Abstract
The results of this study indicate that the maize rhizosphere remains a reservoir for microbial strains with unique beneficial properties. The study sought to provide an indigenous Bacillus strain with a bioprotective potential to alleviate maize fusariosis in South Africa. We selected seven Bacillus isolates (MORWBS1.1, MARBS2.7, VERBS5.5, MOREBS6.3, MOLBS8.5, MOLBS8.6, and NWUMFkBS10.5) with biosuppressive effects against two maize fungal pathogens (Fusarium graminearum and Fusarium culmorum) based on 16S rDNA gene characterization and lipopeptide gene analysis. The PCR analysis revealed that lipopeptide genes encoding the synthesis of iturin, surfactin, and fengycin might be responsible for their antifungal activities. Few of the isolates also showed possible biosurfactant capability, and their susceptibility to known antibiotics is indicative of their eco‐friendly attributes. In addition, in silico genomic analysis of our best isolate (Bacillus velezensis NWUMFkBS10.5) and characterization of its active metabolite with FTIR, NMR, and ESI‐Micro‐Tof MS confirmed the presence of valuable genes clusters and metabolic pathways. The versatile genomic potential of our Bacillus isolate emphasizes the continued relevance of Bacillus spp. in biological management of plant diseases.
Keywords: Bacillus velezensis, ESI‐Micro‐Tof MS, genome, in silico, lipopeptides, plant disease
1. INTRODUCTION
In recent years, the focus has shifted from the use of environmentally harmful plant disease management practices such as the use of chemical pesticides and inorganic fertilizers to the development of ecological friendly approaches that do not pose a public health threat. The use of biological control agents (BCAs) in the management of cereal crop diseases has become popular (Babalola & Glick, 2012; Dimkić et al., 2013; Lugtenberg & Kamilova, 2009). Bacterial species from the genus Bacillus have gained prominent attention due to their tolerance to high temperature, ability to withstand adverse environmental conditions, ability to grow rapidly in liquid media and their ability to produce a large variety of secondary metabolites that have broad antimicrobial capabilities (Santoyo, Orozco‐Mosqueda, & Govindappa, 2012; Sumi, Yang, Yeo, & Hahm, 2014).
Members of the Gram‐positive endospore‐forming Bacillus sp., such as B. amyloliquefaciens and B. subtilis collected from plant parts, have been used in the control of fusariosis in small cereal grains including maize (Bacon & Hinton, 2011; Gond, Bergen, Torres, White, & Kharwar, 2015). Fusariosis in maize, which could manifest as Fusarium head blight (FHB) or Fusarium ear rot (FER) and many more, is caused by fusarium members such as F. graminearum and F. verticillioides. These diseases affect maize production in South Africa and other areas of the world (Boutigny et al., 2012, 2011 ; Summerell & Leslie, 2011). Maize, a staple crop in South Africa, is consumed daily in most households and used in the production of animal feeds (Janse van Rensburg, Mclaren, Flett, & Schoeman, 2015; Lamprecht, Tewoldemedhin, Botha, & Calitz, 2011); therefore, efforts to reduce loss due to preharvest and postharvest contamination by F. graminearum infection have recently gained significant attention (Boutigny et al., 2011; Mngqawa et al., 2016). The presence of mycotoxins zearalenone and deoxynivalenol found in maize grains infected by F. graminearum is also a cause for concern (Wang, Ndoye, Zhang, Li, & Liao, 2011).
Reports show that BCAs used for crop protection perform better in their native geographical regions due to increased survival rate compared to the use of imported commercial BCAs (Abiala, Odebode, Hsu, & Blackwood, 2015; Bardin et al., 2015; Grzywacz, Stevenson, Mushobozi, Belmain, & Wilson, 2014; Pereira, Nesci, Castillo, & Etcheverry, 2010). Our goal in this present work was to select indigenous Bacillus strains from the maize rhizosphere, evaluate their anti‐phytopathogenic potentials in vitro against Fusarium spp., molecularly characterize the Bacillus isolates, and identify the likely mechanisms they employ in their anti‐phytopathogenic activities. Bacillus spp. secrete lipopeptide compounds such as surfactin, fengycin, and iturin that they utilize in antibiosis. The presence of these cyclic lipopeptides in our maize root‐associated strains would be valuable if they are to be considered for in planta studies and subsequently for the management of F. graminearum infections in South Africa.
Strains within the genus have also been reported to synthesize structurally diverse secondary metabolites that exhibit broad‐spectrum antibiotic activities, and the genomic basis for the synthesis of these secondary metabolites has been attributed to the presence of polyketide synthases (PKSs) and non‐ribosomal peptide synthetase (NRPS) in their genomes (Raaijmakers, Bruijn, & Kock, 2006; Roongsawang, Washio, & Morikawa, 2011; Tyc, Song, Dickschat, Vos, & Garbeva, 2017). The amphipathic structure, the hydrophilic peptide portion, and a hydrophobic fatty acid portion of these peptides show resemblances. These peptides also exhibit a cyclic nature due to the linkage of their C‐terminal peptide residue either indirectly to a β‐hydroxy fatty acid or directly to a β‐amino acid (Mnif & Ghribi, 2015; Ongena & Jacques, 2008). These antimicrobial peptides have been isolated, quantified, purified, and characterized using various approaches and techniques that ensure the chemical components responsible for their bioactivity are well understood. The majority of the current approaches employed involve the combination of chromatographic techniques, mass spectrometry, nuclear magnetic resonance (NMR), and Fourier transform infrared spectroscopy (FTIR) (Biniarz, Łukaszewicz, & Janek, 2017; Jasim, Sreelakshmi, Mathew, & Radhakrishnan, 2016).
Reports have shown that expression of biosynthetic genes and secretion of secondary metabolites may be difficult during laboratory culture of potential BCAs due to growth conditions (Laureti et al., 2011). The non‐expression of genes or secretion of secondary metabolites can hinder the identification or detection of the specific metabolite or gene responsible for the antimicrobial activities of a BCA (Michelsen et al., 2015). To fully understand beneficial bacterial species, genomes of multiple independent isolates are required for comparison (Tettelin et al., 2005). Comparing the total repertoire of genes for a group of genomes from close bacterial species is an instrumental approach for the development of novel beneficial compounds and for the functional characterization of important genetic determinants in significant microbial strains (Medini, Donati, Tettelin, Masignani, & Rappuoli, 2005). The bacterial pan‐genome can be defined as the complete repository of genes located in the genome of closely related bacterial species. This includes the “core genome” (genes identified in two or more strains) and the “dispensable genome” (genes peculiar to single strains) (Medini et al., 2005; Rouli, Merhej, Fournier, & Raoult, 2015; Tettelin et al., 2005). The core and dispensable genes are crucial signatures for recognizing species diversity.
Researchers now use a combinational approach such as genome mining, pan‐genome analysis, structural data elucidation, and metabolomic characterization to identify biosynthetic products secreted by important microbes (Dunlap, Bowman, & Schisler, 2013; Van Der Voort et al., 2015). Often, genomic data offer predictions that lead to the detection of novel biosynthetic pathways, genes, and enzymes which then enables experimental isolation, structural elucidation, and chemical characterization of novel compounds (Challis, 2008). The combinational approach ensures that in silico or theoretically predicted biosynthetic products correlate with structurally or chemically identified metabolites (Ziemert, Alanjary, & Weber, 2016). Here, we examined the antimicrobial potential of the lyophilized extract of the secondary metabolites secreted by Bacillus velezensis NWUMFkBS10.5 while employing electro‐spray ionization mass spectrometry (ESI‐Q‐TOF MS), FTIR, and NMR to confirm that its secondary metabolites were functionally active.
Lastly, the genomic information of B. velezensis and other related Bacillus strains having strong potential for the control of phytopathogens including F. graminearum has been made available in recent years (Dunlap et al., 2013; Dunlap, Schisler, Bowman, & Rooney, 2015; Lee et al., 2015; Palazzini, Dunlap, Bowman, & Chulze, 2016; Pan, Li, & Hu, 2017). In light of this, we sequenced the genome of our best isolate NWUMFkBS10.5, to determine its phylogenomic association and its unique antimicrobial trait.
2. EXPERIMENTAL PROCEDURES
2.1. Description of sampling sites and sample collection from rhizosphere
Depending on the width of the maize plot and in no particular order, 20–30 g of rhizospheric soil was collected randomly from four maize rows, 15–25 m, apart in each plot from 10 maize farms in the North West Province of South Africa at harvest time. The geographic location of the sampling sites covers 28,206 km2 area. The temperature ranges between 17°C and 31°C during the summer and between 3°C and 21°C during the winter, with an average rainfall of 360 mm. Harvested maize plants were shaken gently at the roots to manually remove the loosely attached soil. The adhering root soil was considered as the rhizosphere soil, and these samples were pooled for each location. We obtained 10 different soil samples from the different maize plots.
2.2. Differential and selective isolation of Bacillus spp. from rhizosphere sample
Five grams of each soil sample was inoculated in 45 ml of LB (Sigma‐Aldrich L3522) broth and incubated for 16 hr with continuous shaking, at 150 g in an incubator at 35°C after which a calibrated inoculating loop was used to streak on the surface of 20 HiCrome™ (Sigma‐Aldrich) Bacillus commercial agar plates without polymyxin supplement and 10 Bacillus agar with polymyxin supplement (manufacturer protocols). HiCrome Bacillus agar is used for rapid identification of Bacillus spp. from a mixed culture by a chromogenic method. Following manufacturer's directions, distinct colonies were randomly selected from the agar plates of each sample. Grams reaction, oxidase activity, and catalase were performed to presumptively ascertain the genera of the selected isolates. Then, 200 isolates were selected and maintained at −80°C in Luria–Bertani (LB) broth with 15% (v/v) glycerol, and a 15 ml LB broth or agar slant of each isolate was kept at 4°C as working culture.
2.3. In vitro screening for Fusarium‐suppressing isolates
2.3.1. Preliminary antagonistic activity
Fusarium graminearum and Fusarium culmorum were kindly provided by Dr Claire Prigent Combaret (UMR CNRS 5557) Microbial Ecology of Lyon, University Lyon 1, France, and Prof Cristina Cruz, Centre for Ecology, Evolution and Environmental Changes, Faculdade de Ciências da Universidade de Lisboa, Portugal, respectively, and they were maintained on potato dextrose agar (PDA Sigma‐Aldrich P2182) plates. Preliminary detection of the antagonistic activities of the 200 Bacillus isolates against F. graminearum was carried out by multiple confrontation dual culture tests. The protocols of Chen, Chen, Zhang, and Zhu (2014) were slightly modified. A 5‐mm‐diameter plug from an actively a growing (7‐day‐old) mycelial culture of F. graminearum was placed in the center of freshly prepared PDA plates (90 mm). From the 200 Bacillus isolates initially selected, six fresh colonies from 24 hr LB agar culture were circularly streaked (equidistance 1.5 cm) along each PDA plate at a distance of 1.5 cm from the edge of the plate using a sterile inoculating loop. Control plates consisted of F. graminearum placed on PDA alone. The plates were further incubated at 28°C for 7 days. Thereafter, only 11 isolates (BS1.1, BS2.7, BS3.5, BS4.3, BS4.6, BS5.5, BS6.2, BS6.3, BS8.5, BS8.6, and BS10.5) exhibiting strong inhibition were selected for further antifungal confirmatory tests.
2.3.2. Confirmatory in vitro antifungal test
Plates were prepared as described above; however, antagonism was carried out against two fungal pathogens (F. graminearum and F. culmorum) in three conditions. (a) Single loop full of bacterial antagonist streaked at the center of PDA 3 days before both fungal agar plugs were inoculated on opposite sides (condition 1); (b) single loop full of bacterial antagonist streaked at the center of plate while simultaneously inoculating both fungal agar plugs on opposite sides (condition 2); and (c) single loop full of bacterial antagonist streaked 3 days after the fungal agar plugs were inoculated on opposite sides (condition 3). The Bacillus isolate with strong inhibition zones (around their streaks) against the two pathogens in the screening plates was selected for further characterization. The antagonistic effect was determined by measuring the zones of inhibition (mm). The percentage of growth inhibition was calculated using the formula.
where PGI is the percentage of growth inhibition, C1 is the control mycelia area of uninhibited fungi, and C2 is the distance between the bacterial colony and the growing edge of the fungal mycelia. Experiments were repeated three times, and the values were recorded as the means of three replicates.
2.4. Detection of biosurfactant ability
2.4.1. Hemolysis blood agar test
Isolates, BS1.1, BS4.6, BS5.5, BS6.3, BS8.5, BS8.6, and BS10.5, were subjected first to hemolysis test, as isolates with biosurfactant‐producing capability can lyse erythrocytes. A colony loopful of fresh cultures of each isolate or 20 μl of each fresh culture in LB broth was taken and streaked on blood agar plates (HiMedia, India). Plates were incubated from 48 to 72 hr at 37°C (Chakraborty, Chakrabarti, & Das, 2014). The plates were observed for clear zones around the colonies. However, a clearing zone around the bacterial colony on blood agar is not always confirmatory for biosurfactant production (Hazra et al., 2011; Youssef et al., 2004).
2.4.2. Drop collapse test and microplate assay
To determine the production of biosurfactant compounds, a modified “drop collapse test,” applied according to Yanes, Fuente, Altier, and Arias (2012), was conducted. Briefly, each well of a 96‐well plate lid coated with 2 μl of commercial test substances consisting of vegetable oil, motor engine oil, kerosene, hexadecane, and paraffin oil was equilibrated for 2 hr. A cell‐free supernatant from an overnight LB broth culture of each isolate was prepared by filtering through 0.22 μm nitrocellulose membranes (Millipore Corporation, Bedford, MA, USA), and a 5 μl drop of the cell‐free supernatant of each isolate was then placed in the center of the coated well. The result was determined visually after 1 min. If the drop remained beaded, the result was scored as negative, and if the drop collapsed, the result was scored as positive. Water and SDS were used as negative controls of the media. Each treatment was repeated three times. Drop collapse test and microplate assay were also carried out according to the method described by Ben Belgacem et al. (2015). For the microplate assay, 100 μl supernatant was pipetted into a well of 96 microplate and the plate was viewed using a backing sheet of white paper with black grid. A positive result is indicated by the surfactant causing some wetting at the edge of the well and the fluid taking the shape of a diverging lens. The negative result is based on the premise that pure water in a hydrophobic well shows a flat surface, or an optical distortion is observed when surfactants are added to an aqueous solution.
2.5. Extraction of genomic DNA
We further chose seven isolates (BS1.1, BS4.6, BS5.5, BS6.3, BS8.5, BS8.6, and BS10.5) out of the 11 Bacillus isolates, based on their strong antifungal activity, for molecular analysis. Genomic DNA was extracted from overnight culture of the selected isolates using the Zymo Research ZR Soil Microbe DNA Miniprep genomic isolation kit (Epigenetics) following manufacturer's procedure. DNA quantity and quality were assessed with spectroscopic methods using a NanoDrop 1000 (Thermo Scientific, Wilmington, DE, USA), and the DNA was used as the template for polymerase chain reaction (PCR) analysis.
2.6. Detection of lipopeptide genes and molecular characterization of Bacillus isolates
Identification of the selected Bacillus isolates was by 16S rDNA gene sequencing (Garbeva, Veen, & Elsas, 2003), and the presence of lipopeptide genes in the DNA extracts of the Bacillus isolates was determined with a 25 μl reaction mixture containing 1.5–2.5 μg of template DNA; 1 μl of primer, 12.5 μl OneTaq Quick‐Load 2× master mix with standard buffer (New England Biolabs NEB), and 9.5–10.5 μl nuclease‐free water in PCR thermocycler. All the primers utilized in PCR amplification protocols were synthesized by Whitehead Scientific, Integrated DNA Technologies (Supporting Information Table S3). The PCR amplicons were analyzed by electrophoresis in 1% (w/v) agarose gel, and the sizes of the bands were determined using 1‐kb molecular marker. The gel containing 10 μg/ml ethidium bromide (Bio‐Rad) was visualized using a gel documentation system (Gel Doc 2000, Bio‐Rad) to confirm the expected size of the PCR products. NucleoSpin Microbial DNA Purification Kit (Macherey‐Nagel) was used to purify the PCR products which were then sent to Inqaba Biotec (Pretoria, South Africa) for sequencing. 16S rDNA sequences were blast searched on the NCBI GenBank and ENA database (default settings). Aligned sequences were analyzed using MEGA 7.0 software (Tamura et al., 2011), and phylogenetic trees were reconstructed based on the 16S rDNA gene using the neighbor‐joining methods (Saitou & Nei, 1987). Topological robustness was evaluated by bootstrap analysis (Felsenstein, 1985) based on 1,000 replicates.
2.7. Extraction, collection of cell‐free supernatant, and purification of secondary metabolites
From the seven antagonistic subsets, isolate BS10.5 (NWUMFkBS10.5) was the most effective with the highest number of encoding genes, and it was chosen for further analysis. The production and purification of BS10.5 active metabolites were done according to Gond, Bergen, Torres, and White (2015) with slight modification. Cell‐free antimicrobial substances from BS10.5 were collected after the rhizobacteria were grown in 1 L LB broth at 30°C with continuous shaking at 200 g for 72 hr. The cells were harvested by centrifugation at 13,000 g for 15 min, and the culture supernatant was filter sterilized through 0.22 µm nitrocellulose membranes (Millipore Corporation) filters to obtain cell‐free supernatants. About 100 ml of cell‐free supernatant was stored for anti‐pathogen test. Isolate BS10.5 was further grown in 1 L LB broth at 30°C with constant shaking at 200 g for 4 days. After fermentation, the cell filtrate was collected by centrifugation at 6,000 g for 15 min at 4°C, and the supernatant was acid‐precipitated by adjusting to pH 2.0 with 6 M HCl. After an overnight incubation at 4°C, the precipitate was centrifuged at 8,000 g at 4°C for 15 min and the pellet was dissolved in methanol–water (50:50) and then filtered through 0.22 µm PTFE membrane filter to remove larger particles and cell components. The mixture was then concentrated by vacuum evaporator at 45°C and then finally lyophilized.
2.8. Effect of culture‐free supernatant and lyophilized extracts of isolate BS10.5 on bacterial pathogens and Fusarium pathogens
2.8.1. Antibacterial activity
The activity of the cell‐free supernatants was determined by disk diffusion assay. Sterile filter paper disks were impregnated with 60 µl of the cell‐free supernatant of BS10.5, and the disks were placed on a MHA plate previously inoculated with bacterial pathogens (BC = Bacillus cereus ATCC 10876, EF = Enterococcus faecalis ATCC 29212, KP = Klebsiella pneumoniae ATCC 25923, and PA = Pseudomonas aeruginosa ATCC 27853). Control plates included antibiotic disks of norfloxacin (5 μg/disk) and tetracycline (30 μg/disk). The plates were incubated overnight at 25°C and 28°C. Antibacterial activity was observed as inhibition zones around the disk, and the experiment was conducted twice in triplicates.
2.8.2. Antifungal activity
Sterile filter paper disks impregnated with 60 µl of the cell‐free supernatant of BS10.5 were placed at the far edge of a PDA plate, and 5‐mm agar plugs of the Fusarium pathogens (F. graminearum and F. culmorum) were transferred to the opposite edge of the PDA plates. Nystatin (30 μg/disk) was used as control, and after 7 days of incubation and observation of plates at 25°C, zones of inhibition were recorded.
2.9. Dose‐dependent anti‐pathogenic activity of the BS10.5 lyophilized extract
Disk diffusion was employed for the antimicrobial assay following the method described by Chen, Wang, Wang, Hu, and Wang (2010) with modifications. The lyophilized extract was dissolved in phosphate‐buffered saline (PBS) (pH 7.5) to a concentration of 10 mg/ml and serially diluted to varying concentrations of 100, 90, 70, 50, and 30 µg/ml. Sterile disks made from punctured filter papers were then impregnated with 40 µl of the solutions and allowed to dry. Impregnated disks were then placed on the periphery of freshly prepared PDA plate containing a 5‐mm disk of the fungal pathogens in the center or a MHA plate in which 100 µl of overnight cultures of the bacterial pathogens at OD 0.5:600 nm had been spread. Forty microliters of PBS (pH 7.5) was used as a control disk. Antimicrobial activity was observed by the inhibition of microbial growth around the disk. The plates were incubated for 5 days at 28°C for fungal pathogens and 2 days at 28°C for bacterial pathogens (KP, PA, MC, and EF). Each test consisted of three replicates. Recordings were taken if zones of inhibition were spotted from the border of the disk to the perimeter of visible pathogen.
2.10. Effect of the BS10.5 lyophilized extract and commercial fungicides on fungal mycelia growth
Using well diffusion, the activity of the BS10.5 extract powder was also compared to the activity of commercial fungicides using PBS as diluent following a modified protocol of Mousa, Shearer, Limay‐Rios, Zhou, and Raizada (2015). 0.2 g/ml of the lyophilized extract, 10 µg/ml of amphotericin B, and 10 µg/ml of nystatin and triazole (10 µg/ml) were prepared with PBS (pH 7.5). Each mixture was then loaded into each of the wells (made previously with bottom parts of 200‐µl sterile pipette tips) of PDA plates previously inoculated with a 5‐mm agar plug of fungal pathogens. The plates were incubated for 5 days at 28°C, and each plate consisted of three replicates. Recordings were taken if zones of inhibition were spotted from the border of the well to the perimeter of visible pathogen.
2.11. Identification and characterization of BS10.5 bioactive compounds by NMR, FTIR, and ESI‐Q‐TOF MS analysis
2.11.1. Fourier transform infrared spectroscopy
Fourier transform infrared (FTIR) spectroscopy identifies the types of chemical bonds and functional groups in compounds, and it is useful in elucidating the components of an unknown sample. Ten microgram of the lyophilized extract of BS10.5 was analyzed with transform alpha (FTIR) KBr‐integrated spectrometer (Bruker). Spectrum reading was at 400–4,000 wavenumbers (cm−1) with an average of 32 scans. Spectrum was viewed and collated with the OPUS spectroscopy software.
2.11.2. Nuclear magnetic resonance spectroscopy (NMR)
Twenty microgram of the lyophilized extract of BS10.5 was dissolved in 0.5 ml of deuterated DMSO (dimethyl sulfoxide), and 1H and 13C NMR spectrum were acquired on the specific signal assignment. NMR spectra were recorded using a Bruker Avance III 500 MHz spectrometer at room temperature with chemical shifts () recorded against the internal standard tetramethylsilane (TMS).
2.11.3. Mass spectrometry analysis by ESI‐Q‐TOF MS
A high‐resolution mass spectrum was obtained for the lyophilized extract of BS10.5 with an Applied Biosystems 4800 Plus MICRO‐TOF/TOF analyzer (AB Sciex, USA) operated in the positive ion mode with an accelerating voltage of 20 kV, 337 nm nitrogen laser for ionization and α‐cyano‐4‐hydroxycinnamic acid for matrix. Bruker compass data analysis was used to process the mass spectrometry data while molecular weights and formulae were characterized by mass spectrum smart formula tools. The FTIR, NMR, and mass spectrometry analyses were carried out at the Chemical Resource Beneficiation/Laboratory Analytical Services of the North‐West University (Potchefstroom, South Africa).
2.12. Genome sequence of isolate NWUMFkBS10.5
The whole genome of the Bacillus strain NWUMFkBS10.5, (the preferred isolate), was extracted using the Zymo Research ZR Soil Microbe DNA Miniprep Genomic Isolation Kit and sequenced with Illumina MiSeq Reagent Kit v2 microsystem by MR DNA (Shallowater, TX, USA) using their protocols. The Kbase (Arkin et al., 2016) platform was used to check quality of reads (FastQC v.1.0.1), trim reads (Trimmomatic v0.32), and close gaps and remove adaptor sequences (Cutadapt v1.0.1). The reads were also assembled into contigs on the Kbase platform using ARAST v0.0.4 (Velvet and Kb_SPAdes v0.0.9). Contigs from the Kbase were uploaded on the RAST Server version (v) 2.0 (Aziz et al., 2008), PATRICK v3.3.15 (Wattam et al., 2017), and NCBI Prokaryotic Genome Automatic Annotation Pipeline (PGAAP v4.2) (Pruitt, Tatusova, Brown, & Maglott, 2011) for automated annotation. Default parameters were used for the bioinformatics analysis.
2.13. Data mining and in silico analysis of NWUMFkBS10.5 genome
Anti‐smash (4.0.0rc1) and PRISM were used to predict the biosynthetic products and genetic clusters present in BS10.5. A Pan‐genome was created on the Kbase platform to run a comparison of the total genes present in BS10.5 and other established biocontrol Bacillus strains.
2.14. Statistical analysis
A multivariate general linear model was used to analyze treatment means and inhibition rates. Least significant difference test (LSD), Duncan multiple test, and Student–Newman–Keuls (SNK) test were used to compare observed means, pathogen–antagonist relationship, treatment effects, and effect of conditions of inoculation using SPSS statistical software (version 22) at the significance level of 5%.
3. RESULTS
3.1. Presumptive selection and identification of bacterial isolates
All the 200 isolates selected from the HiChrome Bacillus agar according to manufacturer's descriptions (Supporting Information Table S1) were Gram‐positive, oxidase‐positive, and catalase‐positive. Supporting Information Table S2 further describes the location and total number of isolates selected from each site. A large percentage of the isolates showed cultural characteristics that resembled B. subtilis strains because of their light green to green colonies on the agar. Isolate BS10.5 showed a unique cultural characteristic that was not included in manufacturer's identification profile (Figure 1), so, as such, it was not assigned to any of the Bacillus specie. Isolates BS8.5 and BS8.6 showed characteristics resembling strains of B. thuringensis. Isolates BS4.6 and BS4.3 had close resemblance with B. cereus while BS6.3 shared a close resemblance with B. pumilis. Characteristics that resembled B. coagulans were seen in isolates BS1.7, BS2.7, and BS3.5.
Figure 1.
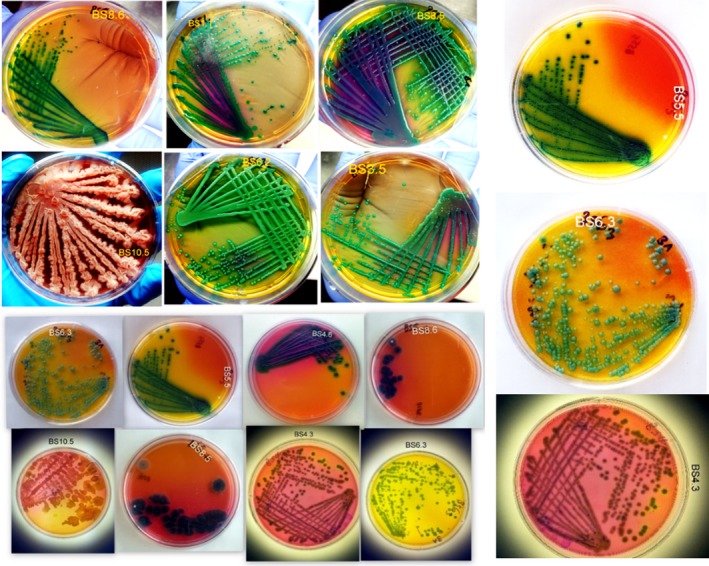
Colonial characteristics and presumptive identification of the isolated Bacillus strains on HiChrome Bacillus agar
3.2. Bacillus inhibition of Fusarium mycelia
Antagonistic activity of the Bacillus isolates against the two Fusarium pathogens showed that lower inhibition rates were seen in condition 1 (antagonist inoculated on PDA 3 days before pathogens) and condition 2 compared to condition 3, where the fungal mycelia plug was inoculated on the PDA 3 days before the antagonists were inoculated (Tables 1 and 2). Overall, F. culmorum was more resistant to the antagonists during condition 1 and condition 2 when compared with susceptibility of F. graminearum during those conditions of treatment. However, the inhibition of F. graminearum was less during condition 3.
Table 1.
Percentage inhibition of Fusarium graminearum mycelia by Bacillus isolates
| Treatment | Condition (% inhibition zone) | Mean | ||
|---|---|---|---|---|
| 1 | 2 | 3 | ||
| BS1.1 | 42.67 | 46.67 | 66.67 | 52.00b |
| BS2.7 | 42.67 | 41.33 | 56.00 | 46.67cd |
| BS3.5 | 45.33 | 41.33 | 68.00 | 51.56b |
| BS4.3 | 40.00 | 38.67 | 44.00 | 40.89f |
| BS4.6 | 42.67 | 38.67 | 44.00 | 41.78ef |
| BS5.5 | 36.00 | 30.67 | 58.67 | 41.78ef |
| BS6.2 | 32.00 | 26.67 | 69.33 | 42.67ef |
| BS6.3 | 36.00 | 33.33 | 65.33 | 44.89de |
| BS8.5 | 45.33 | 46.67 | 54.67 | 48.89bc |
| BS8.6 | 40.00 | 37.33 | 70.67 | 49.33bc |
| BS10.5 | 54.67 | 42.67 | 77.33 | 58.22a |
| Mean | 41.58b | 38.55c | 61.33a | |
| ANOVA | ||||
| Treatment (T) | *** | |||
| Condition (C) | *** | |||
| T × C | *** | |||
Values are means and standard error of four replicates of in vitro antagonistic activity of selected Bacillus isolates against F. graminearum; values having the same letters are not significantly different according to Duncan's least significant difference test at p ≤ 0.05. 1, 2, and 3 represent conditions of inoculation. *** = treatment, conditions and treatment and conditions are significantly different.
Table 2.
Percentage inhibition of Fusarium culmorum mycelia by Bacillus isolates
| Treatment | Condition (% inhibition zone) | Mean | ||
|---|---|---|---|---|
| 1 | 2 | 3 | ||
| BS1.1 | 37.33 | 34.67 | 60.00 | 44.00e |
| BS2.7 | 44.00 | 41.33 | 62.67 | 49.33d |
| BS3.5 | 45.33 | 46.67 | 57.33 | 49.78cd |
| BS4.3 | 45.33 | 50.67 | 70.67 | 55.56ab |
| BS4.6 | 53.33 | 45.33 | 53.33 | 50.67cd |
| BS5.5 | 45.33 | 44.00 | 65.33 | 51.56cd |
| BS6.2 | 37.33 | 36.00 | 46.67 | 40.00f |
| BS6.3 | 56.00 | 50.67 | 69.33 | 58.67a |
| BS8.5 | 52.00 | 54.67 | 62.67 | 56.44a |
| BS8.6 | 50.67 | 57.33 | 50.67 | 52.89bc |
| BS10.5 | 45.33 | 38.67 | 70.67 | 51.56c |
| Mean | 46.55b | 45.45b | 60.85a | |
| ANOVA | ||||
| Treatment (T) | *** | |||
| Condition (C) | *** | |||
| T × C | *** | |||
Values are means and standard error of four replicates of in vitro antagonistic activity of selected Bacillus isolates against F. culmorum; values same having the same letters are not significantly different according to Duncan's least significant difference test at p ≤ 0.05. 1, 2, and 3 represent conditions of inoculation. *** = treatment, conditions and treatment and conditions are significant different.
3.3. Detection of biosurfactant production
The multiple test on the seven isolates for detecting biosurfactant production revealed that four of the isolates (BS1.1, BS3.5, BS8.6, and BS10.5) are biosurfactant producers (Table 3). The detection of biosurfactant production in BS1.1, BS3.5, BS8.6, and BS10.5 further correlates the PCR detection of surfactin genes in these isolates, which is not the case with isolates BS5.5, BS6.3, and BS8.5. The stability of the biosurfactants produced under different growth conditions, however, might need to be determined because the production of biosurfactants is dependent on conducive pH, temperature, and salinity concentration (Rivardo, Turner, Allegrone, Ceri, & Martinotti, 2009).
Table 3.
Test for biosurfactant properties of potential isolates
| Isolates | Test substance | ||||||
|---|---|---|---|---|---|---|---|
| Drop collapse | |||||||
| Blood agara | Microplate assay | Vegetable oilb | Motor engine oilb | Keroseneb | Hexadecaneb | Parafilmb | |
| BS1.1 | ● | ●● | ● | ● | ● | ● | ● |
| BS3.5 | ● | ○ | ● | ● | ● | ○ | ● |
| BS5.5 | ● | ○ | ○ | ○ | ● | ○ | ○ |
| BS6.3 | ● | ○ | ○ | ○ | ○ | ○ | ○ |
| BS8.5 | ● | ○ | ○ | ○ | ○ | ○ | ○ |
| BS8.6 | ● | ●● | ○ | ● | ● | ● | ● |
| BS10.5 | ● | ●● | ● | ● | ● | ● | ● |
| Water | ○ | ○ | ○ | ○ | ○ | ○ | ○ |
| SDS | ● | ●● | ● | ● | ● | ● | ● |
a = hemolysis test: b‐hemolysis (●), no hemolysis (○); b = Drop collapse assay ●: collapse; ○: no collapse; hemolysis test ●: positive; ○: negative; drop collapse test: negative (○), spreading (●), comparable with sds (●); microplate assay: optical distortion comparable with water (○), optical distortion comparable with sds (●●).
3.4. Molecular characterization of Bacillus isolates
The PCR analysis carried out against the seven isolates revealed that each isolates harbored at least one lipopeptide gene. PCR amplification was carried out according to previously established protocols. The targeted genes and primers utilized during the PCR amplification are shown in Supporting Information Table S3. Multiple lipopeptide antibiotic genes were detected in BS10.5 amplicon (Table 4), and all the isolates showed a positive result for the fengycin (Af2‐F) primer. Clusters for surfactin genes were detected in all the isolates using the SrfC primer. However, only three isolates harbored iturin genes, and no amplification was detected using the fengycin (FenD), Ipdc (indole pyruvate decarboxylase), and Acc deaminase primers. Single predominant amplicon bands were produced from the PCR products from the gel electrophoresis (Supporting Information Figure S1). The presence of the detected lipopeptide antibiotics could be responsible for the antagonistic activities of the Bacillus isolates against fungal pathogens.
Table 4.
Genes detected in the antagonistic Bacillus isolates using specific primers sets
| Primer set | Isolates | ||||||
|---|---|---|---|---|---|---|---|
| BS10.5 | BS8.6 | BS8.5 | BS6.3 | BS5.5 | BS4.6 | BS1.1 | |
| Iturin A (ItuD) | ● | ○ | ○ | ● | ○ | ● | ○ |
| 16S (BacF/R1378) | ● | ● | ● | ● | ● | ● | ● |
| Surfactins (As1‐F) | ● | ● | ● | ○ | ● | ○ | ● |
| Fengycins (Af2‐F) | ● | ● | ● | ● | ● | ● | ● |
| Surfactin (SrfC) | ● | ● | ● | ● | ● | ● | ● |
| Surfactin (sfp) | ● | ○ | ○ | ○ | ○ | ○ | ○ |
| Fengycin (FenD) | ○ | ○ | ○ | ○ | ○ | ○ | ○ |
| Bacillomycin D (BamC) | ● | ○ | ○ | ○ | ○ | ○ | ○ |
| Ipdc (Indole Pyruvate decarboxylase) | ○ | ○ | ○ | ○ | ○ | ○ | ○ |
| Acc Deaminase | ○ | ○ | ○ | ○ | ○ | ○ | ○ |
●: (Positive) a PCR amplicon of expected size was seen; ○: negative.
3.5. Phylogenetic analysis
Based on the blast search of the partial 16S rDNA gene sequences and submission to NCBI, GenBank accession numbers were assigned to the seven isolates (Supporting Information Table S4). The blast search correlated with the presumptive identification from the chromogenic agar. Using the aligned MAFFT 16S rDNA sequences, the phylogenetic tree was constructed with MEGA 7.0 software. In addition, the bootstrap support of the genetic relatedness among six of the isolates (BS1.1, BS4.6, BS5.5, BS6.3, BS8.5, BS8.6, and BS10.5) and their nearest neighbors was 100% except isolate BS5.5 that showed 71% close match. In addition, isolates BS1.1, BS5.5, and BS6.3 had no direct clustering with any strain on the tree (Supporting Information Figure S2).
3.6. Antimicrobial activity of cell‐free supernatants
The results of the antimicrobial activity of the cell‐free supernatants showed that the cell‐free culture filtrate from BS10.5 contains strong bioactive substances with inhibitory potentials against fungal and bacterial pathogens (Supporting Information Table S5). The cell‐free filtrates inhibited F. graminearum and B. cereus at the same rate, however, but inhibited K. pneumoniae and P. aeruginosa moderately. In comparison with antibiotics used against the fungal pathogens, the cell‐free supernatant exhibited an inhibition level close to that of nystatin. However, it showed higher inhibitory potential than tetracycline and ciprofloxacin against the bacterial pathogens based on the concentration of the antibiotics used.
3.7. Antipathogenic activity of the BS10.5 lyophilized extract at different concentrations
During the lyophilized extract antipathogenic test, the effect of the extracts decreased relative to an increase in dilution with PBS. Among the bacterial pathogens, EF was the most susceptible to the lipopeptides extracts of BS10.5, while MC was the least susceptible. F. graminearum was more sensitive to the extract concentrations than F. culmorum (Figure 2). At 30 µl, three of the pathogens (PA, MC, and KP) showed no sensitivity.
Figure 2.
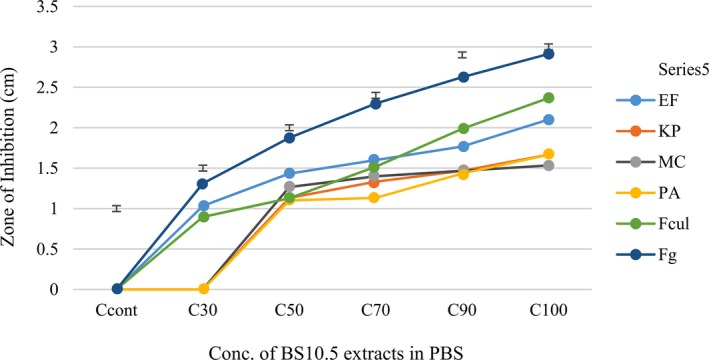
Means of three replicates showing activity of BS10.5 extracts on the microbial pathogens at different concentrations. Values are significantly different according to Duncan's least significant difference test at p ≤ 0.05, and means are significantly different from the control
3.8. Comparative antifungal activity of the BS10.5 lyophilized extract and commercial fungicides
The BS10.5 extract showed a higher inhibition rate against F. graminearum than the commercial fungicides (nystatin and amphotericin) (Figure 3).
Figure 3.
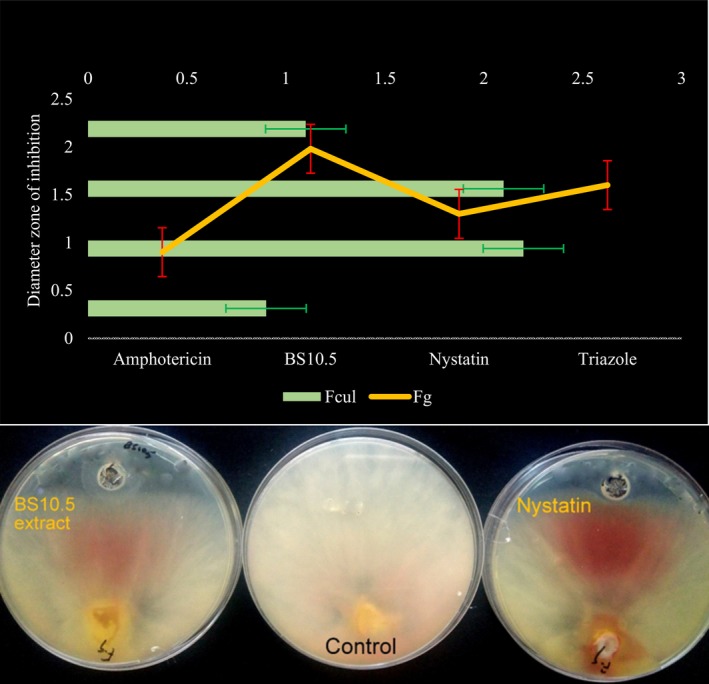
Upper plate: inhibitory effect of the BS10.5 extract (20 mg/ml) and fungicide controls (triazole, amphotericin B, and nystatin) (at concentrations 10 µg/ml, respectively), on the Fusarium graminearum and Fusarium culmorum growth in vitro; Lower plate: inhibitory effect of the BS10.5 extract (20 mg/ml) and nystatin (10 µg/ml), on the F. graminearum. For the experiment, n = 4. Activity of the lyophilized extract and commercial fungicide on fungal growth were significantly different at p ≤ 0.05
3.9. FTIR, NMR, and ESI‐Q‐TOF MS analysis of the bioactive compounds present in extracts of BS10.5
3.9.1. FTIR analysis
The FTIR absorbance showed functional groups corresponding to OH at 3,600–3,500 cm−1, CH stretch at 3,000–2,500 cm−1, NH stretch at 2,500–2,000 cm−1, COO‐ at 1,900–1,500 cm−1, and CC and CN at 1,500–1,000 cm−1 (Supporting Information Figure S3). These wave numbers show characteristics similar to lipopeptides. The stretching and vibration mode of the absorbance is indicative of aliphatic chains, alkyl chains, peptide bonds, and two amide bonds, signifying the presence of a compound with ester and amino groups. This result, which is consistent with the report of Romero et al. (2007), Nam et al. (2015), Rivardo et al. (2009), is indicative of fengycin, surfactin, and iturin moieties.
3.9.2. NMR analysis
The proton analysis of the compound showed NH and OH proton at >8.00 ppm, some –CH3 and CH2 signals at <2.00 ppm and CH2–COO>2.00 ppm but <4.00 ppm (Figure 4). These are suggestive of a peptide backbone of secondary amide, aliphatic chains, and ester linkages, respectively. Chakraborty et al., (2014), gave a similar report from the proton spectra of the lipopeptide extract of B. vallismortis JB201 and B. subtilis SJ301. The signals of the carbon analysis were inconclusive despite several runs (data not shown).
Figure 4.
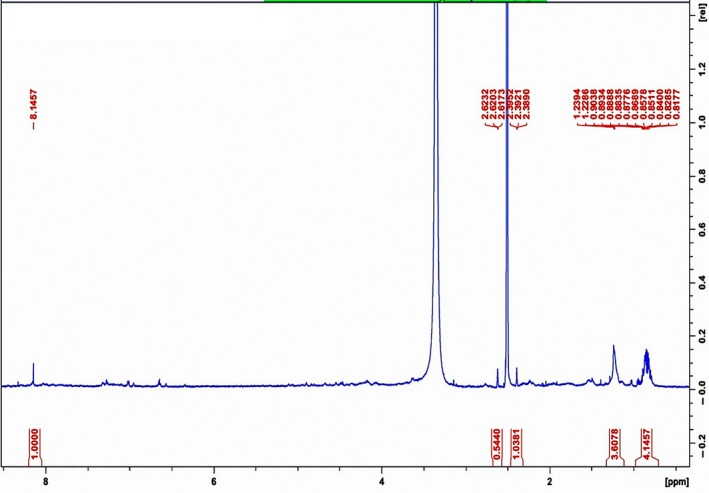
NMR spectrum of BS10.5
3.9.3. ESI‐Q‐TOF MS characterization
The ESI‐Q‐TOF MS characterization showed that the BS10.5 lipopeptide extracts contained antifungal iturin, surfactin, and fengycin families (Figure 5). Isolate BS10.5 had six mass ranges starting from m/z 186.1026, 365.1063, 750.4056, 1,058.6738, 1,477.8184, and 2,095.3363. Mass 1,058.6738 suggests the possible presence of iturin, surfactin, and bacillomycin fragments, while 1,477.8184 represents the mass fragment of fengycin, respectively (Ben Ayed et al., 2014; Jasim et al., 2016; Nam et al., 2015). This conclusion is based on multiple comparison of literature and fragment matches.
Figure 5.
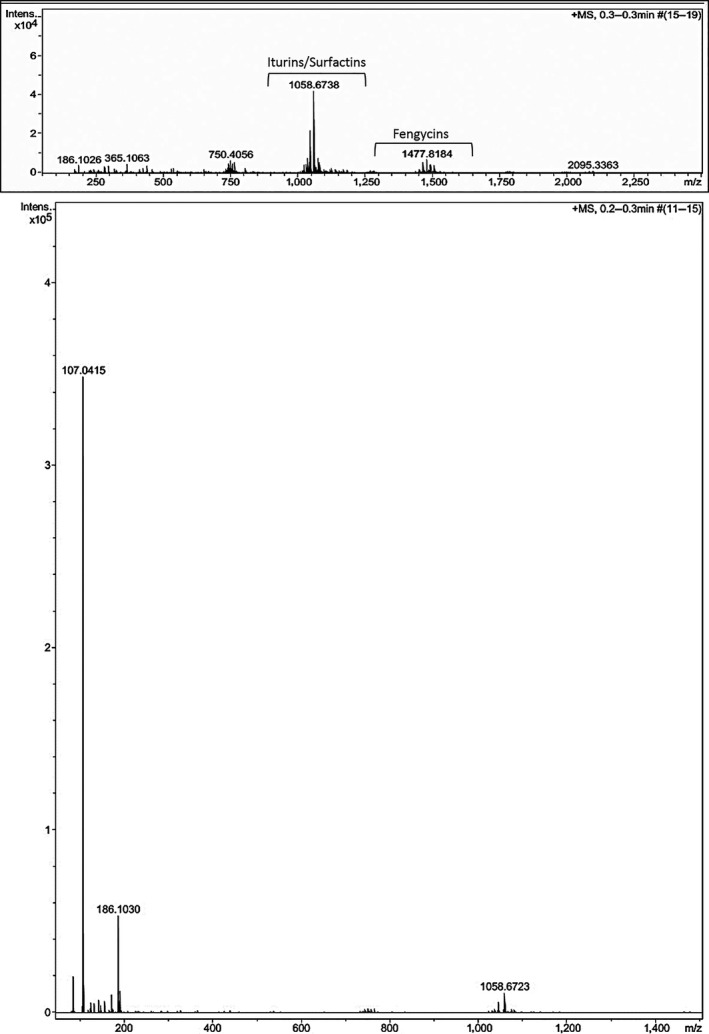
Positive ESI‐Q‐TOF MS spectrum of lipopeptides extract of BS10.5 strain. Clusters of iturin, surfactin, and bacillomycin (m/z 1,058.6738/1,058.6723, 1,058.6740), fengycin (m/z 1,477.8184), and an unidentified (m/z 2,095.3363) molecular ion species are labeled
3.10. Genome sequence data
Isolate BS10.5 was identified as B. velezensis strain NWUMFkBS10.5. The sequencing yielded 7,505,117 clusters and 15,010,234 paired‐end reads with an average read length of 151 bp. After compilation of the data, the genome yielded between 3,905,592 and 3,964,473 bp, and G+C content of 46.4%, showing similarity to known plant growth‐promoting Bacillus spp. (Table 5 and Supporting Information Figure S4). Subsystem information of the isolate was predicted by SEED viewer (Overbeek et al., 2014) (Figure 6). Overview of the secondary metabolite biosynthesis gene clusters with antiSMASH v4.0.0rc1 (Weber et al., 2015) revealed that BS10.5 had 16 gene clusters involved in secondary metabolic activities. The clusters were devoted to the synthesis of antimicrobial peptides such as surfactin, mersacidin, fengycin, and oocydin (Table 6). BS10.5 was labeled non‐pathogenic by Pathogenfinder v1.1 web server (Cosentino, Voldby Larsen, Møller Aarestrup, & Lund, 2013).
Table 5.
Comparison of the Bacillus velezensis NWUMFkBS10.5 genome with other B. velezensis strains
| Attributes | B. velezensis NWUMFkBS10.5 | B. velezensis LM2303 | B. velezensis 157 | B. velezensis LABIM40 | B. velezensis FZB42 | B. velezensis AS43.3 | B. velezensis UCMB5113 | B. velezensis G341 | B. velezensis LS69 | B. velezensis 9912D |
|---|---|---|---|---|---|---|---|---|---|---|
| Size of genome (bp) | 3,964,473 | 3,989,393 | 4,013,317 | 3,972,310 | 3,918,589 | 3,961,291 | 3,889,532 | 4,009,746 | 3,917,761 | 4,206,167 |
| G+C numbers (%) | 46.39 | 46.68 | 40.32 | 46.5% | 46.49 | 46.60 | 46.71 | 46.49 | 46.40 | 46.03 |
| Number of coding sequences | 3,875 | 3,531 | 3,369 | 3,777 | 3,693 | 3,861 | 3,656 | 3,953 | 3,643 | 4,436 |
| Total genes | 3,916 | 3,866 | 3,789 | n.d | 3,421 | 4,037 | n.d | 4,114 | n.d | n.d |
| tRNA | 89 | 86 | 87 | 75 | 89 | 89 | 89 | 95 | 72 | 86 |
| rRNA | 13 | 27 | 27 | 7 | 9 | 29 | 10 | 30 | 7 | 27 |
| Number of RNAs | 93 | n.d | n.d | n.d | 117 | 118 | 182 | 161 | n.d | n.d |
n.d: not documented
Figure 6.
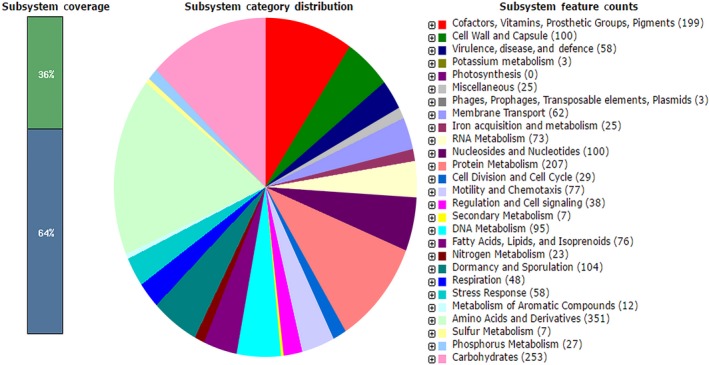
Subsystem summary of the genome Bacillus velezensis NWUMFkBS10.5 predicted by SEED Viewer v2.0. Genomic features are colored according to their functional classification types (Overbeek et al., 2014)
Table 6.
Description and location of BGC in NWUMFkBS10.5 identified in silico
| Cluster Identity (ID) | Type | Position/Region | Numbers of BGC | Most Similar BGC predicted | Percentage similarity |
|---|---|---|---|---|---|
| Cluster 1 (218.374_ID_10360) | Terpene | 42,380–64,263 | 1 | Undefined | — |
| Cluster 2 (237.089_ID_10346) | Otherks | 65,373–106,617 | 1 | Butirosin | 7% of genes similar |
| Cluster 3 (237.089_ID_10346) | Terpene | 189,395–210,135 | 1 | Undefined | — |
| Cluster 4 (237.089_ID_10346) | Transatpks | 493,558–579,439 | 5 | Macrolactin | 100% of genes similar |
| Cluster 5 (237.089_ID_10346) | Transatpks—Nrps | 808,101–910,775 | 10 | Bacillaene | 100% of genes similar |
| Cluster 6 (237.089_ID_10346) | Transatpks—Nrps | 973,335 –1,061,819 | 10 | Fengycin | 86% of genes similar |
| Cluster 7 (228.907_ID_10356) | Transatpks | 1–45,825 | 10 | Difficidin | 53% of genes similar |
| Cluster 8 (228.907_ID_10356) | T3pks | 161,376–202,527 | 1 | Undefined | — |
| Cluster 9 (242.696_ID_1037) | Nrps | 1–10,331 | 1 | Undefined | — |
| Cluster 10 (242.696_ID_1037) | Nrps | 1–14,180 | 2 | Fengycin | 13% of genes similar |
| Cluster 11 (232.565_ID_10370) | Transatpks | 1–23,720 | 7 | Difficidin | 26% of genes similar |
| Cluster 12 (246.163_ID_10350) | Nrps | 516,770–569,433 | 1 | undefined | — |
| Cluster13 (263.57_ID_10354) | Nrps | 1–25,748 | 3 | Surfactin | 47% of genes similar |
| Cluster 14 (265.136_ID_10348) | Bacteriocin—Nrps | 1,981–68,772 | 10 | Bacillibactin | 100% of genes similar |
| Cluster 15 (265.136_ID_10348) | Other | 574,581–615,999 | 6 | Bacilysin | 100% of genes similar |
| Cluster 16 (265.136_ID_10348) | Lantipeptide | 766,502–789,690 | 1 | Mersacidin | 90% of genes similar |
3.11. Insights from exploration and in silico mining of B. velezensis NWUMFkBS10.5 genome
The in silico analyses revealed that the NWUMFkBS10.5 genome has 16 clusters which harbored over 76 homologous biosynthetic gene clusters (BGC). The genome has four nonribosomal peptide synthetases (NRPSs), two terpenes, three trans‐AT PKS (transatpks), a single lantipeptide, a type 3 polyketide synthetase (T3pks), two trans‐AT PKS nonribosomal peptide synthetases, a single bacteriocin nonribosomal peptide synthetase cluster and two unidentified ketide synthetases (Table 6 and Supporting Information Figure S5a–k). In Supporting Information Figure S5a–k, the colors symbolize different functional gene types: blue (transport‐related genes), green (biosynthetic genes), red (regulatory genes), and gray (additional genes). In the antismash data, the highest number of biosynthetic genes was found in clusters 5, 6, 7, and 14 (10 each), whereas the PRISM result predicted 18 clusters in the NWUMFkBS10.5 genome. In Table 7, the functions of the predicted biosynthetic compounds are given.
Table 7.
Overview of the biosynthetic compounds predicted in the NWUMFkBS10.5 genome
| Biosynthetic compound | Cluster located | Biomedical/Biocontrol function | Reference |
|---|---|---|---|
| Mycosubtilin | 6 | Antifungal, hemolytic, and limited antibacterial activity | Duitman et al. (2007), Leclère et al. (2005) |
| Fengycin and Plipastatin | 6 | Broad‐spectrum antifungal and antitumoral agent | Cochrane and Vederas (2016), Vanittanakom, Loeffler, Koch, and Jung (1986) |
| Surfactin | 13 | Antibacterial, antifungal, antiviral, antimycoplasma, antitumoral, insecticidal, anticoagulant activities, and enzyme inhibitors | Mnif and Ghribi (2015), Ongena and Jacques (2008) |
| Iturin | 6 | Antibacterial and antifungal activity | Dunlap et al. (2013), Ongena and Jacques (2008) |
| Polymyxin | 6 | Antibacterial, antifungal, and Immuno‐modulating activity | Cochrane et al. (2016) |
| Sessilin | 10 | Antifungal activity | D’Aes et al. (2011), Olorunleke, Hua, Kieu, Ma, and Höfte (2015) |
| Bananamides | 10 | Unspecified | Nguyen et al. (2016) |
| Cichopeptin | 10 | Limited information | Huang et al. (2015) |
| Viscosin | 10 | Biosurfactant | Alsohim et al. (2014), Bonnichsen et al. (2015) |
| Taiwachelin, tolaasinand, orfamide | 10 | Iron chelation, therapeutic peptide, insecticidal biosurfactant, and elicitor of induced systemic resistance | Andolfi, Cimmino, Cantore, Iacobellis, and Evidente (2008), Jang et al. (2013), Kreutzer, Kage, and Nett (2012), Ma, Ongena, and Höfte (2017) |
| Mersacidin | 16 | Antibacterial | Abriouel, Franz, Ben Omar, and Gálvez (2011) |
| Bacitracin and bacilysin | 15 | Limited use as animal growth promoter, topical antibiotic, antibacterial, and antifungal | Mousa and Raizada (2015), Phillips (1999) |
| Bacillibactin, paenibactin, griseobactin, heterobactin, mirubactin, myxochelin, vanchrobactin | 14 | Iron chelation and anticancer agent | Balado, Osorio, and Lemos (2008), Giessen et al. (2012), Patzer and Braun (2010), Sandy et al. (2010), Wen et al. (2011) |
| Amylocyclicin | 1 | Antibacterial and plant growth promoter | Scholz et al. (2014) |
| Lichenysin | 13 | Biosurfactant | Grangemard, Bonmatin, Bernillon, Das, and Peypoux (1999) |
| Basiliskamides | 13 | Antifungal | Theodore et al. (2014) |
| Difficidin | 11, 7, 4 | Broad‐spectrum antibacterial compound | Chen et al. (2009), Mousa and Raizada (2015) |
| Kalimantacin/batumin and oocydin A | 11, 7, 5 | Antibacterial and antifungal haterumalide | Matilla, Leeper, and Salmond (2015), Mattheus et al. (2010), Tokunaga et al., (1996) |
| Sorangicin | 11, 7 | Antibacterial macrolide antibiotic | Campbell et al. (2005) |
| Elansolid | 7 | Antibiotic (Bactericidal) | Steinmetz et al. (2011) |
| Myxovirescin | 7, 5 | Antibiotic (Bactericidal) | Xiao, Gerth, Müller, and Wall (2012) |
| Phormidolide, thiomarinol, and mupirocin | 5 | Antitumor agent and antibacterial metabolite/clinical antibiotic | Mousa and Raizada (2015), Murphy et al. (2014) |
| Paenilarvins, tridecaptin, and paenibacterin | 6 | Antifungal, antitumor agent, and antibiotic (Bactericidal) | Cochrane et al. (2016), Hertlein et al. (2016), Huang, Guo, and Yousef (2014), Huang and Yousef (2014) |
3.12. Whole genome nucleotide/NCBI biosynthetic gene blast and Pangenome comparison
Firstly, we carried out a genome blast search of NWUMFkBS10.5 to determine its genetic relatedness, and then, we blast searched the genome against selected lipopeptide gene clusters and constructed the phylogenetic trees (Figure 7 and Supporting Information Figure S6). Thirdly, the pangenome comparisons of four established agriculturally and industrially relevant Bacillus strains with NWUMFkBS10.5 strains confirmed the distinctive status of BS10.5 and its genetic relatedness (Figure 8).
Figure 7.
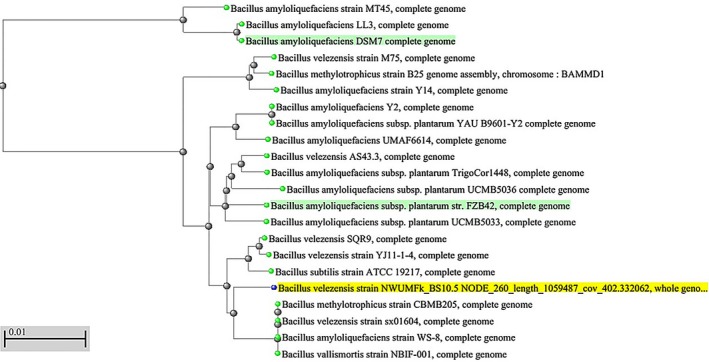
Neighbor‐joining phylogenetic tree from pangenomic sequence of closely related Bacillus velezensis strains. Bar, 0.01 substitutions per nucleotide position. Reference strains highlighted green. Strain B. velezensis NWUMFkBS10.5 is highlighted yellow
Figure 8.
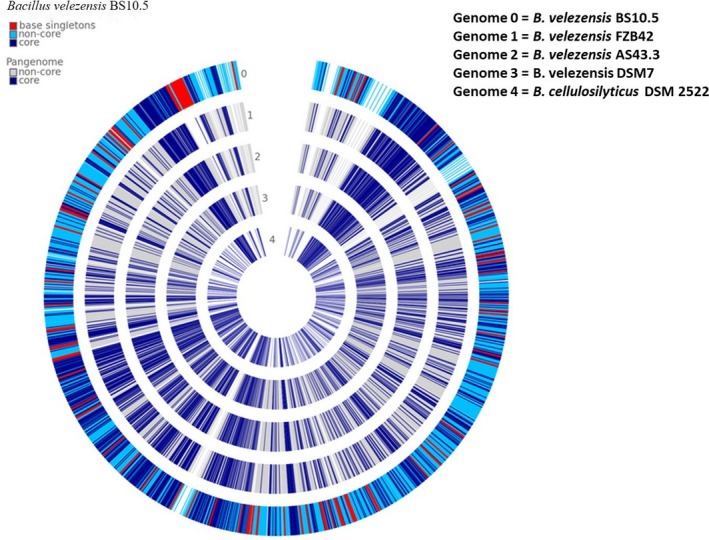
Pangenomic atlas of Bacillus velezensis NWUMFkBS10.5, other closely related B. veleznesis strains, and an out group B. cellulosilyticus DSM 2522 (genome 4). The similarities and dissimilarities in their core and non‐core genome are indicated as predicted in the left key of the figure. Peculiar genes in strain B. velezensis NWUMFkBS10.5 are indicated in its ring as light blue arcs
4. DISCUSSION
When in search of beneficial microbial strains for commercialization, key screening approaches and sequential steps have been identified that lead to selecting robust and effective candidates (Köhl, Postma, Nicot, Ruocco, & Blum, 2011). A comprehensive in vitro analysis starting from the culturing stage to molecular identification of action mechanisms and backed up by in silico genome exploration of potential candidates provides strains with better in planta viability and efficacy (Adeniji & Babalola, 2018). In this study, out of the 200 isolates initially screened for antagonism against the fusarium pathogens, 11 isolates showed consistent antagonism and among 11 isolates, isolate BS10.5 exhibited the strongest and most consistent antagonism considering the conditions. Applying biocontrol agents as a bioprotective coating prior to planting of crop seedlings has been demonstrated in previous reports (Yang et al., 2015). The antifungal result of this study demonstrates that our Bacillus isolates are candidates for preplanting and post‐harvest bioprotective inoculants. The reduction in fungal mycelia growth (Tables 1 and 2) shows they could protect plants against the onset of fusariosis. Members of the Bacillus sp. exhibit remarkable biocontrol activity against phytopathogens due to the production of the lipopeptide group of antibiotics (surfactin, fengycin, bacillomycin, and iturin) (Guo et al., 2014; Vitullo, Pietro, Romano, Lanzotti, & Lima, 2012). We detected these groups in some of our Bacillus isolates.
Production of biosurfactants has been reported for many Bacillus strains (Chen, Zhang, Fu, Li, & Wang, 2016; Plaza, Chojniak, Rudnicka, Paraszkiewicz, & Bernat, 2015). The four biosurfactant producers (BS1.1, BS3.5, BS8.6, and BS10.5) identified in the study have potential agricultural, industrial, biological, and medical applications (Table 3). Biosurfactants are biologically surface‐active agents with both lipophilic and hydrophilic moieties (Kosaric, Gray, & Cairns, 1987). They act by decreasing the surface and interfacial tension between individual molecules in contact, forming selective ionic pores in membrane bilayers. With biosurfactants having antiviral, antibacterial, hemolytic, antifungal, and anti‐insecticidal properties (Mnif & Ghribi, 2015), the antimicrobial mechanisms of these surfactant producers are further worth exploiting.
Culture‐free extracts from B. velezensis antagonists have been used in bioprotection assays through several modes of application such as seed treatments and pour plate mixtures (Cao et al., 2018). These culture‐free extracts are of beneficial importance when produced in sufficient volume during fermentation processes. The supernatant contains diverse metabolites such as indole‐acetic acid (IAA), antibiotics, lytic enzymes, iron chelating compounds, and hormones depending on the microbial growth phase and culture conditions (Ahmad, Ahmad, & Khan, 2008; Chen et al., 2014). The secretion and accumulation of surfactin occur at the logarithmic phase while the synthesis of iturin occurs during the stationary phase of growth (Leclère et al., 2005). The antimicrobial activity shown by the cell‐free supernatants of isolate BS10.5 suggests that the secondary metabolite of the isolate can also be of immense industrial benefit when purified in sufficient quantity.
Having employed a dose‐dependent assay to determine the concentrations at which the pathogens were most sensitive to BS10.5 extracts, we report that the growth of the pathogens was retarded relative to increased concentration of BS10.5 extract. At 30 µl, three of the pathogens (PA, MC, and KP) showed no sensitivity. This gives an indication of the lowest possible concentration at which the pathogens will remain susceptible. Dose‐dependent effects of lipopeptides have been reported elsewhere (Falardeau, Wise, Novitsky, & Avis, 2013). A detailed assay might however be needed to confirm the minimum inhibitory or cidal dose of the extract. Commercial fungicides containing nystatins, triazoles, and amphotericins have been used as controls to compare sensitivity of Fusarium to fungal antibiotics (Arutchelvi & Doble, 2010; Girija, Duraipandiyan, Kuppusamy, Gajendran, & Rajagopal, 2014), and Fusarium spp. are less susceptible to amphotericin than to nystatins and triazoles (Deepak & Jayapradha, 2015). Our lipopeptide extract showed excellent inhibitory activity in comparison with the fungicides we utilized in this study. Our results correlate with other reports showing comparative multi‐antipathogenic effects of crude, partially purified, or purified extracts of bacterial antagonists (Chen et al., 2010; Mousa et al., 2015). The action mechanisms of microbial lipopeptides reduce the possibility of microbes developing resistance to them, while conventional antibiotics are prone to resistance.
The FTIR, NMR, and ESI‐Q‐TOF MS results provide further evidence that the lipopeptides (surfactin, iturin, and fengycin) prominently reported in Bacillus spp. are responsible for the biocontrol activity of NWUMFkBS10.5. Reports showing non‐synergistic and synergistic activity of lipopeptides against Fusarium members are available (Koumoutsi et al., 2004; Xu et al., 2013), and Bacillus spp. with conserved genes for producing multiple lipopeptides are not rare (Jemil et al., 2017). BS10.5 harbored multiple lipopeptide antibiotic genes. The ESI‐MS was effective in detecting the presence of these compounds, but we could not speculate what compound produced the signal at m/z 2,095.3363. Furthermore, except for ericin S with 3,351.543 m/z (Palazzini et al., 2016), high signals of this range have been rarely reported for Bacillus lipopeptides. Fragment spectra of surfactin and its homologs (C13, 14, and 15) are mostly detected at 1,030.8, 1,044.8, 1,046.8, 1,058.8, 1,060.8, and 1,074.8, that of bacillomycin (C14, 15, 16, and 17) at 1,031.7, 1,045.7, 1,053.7, 1,059.7, 1,067.7, 1,069.7, 1,081.7, 1,083.7, 1,097.7, 1,095.7, 1,111.7, 1,527.8, 1,529.9, and 1,543.8, those of fengycins (C15, 16, and 17) at 1,449.9, 1,463.9, 1,471.9, 1,477.9, 1,485.9, 1,487.9, 1,491.8, 1,499.9, 1,501.9, 1,505.8, 1,513.9, and 1,515.9 (Koumoutsi et al., 2004). The majority of these lipopeptides have a large variety of isoforms which sometimes present challenges in distinguishing them (Chen et al., 2014). Our aim in this study was not to resolve each lipopeptide type into its specific isoforms, but to detect the different groups of lipopeptides present in the metabolite and confirm that the gene clusters detected are metabolically functional.
The presence of surfactin, bacillomycin, iturin D, and fengycin D biosynthetic genes, detected in the BS10.5 amplicons during PCR analysis, was confirmed from the WGS data. The BS10.5 genome analysis showed similarity identity above 85% for some biosynthetic genes clusters (BGC). Technically, BGC or biosynthetic genes are considered to be present when the similarity index is 65% and above (Van Der Voort et al., 2015). However, our report here also shows that a low percentage similarity of a BGC does not signify the absence of the predicted BGC in the genome being analyzed and this correlates with our identification of iturin gene during PCR, despite its predicted similarity identity in cluster 6 (BGC0001098) being below 65%. Coding region specific for fengycin is predicted to be present in cluster 6 at 86%, bacillomycin at 66%, and iturin at 53% similarity (Table 6). These genes were also detected by specific primers during the PCR‐gel electrophoresis and ESI‐Q‐TOF MS analysis.
The B. velezensis sp. is categorized as heterotypic synonyms of B. amyloliquefaciens subsp. plantarum FZB42T, B. methylotrophicus KACC 13015T, and B. oryzicola KACC 18228 based on DNA‐DNA hybridization values >84% (Dunlap, Kim, Kwon, & Rooney, 2016; Fan, Blom, Klenk, & Borriss, 2017). The in silico result showed that our B. velezensis NWUMFkBS10.5 strain closely shares some phenotypic and genotypic traits with several of the commercially established plant growth‐promoting strains within the Bacillus genus (Table 6, Supporting Information Figures S4 and S7a,b). We avoided relying only on the structural and functional metabolic capacity of well‐studied or representative B. velezensis strains to deduce the metabolic capacity of NWUMFkBS10.5. The approach has been reported to have limitations, because experimental data have shown that in some species, new genes are discovered after comparing the genomes of newly sequenced strains and reference strains (Medini et al., 2005).
We applied the pangenomic analysis to locate genes that were responsible for metabolic activity and genes that were dispensable to the survival of NWUMFkBS10.5. The analysis showed that the strain shared core resemblance with the other analyzed Bacillus isolates. Additionally, NWUMFkBS10.5 genome had five undefined BGC clusters (1, 3, 8, 9, and 12) (Table 6) and did not contain amylolysin unlike other B. velezensis strains (Liu et al., 2017; Pan et al., 2017). In Table 5 and Figure 7a, our B. velezensis NWUMFkBS10.5 genome did not show close phylogenomic clustering with the popular industrially and agriculturally applied B. velezensis strains (B. velezensis FZB42, AS43.3, UCMB5033, DSM7, Trigorcor 1448, and M75). The iturin cluster was predictably found in cluster 6, subcluster BGC0001098 (Supporting Information Figure S5h) of NWUMFkBS10.5, even though the NWUMFkBS10.5 blast did not branch directly with any of the iturin operons (Supporting Information Figure S6). It, however, contained putative gene clusters encoding sporulation, biofilm formation, and antibiotic synthesis, (macrolactin, bacilysin, bacillibactin, surfactin, bacillopeptin, bacillaene, iturin, fengycin, and difficidin) that could also be found in these strains (Pandin et al., 2018) (Tables 6 and 7). Gene clusters for amylocyclicin (detected in B. velezensis FZB42 and B. velezensis LS69) (Liu et al., 2017; Scholz et al., 2014), butirosin (detected in B. velezensis LM2303) (Chen, 2017), ericin (detected in B. velezensis RC 218) (Palazzini et al., 2016), and lignin degradation (Chen et al., 2018) were located in NWUMFkBS10.5 genome.
Remarkably, biosynthetic clusters predicted to synthesize compounds such as sessilin, bananamides, cichopeptin, taiwachelin, tolaasin, basiliskamides, kalimantacin, and mersacidin were however identified in strain NWUMFkBS10.5 (Table 6). These compounds are rarely documented for B. velezensis strains, and this suggests possible evolutionary pressure for the development of competitive antimicrobials among the B. velezensis group. Our goal in this study was to provide a bioprotective Bacillus strain for maize against the onset of fusariosis. B. velezensis NWUMFkBS10.5 was prudentially selected because it harbored functional and metabolically active gene clusters responsible for synthesizing diverse beneficial compounds. To date, no standard effective chemical fungicide exists for the treatment of fusariosis in South Africa. To the best of our knowledge, this is a rare report describing the genomic and biocontrol potential of a native B. velezensis strain peculiar to Africa. Although we did not corroborate our results with in planta experiments in this study, the exhaustive nature of this combinatorial in vitro study affirms the in planta viability of B. velezensis NWUMFkBS10.5. Lastly, the information gathered on NWUMFkBS10.5 will be valuable for its biotechnological manipulation and its probable development into a biofungicide in South Africa.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interests.
AUTHORS CONTRIBUTION
AAA designed and performed the in vitro assays/experiments, molecular analysis, in silico genome analysis, analyzed the data, and drafted the manuscript. OSA contributed to the chemical characterization and its data interpretation. OOB participated in the experimental design, supervised the work, and provided funding. All authors contributed to the drafts, final version of the paper, and approved submission.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
Supporting information
ACKNOWLEDGMENT
This work is based on the research supported by National Research Foundation, South Africa (Grant UID: 81192).
Adeniji AA, Aremu OS, Babalola OO. Selecting lipopeptide‐producing, Fusarium‐suppressing Bacillus spp.: Metabolomic and genomic probing of Bacillus velezensis NWUMFkBS10.5. MicrobiologyOpen. 2019;8:e742 10.1002/mbo3.742
[Correction added on 15 November 2018, after first online publication: in section 2.3.2, the formula has been corrected in this version].
DATA ACCESSIBILITY
The accession numbers of the 16S rDNA nucleotide sequences have been deposited at the NCBI GenBank (Supporting Information Table S4). Whole Genome Shotgun project is deposited at DDBJ/ENA/GenBank under the accession NZ_NITU01000037.1. The version described in this chapter is version NZ_NITU01000037.1 (GCF_002204665.1). The BioProject and BioSample designation for this project are PRJNA388288 and SAMN07174738. The datasets generated during and/or analyzed during the current study are available on request.
REFERENCES
- Abiala, M. , Odebode, A. , Hsu, S. , & Blackwood, C. (2015). Phytobeneficial properties of bacteria isolated from the rhizosphere of maize in southwestern Nigerian soils. Applied and Environmental Microbiology, 81, 4736–4743. 10.1128/AEM.00570-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abriouel, H. , Franz, C. M. , Ben Omar, N. & Gálvez, A. (2011). Diversity and applications of Bacillus bacteriocins. FEMS Microbiology Reviews, 35, 201–232. [DOI] [PubMed] [Google Scholar]
- Adeniji, A. A. , & Babalola, O. O. (2018). Tackling maize fusariosis: In search of Fusarium graminearum biosuppressors. Archives of Microbiology, 200, 1239–1255. 10.1007/s00203-018-1542-y [DOI] [PubMed] [Google Scholar]
- Ahmad, F. , Ahmad, I. , & Khan, M. S. (2008). Screening of free‐living rhizospheric bacteria for their multiple plant growth promoting activities. Microbiological Research, 163, 173–181. 10.1016/j.micres.2006.04.001 [DOI] [PubMed] [Google Scholar]
- Alsohim, A. S. , Taylor, T. B. , Barrett, G. A. , Gallie, J. , Zhang, X.‐X. , Altamirano‐Junqueira, A. E. , … Jackson, R. W. (2014). The biosurfactant viscosin produced by Pseudomonas fluorescens SBW25 aids spreading motility and plant growth promotion. Environmental Microbiology, 16, 2267–2281. [DOI] [PubMed] [Google Scholar]
- Andolfi, A. , Cimmino, A. , Cantore, P. L. , Iacobellis, N. S. , & Evidente, A. (2008). Bioactive and structural metabolites of Pseudomonas and Burkholderia species causal agents of cultivated mushrooms diseases. Perspectives in Medicinal Chemistry, 2, 81–112. [PMC free article] [PubMed] [Google Scholar]
- Arkin, A. P. , Stevens, R. L. , Cottingham, R. W. , Maslov, S. , Henry, C. S. , Dehal, P. , … Canon, S. (2016). The DOE systems biology knowledgebase (KBase). bioRxiv, 096354. [Google Scholar]
- Arutchelvi, J. , & Doble, M. (2010). Characterization of glycolipid biosurfactant from Pseudomonas aeruginosa CPCL isolated from petroleum‐contaminated soil. Letters in Applied Microbiology, 51, 75–82. 10.1111/j.1472-765X.2010.02858.x [DOI] [PubMed] [Google Scholar]
- Aziz, R. K. , Bartels, D. , Best, A. A. , Dejongh, M. , Disz, T. , Edwards, R. A. , … Zagnitko, O. (2008). The RAST server: Rapid annotations using subsystems technology. BMC Genomics, 9, 75 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babalola, O. O. , & Glick, B. R. (2012). Indigenous African agriculture and plant associated microbes: Current practice and future transgenic prospects. Scientific Research and Essays, 7, 2431–2439. [Google Scholar]
- Bacon, C. W. , & Hinton, D. M. (2011). In planta reduction of maize seedling stalk lesions by the bacterial endophyte Bacillus mojavensis . Canadian Journal of Microbiology, 57, 485–492. [DOI] [PubMed] [Google Scholar]
- Balado, M. , Osorio, C. R. , & Lemos, M. L. (2008). Biosynthetic and regulatory elements involved in the production of the siderophore vanchrobactin in Vibrio anguillarum . Microbiology, 154, 1400–1413. 10.1099/mic.0.2008/016618-0 [DOI] [PubMed] [Google Scholar]
- Bardin, M. , Ajouz, S. , Comby, M. , Lopez‐Ferber, M. , Graillot, B. , Siegwart, M. , & Nicot, P. C. (2015). Is the efficacy of biological control against plant diseases likely to be more durable than that of chemical pesticides? Frontiers in Plant Science, 6, 566 10.3389/fpls.2015.00566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Ayed, H. , Hmidet, N. , Béchet, M. , Chollet, M. , Chataigné, G. , Leclère, V. , … Nasri, M. (2014). Identification and biochemical characteristics of lipopeptides from Bacillus mojavensis A21. Process Biochemistry, 49, 1699–1707. 10.1016/j.procbio.2014.07.001 [DOI] [Google Scholar]
- Ben Belgacem, Z. , Bijttebier, S. , Verreth, C. , Voorspoels, S. , Van De Voorde, I. , Aerts, G. , … Lievens, B. (2015). Biosurfactant production by Pseudomonas strains isolated from floral nectar. Journal of Applied Microbiology, 118, 1370–1384. [DOI] [PubMed] [Google Scholar]
- Biniarz, P. , Łukaszewicz, M. , & Janek, T. (2017). Screening concepts, characterization and structural analysis of microbial‐derived bioactive lipopeptides: A review. Critical Reviews in Biotechnology, 37, 393–410. 10.3109/07388551.2016.1163324 [DOI] [PubMed] [Google Scholar]
- Bonnichsen, L. , Bygvraa Svenningsen, N. , Rybtke, M. , De Bruijn, I. , Raaijmakers, J. M. , Tolker‐Nielsen, T. , & Nybroe, O. (2015). Lipopeptide biosurfactant viscosin enhances dispersal of Pseudomonas fluorescens SBW25 biofilms. Microbiology, 161, 2289–2297. 10.1099/mic.0.000191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutigny, A. L. , Beukes, I. , Small, I. , Zühlke, S. , Spiteller, M. , Van Rensburg, B. J. , … Viljoen, A. (2012). Quantitative detection of Fusarium pathogens and their mycotoxins in South African maize. Plant Pathology, 61, 522–531. 10.1111/j.1365-3059.2011.02544.x [DOI] [Google Scholar]
- Boutigny, A. L. , Ward, T. J. , Van Coller, G. J. , Flett, B. , Lamprecht, S. C. , O’Donnell, K. , & Viljoen, A. (2011). Analysis of the Fusarium graminearum species complex from wheat, barley and maize in South Africa provides evidence of species‐specific differences in host preference. Fungal Genetics and Biology, 48, 914–920. 10.1016/j.fgb.2011.05.005 [DOI] [PubMed] [Google Scholar]
- Campbell, E. A. , Pavlova, O. , Zenkin, N. , Leon, F. , Irschik, H. , Jansen, R. , … Darst, S. A. (2005). Structural, functional, and genetic analysis of sorangicin inhibition of bacterial RNA polymerase. EMBO Journal, 24, 674–682. 10.1038/sj.emboj.7600499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, Y. , Pi, H. , Chandrangsu, P. , Li, Y. , Wang, Y. , Zhou, H. , … Cai, Y. (2018). Antagonism of two plant‐growth promoting Bacillus velezensis isolates against Ralstonia solanacearum and Fusarium oxysporum . Scientific Reports, 8, 4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty, J. , Chakrabarti, S. , & Das, S. (2014). Characterization and antimicrobial properties of lipopeptide biosurfactants produced by Bacillus subtilis SJ301 and Bacillus vallismortis JB201. Applied Biochemistry and Microbiology, 50, 609–618. 10.1134/S0003683814060039 [DOI] [Google Scholar]
- Challis, G. L. (2008). Mining microbial genomes for new natural products and biosynthetic pathways. Microbiology, 154, 1555–1569. 10.1099/mic.0.2008/018523-0 [DOI] [PubMed] [Google Scholar]
- Chen, L. (2017). Complete genome sequence of Bacillus velezensis LM2303, a biocontrol strain isolated from the dung of wild yak inhabited Qinghai‐Tibet plateau. Journal of Biotechnology, 251, 124–127. 10.1016/j.jbiotec.2017.04.034 [DOI] [PubMed] [Google Scholar]
- Chen, H. , Chen, Z. , Zhang, Y. , & Zhu, W. (2014). Identification of antifungal peptides from Bacillus subtilis BS‐918. Analytical Letters, 47, 2891–2899. [Google Scholar]
- Chen, L. , Gu, W. , Xu, H.‐Y. , Yang, G.‐L. , Shan, X.‐F. , Chen, G. , … Qian, A.‐D. (2018). Comparative genome analysis of Bacillus velezensis reveals a potential for degrading lignocellulosic biomass. 3 Biotech, 8, 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X.‐H. , Scholz, R. , Borriss, M. , Junge, H. , Mögel, G. , Kunz, S. , & Borriss, R. (2009). Difficidin and bacilysin produced by plant‐associated Bacillus amyloliquefaciens are efficient in controlling fire blight disease. Journal of Biotechnology, 140, 38–44. 10.1016/j.jbiotec.2008.10.015 [DOI] [PubMed] [Google Scholar]
- Chen, L. , Wang, N. , Wang, X. , Hu, J. , & Wang, S. (2010). Characterization of two anti‐fungal lipopeptides produced by Bacillus amyloliquefaciens SH‐B10. Bioresource Technology, 101, 8822–8827. 10.1016/j.biortech.2010.06.054 [DOI] [PubMed] [Google Scholar]
- Chen, X. , Zhang, Y. , Fu, X. , Li, Y. , & Wang, Q. (2016). Isolation and characterization of Bacillus amyloliquefaciens PG12 for the biological control of apple ring rot. Postharvest Biology and Technology, 115, 113–121. 10.1016/j.postharvbio.2015.12.021 [DOI] [Google Scholar]
- Cochrane, S. A. , Findlay, B. , Bakhtiary, A. , Acedo, J. Z. , Rodriguez‐Lopez, E. M. , Mercier, P. , & Vederas, J. C. (2016). Antimicrobial lipopeptide tridecaptin A1 selectively binds to Gram‐negative lipid II. Proceedings of the National Academy of Sciences USA, 113, 11561–11566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochrane, S. A. , & Vederas, J. C. (2016). Lipopeptides from Bacillus and Paenibacillus spp.: A gold mine of antibiotic candidates. Medicinal Research Reviews, 36, 4–31. 10.1002/med.21321 [DOI] [PubMed] [Google Scholar]
- Cosentino, S. , Voldby Larsen, M. , Møller Aarestrup, F. , & Lund, O. (2013). PathogenFinder – Distinguishing friend from foe using bacterial whole genome sequence data. PLoS One, 8, e77302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’aes, J. , Hua, G. K. H. , DeMaeyer, K. , Pannecoucque, J. , Forrez, I. , Ongena, M. , … Höfte, M. (2011). Biological control of rhizoctonia root rot on bean by phenazine and cyclic lipopeptide‐producing Pseudomonas cmr12a. Phytopathology, 101, 996–1004. [DOI] [PubMed] [Google Scholar]
- Deepak, R. , & Jayapradha, R. (2015). Lipopeptide biosurfactant from Bacillus thuringiensis pak2310: A potential antagonist against Fusarium oxysporum . Journal De Mycologie Médicale/Journal of Medical Mycology, 25, e15–e24. 10.1016/j.mycmed.2014.10.011 [DOI] [PubMed] [Google Scholar]
- Dimkić, I. , Živković, S. , Berić, T. , Ivanović, Ž. , Gavrilović, V. , Stanković, S. , & Fira, D. (2013). Characterization and evaluation of two Bacillus strains, SS‐12.6 and SS‐13.1, as potential agents for the control of phytopathogenic bacteria and fungi. Biological Control, 65, 312–321. 10.1016/j.biocontrol.2013.03.012 [DOI] [Google Scholar]
- Duitman, E. H. , Wyczawski, D. , Boven, L. G. , Venema, G. , Kuipers, O. P. , & Hamoen, L. W. (2007). Novel methods for genetic transformation of natural Bacillus subtilis isolates used to study the regulation of the mycosubtilin and surfactin synthetases. Applied and Environmental Microbiology, 73, 3490–3496. 10.1128/AEM.02751-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap, C. A. , Bowman, M. J. , & Schisler, D. A. (2013). Genomic analysis and secondary metabolite production in Bacillus amyloliquefaciens AS 43.3: A biocontrol antagonist of Fusarium head blight. Biological Control, 64, 166–175. 10.1016/j.biocontrol.2012.11.002 [DOI] [Google Scholar]
- Dunlap, C. A. , Kim, S.‐J. , Kwon, S.‐W. , & Rooney, A. P. (2016). Bacillus velezensis is not a later heterotypic synonym of Bacillus amyloliquefaciens; Bacillus methylotrophicus, Bacillus amyloliquefaciens subsp. plantarum and ‘Bacillus oryzicola’ are later heterotypic synonyms of Bacillus velezensis based on phylogenomics. International Journal of Systematic and Evolutionary Microbiology, 66, 1212–1217. [DOI] [PubMed] [Google Scholar]
- Dunlap, C. A. , Schisler, D. A. , Bowman, M. J. , & Rooney, A. P. (2015). Genomic analysis of Bacillus subtilis OH 131.1 and co‐culturing with Cryptococcus flavescens for control of Fusarium head blight. Plant Gene, 2, 1–9. 10.1016/j.plgene.2015.03.002 [DOI] [Google Scholar]
- Falardeau, J. , Wise, C. , Novitsky, L. , & Avis, T. J. (2013). Ecological and mechanistic insights into the direct and indirect antimicrobial properties of Bacillus subtilis lipopeptides on plant pathogens. Journal of Chemical Ecology, 39, 869–878. 10.1007/s10886-013-0319-7 [DOI] [PubMed] [Google Scholar]
- Fan, B. , Blom, J. , Klenk, H. P. , & Borriss, R. (2017). Bacillus amyloliquefaciens, Bacillus velezensis, and Bacillus siamensis form an “Operational Group B. amyloliquefaciens” within the B. subtilis species complex. Frontiers in Microbiology, 8, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein, J. (1985). Confidence limits on phylogenies: An approach using the bootstrap. Evolution, 39, 783–791. 10.1111/j.1558-5646.1985.tb00420.x [DOI] [PubMed] [Google Scholar]
- Garbeva, P. , Van Veen, J. A. , & Van Elsas, J. D. (2003). Predominant Bacillus spp. in agricultural soil under different management regimes detected via PCR‐DGGE. Microbial Ecology, 45, 302–316. 10.1007/s00248-002-2034-8 [DOI] [PubMed] [Google Scholar]
- Giessen, T. W. , Franke, K. B. , Knappe, T. A. , Kraas, F. I. , Bosello, M. , Xie, X. , … Marahiel, M. A. (2012). Isolation, structure elucidation, and biosynthesis of an unusual hydroxamic acid ester‐containing siderophore from Actinosynnema mirum . Journal of Natural Products, 75, 905–914. [DOI] [PubMed] [Google Scholar]
- Girija, S. , Duraipandiyan, V. , Kuppusamy, P. S. , Gajendran, H. , & Rajagopal, R. (2014). Chromatographic characterization and GC‐MS evaluation of the bioactive constituents with antimicrobial potential from the pigmented ink of Loligo duvauceli. International Scholarly Research Notices, 2014. [DOI] [PMC free article] [PubMed]
- Gond, S. K. , Bergen, M. S. , Torres, M. S. , & White, J. F. Jr (2015). Endophytic Bacillus spp. produce antifungal lipopeptides and induce host defence gene expression in maize. Microbiological Research, 172, 79–87. [DOI] [PubMed] [Google Scholar]
- Gond, S. K. , Bergen, M. S. , Torres, M. S. , White, J. F. Jr , & Kharwar, R. N. (2015). Effect of bacterial endophyte on expression of defense genes in Indian popcorn against Fusarium moniliforme . Symbiosis, 66, 133–140. [Google Scholar]
- Grangemard, I. , Bonmatin, J.‐M. , Bernillon, J. , Das, B. C. , & Peypoux, F. (1999). Lichenysins G, a novel family of lipopeptide biosurfactants from Bacillus licheniformis IM 1307. Journal of Antibiotics, 52, 363–373. [DOI] [PubMed] [Google Scholar]
- Grzywacz, D. , Stevenson, P. C. , Mushobozi, W. L. , Belmain, S. , & Wilson, K. (2014). The use of indigenous ecological resources for pest control in Africa. Food Security, 6, 71–86. 10.1007/s12571-013-0313-5 [DOI] [Google Scholar]
- Guo, Q. , Dong, W. , Li, S. , Lu, X. , Wang, P. , Zhang, X. , … Ma, P. (2014). Fengycin produced by Bacillus subtilis NCD‐2 plays a major role in biocontrol of cotton seedling damping‐off disease. Microbiological Research, 169, 533–540. 10.1016/j.micres.2013.12.001 [DOI] [PubMed] [Google Scholar]
- Hazra, C. , Kundu, D. , Ghosh, P. , Joshi, S. , Dandi, N. , & Chaudhari, A. (2011). Screening and identification of Pseudomonas aeruginosa AB4 for improved production, characterization and application of a glycolipid biosurfactant using low‐cost agro‐based raw materials. Journal of Chemical Technology and Biotechnology, 86, 185–198. 10.1002/jctb.2480 [DOI] [Google Scholar]
- Hertlein, G. , Seiffert, M. , Gensel, S. , Garcia‐Gonzalez, E. , Ebeling, J. , Skobalj, R. , … Genersch, E. (2016). Biological role of paenilarvins, iturin‐like lipopeptide secondary metabolites produced by the honey bee pathogen Paenibacillus larvae . PLoS One, 11, e0164656 10.1371/journal.pone.0164656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, E. , Guo, Y. , & Yousef, A. E. (2014). Biosynthesis of the new broad‐spectrum lipopeptide antibiotic paenibacterin in Paenibacillus thiaminolyticus OSY‐SE. Research in Microbiology, 165, 243–251. 10.1016/j.resmic.2014.02.002 [DOI] [PubMed] [Google Scholar]
- Huang, C.‐J. , Pauwelyn, E. , Ongena, M. , Debois, D. , Leclère, V. , Jacques, P. , … Höfte, M. (2015). Characterization of cichopeptins, new phytotoxic cyclic lipodepsipeptides produced by Pseudomonas cichorii sf1‐54 and their role in bacterial midrib rot disease of lettuce. Molecular Plant‐Microbe Interactions, 28, 1009–1022. [DOI] [PubMed] [Google Scholar]
- Huang, E. , & Yousef, A. E. (2014). Paenibacterin, a novel broad‐spectrum lipopeptide antibiotic, neutralises endotoxins and promotes survival in a murine model of Pseudomonas aeruginosa‐induced sepsis. International Journal of Antimicrobial Agents, 44, 74–77. 10.1016/j.ijantimicag.2014.02.018 [DOI] [PubMed] [Google Scholar]
- Jang, J. Y. , Yang, S. Y. , Kim, Y. C. , Lee, C. W. , Park, M. S. , Kim, J. C. , & Kim, I. S. (2013). Identification of orfamide A as an insecticidal metabolite produced by Pseudomonas protegens F6. Journal of Agricultural and Food Chemistry, 61, 6786–6791. 10.1021/jf401218w [DOI] [PubMed] [Google Scholar]
- Janse Van Rensburg, B. , Mclaren, N. W. , Flett, B. C. , & Schoeman, A. (2015). Fumonisin producing Fusarium spp. and fumonisin contamination in commercial South African maize. European Journal of Plant Pathology, 141, 491–504. 10.1007/s10658-014-0558-7 [DOI] [Google Scholar]
- Jasim, B. , Sreelakshmi, K. , Mathew, J. , & Radhakrishnan, E. (2016). Surfactin, iturin, and fengycin biosynthesis by endophytic Bacillus sp. from Bacopa monnieri . Microbial Ecology, 72, 106–119. [DOI] [PubMed] [Google Scholar]
- Jemil, N. , Manresa, A. , Rabanal, F. , Ben Ayed, H. , Hmidet, N. , & Nasri, M. (2017). Structural characterization and identification of cyclic lipopeptides produced by Bacillus methylotrophicus DCS1 strain. Journal of Chromatography B, 1060, 374–386. 10.1016/j.jchromb.2017.06.013 [DOI] [PubMed] [Google Scholar]
- Köhl, J. , Postma, J. , Nicot, P. , Ruocco, M. , & Blum, B. (2011). Stepwise screening of microorganisms for commercial use in biological control of plant‐pathogenic fungi and bacteria. Biological Control, 57, 1–12. 10.1016/j.biocontrol.2010.12.004 [DOI] [Google Scholar]
- Kosaric, N. , Gray, N. , & Cairns, W. (1987). Biotechnology and the surfactant industry. Biosurfactants and Biotechnology, 25, 1–19. [Google Scholar]
- Koumoutsi, A. , Chen, X. H. , Henne, A. , Liesegang, H. , Hitzeroth, G. , Franke, P. , … Borriss, R. (2004). Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42. Journal of Bacteriology, 186, 1084–1096. 10.1128/JB.186.4.1084-1096.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreutzer, M. F. , Kage, H. , & Nett, M. (2012). Structure and biosynthetic assembly of cupriachelin, a photoreactive siderophore from the bioplastic producer Cupriavidus necator H16. Journal of the American Chemical Society, 134, 5415–5422. 10.1021/ja300620z [DOI] [PubMed] [Google Scholar]
- Lamprecht, S. C. , Tewoldemedhin, Y. T. , Botha, W. J. , & Calitz, F. J. (2011). Fusarium graminearum Species Complex associated with maize crowns and roots in the Kwazulu‐Natal province of South Africa. Plant Disease, 95, 1153–1158. [DOI] [PubMed] [Google Scholar]
- Laureti, L. , Song, L. , Huang, S. , Corre, C. , Leblond, P. , Challis, G. L. , & Aigle, B. (2011). Identification of a bioactive 51‐membered macrolide complex by activation of a silent polyketide synthase in Streptomyces ambofaciens . Proceedings of the National Academy of Sciences USA, 108, 6258–6263. 10.1073/pnas.1019077108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclère, V. , Béchet, M. , Adam, A. , Guez, J.‐S. , Wathelet, B. , Ongena, M. , … Jacques, P. (2005). Mycosubtilin overproduction by Bacillus subtilis BBG100 enhances the organism's antagonistic and biocontrol activities. Applied and Environmental Microbiology, 71, 4577–4584. 10.1128/AEM.71.8.4577-4584.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H. H. , Park, J. , Lim, J. Y. , Kim, H. , Choi, G. J. , Kim, J.‐C. , & Seo, Y.‐S. (2015). Complete genome sequence of Bacillus velezensis G341, a strain with a broad inhibitory spectrum against plant pathogens. Journal of Biotechnology, 211, 97–98. 10.1016/j.jbiotec.2015.07.005 [DOI] [PubMed] [Google Scholar]
- Liu, G. , Kong, Y. , Fan, Y. , Geng, C. , Peng, D. , & Sun, M. (2017). Whole‐genome sequencing of Bacillus velezensis LS69, a strain with a broad inhibitory spectrum against pathogenic bacteria. Journal of Biotechnology, 249, 20–24. 10.1016/j.jbiotec.2017.03.018 [DOI] [PubMed] [Google Scholar]
- Lugtenberg, B. , & Kamilova, F. (2009). Plant‐growth‐promoting rhizobacteria. Annual Review of Microbiology, 63, 541–556. 10.1146/annurev.micro.62.081307.162918 [DOI] [PubMed] [Google Scholar]
- Ma, Z. , Ongena, M. , & Höfte, M. (2017). The cyclic lipopeptide orfamide induces systemic resistance in rice to Cochliobolus miyabeanus but not to Magnaporthe oryzae . Plant Cell Reports, 36, 1731–1746. 10.1007/s00299-017-2187-z [DOI] [PubMed] [Google Scholar]
- Matilla, M. A. , Leeper, F. J. , & Salmond, G. P. C. (2015). Biosynthesis of the antifungal haterumalide, oocydin A, in Serratia, and its regulation by quorum sensing, RpoS and Hfq. Environmental Microbiology, 17, 2993–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattheus, W. , Gao, L.‐J. , Herdewijn, P. , Landuyt, B. , Verhaegen, J. , Masschelein, J. , … Lavigne, R. (2010). Isolation and purification of a new kalimantacin/batumin‐related polyketide antibiotic and elucidation of its biosynthesis gene cluster. Chemistry & Biology, 17, 149–159. 10.1016/j.chembiol.2010.01.014 [DOI] [PubMed] [Google Scholar]
- Medini, D. , Donati, C. , Tettelin, H. , Masignani, V. , & Rappuoli, R. (2005). The microbial pan‐genome. Current Opinion in Genetics & Development, 15, 589–594. 10.1016/j.gde.2005.09.006 [DOI] [PubMed] [Google Scholar]
- Michelsen, C. F. , Watrous, J. , Glaring, M. A. , Kersten, R. , Koyama, N. , Dorrestein, P. C. , & Stougaard, P. (2015). Nonribosomal peptides, key biocontrol components for Pseudomonas fluorescens In5, isolated from a greenlandic suppressive soil. Mbio, 6, e00079–e115. 10.1128/mBio.00079-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mngqawa, P. , Shephard, G. S. , Green, I. R. , Ngobeni, S. H. , De Rijk, T. C. , & Katerere, D. R. (2016). Mycotoxin contamination of home‐grown maize in rural northern South Africa (Limpopo and Mpumalanga Provinces). Food Additives & Contaminants: Part B, 9, 38–45. 10.1080/19393210.2015.1121928 [DOI] [PubMed] [Google Scholar]
- Mnif, I. , & Ghribi, D. (2015). Review lipopeptides biosurfactants: Mean classes and new insights for industrial, biomedical, and environmental applications. Peptide Science, 104, 129–147. 10.1002/bip.22630 [DOI] [PubMed] [Google Scholar]
- Mousa, W. K. , & Raizada, M. N. (2015). Biodiversity of genes encoding anti‐microbial traits within plant associated microbes. Frontiers in Plant Science, 6, 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousa, W. K. , Shearer, C. R. , Limay‐Rios, V. , Zhou, T. , & Raizada, M. N. (2015). Bacterial endophytes from wild maize suppress Fusarium graminearum in modern maize and inhibit mycotoxin accumulation. Frontiers in Plant Science, 6, 805 10.3389/fpls.2015.00805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, A. C. , Gao, S.‐S. , Han, L.‐C. , Carobene, S. , Fukuda, D. , Song, Z. , … Crump, M. P. (2014). Biosynthesis of thiomarinol A and related metabolites of Pseudoalteromonas sp. SANK 73390. Chemical Science, 5, 397–402. 10.1039/C3SC52281D [DOI] [Google Scholar]
- Nam, J. , Jung, M. Y. , Kim, P. I. , Lee, H. B. , Kim, S. W. , & Lee, C. W. (2015). Structural characterization and temperature‐dependent production of C17‐fengycin B derived from Bacillus amyloliquefaciens subsp. plantarum BC32‐1. Biotechnology and Bioprocess Engineering, 20, 708–713. 10.1007/s12257-015-0350-3 [DOI] [Google Scholar]
- Nguyen, D. D. , Melnik, A. V. , Koyama, N. , Lu, X. , Schorn, M. , Fang, J. , … Dorrestein, P. C. (2016). Indexing the Pseudomonas specialized metabolome enabled the discovery of poaeamide B and the bananamides. NatureMicrobiology, 2, 16197–16197. 10.1038/nmicrobiol.2016.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olorunleke, F. E. , Hua, G. K. H. , Kieu, N. P. , Ma, Z. , & Höfte, M. (2015). Interplay between orfamides, sessilins and phenazines in the control of Rhizoctonia diseases by Pseudomonas sp. CMR12a. Environmental Microbiology Reports, 7, 774–781. [DOI] [PubMed] [Google Scholar]
- Ongena, M. , & Jacques, P. (2008). Bacillus lipopeptides: Versatile weapons for plant disease biocontrol. Trends in Microbiology, 16(3), 115–125. 10.1016/j.tim.2007.12.009 [DOI] [PubMed] [Google Scholar]
- Overbeek, R. , Olson, R. , Pusch, G. D. , Olsen, G. J. , Davis, J. J. , & Disz, T. (2014). The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Research, 42, D206–D214. 10.1093/nar/gkt1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzini, J. M. , Dunlap, C. A. , Bowman, M. J. , & Chulze, S. N. (2016). Bacillus velezensis RC 218 as a biocontrol agent to reduce Fusarium head blight and deoxynivalenol accumulation: Genome sequencing and secondary metabolite cluster profiles. Microbiological Research, 192, 30–36. 10.1016/j.micres.2016.06.002 [DOI] [PubMed] [Google Scholar]
- Pan, H. Q. , Li, Q. L. , & Hu, J. C. (2017). The complete genome sequence of Bacillus velezensis 9912D reveals its biocontrol mechanism as a novel commercial biological fungicide agent. Journal of Biotechnology, 247, 25–28. 10.1016/j.jbiotec.2017.02.022 [DOI] [PubMed] [Google Scholar]
- Pandin, C. , Le Coq, D. , Deschamps, J. , Vedie, R. , Rousseau, T. , Aymerich, S. , & Briandet, R. (2018). Complete genome sequence of Bacillus velezensis QST713: A biocontrol agent that protects Agaricus bisporus crops against the green mould disease. Journal of Biotechnology, 278, 10–19. 10.1016/j.jbiotec.2018.04.014 [DOI] [PubMed] [Google Scholar]
- Patzer, S. I. , & Braun, V. (2010). Gene cluster involved in the biosynthesis of griseobactin, a catechol‐peptide siderophore of Streptomyces sp. ATCC 700974. Journal of Bacteriology, 192, 426–435. 10.1128/JB.01250-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, P. , Nesci, A. , Castillo, C. , & Etcheverry, M. (2010). Impact of bacterial biological control agents on fumonisin B1 content and Fusarium verticillioides infection of field‐grown maize. Biological Control, 53, 258–266. 10.1016/j.biocontrol.2010.02.001 [DOI] [Google Scholar]
- Phillips, I. (1999). The use of bacitracin as a growth promoter in animals produces no risk to human health. Journal of Antimicrobial Chemotherapy, 44, 725–728. 10.1093/jac/44.6.725 [DOI] [PubMed] [Google Scholar]
- Plaza, G. , Chojniak, J. , Rudnicka, K. , Paraszkiewicz, K. , & Bernat, P. (2015). Detection of biosurfactants in Bacillus species: Genes and products identification. Journal of Applied Microbiology, 119, 1023–1034. [DOI] [PubMed] [Google Scholar]
- Pruitt, K. D. , Tatusova, T. , Brown, G. R. , & Maglott, D. R. (2011). NCBI Reference Sequences (RefSeq): Current status, new features and genome annotation policy. Nucleic Acids Research, 40, D130–D135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers, J. M. , Bruijn, I. , & Kock, M. J. (2006). Cyclic lipopeptide production by plant‐associated Pseudomonas spp.: diversity, activity, biosynthesis, and regulation. Molecular Plant‐Microbe Interactions, 19, 699–710. [DOI] [PubMed] [Google Scholar]
- Rivardo, F. , Turner, R. J. , Allegrone, G. , Ceri, H. , & Martinotti, M. G. (2009). Anti‐adhesion activity of two biosurfactants produced by Bacillus spp. prevents biofilm formation of human bacterial pathogens. Applied Microbiology and Biotechnology, 83, 541–553. [DOI] [PubMed] [Google Scholar]
- Romero, D. , De Vicente, A. , Rakotoaly, R. H. , Dufour, S. E. , Veening, J.‐W. , Arrebola, E. , … Pérez‐García, A. (2007). The Iturin and Fengycin families of lipopeptides are key factors in antagonism of Bacillus subtilis toward Podosphaera fusca . Molecular Plant‐Microbe Interactions, 20, 430–440. [DOI] [PubMed] [Google Scholar]
- Roongsawang, N. , Washio, K. , & Morikawa, M. (2011). Diversity of nonribosomal peptide synthetases involved in the biosynthesis of lipopeptide biosurfactants. International Journal of Molecular Sciences, 12, 141–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouli, L. , Merhej, V. , Fournier, P. E. , & Raoult, D. (2015). The bacterial pangenome as a new tool for analysing pathogenic bacteria. New Microbes and New Infections, 7, 72–85. 10.1016/j.nmni.2015.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou, N. , & Nei, M. (1987). The neighbor‐joining method: A new method for reconstructing phylogenetic trees. Molecular Biology and Evolution, 4, 406–425. [DOI] [PubMed] [Google Scholar]
- Sandy, M. , Han, A. , Blunt, J. , Munro, M. , Haygood, M. , & Butler, A. (2010). Vanchrobactin and anguibactin siderophores produced by Vibrio sp. DS40M4. Journal of Natural Products, 73, 1038–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoyo, G. , Orozco‐Mosqueda, M. D. C. , & Govindappa, M. (2012). Mechanisms of biocontrol and plant growth‐promoting activity in soil bacterial species of Bacillus and Pseudomonas: A review. Biocontrol Science and Technology, 22, 855–872. [Google Scholar]
- Scholz, R. , Vater, J. , Budiharjo, A. , Wang, Z. , He, Y. , Dietel, K. , … Borriss, R. (2014). Amylocyclicin, a novel circular bacteriocin produced by Bacillus amyloliquefaciens FZB42. Journal of Bacteriology, 196, 1842–1852. 10.1128/JB.01474-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz, H. , Gerth, K. , Jansen, R. , Schläger, N. , Dehn, R. , Reinecke, S. , … Müller, R. (2011). Elansolid A, a unique macrolide antibiotic from Chitinophaga sancti isolated as two stable atropisomers. Angewandte Chemie International Edition, 50, 532–536. 10.1002/anie.201005226 [DOI] [PubMed] [Google Scholar]
- Sumi, C. D. , Yang, B. W. , Yeo, I. C. , & Hahm, Y. T. (2014). Antimicrobial peptides of the genus Bacillus: A new era for antibiotics. Canadian Journal of Microbiology, 61, 93–103. [DOI] [PubMed] [Google Scholar]
- Summerell, B. A. , & Leslie, J. F. (2011). Fifty years of Fusarium: How could nine species have ever been enough? Fungal Diversity, 50, 135–144. 10.1007/s13225-011-0132-y [DOI] [Google Scholar]
- Tamura, K. , Peterson, D. , Peterson, N. , Stecher, G. , Nei, M. , & Kumar, S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28, 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin, H. , Masignani, V. , Cieslewicz, M. J. , Donati, C. , Medini, D. , Ward, N. l. , … Fraser, C. M. (2005). Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: Implications for the microbial "pan‐genome". Proceedings of the National Academy of Sciences USA, 102, 13950–13955. 10.1073/pnas.0506758102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodore, C. M. , Stamps, B. W. , King, J. B. , Price, L. S. L. , Powell, D. R. , Stevenson, B. S. , & Cichewicz, R. H. (2014). Genomic and metabolomic insights into the natural product biosynthetic diversity of a feral‐hog‐associated Brevibacillus laterosporus strain. PLoS One, 9, e90124 10.1371/journal.pone.0090124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga, T. , Kamigiri, K. , Orita, M. , Nishikawa, T. , Shimizu, M. , & Kaniwa, H. (1996). Kalimantacin A, B, and C, novel antibiotics produced by Alcaligenes sp. YL‐02632S. Journal of Antibiotics, 49, 140–144. [DOI] [PubMed] [Google Scholar]
- Tyc, O. , Song, C. , Dickschat, J. S. , Vos, M. , & Garbeva, P. (2017). The ecological role of volatile and soluble secondary metabolites produced by soil bacteria. Trends in Microbiology, 25, 280–292. 10.1016/j.tim.2016.12.002 [DOI] [PubMed] [Google Scholar]
- Van Der Voort, M. , Meijer, H. J. G. , Schmidt, Y. , Watrous, J. , Dekkers, E. , Mendes, R. , … Raaijmakers, J. M. (2015). Genome mining and metabolic profiling of the rhizosphere bacterium Pseudomonas sp. SH‐C52 for antimicrobial compounds. Frontiers in Microbiology, 6, 693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanittanakom, N. , Loeffler, W. , Koch, U. , & Jung, G. (1986). Fengycin‐A novel antifungal lipopeptide antibiotic produced by Bacillus subtilis F‐29‐3. Journal of Antibiotics, 39, 888–901. 10.7164/antibiotics.39.888 [DOI] [PubMed] [Google Scholar]
- Vitullo, D. , Di Pietro, A. , Romano, A. , Lanzotti, V. , & Lima, G. (2012). Role of new bacterial surfactins in the antifungal interaction between Bacillus amyloliquefaciens and Fusarium oxysporum . Plant Pathology, 61, 689–699. 10.1111/j.1365-3059.2011.02561.x [DOI] [Google Scholar]
- Wang, J.‐H. , Ndoye, M. , Zhang, J.‐B. , Li, H.‐P. , & Liao, Y.‐C. (2011). Population structure and genetic diversity of the Fusarium graminearum species complex. Toxins, 3, 1020–1037. 10.3390/toxins3081020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattam, A. R. , Davis, J. J. , Assaf, R. , Boisvert, S. , Brettin, T. , Bun, C. , … Stevens, R. L. (2017). Improvements to PATRIC, the all‐bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Research, 45, D535–D542. 10.1093/nar/gkw1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, T. , Blin, K. , Duddela, S. , Krug, D. , Kim, H. U. , Bruccoleri, R. , … Medema, M. H. (2015). antiSMASH 3.0—A comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Research, 43, W237–W243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, Y. , Wu, X. , Teng, Y. , Qian, C. , Zhan, Z. , Zhao, Y. , & Li, O. (2011). Identification and analysis of the gene cluster involved in biosynthesis of paenibactin, a catecholate siderophore produced by Paenibacillus elgii B69. Environmental Microbiology, 13, 2726–2737. 10.1111/j.1462-2920.2011.02542.x [DOI] [PubMed] [Google Scholar]
- Xiao, Y. , Gerth, K. , Müller, R. , & Wall, D. (2012). Myxobacterium‐produced antibiotic ta (myxovirescin) inhibits type ii signal peptidase. Antimicrobial Agents and Chemotherapy, 56, 2014–2021. 10.1128/AAC.06148-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Z. H. , Shao, J. H. , Li, B. , Yan, X. , Shen, Q. R. , & Zhang, R. F. (2013). Contribution of bacillomycin D in Bacillus amyloliquefaciens SQR9 to antifungal activity and biofilm formation. Applied and Environmental Microbiology, 79, 808–815. 10.1128/AEM.02645-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanes, M. L. , De La Fuente, L. , Altier, N. , & Arias, A. (2012). Characterization of native fluorescent Pseudomonas isolates associated with alfalfa roots in Uruguayan agroecosystems. Biological Control, 63, 287–295. 10.1016/j.biocontrol.2012.08.006 [DOI] [Google Scholar]
- Yang, L. , Quan, X. , Xue, B. , Goodwin, P. H. , Lu, S. , Wang, J. , … Wu, C. (2015). Isolation and identification of Bacillus subtilis strain YB‐05 and its antifungal substances showing antagonism against Gaeumannomyces graminis var. tritici . Biological Control, 85, 52–58. 10.1016/j.biocontrol.2014.12.010 [DOI] [Google Scholar]
- Youssef, N. H. , Duncan, K. E. , Nagle, D. P. , Savage, K. N. , Knapp, R. M. , & Mcinerney, M. J. (2004). Comparison of methods to detect biosurfactant production by diverse microorganisms. Journal of Microbiological Methods, 56, 339–347. 10.1016/j.mimet.2003.11.001 [DOI] [PubMed] [Google Scholar]
- Ziemert, N. , Alanjary, M. , & Weber, T. (2016). The evolution of genome mining in microbes – A review. Natural Product Reports, 33, 988–1005. 10.1039/C6NP00025H [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession numbers of the 16S rDNA nucleotide sequences have been deposited at the NCBI GenBank (Supporting Information Table S4). Whole Genome Shotgun project is deposited at DDBJ/ENA/GenBank under the accession NZ_NITU01000037.1. The version described in this chapter is version NZ_NITU01000037.1 (GCF_002204665.1). The BioProject and BioSample designation for this project are PRJNA388288 and SAMN07174738. The datasets generated during and/or analyzed during the current study are available on request.


