Abstract
Background
Certain tumors rely heavily on their DNA repair capability to survive the DNA damage induced by chemotherapeutic agents. Therefore, it is important to monitor the dynamics of DNA repair in patient samples during the course of their treatment, in order to determine whether a particular drug regimen perturbs the DNA repair networks in cancer cells and provides therapeutic benefits. Quantitative measurement of proteins and/or their posttranslational modification(s) at DNA double strand breaks (DSBs) induced by laser microirradiation provides an applicable diagnostic approach to examine DNA repair and its dynamics. However, its use is restricted to adherent cell lines and not employed in suspension tumor cells that include the many hematological malignancies.
Methods
Here, we report the development of an assay to laser micro-irradiate and quantitatively measure DNA repair transactions at DSB sites in normal mononuclear cells and a variety of suspension leukemia and lymphoma cells including primary patient samples.
Findings
We show that global changes in the H3K27me3-ac switch modulated by inhibitors of Class I HDACs, EZH2 methyltransferase and (or) H3K27me3 demethylases do not reflect the dynamic changes in H3K27me3 that occur at double-strand break sites during DNA repair.
Interpretation
Results from our mechanistic studies and proof-of-principle data with patient samples together show the effectiveness of using the modified micro-laser-based assay to examine DNA repair directly in suspension cancer cells, and has important clinical implications by serving as a valuable tool to assess drug efficacies in hematological cancer cells that grow in suspension.
Keywords: Histone deacetylases, Chromatin modifiers, Laser damage, DNA repair, Cancer, Hematological cells
Research in context.
Evidence before this study
Investigations into DNA repair dynamics at DNA break sites using microlaser-induced DNA damage has provided valuable insights into various facets of the DNA repair process. However, the use of this powerful technique was restricted to solid tumors or adherent cells, and not employed in liquid tumors or suspension cells that include the many hematological cancers. We have previously used the microlaser technique to demonstrate the effects of histone deacetylase (HDAC) inhibitors on DNA repair in leukemia and lymphoma cells.
Added value of this study
We present here a detailed methodology to study DNA repair at microlaser-induced double strand breaks in a variety of suspension hematological cells that include normal blood cells and liquid tumors. We show that the microlaser technique can be used as an assay to examine the dynamics of DNA repair in hematological cancer cells. Using the optimized methodology, we provide insights into the mechanisms that regulate H3K27me3 epigenetic mark at double-strand break sites during DNA repair in hematological cancer cells. Overall, this study is of value to the HDACs, DNA repair, epigenetics and chromatin fields, and it also has an important application in the field of cancer therapeutics—by serving as a diagnostic tool to evaluate treatment efficacy in either preclinical or clinical setting.
Implications of all the available evidence
Using the refined microlaser technique and small molecule inhibitors, we demonstrate the mechanistic interplay between epigenetic enzymes—HDAC1,2, EZH2 H3K27 methyltransferase and H3K27me3 demethylases—during DNA repair in suspension cancer cells. Therefore, findings from our study show that the microlaser technique can be used to examine epigenetic and other regulatory mechanisms that occur during DNA repair, directly in hematological cells. Thus, obviating the need to extrapolate results obtained from adherent cancer cells to liquid tumors. Moreover, the methodology described here provides a diagnostic test in clinics, to evaluate the efficacies of drugs, especially those targeting aberrant DNA repair, and guide treatment plans with mono- or combination therapies for hematological cancers.
Alt-text: Unlabelled Box
1. Introduction
Many new components of the DNA damage response were identified in the past two decades, which include proteins that function as DNA damage sensors or transducers, or those that act in the actual DNA repair. Because the majority of these factors act primarily at the sites of DNA breaks, they can be detected using immunofluorescence (IF) as coalesced structures called foci after exposure to DNA damaging agents. However, this standard IF approach fails to detect proteins that bind transiently to DNA break sites, and it does not permit the quantitative measurement of the spreading of DNA damage signal and the temporal examination of the assembly/disassembly of repair proteins at break sites. These limitations of the standard IF approach were overcome by the development of localized irradiation technique using micro-laser-fitted microscopes and its combination with immunofluorescence. Moreover, laser micro-irradiation has enabled the induction of breaks at discrete regions within the nucleus and has allowed the assessment of DNA repair dynamics in real time. Using the laser micro-irradiation technique, novel roles for several chromatin modifiers and remodelers at the sites of DNA damage were elucidated [[1], [2], [3]]. While this versatile approach has contributed significantly to the DNA repair field, its use has been restricted to adherent cells, particularly HeLa and U2OS cell lines [[3], [4], [5]], and it was not optimized for use in ‘liquid tumors’ or suspension cells that include hematological malignancies, such as, leukemia and lymphoma.
Misregulated DNA repair contributes to the pathogenesis of and/or treatment resistance in many hematological cancers [6,7]. An underlying aberrant or altered epigenetic mechanism, such as histone post-translational modifications, contributes to the poor prognosis in many of these cancers. For instance, changes in histone H3 lysine-27 trimethylation (H3K27me3) are associated with poor prognosis in lymphoid malignancies [[8], [9], [10]]. Overexpression of EZH2 H3K27 methyltransferase occurs in many lymphoid malignancies including Burkitt lymphoma, follicular lymphoma, diffuse large B-cell lymphoma (DLBCL), mantle cell lymphoma and multiple myeloma, and EZH2 gain-of-function mutation is observed in germinal center-derived DLBCL and follicular lymphomas [11,12]. UTX, the demethylase for H3K27, is mutated in multiple hematological malignancies [13] and its inhibition is proposed as a potential epigenetic therapy for T-cell acute lymphoblastic leukemia [14]. EZH2 and H3K27me3 are linked to DNA double-strand break repair [15,16]. The H3K27 residue undergoes mutually exclusive acetylation (ac) and methylation modifications. We developed a method to laser micro-irradiate suspension cells, and using this modified approach, we previously reported a novel functional relationship between epigenetic enzymes, histone deacetylase 1 and 2 (HDAC1,2) and the EZH2 H3K27 methyltransferase, regulating the H3K27me3-ac switch during DNA repair in suspension EZH2 gain-of-function mutant DLBCL cells [3]. Translating the findings from the micro-laser-based assay, we reported that histone deacetylase (HDAC) inhibitors either in isolation or in combination with standard chemotherapies impair DNA repair, reset the altered H3K27me3-ac epigenetic switch and provide therapeutic benefits in the EZH2 gain-of-function (GOF) mutant DLBCL cells [3]. Therefore, based on this published study and given the importance of DNA repair in hematological malignancies and their drug response [6,7,17], it has become essential to develop a methodology to track and measure DNA repair dynamics in suspension cancer cells, which in turn could greatly enhance our ability to treat these cancers in an effective manner.
Here, we now present a detailed methodology to micro-irradiate a variety of suspension cancer cells including those obtained from primary patients with hematological cancers, and to detect DNA repair proteins or protein post-translational modifications at the sites of laser-induced DNA damage directly in suspension cells. We further highlight the use of the modified micro-laser technique for mechanistic studies by demonstrating the effects of small molecule inhibitors of HDACs, EZH2 or UTX on the deposition of H3K27me3 mark at DNA break sites in suspension cells. Overall, the micro-laser technique described here provides a powerful system to further our fundamental understanding of DNA repair mechanisms in suspension cancer cells, while advancing the clinical applicability of the DNA repair-based assays as biomarkers for cancer treatment.
2. Materials used in the study
2.1. Cell culture
Human lymphoma cells were maintained in culture medium in an exponentially growing state in a 37 °C, 5% CO2 incubator. Culture medium was prepared by adding 20% heat inactivated fetal bovine serum (FBS), 1% Penicillin-Streptomycin, and 1% L-Glutamine to RPMI 1640. Primary lymphoma patient cells and normal peripheral blood mononuclear cells were maintained in Mononuclear Cell Medium purchased from PromoCell. The patient sample culture was carried out in accordance with the University of Utah IRB guidelines. The samples are de-identified samples and hence we do not have access to the patient history. SupB15 leukemia cells were cultured in IMDM supplemented with 20% fetal bovine serum and penicillin-streptomycin.
2.2. Reagents used
Karpas-422 DLBCL cell line (Sigma, 06101702-1VL), SupB15 leukemia cell line (ATCC, CRL-1929), RPMI 1640 Medium (Caisson, RPL09-500ML), Fetal Bovine Serum (HyClone, SH30910.03), L-Glutamine (Corning, 25-005-Cl), NaHCO3 (Sigma, S4019), NaOH Pellets (Mallinckrodt Analytical, 7708-10), Cell-Tak Cell and Tissue Adhesive (Corning, 354240), HCl (Fisher, A144-212), NaCl (Fisher, S671-3), KCl (Fisher, P217-500), Na2HPO4 (Fisher, S374-1), KH2PO4 (Fisher, P285-500), Hoechst 33342 Solution in Water (Thermo, H3570), Formalin 1:10 (Fisher, 23-245-685), Triton X-100 (Sigma, X100-1L), Normal Goat Serum (Sigma, G9023-10ML), Mouse α-γH2AX (Millipore, 05-636), Rabbit α-γH2AX (Cell Signaling, 2577), Rabbit α H3K27me3 (Cell Signaling, 9733), Mouse α-pS1981-ATM (Rockland, 200-301-400S), Rabbit α-Cleaved Caspase-3 (Cell Signaling, 9661), Goat α Mouse Alexa 546 (Thermo-Life Technologies, A11030), Goat α Rabbit Alexa 488 (Thermo-Life Technologies, A11034), Goat α Mouse Alexa 488 (Thermo-Life Technologies, A11029), Goat α Rabbit Alexa 546 (Thermo-Life Technologies, A11035), Fluoromount-G (Southern Biotech, 0100–01). Sodium Bicarbonate Solution: Dissolve 0.42 g of NaHCO3 in 50 mL of sterile H2O to make a 0.1 M solution and add HCl to bring solution to pH 8. Cell Tak Solution: To prepare 100 μl of solution, 1.25 μl 1 M NaOH and 2.5 μl Cell-Tak was added to 96.25 μl 0.1 M NaHCO3 to obtain final concentrations of 12.5 mM NaOH and 5 μg Cell-Tak in 0.1 M NaHCO3.
2.3. Preparation of chromatin
Chromatin fractions from DLBCL and Pre-B-ALL cells were made as described previously [24]. Briefly, 3-4 × 106 cells were pelleted and resuspended in buffer A containing a mixture of 10 mM HEPES (pH 7.9), 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM dithiothreitol and protease inhibitor cocktail). Triton-X-100 was added to the samples at a final concentration of 0.1% and incubated on ice for 8 min. Nuclei were collected by centrifugation at 8000 rpm for 5 min at 4 °C and lysed on ice for 30 min in buffer B (3 mM EDTA, 0.2 mM EGTA, 1 mM dithiothreitol and protease inhibitor cocktail). The chromatin fraction was separated from the soluble fraction by centrifugation at 10,000 rpm at 4 °C for 5 min. The supernatant was discarded and the chromatin pellet was resuspended in RIPA buffer with protease inhibitor and sonicated prior to immunoblotting.
2.4. Transient attachment of suspension cells to chamber slides
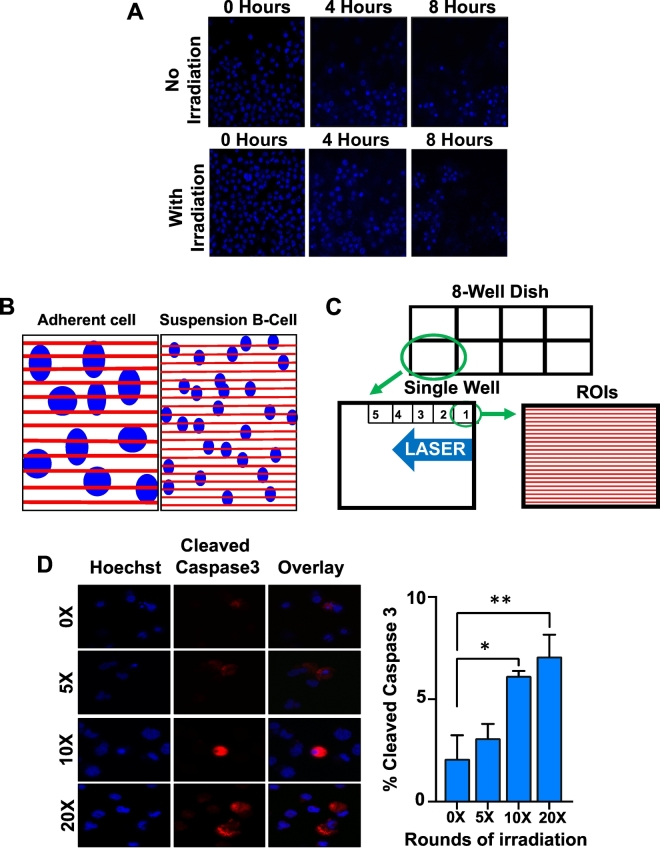
The Corning™ Cell-Tak reagent is a solution derived from the ‘polyphenolic proteins’ secreted by the marine mussel that enable it to anchor itself onto solid structures [42]. The Cell-Tak reagent acts as a cell adhesive, and therefore can be used to attach the suspension leukemia or lymphoma cancer cells onto the chamber dish. The Cell-Tak reagent was diluted in a solution of NaHCO3 and NaOH outlined above. We coated the chamber dish wells with this solution (100 μl/cm3/well of an 8-well chamber) and left the chamber undisturbed in the tissue culture hood at room temperature for 30 min. Following this incubation, the solution was removed and wells were washed with sterile water prior to adding the cells. During the last 10 min of this 30-min coating period, cells that were either untreated or pre-treated with drugs were centrifuged (4000 rpm, 5 min) and re-suspended in 300 μl fresh culture media before adding them to the Cell-Tak-treated wells. Cells were then allowed to settle for 15–20 min in a 5% CO2 incubator at 37 °C. Following the attachment of cells to the dish and prior to micro-irradiation, Hoechst 33342 solution was added to a final concentration of 55 μM to stain DNA to identify the nucleus. We found that the timing between plating of cells and laser micro-irradiation is critical for the successful implementation of this protocol. Imaging of live cells with or without laser micro-irradiation showed that cells get washed off 4–8 h after their initial adhesion to the Cell-Tak-treated chamber (Fig. 1A). Therefore, it is important to perform micro-irradiation immediately following a 15–20 min incubation of cells in the Cell-Tak coated culture dishes.
Fig. 1.
A methodology to make suspension lymphoma cells adhere to culture dish. A. Karpas-422 (EZH2 GOF DLBCL) cells were adhered to the dish using the Cell-Tak reagent and certain regions in the dish were micro-irradiated as described in the main text. Cells were imaged every 30 min for 8 h. The Cell-Tak reagent stopped working within 8 h in most regions of the dish regardless of the damage. In regions where cells adhered all 8 h, there was a little morphological change indicating that the damage was not sufficient to cause cell death. B. A comparison of ROIs (red lines) between adherent and suspension cell lines. C. A zoomed view of a single well to demonstrate irradiation of 2–5 fields of view within a span of 5 min. 24 ROIs per field of view is depicted in the fig. D. Cells were micro-irradiated for 0, 5, 10 or 20 rounds of irradiation prior to immunostaining with anti-Cleaved Caspase-3. Two-tailed t-test was performed. *p = .027, **p = .034 (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.5. Laser micro-irradiation
For micro-irradiation, we used a 405 nm solid-state laser, and a 60× oil-immersion Plan Apo,1.4 numerical aperture (NA) objective with a pinhole of 1.8 airy units (AU) on a Nikon A1R (or Nikon A1) confocal microscope that was controlled using the Nikon Instruments Software (NIS-Elements). During irradiation, the microscope was programmed to utilize the laser to irradiate specific regions in the well. The areas targeted for laser irradiation are termed as regions of interest (ROIs) in the NIS-Elements software (Fig. 1B). Because B-cells (and their nuclei) are smaller in size compared to the adherent cells (such as HeLa or U2OS), we chose to use 24 ROIs for irradiation as opposed to 12 ROIs used for HeLa or U2OS cells, in order to irradiate the maximum number of B-cells (Fig. 1B–C).
Utilizing the NIS-Elements software, we created target areas using the ROI manager by selecting the “rectangle ROI” option and drawing rectangles across the field of view. We then duplicated the rectangle until there were 24 ROIs spanning the region (Fig. 1B), with each ROI measuring 208 × 1.7 μm2. When creating these ROIs, it was important to ensure that they were labeled as “stimulation ROIs” in order for the laser to recognize them as areas to be irradiated. We set the 405 nm laser to irradiate the 24 ROIs spanning the field under view. Each well was irradiated over a period of five minutes and this time period included moving to each field of view within the same well. We focused on a corner of the well and then worked our way across the edge to irradiate 2–3 fields of view (Fig. 1C). We found that the ~2 min 5× irradiation cycle in addition to focusing on the next field of view allowed for 2–3 fields to be irradiated in the allotted 5 min. Following the 5 min of micro-irradiation, cells in each well were allowed to recover for an additional 10 min before fixing them with 3.7% formalin for 10 min.
2.6. Immunofluorescence
Immunofluorescence following micro irradiation was performed according the protocol described previously except that 0.1% instead of 0.5% Triton-X-100 was used [43]. The images were overlaid using the NIS elements software.
2.7. Statistical analyses
At least 100 cells with γH2AX lines per experiment were used to determine if H3K27me3 co-localizes with γH2AX and each experiment had at least three biological replicates. Averages and standard errors from three or more biological replicates were determined. All the p-values were calculated using the student's two tailed t-test.
3. Results
3.1. Detection of DSB tracks in suspension cancer cells
Having optimized the conditions to immobilize suspension cells onto chamber slides using the Cell-Tak reagent (see methods section for details), we then determined the correct laser settings to induce double strand breaks. We used a wide range of laser power and found that the damage lines or tracks are most distinct when a 100% power of irradiation was used. We further validated the suitability of this high-power setting by micro-irradiating immobilized B-cells and performing time-lapse live cell imaging. A number of regions were subjected to micro-irradiation and each region was imaged every half hour for 8 h. We also performed multiple cycles of irradiation to identify the optimal dose of DNA damage for suspension cells. One cycle includes one round of irradiation of all the regions of interest (ROIs), and five cycles of 24 ROIs refers to zapping each of the 24 micro-lasered lines five times for a total of 120 irradiations over a particular field of view. No significant changes in cell morphology were observed in the irradiated regions under these conditions (Fig. 1D). We also performed immunostaining of laser irradiated cells to detect the cleaved form of caspase-3 (an apoptosis marker), which showed that cell viability was not compromised when laser irradiation was performed for 5 cycles, but a statistically significant number of cells undergoing active apoptosis were observed when irradiation was done for ≥10 cycles (Fig. 1D).
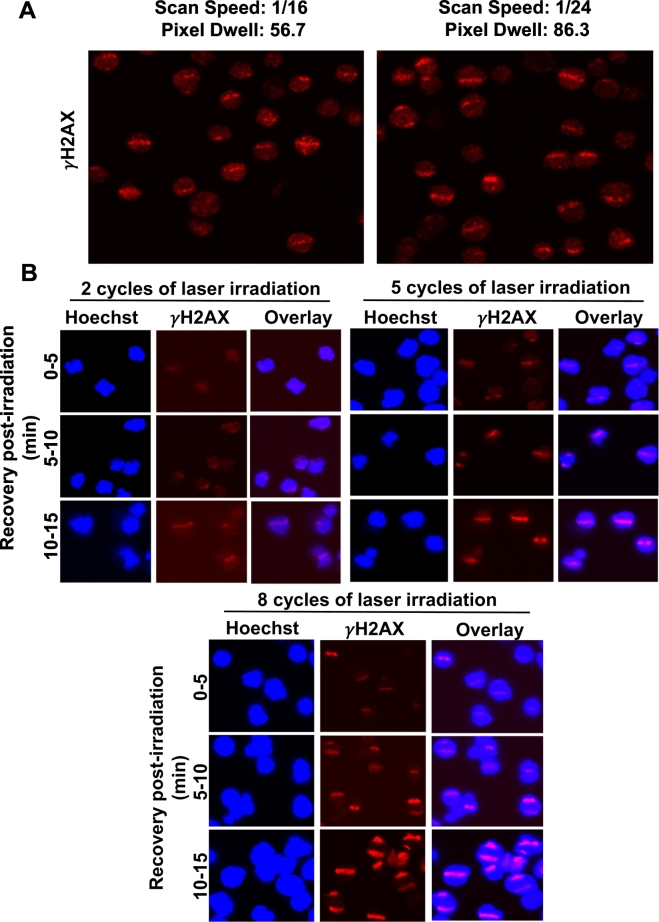
γH2AX or H2AX serine-139 phosphorylation is the gold standard for detecting DNA damage [18]. We manipulated the scan speed (that is, speed at which one field of view is imaged or irradiated) and pixel dwell (which is inversely related to the scan speed; a higher scan speed refers to a lower laser dwell time), in order to further examine if changing the irradiation intensity affected the formation of γH2AX lines. Our results showed that the settings detailed above can effectively cause laser-induced damage lines at both scan speeds and pixel dwell time (Fig. 2A). We also irradiated the same field of view with increasing number of cycles (2–8 cycles) and found that increasing the cycle number magnifies the spreading of the damage signal, as evidenced by the broadening of the γH2AX tracks with increasing cycle numbers (Fig. 2B). An increased number of cycles may be optimal when using an older laser, as overall laser power can decrease over time with usage of the microscope. We measured the time for γH2AX tracks to be formed or seen after laser irradiation. We find that it takes 5–10 min after laser irradiation to detect the γH2AX-labeled tracks (Fig. 2B). In our standard γH2AX immunofluorescence assays, we see that it can take >24 h for the disappearance of radiation-induced γH2AX foci (Supplementary Fig. 1). This poses a technical impediment to measure dephosphorylation of γH2AX in our micro-laser assays, as the suspension cells start to detach from the assay plate after 4–8 h following Cell-Tak-mediated adherence (Fig. 1A).
Fig. 2.
Optimization of the irradiation cycles for B-cells. A. Representative images of micro-irradiation at different scan speeds, illustrating the use of different laser intensities in Karpas-422 (EZH2 GOF DLBCL) cells. Localization of γH2AX immunofluorescence to the damage lines is shown in this fig. B. SUDHL4 cells were irradiated for increasing numbers of cycles and allowed to recover for the indicated time periods: 0–5 min, cells were fixed between 0 and 5 min post start of irradiation. That is, when irradiating for 8 cycles it takes over 3 min to irradiate all 24 lines for 8 times each and therefore the closest to no recovery possible is approximately 3–4 min post start of irradiation. Cells were then stained with anti-γH2AX antibody to evaluate the appearance and intensity of laser-induced damage.
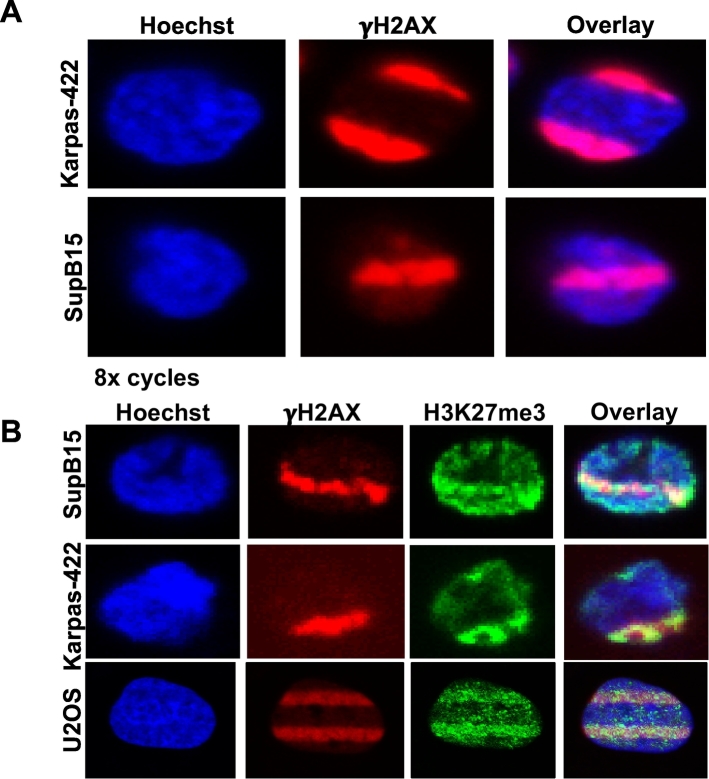
The DNA damage tracks were observed in both lymphoma (Karpas-422) as well as leukemia (SupB15) cells (Fig. 3A), demonstrating the feasibility and effectiveness of the technique across different cell types. In addition, the settings that were used in SupB15 and Karpas-422 cells also created γH2AX damage and H3K27me3 stripes in adherent U2OS cells, except that we used 12 ROIs instead of 24 ROIs and did not use the Cell-Tak reagent for U2OS cells (Fig. 3B). Therefore, comparison of repair dynamics in suspension as well as adherent cells can also be made using our newly developed technology (Fig. 3B).
Fig. 3.
Detection of γH2AX and H3K27me3 at DSB sites in SupB15 and Karpas-422 cells. A. Karpas-422 (DLBCL) and SupB15 (Ph + Pre-B-ALL) cells were irradiated for 8 cycles and γH2AX immunofluorescence was performed. B. Indicated cell lines were irradiated for 5 cycles and allowed to recover for 10–15 min. Immunofluorescence was performed with antibodies recognizing γH2AX or H3K27me3. Magnified images for Karpas-422 and SupB15 cells are shown to facilitate better visualization of the laser-induced DNA damage tracks.
3.2. Detection of DSB proteins at micro-laser damage sites in DLBCL lines as well as in primary DLBCL patient samples
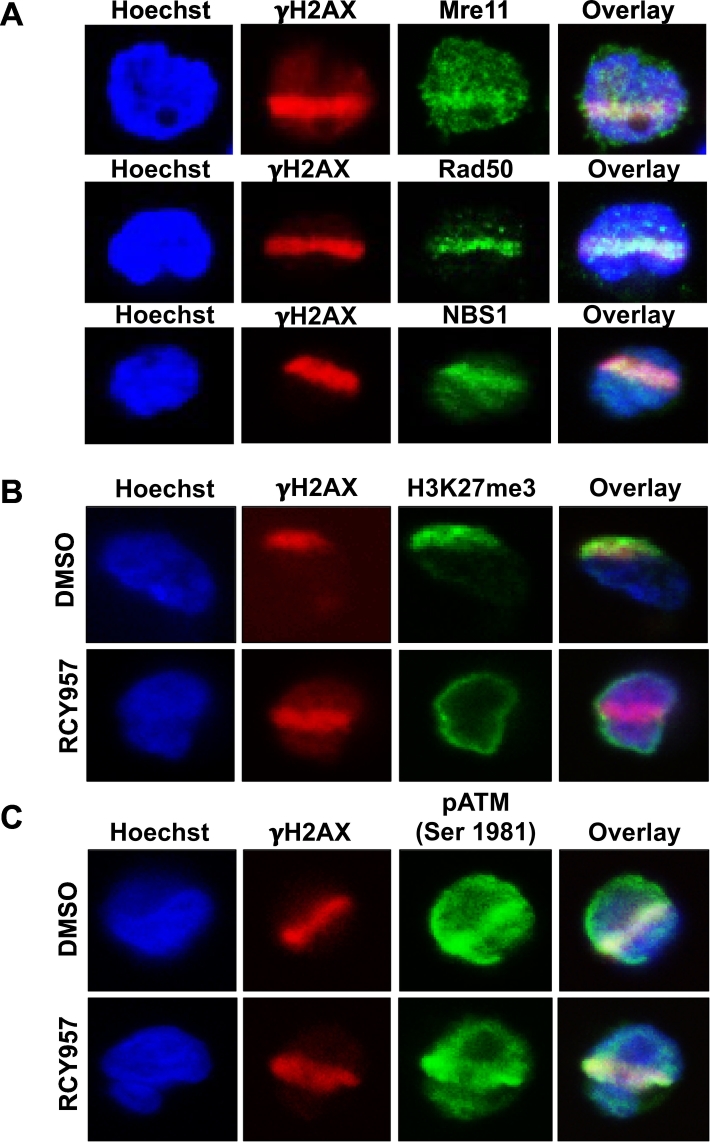
To detect proteins at laser-induced DNA breaks following immunofluorescence, imaging was done in various Z-stacks to determine whether a protein of interest co-localizes with γH2AX containing damage lines in different planes. Cells were imaged by Z-stacks utilizing a 60× Plan Apo objective on a Nikon A1R (or Nikon A1) confocal microscope. The software was set to perform imaging at increments of 0.5 μm to result in an average of 5–15 steps per Z-stack. In order to visualize the nucleus, the protein-of-interest and γH2AX, three channels were selected for imaging: 405 nm (blue), 488 nm (green), and 561 nm (red), respectively. Using the selected stack, the number of γH2AX lines within a focal plane were counted. Only lines for the protein-of-interest that co-localized with the γH2AX line were counted as a true DNA damage signal. Approximately, 100–300 cells with γH2AX lines per treatment were counted and the experiment was repeated at least three times in order to obtain statistically significant data. To confirm that our laser settings induced DNA double strand breaks (DSBs), we examined DNA repair proteins that localize to DSB sites and not to single-strand DNA breaks, namely, components of the MRN complex (Mre11-Rad50-Nbs1) [19]. Our results showed an efficient localization of Mre11, Rad50 and Nbs1 to the laser-induced break sites (Fig. 4A), verifying that the micro-laser technique that we have optimized for blood-related or hematological cancers creates double-strand DNA breaks.
Fig. 4.
Localization of DSB repair proteins and histone H3K27me3 mark to γH2AX tracks in DLBCL cells. A. Karpas-422 (EZH2 GOF DLBCL) cells were micro-irradiated as described in the main text and stained with anti-γH2AX along with anti-Mre11, Rad50 or NBS1. B. Karpas-422 DLBCL cells were micro-irradiated as described in the main text and stained with anti-γH2AX and H3K27me3. A reduction in H3K27me3 is seen following treatment of cells with HDAC1,2 inhibitor (RCY957). C. Karpas-422 DLBCL cells were micro-irradiated and stained with anti-γH2AX and phospho-ATM. No change in phospho-ATM localization was observed following treatment of Karpas-422 cells with HDAC1,2-selective inhibitor (RCY957).
ATM is a key regulator of DNA damage response [20]. During DNA repair, ATM is phosphorylated at Serine-1981 in response to DSB induction, and ATM phosphorylation dissociates the inactive dimers to active monomers [21,22]. We then examined whether our optimized protocol can detect post-translational modification(s) of DNA repair factors as effectively as histone modifications. Given the link we established between HDAC1,2 activity and DSB repair [3,[23], [24], [25]], we examined whether HDAC1,2 inhibition affects phospho-ATM levels at DSB sites in EZH2 gain-of-function mutant DLBCL cells. Control DMSO-treated cells or the HDAC1,2 inhibitor (RCY957) treated cells were co-stained with antibodies that detect H3K27me3 or phospho-ATM along with γH2AX. Our data showed that HDAC1,2 inhibition decreases H3K27me3 at DSB sites in agreement with our previous results [3], and that HDAC1,2 inhibition does not affect phospho-ATM at DSB sites (Fig. 4B-C). These findings together indicate that HDAC1,2 inhibition impairs EZH2-mediated but not ATM-dependent DNA repair signaling. Collectively, our results (as shown in Fig. 4A-C) demonstrate that the micro-laser technique we optimized can successfully detect proteins and their post-translational modifications at DSBs in suspension cells.
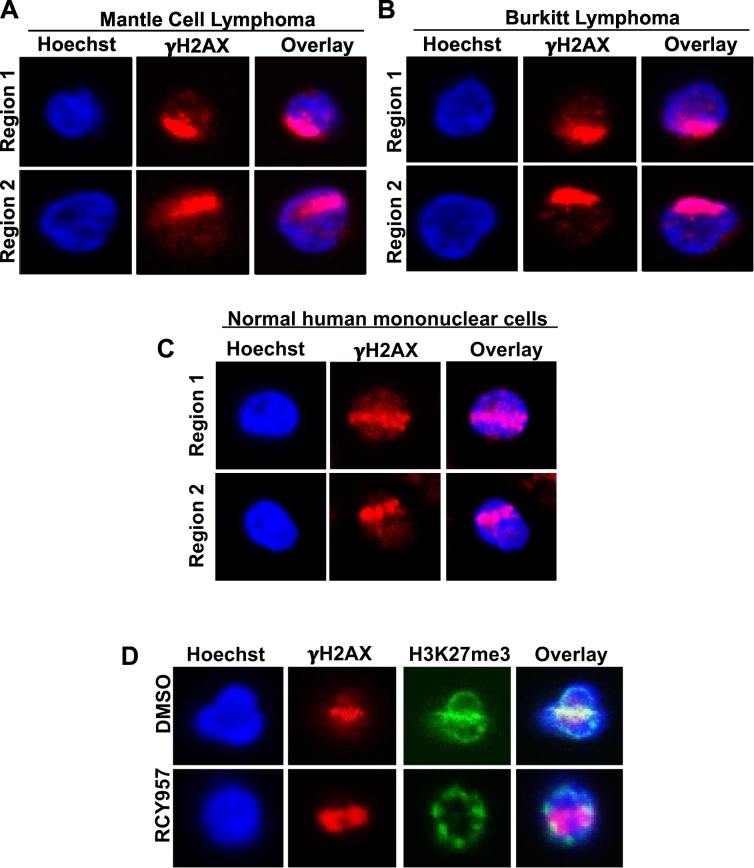
We used the settings detailed above to successfully irradiate and detect γH2AX tracks in primary mantle cell lymphoma (Fig. 5A), primary Burkitt lymphoma mononuclear cells (Fig. 5B), and in normal human peripheral blood mononuclear cells (PBMCs) (Fig. 5C). Importantly, we found that HDAC1,2 inhibition reduced H3K27me3 in primary mantle cell lymphoma (Fig. 5D), similar to that seen in EZH2 gain-of-function mutant DLBCL cells (Fig. 4B) [3]. Therefore, regulation of H3K27me3 by HDAC1,2 appears to be a general phenomenon seen in diverse cell lines and also occurs in primary patient samples. More importantly, these results demonstrate that our micro-laser technique can be applied to monitor DNA repair dynamics directly in normal PBMCs and in primary patient suspension cancer cells.
Fig. 5.
The utility of micro-irradiation technique in primary patient lymphoma samples. A and B. Representative images of laser-induced DNA damage in primary lymphoma patient cells. Mononuclear cells from mantle cell and Burkitt's lymphoma patients were cultured in vitro and irradiated with the same settings that were used for Karpas-422 cells. Localization of γH2AX to the laser damage lines is shown in this fig. C. Normal mononuclear cells from a healthy individual were micro-irradiated and γH2AX containing DSB regions were detected using the protocol described for lymphoma and leukemia cells as described in the main text. D. Representative images of primary mantle cell lymphoma cells following micro-irradiation and staining with anti-γH2AX and H3K27me3 are shown. A reduction in H3K27me3 is seen following treatment of lymphoma cells with HDAC1,2 inhibitor (RCY957) for 24 h.
3.3. Use of the optimized micro-laser technique to examine H3K27me3 during DNA repair in suspension DLBCL cells following small molecule inhibition of epigenetic modifiers
Next, we employed our modified micro-laser approach to determine the changes in the DNA repair-associated H3K27me3 at DSBs following treatment of suspension diffuse large B-cell lymphoma cells with small molecule inhibitors of enzymes that regulate this epigenetic mark. We chose two different DLBCL cell lines for the study: SUDHL4, a germinal center-derived diffuse large B-cell lymphoma (GC-DLBCL) line with a gain-of-function mutation in EZH2 [3] and SUDHL2, an activated B-cell-derived diffuse large B-cell lymphoma (ABC-DLBCL) line with wild-type EZH2 and expressing a truncated mutant form of the p300 histone acetyltransferase (HAT) that catalyzes histone H3K27 acetylation [26]. We treated these lymphoma lines with small molecule inhibitors of three different epigenetic enzymes: 1) DZNep that degrades the PRC2 complex that contains the EZH2 H3K27 methyltransferase [27]; 2) GSKJ4, an inhibitor of JMJD3/UTX H3K27 demethylases [28] [29,30]; and 3) RCY957, an HDAC1,2 selective inhibitor that could indirectly impact H3K27me3 via regulating H3K27 acetylation [3].
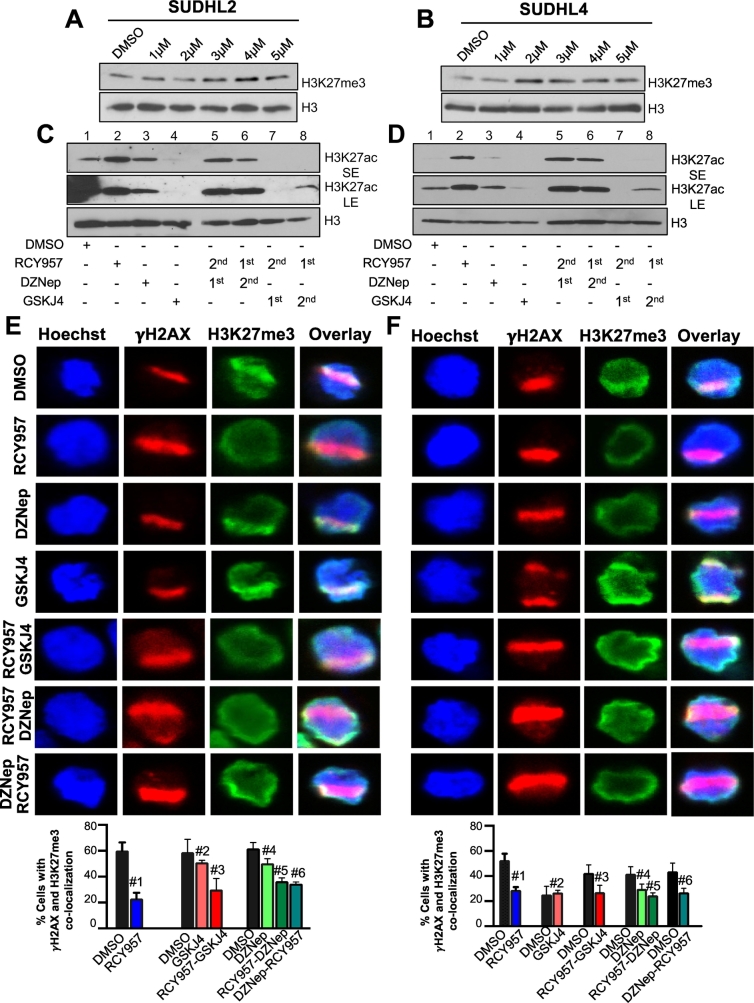
We first examined the effects of these small molecule inhibitor treatments on bulk H3K27me3 and H3K27ac levels using immunoblotting. A dose dependent increase in bulk H3K27me3 levels was observed upon treatment of SUDHL2 or SUDHL4 cells with GSKJ4 (Fig. 6A–B), confirming the inhibition of H3K27 demethylases by this compound. We previously showed that treatment of SUDHL4 cells with HDAC1,2 inhibitor RCY957 or EZH2 inhibitor DZNep does not affect global H3K27me3 levels [3]. A robust increase in H3K27ac was obtained in both SUDHL4 and SUDHL2 cells following treatment with RCY957, demonstrating its ability to inhibit the deacetylase activities of HDAC1,2 (Fig. 6C–D, compare lanes 1–2). We find that SUDHL2 cells possess H3K27ac even in the absence of a functional p300 acetyltransferase, suggesting that the homologous CBP acetyltransferase might be catalyzing H3K27ac (Fig. 6C). A subtle increase in H3K27ac was observed following treatment with DZNep (Fig. 6C-D, compare lanes 1–3), which could be ascribed to an increased access to the substrate H3K27 residue by acetyltransferases as a result of the reported DZNep-mediated disruption of the EZH2-containing PRC2 complex [27]. A decrease in bulk H3K27ac was observed following treatment with GSKJ4 (Fig. 6C–D, compare lanes 1–4), which correlates well with the increase in bulk H3K27me3 levels upon inhibiting the H3K27 demethylases (Fig. 6A–B). Pre-treatment of SUDHL2 or SUDHL4 cells with DZNep or addition of DZNep to RCY957-treated cells led to an accumulation of H3K27ac mark globally (Fig. 6C–D), further demonstrating an increased access to the substrate by acetyltransferases and inhibition of HDAC1,2 activities. Pre-treatment of SUDHL2 or SUDHL4 cells with the H3K27 demethylase inhibitor GSKJ4 prior to HDAC1,2 inhibition prevented the accumulation of H3K27ac (Fig. 6C-D, lane 7), suggesting an acquisition of a ‘saturated’ methylated state at H3K27 globally that effectively blocks acetylation. Addition of GSKJ4 to cells pretreated with RCY957 also prevented the accumulation of H3K27ac (Fig. 6C–D, lane 8), showing that the effects of RCY957 are reversible. Taken together, these results show the effectiveness of the small molecules in inhibiting the enzymes involved in establishing the H3K27me3-ac epigenetic switch in DLBCL cells.
Fig. 6.
The effect of HDAC, EZH2 and UTX/JMJD3 inhibitors on H3K27ac and H3K27me3 levels in DLBCL cells. A-B. SUDHL2 or SUDHL4 cells were treated with increasing concentrations of GSKJ4 (1–5 μM) for 12 h. Immunoblotting were performed with whole cell lysates obtained from control and treated cells. Blots were probed with anti-H3K27 trimethyl (me3) or anti-H3 antibody. C-D. SUDHL2 or SUDHL4 cells were treated for 12 h with single drugs, 2 μM RCY957, 0.5 μM DZNep or 5 μM GSKJ4 or treated with a single drug for 12 h, washed with PBS and treated with the second drug for an additional 12 h. Following indicated treatments, the chromatin fractions were prepared and immunoblotting was performed. Blots were probed with either anti-H3K27 acetyl (ac) or anti-H3 antibody. SE or LE, short or long exposure, respectively. E-F. SUDHL2 or SUDHL4 cells were treated for 24 h with single drugs RCY957, GSKJ4 or DZNep. Dual treatments were performed for 12 h with the first drug followed by washing of cells and treatment with the second drug for an additional 12 h. Following micro-irradiation, cells were stained with anti-γH2AX and H3K27me3. The overlay of γH2AX and H3K27me3 signals with the hoechst stained nucleus is shown in this fig. E. The p values calculated using the two tailed t-test are as follows: SUDHL2 cell line: DMSO vs 957: 0.0098 (#1), DMSO vs GSKJ4: 0.5243 (#2), DMSO vs 957 then GSKJ4: 0.164 (#3), DMSO vs DZNep: 0.107 (#4), DMSO vs 957 then DZNep: 0.0016 (#5) and DMSO vs DZNep then RCY957: 0.003 (#6). F. SUDHL4 cell line: DMSO vs 957: 0.0148 (#1), DMSO vs GSKJ4: 0.842 (#2), DMSO vs 957 then GSKJ4: 0.156 (#3), DMSO vs DZNep: 0.0303 (#4) and DMSO vs 957 then DZNep: 0.0212 (#5) and DMSO vs DZNep then RCY957:0.05 (#6). The images were overlaid using the NIS elements software.
We then tested the effects of these small molecule inhibitors on cell viability and the cell cycle progression of SUDHL2 and SUDHL4 lines after different treatment times. The HDAC1,2 inhibitor (RCY957) or EZH2 inhibitor (DZNep) treatment showed no effect on cell viability or the cell cycle phases after 24 h treatment (Supplemental Fig. 2), but increased the sub-G1 or dead cell population after 72 h treatment (Supplemental Fig. 3), which agrees well with our previous study [3]. The H3K27 demethylase inhibitor GSKJ4 alone or when added before or after RCY957 caused death in SUDHL2 and SUDHL4 cells after 12 h treatment (Supplemental Fig. 2), and resulted in a robust cell death after 72 h treatment (Supplemental Fig. 3). Surprisingly, SUDHL4 cells expressing the hyperactive mutant EZH2 and containing elevated levels of H3K27me3 were more sensitive to GSKJ4 treatment than SUDHL2 cells expressing wild-type EZH2 (Supplemental Figs. 2 and 3). This result reveals for the first time that increasing the already high H3K27me3 levels by inhibiting H3K27 demethylation in the EZH2 gain-of-function mutant SUDHL4 cells can also be detrimental. Collectively, results from these cell cycle assays suggest that altering the levels of H3K27me3 and H3K27ac compromises the viability of SUDHL2 and SUDHL4 DLBCL cells.
We next used our optimized micro-laser assay to examine the effects of these small molecule inhibitors on H3K27me3 at DSB sites in the suspension DLBCL cells. For this assay, we chose treatment time points where the inhibitors showed no significant induction of cell death. Therefore, we used the 24 h treatment time-point for HDAC1,2 inhibitor (RCY957) or EZH2 inhibitor (DZNep) and the 12 h time-point for the H3K27 demethylase inhibitor (GSKJ4), (Supplemental Figs. 2 and 4). While treatment with GSKJ4 increased global H3K27me3 levels (Fig. 6A-B), an increase in H3K27me3 was not evident at the micro-laser-induced DSBs (Fig. 6E) and the difference was statistically insignificant between DMSO and GSKJ4 treatments in SUDHL2 (p = .5243) or SUDHL4 cells (p = .842). This finding indicates to a limitation of the micro-laser assays in general, wherein increased accumulation of a preexisting factor or protein post-translational modification at DSBs are not easily measurable. Interestingly, DZNep treatment caused a very modest reduction in H3K27me3 at DSB sites in SUDHL2 cells (p = .107), but showed greater reduction in SUDHL4 cells (p = .0303) (Fig. 6E-F), alluding to a likely higher sensitivity of the EZH2 hyperactive mutant-containing PRC2 complex in the latter cell line to the DZNep-mediated disassembly. While HDAC1,2 inhibition by RCY957 robustly increases global H3K27ac (Fig. 6C–D), it does not reduce global H3K27me3 [3]. However, H3K27me3 deposition at DSBs is severely reduced following RCY957 treatment in both SUDHL2 (p = .0098) and SUDHL4 (p = .0148) cells (Fig. 6E–F), suggesting that increased H3K27ac as a result of HDAC1,2 inhibition blocks the active deposition of EZH2-catalyzed H3K27me3 at DSBs during DNA repair. We were unable to reliably assess H3K27ac at DSB sites as it exhibited a pan nuclear staining pattern in immunofluorescence (data not shown). Nevertheless, our results show that the micro-laser assay can be effectively used to detect and measure factors that are actively recruited to or deposited at DSBs during DNA repair, such as, the MRN complex and phospho-ATM (Fig. 4A–C) in addition to H3K27me3. Moreover, these results also demonstrate that the micro-laser assay can provide valuable insights into the mode-of-action of small molecule inhibitors at DSBs during DNA repair.
We also used our optimized micro-laser assay to examine the effects of sequential inhibitor treatments on H3K27me3 at DSBs. Pretreatment with RCY957 did not significantly impact the effects of the H3K27 demethylase inhibitor GSKJ4 on H3K27me3 at DSBs in either SUDHL2 (p = .164) or SUDHL4 (p = .156) cell lines (Fig. 6E–F). Because of its high cytotoxicity (Supplemental Figs. 2 and 3), we were unable to examine the effects of pretreatment with GSKJ4. However, pretreatment of SUDHL2 or SUDHL4 cells with RCY957 before DZNep or vice versa, both caused a statistically significant decrease in H3K27me3 when compared to DZNep treatment alone (p < .05) (Fig. 6E–F). Based on this observation, we tested the effects of RCY957 and DZNep combination treatments on cell viability. Our results showed a robust cell death induced by the combination or sequential treatments when compared to the single agent treatments (Supplemental Fig. 3) [3]. This result suggests that the HDAC1,2 and EZH2 inhibitors can possibly serve as a mechanism-based combination therapeutic strategy for EZH2 gain-of-function DLBCL. Importantly, these results strongly support the use of micro-laser assay to guide treatment plans in the clinic.
4. Discussion
Here we report the development and application of the micro-laser DNA repair assay that can now be used for liquid tumor cells. We report extensive optimizations for this assay that include determining the adherence strategy, the cell density, the laser power and microscope settings for successful micro-irradiation and imaging of suspension lymphoma and leukemia cells (Fig. 1, Fig. 2, Fig. 3). We have confirmed that the optimized settings and conditions do not compromise cell viability (Fig. 1D).
We recently reported that inhibition of Class I family histone deacetylases HDAC1 and HDAC2 (HDAC1,2) reduces the amount of DNA break-induced silencing in cis (DISC) during DSB repair, a phenomenon that normally affects several kilobases flanking DSBs [25]. DISC is regulated by two histone modifications belonging to the EZH2 repair pathway: H3K27me3 installed by EZH2, and the PBAF nucleosome remodeler complex controlled H2AK119 monoubiquitination (H2AK119ub1), another transcriptional repression mark that acts downstream of EZH2 during DNA repair [1,16,25]. H3K27 acetylation (ac) acts as a positive epigenetic signal during transcription [31]. Acetylation blocks methylation at H3K27 residue, and thus creating an epigenetic switch between these two signaling states. Consistent with this model, using the micro-laser technique, we have found that HDAC1,2 inhibition decreases H3K27me3 at DSBs during DNA repair and increases global levels of H3K27ac in DLBCL cells [3] (Fig. 6). Thus, using our optimized micro-laser technique, we have made a novel link between HDAC1,2 and EZH2 in DNA repair for the first time in DLBCL cells [3]. Overall, our optimized microlaser technique enables determining the recruitment of factors including chromatin modifiers or remodelers to DSB sites, and uncovering their functions in DNA repair, directly in liquid tumor cells.
DLBCL is the most common disease entity of B-cell non-Hodgkin lymphoma (NHL) in adults, and has one of the highest mortality rates [32]. DLBCL subtypes, GC and ABC, originate from germinal center B-cells or activated B-cells, respectively [33]. While >50% of DLBCL patients are cured with the available multi-agent chemo, radio- or immune-therapies, ~30–40% of patients develop a relapsed or refractory disease [34] and many respond poorly to a second line therapy using high-dose chemotherapy and autologous stem cell transplantation [[35], [36], [37]]. Therefore, there is a compelling need for new and effective therapeutic strategies and prognostic biomarkers. Sequencing of lymph node biopsies from DLBCL patients has identified recurrent somatic mutations in the polycomb group proto-oncogene EZH2 [38], occurring in ~22% of GC-DLBCL subtype. These are gain-of-function (GOF) mutations within the epigenetic modifier EZH2's catalytic SET domain resulting in a hyperactive enzyme producing high levels of H3K27 trimethylation (K27me3) [8,39]. EZH2 GOF GC-DLBCL cells with aberrantly elevated H3K27me3 are chemoresistant to DNA damaging agents such as doxorubicin compared to GC-DLBCL with wild-type EZH2 [40].
Small molecule inhibitors of EZH2 have reached clinical trials for these refractory EZH2GOF B-cell lymphomas. In addition to EZH2 inhibitors, we have shown a novel strategy of targeting the EZH2 gain-of-function mutant DLBCL using HDAC1,2 selective inhibitors [3]. We reported that HDAC1,2 inhibitor decreases the survival advantages promoted by the EZH2 GOF mutation in DLBCL cells. In addition to EZH2, H3K27 demethylase JMJD3 or KDM6B is overexpressed in germinal center-derived DLBCL and this high expression of JMJD3 is also correlated with poor prognosis [41]. Here, we have now shown that GSKJ4, an inhibitor of H3K27me3 demethylase, causes rapid and robust apoptosis in GC-DLBCL cells (Supplementary Figs. 2 and 3). Interestingly, the EZH2 gain-of-function mutant DLBCL cells with high H3K27me3 are more sensitive to GSKJ4 treatment than DLBCL cells with wild-type EZH2 (Supplemental Figs. 2 and 3). This finding along with those obtained using EZH2 and HDAC1,2 inhibitors together allude to a model where maintaining the H3K27me3-H3K27ac balance is critical for DNA repair capabilities and survival of certain subset of cancer cells. The use of laser-based repair assay optimized here for hematological liquid cancers fills a major void that currently exist between assessing global histone modifications and chromatin dynamics that occur globally and those that occur specifically at DSB sites in liquid tumors. Because the suspension cells remain attached to the Cell-Tak-coated culture dish for a limited time (4–8 h), one limitation of the assay is the inability to measure γH2AX dephosphorylation and attenuation of DNA damage signal, which take several hours to be completed (Supplemental Fig. 1).
The era of using the same treatment plan for all DLBCLs is fading away as personalized medicine approaches gain ground. As illustrated by our data, the technique presented here could lend itself to important clinical applications as we demonstrate the applicability of this technique in primary lymphoma patient sample cultures. We performed micro-irradiation in two different primary patient lymphoma samples and were able to detect γH2AX and/or H3K27me3 at the DSB laser stripes (Fig. 5A–B). We could also observe a decrease in H3K27me3 at the DSB laser tracks following HDAC1,2 inhibitor treatment in a primary DLBCL patient sample (Fig. 5D). Thus, our data demonstrate an important and direct applicability of our optimized micro-laser technique to predict the response of a blood cancer patient to chemotherapy and guide the manipulation of the drug dosage or regimen for subsequent treatments using bone marrow aspirates. The optimized micro-irradiation followed by immunofluorescence described here can be used to assess the DNA repair efficiency in patients recruited to clinical trials, as the standard immunofluorescence technique to examine epigenetic histone marks or DNA damage markers would not inform us about the rapid DNA repair events that occurr at DSB sites. In summary, the method described here is not only useful to decipher the mechanisms of action of novel drugs, but can also provide a valuable diagnostic tool that could be used to devise mechanism-based therapeutic strategies.
Acknowledgments
Acknowledgements
This work was supported by National Institute of Health/National Cancer Institute R01 grant (1R011CA188520) to SB. We thank the HCI cell bank for the patient samples. Normal mononuclear cells were isolated from the blood collected from Drs. Srividya Bhaskara and Mahesh Chandrasekharan. We thank Drs. Simon Jones and Matt Jarpe for the HDAC inhibitor. We thank Dr. Mahesh Chandrasekharan for his comments on the manuscript. We thank Shaistah Din for her technical assistance with standard immunofluorescence. We also thank the University of Utah cell imaging core services.
Conflict of interest
The authors have no competing financial interests.
Author contributions
DJ and GSSB optimized the laser conditions for hematological cells. DJ, GSSB and SB designed experiments and critically evaluated the data. DJ, GSSB and KC performed the experiments described in this study. DJ and SB wrote the manuscript. SB obtained funding for this work.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.03.083.
Appendix A. Supplementary data
Supplementary Fig. 1 SUDHL4 cells were irradiated with a 5Gy dose using a RadSource X-ray irradiation cabinet. Cells were allowed to recover for the indicated time points prior to fixation and immunofluorescence with γH2AX was performed.
Supplementary Fig. 2 A and B. SUDHL2 or SUDHL4 cells were treated for 24 h with 2 μM RCY957, 5 μM GSKJ4 or 0.5 μM DZnep prior to cell cycle analysis. Sequential treatments of 2 μM RCY957 then 5 μM GSKJ4 were performed for 12 h per treatment. Cells were treated with RCY957 for 12 h, then drug was removed and GSKJ4 was added for an additional 12 h. All the treatments were done for a total of 24 h. After 24 h total treatment, cells were fixed in 70% ethanol and stained with propidium iodide for cell cycle analyses.
Supplementary Fig. 3 A and B. SUDHL4 or SUDHL2 cells were treated with 2 μM RCY957, 5 μM GSKJ4 or 0.5 μM DZNep for 72 h and cell cycle analysis of propidium iodide stained cells was done.
Supplementary Fig. 4 A and B. SUDHL4 or SUDHL2 cells were treated with 2.5 or 5 μM GSKJ4 for 12 h and cell cycle analysis of propidium iodide stained cells was done.
References
- 1.Kakarougkas A., Ismail A., Chambers A.L., Riballo E., Herbert A.D., Kunzel J. Requirement for PBAF in transcriptional repression and repair at DNA breaks in actively transcribed regions of chromatin. Mol Cell. 2014;55(5):723–732. doi: 10.1016/j.molcel.2014.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Toiber D., Erdel F., Bouazoune K., Silberman D.M., Zhong L., Mulligan P. SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol Cell. 2013;51(4):454–468. doi: 10.1016/j.molcel.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson D.P., Spitz G.S., Tharkar S., Quayle S.N., Shearstone J.R., Jones S. HDAC1,2 inhibition impairs EZH2- and BBAP-mediated DNA repair to overcome chemoresistance in EZH2 gain-of-function mutant diffuse large B-cell lymphoma. Oncotarget. 2015;6(7):4863–4887. doi: 10.18632/oncotarget.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baldeyron C., Soria G., Roche D., Cook A.J., Almouzni G. HP1alpha recruitment to DNA damage by p150CAF-1 promotes homologous recombination repair. J Cell Biol. 2011;193(1):81–95. doi: 10.1083/jcb.201101030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chou D.M., Adamson B., Dephoure N.E., Tan X., Nottke A.C., Hurov K.E. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc Natl Acad Sci U S A. 2010;107(43):18475–18480. doi: 10.1073/pnas.1012946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cottini F., Hideshima T., Suzuki R., Tai Y.T., Bianchini G., Richardson P.G. Synthetic lethal approaches exploiting DNA damage in aggressive myeloma. Cancer Discov. 2015;5(9):972–987. doi: 10.1158/2159-8290.CD-14-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Economopoulou P., Pappa V., Papageorgiou S., Dervenoulas J., Economopoulos T. Abnormalities of DNA repair mechanisms in common hematological malignancies. Leuk Lymphoma. 2011;52(4):567–582. doi: 10.3109/10428194.2010.551155. [DOI] [PubMed] [Google Scholar]
- 8.Kim K.H., Roberts C.W. Targeting EZH2 in cancer. Nat Med. 2016;22(2):128–134. doi: 10.1038/nm.4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ezponda T., Licht J.D. Molecular pathways: deregulation of histone h3 lysine 27 methylation in cancer-different paths, same destination. Clin Cancer Res. 2014;20(19):5001–5008. doi: 10.1158/1078-0432.CCR-13-2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Safaei S., Baradaran B., Hagh M.F., Alivand M.R., Talebi M., Gharibi T. Double sword role of EZH2 in leukemia. Biomed Pharmacother. 2018;98:626–635. doi: 10.1016/j.biopha.2017.12.059. [DOI] [PubMed] [Google Scholar]
- 11.Herviou L., Cavalli G., Cartron G., Klein B., Moreaux J. EZH2 in normal hematopoiesis and hematological malignancies. Oncotarget. 2016;7(3):2284–2296. doi: 10.18632/oncotarget.6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu B., Konze K.D., Jin J., Wang G.G. Targeting EZH2 and PRC2 dependence as novel anticancer therapy. Exp Hematol. 2015;43(8):698–712. doi: 10.1016/j.exphem.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Popovic R., Shah M.Y., Licht J.D. Epigenetic therapy of hematological malignancies: where are we now? Ther Adv Hematol. 2013;4(2):81–91. doi: 10.1177/2040620712466864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benyoucef A., Palii C.G., Wang C., Porter C.J., Chu A., Dai F. UTX inhibition as selective epigenetic therapy against TAL1-driven T-cell acute lymphoblastic leukemia. Genes Dev. 2016;30(5):508–521. doi: 10.1101/gad.276790.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campbell S., Ismail I.H., Young L.C., Poirier G.G., Hendzel M.J. Polycomb repressive complex 2 contributes to DNA double-strand break repair. Cell Cycle. 2013;12(16):2675–2683. doi: 10.4161/cc.25795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vissers J.H., van Lohuizen M., Citterio E. The emerging role of Polycomb repressors in the response to DNA damage. J Cell Sci. 2012;125:3939–3948. doi: 10.1242/jcs.107375. Pt 17. [DOI] [PubMed] [Google Scholar]
- 17.Fong C.Y., Morison J., Dawson M.A. Epigenetics in the hematologic malignancies. Haematologica. 2014;99(12):1772–1783. doi: 10.3324/haematol.2013.092007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharma A., Singh K., Almasan A. Histone H2AX phosphorylation: a marker for DNA damage. Methods Mol Biol. 2012;920:613–626. doi: 10.1007/978-1-61779-998-3_40. [DOI] [PubMed] [Google Scholar]
- 19.Carney J.P., Maser R.S., Olivares H., Davis E.M., Le Beau M., Yates J.R., 3rd The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93(3):477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 20.Marechal A., Zou L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb Perspect Biol. 2013;5(9) doi: 10.1101/cshperspect.a012716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kozlov S.V., Graham M.E., Jakob B., Tobias F., Kijas A.W., Tanuji M. Autophosphorylation and ATM activation: additional sites add to the complexity. J Biol Chem. 2011;286(11):9107–9119. doi: 10.1074/jbc.M110.204065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lavin M.F., Delia D., Chessa L. ATM and the DNA damage response. Workshop on ataxia-telangiectasia and related syndromes. EMBO Rep. 2006;7(2):154–160. doi: 10.1038/sj.embor.7400629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhaskara S. Histone deacetylases 1 and 2 regulate DNA replication and DNA repair: potential targets for genome stability-mechanism-based therapeutics for a subset of cancers. Cell Cycle. 2015;14(12):1779–1785. doi: 10.1080/15384101.2015.1042634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhaskara S., Jacques V., Rusche J.R., Olson E.N., Cairns B.R., Chandrasekharan M.B. Histone deacetylases 1 and 2 maintain S-phase chromatin and DNA replication fork progression. Epigenetics Chromatin. 2013;6(1):27. doi: 10.1186/1756-8935-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tharkar-Promod S., Johnson D.P., Bennett S.E., Dennis E.M., Banowsky B.G., Jones S.S. HDAC1,2 inhibition and doxorubicin impair Mre11-dependent DNA repair and DISC to override BCR-ABL1-driven DSB repair in Philadelphia chromosome-positive B-cell precursor acute lymphoblastic leukemia. Leukemia. 2018;32(1):49–60. doi: 10.1038/leu.2017.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haery L., Lugo-Pico J.G., Henry R.A., Andrews A.J., Gilmore T.D. Histone acetyltransferase-deficient p300 mutants in diffuse large B cell lymphoma have altered transcriptional regulatory activities and are required for optimal cell growth. Mol Cancer. 2014;13:29. doi: 10.1186/1476-4598-13-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fiskus W., Wang Y., Sreekumar A., Buckley K.M., Shi H., Jillella A. Combined epigenetic therapy with the histone methyltransferase EZH2 inhibitor 3-deazaneplanocin a and the histone deacetylase inhibitor panobinostat against human AML cells. Blood. 2009;114(13):2733–2743. doi: 10.1182/blood-2009-03-213496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heinemann B., Nielsen J.M., Hudlebusch H.R., Lees M.J., Larsen D.V., Boesen T. Inhibition of demethylases by GSK-J1/J4. Nature. 2014;514(7520):E1–E2. doi: 10.1038/nature13688. [DOI] [PubMed] [Google Scholar]
- 29.Sui A., Xu Y., Li Y., Hu Q., Wang Z., Zhang H. The pharmacological role of histone demethylase JMJD3 inhibitor GSK-J4 on glioma cells. Oncotarget. 2017;8(40):68591–68598. doi: 10.18632/oncotarget.19793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y., Zhang M., Sheng M., Zhang P., Chen Z., Xing W. Therapeutic potential of GSK-J4, a histone demethylase KDM6B/JMJD3 inhibitor, for acute myeloid leukemia. J Cancer Res Clin Oncol. 2018;144(6):1065–1077. doi: 10.1007/s00432-018-2631-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 32.Pasqualucci L., Dalla-Favera R. The genetic landscape of diffuse large B-cell lymphoma. Semin Hematol. 2015;52(2):67–76. doi: 10.1053/j.seminhematol.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Basso K., Dalla-Favera R. Germinal centres and B cell lymphomagenesis. Nat Rev Immunol. 2015;15(3):172–184. doi: 10.1038/nri3814. [DOI] [PubMed] [Google Scholar]
- 34.Raut L.S., Chakrabarti P.P. Management of relapsed-refractory diffuse large B cell lymphoma. South Asian J Cancer. 2014;3(1):66–70. doi: 10.4103/2278-330X.126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Verdonck L.F., Dekker A.W., de Gast G.C., van Kempen M.L., Lokhorst H.M., Nieuwenhuis H.K. Salvage therapy with ProMACE-MOPP followed by intensive chemoradiotherapy and autologous bone marrow transplantation for patients with non-Hodgkin's lymphoma who failed to respond to first-line CHOP. J Clin Oncol. 1992;10(12):1949–1954. doi: 10.1200/JCO.1992.10.12.1949. [DOI] [PubMed] [Google Scholar]
- 36.Kewalramani T., Zelenetz A.D., Hedrick E.E., Donnelly G.B., Hunte S., Priovolos A.C. High-dose chemoradiotherapy and autologous stem cell transplantation for patients with primary refractory aggressive non-Hodgkin lymphoma: an intention-to-treat analysis. Blood. 2000;96(7):2399–2404. [PubMed] [Google Scholar]
- 37.Elstrom R.L., Martin P., Ostrow K., Barrientos J., Chadburn A., Furman R. Response to second-line therapy defines the potential for cure in patients with recurrent diffuse large B-cell lymphoma: implications for the development of novel therapeutic strategies. Clin Lymphoma Myeloma Leuk. 2010;10(3):192–196. doi: 10.3816/CLML.2010.n.030. [DOI] [PubMed] [Google Scholar]
- 38.Morin R.D., Johnson N.A., Severson T.M., Mungall A.J., An J., Goya R. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42(2):181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCabe M.T., Graves A.P., Ganji G., Diaz E., Halsey W.S., Jiang Y. Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27) Proc Natl Acad Sci U S A. 2012;109(8):2989–2994. doi: 10.1073/pnas.1116418109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clozel T., Yang S., Elstrom R.L., Tam W., Martin P., Kormaksson M. Mechanism-based epigenetic chemosensitization therapy of diffuse large B-cell lymphoma. Cancer Discov. 2013;3(9):1002–1019. doi: 10.1158/2159-8290.CD-13-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mathur R., Sehgal L., Havranek O., Kohrer S., Khashab T., Jain N. Inhibition of demethylase KDM6B sensitizes diffuse large B-cell lymphoma to chemotherapeutic drugs. Haematologica. 2017;102(2):373–380. doi: 10.3324/haematol.2016.144964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waite J.H., Tanzer M.L. Polyphenolic substance of Mytilus edulis: novel adhesive containing L-Dopa and Hydroxyproline. Science. 1981;212(4498):1038–1040. doi: 10.1126/science.212.4498.1038. [DOI] [PubMed] [Google Scholar]
- 43.Bhaskara S., Chyla B.J., Amann J.M., Knutson S.K., Cortez D., Sun Z.W. Deletion of histone deacetylase 3 reveals critical roles in S phase progression and DNA damage control. Mol Cell. 2008;30(1):61–72. doi: 10.1016/j.molcel.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1 SUDHL4 cells were irradiated with a 5Gy dose using a RadSource X-ray irradiation cabinet. Cells were allowed to recover for the indicated time points prior to fixation and immunofluorescence with γH2AX was performed.
Supplementary Fig. 2 A and B. SUDHL2 or SUDHL4 cells were treated for 24 h with 2 μM RCY957, 5 μM GSKJ4 or 0.5 μM DZnep prior to cell cycle analysis. Sequential treatments of 2 μM RCY957 then 5 μM GSKJ4 were performed for 12 h per treatment. Cells were treated with RCY957 for 12 h, then drug was removed and GSKJ4 was added for an additional 12 h. All the treatments were done for a total of 24 h. After 24 h total treatment, cells were fixed in 70% ethanol and stained with propidium iodide for cell cycle analyses.
Supplementary Fig. 3 A and B. SUDHL4 or SUDHL2 cells were treated with 2 μM RCY957, 5 μM GSKJ4 or 0.5 μM DZNep for 72 h and cell cycle analysis of propidium iodide stained cells was done.
Supplementary Fig. 4 A and B. SUDHL4 or SUDHL2 cells were treated with 2.5 or 5 μM GSKJ4 for 12 h and cell cycle analysis of propidium iodide stained cells was done.