.83–80.95) | 0.072 |
Abbreviations: HER2, human epidermal growth factor receptor-2; mTBI, molecular tumor burden index; PFS, progression-free survival; OS, overall survival; HR, hazard ratio; CI, confidence interval.
p ≤ 0.05
4. Discussion
Monitoring tumor load and analyzing drug-resistant genes with ctDNA has been previously reported in colorectal cancer, lung cancer, breast cancer, and other solid tumors [[36], [37], [38], [39]]. Spatiotemporally, clinical and biological heterogeneity are key challenges for detection methods based on a single gene or a few mutations. A targeted capture sequencing and clone structure reconstructing approach was used to understand clonal evolution in ctDNA. We applied this method in 173 ctDNA samples from 39 AGC patients to monitor serial changes and analyze the mechanism involved in trastuzumab resistance. The mTBI, calculated from mean VAF of clonal mutations, achieved high patient coverage and sensitivity for broad clinical applicability in predicting disease progression. Higher mTBI values were demonstrated to be a risk factor for PFS. Clonal evolution during treatment demonstrated the probability of multiple resistance mechanisms for trastuzumab in AGC. These results highlight the advantages of using ctDNA as a biomarker to detect progression with an increased lead time, with unclear survival advantage.
Detecting ctDNA with a single hotspot gene, such as TP53, is challenged by the limited detectability coverage in cancer patients (50–70%). In our study, combinations of targeted capture sequencing and clone analysis have improved patient coverage to 100% and enabled the detection rate of increasing mTBI prior to clinical progression, with a mean lead time of 18 weeks. These findings, along with drug-resistant gene analysis and the development of therapeutic strategies (e.g., targeted and immunological therapy), may inform future trial designs based on earlier interventions in advanced or early disease directed by ctDNA [23,40]. Further work will characterize whether ctDNA analyses may be used to stratify patients who are at high risk for recurrence, with the goal of sparing low-risk patients from the toxicities of unnecessary systemic therapies.
The high heterogeneity of histological and morphological characteristics of AGC makes clinical classification and lesion measurements difficult to accurately assess [41,42]. Molecular classification, including HER2 status and mTBI, performed at baseline and used as a prognostic and predictive factor in AGC, can be meaningful. The abundance of clonal mutations has been demonstrated to be related to tumor size in early-stage lung cancer [43]. A higher amount of pre-operative ctDNA, likely related to large primary lesions or occult metastases, is associated with worse prognosis in operable colorectal cancer [44]. To our knowledge, this report is the first to show that the mTBI of pretreatment ctDNA is a potential factor to predict treatment outcome in AGC. Our report suggests that patients with higher mTBI are at high risk for earlier progression and require closer response monitoring. Moreover, enrolling cohorts of patients with higher mTBI into clinical trials may enrich for higher risk patients.
Our study analyzed clonal temporal evolution in ctDNA and observed expanding clones that are likely related to drug resistance to trastuzumab. Most of these mutation clones were enriched in the MAPK and RAS pathways, and were consistent with a previous report in patients on trastuzumab with late-stage breast cancer [45,46]. A recent study published in Gut has explored the efficacy of predictive markers, and demonstrates the substantial contributions of PIK3CA/R1/C3 and ERBB2/4 mutations substantial contributions to trastuzumab treatment, resulting and resulted in a poor progression-free survival of HER2 + mGC patients [47]. However, there have been limited studies focused on trastuzumab resistance in AGC. As already shown in our results, CNVs in the resistance resistant genes EGFR and MET CNV were identified in AGC patients, suggesting the possibility of combining of trastuzumab with other targeted agents. Of even more interest, we observed that the mean VAF of clonal mutations, defined as molecular tumor burden index (mTBI), could help standardize the applicability and accuracy of ctDNA in monitoring tumor burden in highly heterogeneous gastric cancer. This result underlines the heterogeneity of gastric cancer, and indicates challenges in drug research and therapeutic strategy.
One false-negative patient with malignant abdominal effusion was diagnosed as having progressive disease. This result may be due to the fact that ctDNA from peritoneal metastasis can lead to variable shedding and causing a low abundance of ctDNA in peripheral blood [47]. We also sequenced paired ctDNA from paired peripheral blood and ascites and found a significantly increased abundance in ascites compared with that in peripheral blood (36% versus 11.6%,P < 10−5; data not shown). In patients with malignant ascites, ctDNA extracted from ascites might be an appropriate supplement to serial monitoring.
5. Conclusions
Despite the relatively large number of samples and long follow-up time in our study, our conclusions may be affected by the small number of patients. Large-scale studies of ctDNA will be required to validate the parameters of mTBI. For expanding clones, a series of biological functional experiments will need to be designed to verify whether advantageous mutations lead to drug resistance. In addition, certain technical improvements are needed, such as the inclusion of more SNP locations in the panel, to increase the sensitivity of CNV detection [48].
Mounting data suggests that ctDNA may have the capacity to track resistance before conventional imaging modalities. Whether this is clinically meaningful is an active area of investigation. In high-heterogeneity cancers, targeted capture sequencing has several advantages over single-gene sequencing. Understanding the clonal population structure in cancer by investigating expanding mutations in ctDNA during treatment is a promising approach for uncovering drug resistance mechanisms. Although our study included only a small cohort of patients with AGC, our study highlights the potential utility of mTBI for the identification of high-risk groups and tracking the progression of cancer.
The following are the supplementary data related to this article
Targeted region by pan-cancel panel sequencing.
Summary of panel sequencing of all samples.
Mutations detected in tissue and baseline plasma samples.
Summary of clone analysis in serial ctDNA from patients received chemotherapy plus transtuzumab.
Expanding mutations identified in ctDNA at disease progression.
Clinical characteristics of all patients.
Supplementary material
Acknowledgments
Acknowledgements
We are grateful to the patients, physicians, and pathologists at the Department of GI Oncology, the Fifth Medical Center, General Hospital of PLA who contributed patient material.
Funding
This work was supported by grants from the National International Cooperation Grant (to J.X.; Project No. 2014DFB33160).
Declarations of interests
The authors have no potential conflicts of interest to disclose.
Authors' contributions
J.X. and X.Y. designed the study. Y·W., L.C., and R.J. did the literature search. J.X., Y.X., Y.W. and L.C. wrote and revised the study protocol. Y.W., R.J., C.Z., R.L., and Y.Z. collected samples and clinical data. J.X., X.Y., Y.W, L.C., R.J., R.C., X.X., A.B., H.H. and Y.G. participated in the analysis and interpretation of data. Y.W., L.C. and J.X. developed an early draft. All authors contributed to the drafting and revision of the manuscript.
Contributor Information
Xin Yi, Email: yix@geneplus.org.cn.
Jianming Xu, Email: jmxu2003@yahoo.com.
References
- 1.Watanabe H., Okada M., Kaji Y., Satouchi M., Sato Y., Yamabe Y. New response evaluation criteria in solid tumours-revised RECIST guideline (version 1.1) Gan To Kagaku Ryoho. 2009;36(13):2495–2501. [PubMed] [Google Scholar]
- 2.Su B.B., Shi H., Wan J. Role of serum carcinoembryonic antigen in the detection of colorectal cancer before and after surgical resection. World J Gastroenterol. 2012;18(17):2121–2126. doi: 10.3748/wjg.v18.i17.2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruibal Morell A. CEA serum levels in non-neoplastic disease. Int J Biol Markers. 1992;7(3):160–166. doi: 10.1177/172460089200700307. [DOI] [PubMed] [Google Scholar]
- 4.Tempero M.A., Uchida E., Takasaki H., Burnett D.A., Steplewski Z., Pour P.M. Relationship of carbohydrate antigen 19-9 and Lewis antigens in pancreatic cancer. Cancer Res. 1987;47(20):5501–5503. [PubMed] [Google Scholar]
- 5.Shimada H., Noie T., Ohashi M., Oba K., Takahashi Y. Clinical significance of serum tumor markers for gastric cancer: a systematic review of literature by the task force of the Japanese gastric Cancer association. Gastric Cancer. 2014;17(1):26–33. doi: 10.1007/s10120-013-0259-5. [DOI] [PubMed] [Google Scholar]
- 6.Thibault I., Chang E.L., Sheehan J., Ahluwalia M.S., Guckenberger M., Sohn M.J. Response assessment after stereotactic body radiotherapy for spinal metastasis: a report from the SPIne response assessment in Neuro-oncology (SPINO) group. Lancet Oncol. 2015;16(16):e595–e603. doi: 10.1016/S1470-2045(15)00166-7. [DOI] [PubMed] [Google Scholar]
- 7.Albert J.M. Radiation risk from CT: implications for cancer screening. AJR Am J Roentgenol. 2013;201(1):W81–W87. doi: 10.2214/AJR.12.9226. [DOI] [PubMed] [Google Scholar]
- 8.Siravegna G., Marsoni S., Siena S., Bardelli A. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol. 2017;14:531–548. doi: 10.1038/nrclinonc.2017.14. [DOI] [PubMed] [Google Scholar]
- 9.Crowley E., Di Nicolantonio F., Loupakis F., Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. 2013;10(8):472–484. doi: 10.1038/nrclinonc.2013.110. [DOI] [PubMed] [Google Scholar]
- 10.Yi X., Ma J., Guan Y., Chen R., Yang L., Xia X. The feasibility of using mutation detection in ctDNA to assess tumor dynamics. Int J Cancer. 2017;140:2642–2647. doi: 10.1002/ijc.30620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poruk K.E., Firpo M.A., Adler D.G., Mulvihill S.J. Screening for pancreatic cancer: why, how, and who? Ann Surg. 2013;257(1):17–26. doi: 10.1097/SLA.0b013e31825ffbfb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loman N., Saal L.H. The state of the art in prediction of breast cancer relapse using cell-free circulating tumor DNA liquid biopsies. Ann Transl Med. 2016;4(Suppl. 1):S68. doi: 10.21037/atm.2016.10.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perdigones N., Murtaza M. Capturing tumor heterogeneity and clonal evolution in solid cancers using circulating tumor DNA analysis. Pharmacol Ther. 2017;174:22–26. doi: 10.1016/j.pharmthera.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 14.Tie J., Wang Y., Tomasetti C., Li L., Springer S., Kinde I. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med. 2016;8(346):346ra92. doi: 10.1126/scitranslmed.aaf6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dawson S.J., Tsui D.W., Murtaza M., Biggs H., Rueda O.M., Chin S.F. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368(13):1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 16.Reinert T., Scholer L.V., Thomsen R., Tobiasen H., Vang S., Nordentoft I. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut. 2016;65(4):625–634. doi: 10.1136/gutjnl-2014-308859. [DOI] [PubMed] [Google Scholar]
- 17.Garcia-Murillas I., Schiavon G., Weigelt B., Ng C., Hrebien S., Cutts R.J. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med. 2015;7(302):302ra133. doi: 10.1126/scitranslmed.aab0021. [DOI] [PubMed] [Google Scholar]
- 18.Leary R.J., Kinde I., Diehl F., Schmidt K., Clouser C., Duncan C. Development of personalized tumor biomarkers using massively parallel sequencing. Sci Transl Med. 2010;2(20):20ra14. doi: 10.1126/scitranslmed.3000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacoby M.A., Duncavage E.J., Walter M.J. Implications of tumor clonal heterogeneity in the era of next-generation sequencing. Trends Cancer. 2015;1(4):231–241. doi: 10.1016/j.trecan.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 20.Newman A.M., Bratman S.V., To J., Wynne J.F., Eclov N.C., Modlin L.A. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20(5):548–554. doi: 10.1038/nm.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burrell R.A., Swanton C. Re-evaluating clonal dominance in Cancer evolution. Trends Cancer. 2016;2(5):263–276. doi: 10.1016/j.trecan.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 22.McGranahan N., Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell. 2017;168(4):613–628. doi: 10.1016/j.cell.2017.01.018. [DOI] [PubMed] [Google Scholar]
- 23.Wan J.C., Massie C., Garcia-Corbacho J., Mouliere F., Brenton J.D., Caldas C. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17(4):223–238. doi: 10.1038/nrc.2017.7. [DOI] [PubMed] [Google Scholar]
- 24.Nong J., Gong Y., Guan Y., Yi X., Yi Y., Chang L. Circulating tumor DNA analysis depicts subclonal architecture and genomic evolution of small cell lung cancer. Nat Commun. 2018;9(1):3114. doi: 10.1038/s41467-018-05327-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J., Vasaikar S., Shi Z., Greer M., Zhang B. WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 2017;45:W130–W137. doi: 10.1093/nar/gkx356. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roth A., Khattra J., Yap D., Wan A., Laks E., Biele J. PyClone: statistical inference of clonal population structure in cancer. Nat Methods. 2014;11(4):396–398. doi: 10.1038/nmeth.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bardelli A., Pantel K. Liquid biopsies, what we do not know (yet) Cancer Cell. 2017;31(2):172–179. doi: 10.1016/j.ccell.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 28.Network CGAR Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513(7517):202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang K., Kan J., Yuen S.T., Shi S.T., Chu K.M., Law S. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat Genet. 2011;43(12):1219–1223. doi: 10.1038/ng.982. [DOI] [PubMed] [Google Scholar]
- 30.Kakiuchi M., Nishizawa T., Ueda H., Gotoh K., Tanaka A., Hayashi A. Recurrent gain-of-function mutations of RHOA in diffuse-type gastric carcinoma. Nat Genet. 2014;46(6):583–587. doi: 10.1038/ng.2984. [DOI] [PubMed] [Google Scholar]
- 31.Wang K., Yuen S.T., Xu J., Lee S.P., Yan H.H., Shi S.T. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 2014;46(6):573–582. doi: 10.1038/ng.2983. [DOI] [PubMed] [Google Scholar]
- 32.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 33.Adzhubei I., Jordan D.M., Sunyaev S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013 doi: 10.1002/0471142905.hg0720s76. [Chapter 7, Unit 7.20] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nahta R., Yu D., Hung M.C., Hortobagyi G.N., Esteva F.J. Mechanisms of disease: understanding resistance to HER2-targeted therapy in human breast cancer. Nat Clin Pract Oncol. 2006;3(5):269–280. doi: 10.1038/ncponc0509. [DOI] [PubMed] [Google Scholar]
- 35.Shattuck D.L., Miller J.K., Carraway K.L., 3rd, Sweeney C. Met receptor contributes to trastuzumab resistance of Her2-overexpressing breast cancer cells. Cancer Res. 2008;68(5):1471–1477. doi: 10.1158/0008-5472.CAN-07-5962. [DOI] [PubMed] [Google Scholar]
- 36.Piotrowska Z., Niederst M.J., Karlovich C.A., Wakelee H.A., Neal J.W., Mino-Kenudson M. Heterogeneity underlies the emergence of EGFRT790 wild-type clones following treatment of T790M-positive cancers with a third-generation EGFR inhibitor. Cancer Discov. 2015;5(7):713–722. doi: 10.1158/2159-8290.CD-15-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schiavon G., Hrebien S., Garcia-Murillas I., Cutts R.J., Pearson A., Tarazona N. Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci Transl Med. 2015;7(313):313ra182. doi: 10.1126/scitranslmed.aac7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Russo M., Siravegna G., Blaszkowsky L.S., Corti G., Crisafulli G., Ahronian L.G. Tumor heterogeneity and lesion-specific response to targeted therapy in colorectal Cancer. Cancer Discov. 2016;6(2):147–153. doi: 10.1158/2159-8290.CD-15-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murtaza M., Dawson S.J., Tsui D.W., Gale D., Forshew T., Piskorz A.M. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497(7447):108–112. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 40.Willyard C. Cancer therapy: an evolved approach. Nature. 2016;532(7598):166–168. doi: 10.1038/532166a. [DOI] [PubMed] [Google Scholar]
- 41.Caldas C., Carneiro F., Lynch H., Yokota J., Wiesner G., Powell S. Familial gastric cancer: overview and guidelines for management*. J Med Genet. 1999;36(12):873–880. [PMC free article] [PubMed] [Google Scholar]
- 42.Chung Y.J., Kim K.M., Choi J.R., Choi S.W., Rhyu M.G. Relationship between intratumor histological heterogeneity and genetic abnormalities in gastric carcinoma with microsatellite instability. Int J Cancer. 1999;82(6):782–788. doi: 10.1002/(sici)1097-0215(19990909)82:6<782::aid-ijc2>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 43.Abbosh C., Birkbak N.J., Wilson G.A., Jamal-Hanjani M., Constantin T., Salari R. Phylogenetic ctDNA analysis depicts early stage lung cancer evolution. Nature. 2017;545:446–451. doi: 10.1038/nature22364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Phallen J., Sausen M., Adleff V., Leal A., Hruban C., White J. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med. 2017;9(403) doi: 10.1126/scitranslmed.aan2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu Y., Zi X., Zhao Y., Mascarenhas D., Pollak M. Insulin-like growth factor-I receptor signaling and resistance to trastuzumab (Herceptin) J Natl Cancer Inst. 2001;93(24):1852–1857. doi: 10.1093/jnci/93.24.1852. [DOI] [PubMed] [Google Scholar]
- 46.Valabrega G., Montemurro F., Aglietta M. Trastuzumab: mechanism of action, resistance and future perspectives in HER2-overexpressing breast cancer. Ann Oncol. 2007;18(6):977–984. doi: 10.1093/annonc/mdl475. [DOI] [PubMed] [Google Scholar]
- 47.Husain H., Nykin D., Bui N., Quan D., Gomez G., Woodward B. Cell-free DNA from ascites and pleural effusions: molecular insights into genomic aberrations and disease biology. Mol Cancer Ther. 2017;16(5):948–955. doi: 10.1158/1535-7163.MCT-16-0436. [DOI] [PubMed] [Google Scholar]
- 48.Gambin T., Akdemir Z.C., Yuan B., Gu S., Chiang T., Carvalho C.M.B. Homozygous and hemizygous CNV detection from exome sequencing data in a Mendelian disease cohort. Nucleic Acids Res. 2017;45(4):1633–1648. doi: 10.1093/nar/gkw1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Targeted region by pan-cancel panel sequencing.
Summary of panel sequencing of all samples.
Mutations detected in tissue and baseline plasma samples.
Summary of clone analysis in serial ctDNA from patients received chemotherapy plus transtuzumab.
Expanding mutations identified in ctDNA at disease progression.
Clinical characteristics of all patients.
Supplementary material