Abstract
Polyhydroxyalkanoates (PHAs) are biodegradable plastics that can be produced by some methanotrophic organisms such as those of the genus Methylocystis. This allows the conversion of a detrimental greenhouse gas into an environmentally friendly high added‐value bioproduct. This study presents the genome sequence of Methylocystis hirsuta CSC1 (a high yield PHB producer). The genome comprises 4,213,043 bp in 4 contigs, with the largest contig being 3,776,027 bp long. Two of the other contigs are likely to correspond to large size plasmids. A total of 4,664 coding sequences were annotated, revealing a PHA production cluster, two distinct particulate methane monooxygenases with active catalytic sites, as well as a nitrogen fixation operon and a partial denitrification pathway.
Keywords: genome, methane monooxygenases, methanotrophs, methylocystis, polyhydroxyalkanoates
1. INTRODUCTION
Anthropogenic emissions of methane currently account for up to 30% of the global emissions of greenhouse gases (considering that methane has a 25 times greater global warming potential than CO2; Desai & Harvey, 2017). Despite the fact that methane can be used as an industrial energy source via combustion at concentrations higher than 20%, more than 56% of its emissions have concentrations lower than 5% (Lebrero et al., 2016). Biological methane abatement is a very attractive alternative to treat diluted methane emissions based on its high effectiveness and environmentally friendliness. In addition, biological methane abatement can be coupled to the production of added‐value compounds. Polyhydroxyalkanoates (PHAs) are intracellular biopolyesters produced under nutrient‐limiting conditions by a wide range of methane‐consuming organisms (Pieja, Morse, & Cal, 2017).
Methanotrophs are organisms able to use methane as the sole energy and carbon source, some of them use methane exclusively and others are facultative methanotrophs, able to grow also in other carbon sources. We focus here on bacterial methanotrophic species using oxygen as electron acceptor. Even if anaerobic methane oxidation can also occur coupled to sulfate, nitrate and nitrite reduction, this phenomenon plays a minor ecological role compared to aerobic oxidation. Aerobic methanotrophic bacteria are usually classified into type I and type II methanotrophs, which are different in their membrane arrangement, fatty acid composition, and methane assimilation pathways (Hanson & Hanson, 1996). Type II methanotrophs such as Methylocystis, Methylosinus, and Methylocella are considered the main methanotrophic PHA‐synthesizing genera. For instance, Methylocystis hirsuta has been shown to accumulate PHB up to 45% of its total biomass (García‐Pérez et al., 2018), which makes it a very interesting cell factory. This value is higher than those previously reported for other methanotrophs (Pieja, Rostowski, & Criddle, 2011), including its close relative Methylocystis sp. SC2.
2. MATERIALS AND METHODS
The strain M. hirsuta CSC1, which was obtained from DSMZ (DSM no. 18500) (Lidner et al., 2007), was cultured in 1,250 ml gas‐tight bottles containing 210 ml of a mineral salt medium (Mokhtari‐Hosseini et al., 2009). Methane was then supplied at an initial headspace concentration of 195 ± 7 g/m3 (under O2 sufficient conditions), and the bottles were incubated in an orbital shaker (MaxQ 4000; Thermo Scientific, USA) at 30°C and 200 rpm for a week. Biomass was centrifuged, and sequencing libraries were prepared (after checking the purity of the culture by 16S sequencing) using the protocol for multiplexed microbial SMRTbell libraries for the PacBio Sequel System. The genetic material was fragmented and selected to have a size close to 20 kb. The final size distribution was checked using AATI Femto Pulse. The library was sequenced using the platform Sequel from PacBio, with an acquisition time of 10 hr. The sequenced reads were assembled using HGAP 4.0 (Chin et al., 2013).
3. RESULTS
The assembly's results are summarized in Table 1.
Table 1.
Genome statistics of Methylocystis hirsuta CSC1
| Genome feature | Value |
|---|---|
| Size (bp) | 4,213,043 |
| Contigs | 4 |
| N50 (bp) | 3,776,027 |
| L50 | 1 |
| GC content (%) | 62.4 |
| Coding sequences | 4,664 |
| Number of RNAs | 50 |
A phylogenetic analysis was carried out using JSpeciesWS (Richter, Rosselló‐Móra, Glöckner, & Peplies, 2015), which calculates the average nucleotide identity (ANI) comparing all shared orthologous protein‐coding genes of two genomes (Richter & Rosselló‐Móra, 2009). The phylogenetic tree (Figure 1) containing the 10 closest species identified by JSpeciesWS was built using the function dendrogram from SciPy after defining the distance between species as 100 minus their ANI value.
Figure 1.
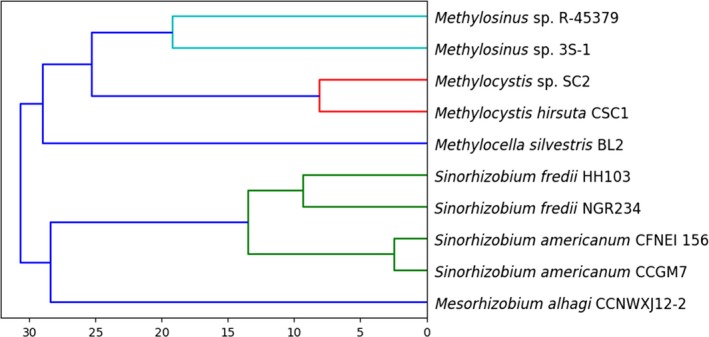
Phylogenetic relations of Methylocystis hirsuta CSC1. The x‐axis represents 100 minus the average nucleotide identity among species. The closest related species was Methylocystis sp. CS2. Methylosinus appeared to be the closest genus to Methylocystis
The genome was annotated using the NCBI Prokaryotic Genome annotation pipeline (Tatusova et al., 2016). A general summary of the biological functions coded in the genome was obtained using RAST (Overbeek et al., 2014). Approximately, 25% of the annotated genes corresponded to RAST subsystems (Table 2).
Table 2.
Biological subsystem distribution of annotated genes in Methylocystis hirsuta CSC1
| Code | Description | Value | Percentage |
|---|---|---|---|
| A | Cofactors, vitamins, prosthetic groups, pigments | 143 | 8.8 |
| B | Cell wall and capsule | 47 | 2.9 |
| C | Virulence, disease and defense | 77 | 4.7 |
| D | Potassium metabolism | 7 | 0.4 |
| E | Miscellaneous | 37 | 2.3 |
| F | Phages, prophages, transposable elements, plasmids | 41 | 2.5 |
| G | Membrane transport | 75 | 4.6 |
| H | Iron metabolism | 0 | 0 |
| I | RNA metabolism | 49 | 3 |
| J | Nucleosides and nucleotides | 51 | 3.1 |
| K | Protein metabolism | 187 | 11.5 |
| L | Cell division and cell cycle | 0 | 0 |
| M | Motility and chemotaxis | 0 | 0 |
| N | Regulation and cell signaling | 34 | 2.1 |
| O | Secondary metabolism | 7 | 0.4 |
| P | DNA metabolism | 76 | 4.7 |
| Q | Fatty acids, lipids, and isoprenoids | 68 | 4.2 |
| R | Nitrogen metabolism | 39 | 2.4 |
| S | Dormancy and sporulation | 1 | 0.1 |
| T | Respiration | 126 | 7.7 |
| U | Stress response | 70 | 4.3 |
| V | Metabolism of aromatic compounds | 13 | 0.8 |
| W | Amino acids and derivatives | 243 | 14.9 |
| X | Sulfur metabolism | 13 | 0.8 |
| Y | Phosphorous metabolism | 17 | 1 |
| Z | Carbohydrates | 184 | 11.3 |
Despite the presence of plasmids has not been assessed experimentally, a cluster of three plasmid replication genes (RepA, RepB and RepC) was detected in the fourth contig (which is 158,363 bases long). This suggests that this contig could correspond to a large plasmid. The locus tags of the genes forming this cluster of plasmid replication genes are as follows: D1030_20715, D1030_20710, and D1030_20705. The fourth contig also contains several repeat regions that could be binding sites for the plasmid‐encoded repeat proteins (Sekine et al., 2006). The third contig (260,028 bp) contains also two plasmid replication genes (RepA and RepC) separated by a protein that could not be annotated and has repeated regions in their proximity, which suggests the possibility of this contig being also a large size plasmid. The chromosome also contains two genes annotated as RepA proteins and two other putative RepC proteins. Overall, it appears very likely that M. hirsuta is able to sustain plasmid replication. The closest strain Methylocystis sp. SC2 does contain two large plasmids.
In order to identify gene clusters involved in the synthesis of secondary metabolites, the platform antiSMASH 4.0 was used (Blin et al., 2017). The two clusters showing higher similarity (to known clusters) were a PHA biosynthetic gene cluster and an enterobactin biosynthetic cluster (Table 3). The PHA biosynthetic cluster (Figure 2a) contains the genes phbA, phbB and the regulator phaR. The gene involved in the last step of PHB biosynthesis (D1030_08315) is outside of this cluster. The same arrangement of phbA, phbB, and phaR is observed in the strain Methylocystis sp. SC2 (Dam, Dam, Kube, Reinhardt, & Liesack, 2012). On the other hand, enterobactin is a chelating agent that has a strong affinity for iron and is secreted to the environment to improve iron assimilation. A cluster of eight genes involved in the synthesis of enterobactin from chorismate was found. This cluster has the same architecture in Methylocystis sp. SC2, which suggests that these organisms are able to uptake iron with high efficiency.
Table 3.
Enterobactin biosynthesis and transport genes in Methylocystis hirsuta CSC1
| Locus tag | Genomic coordinates | Annotation |
|---|---|---|
| D1030_04505 | 893002–894342 (+) | Enterobactin esterase |
| D1030_04515 | 894639–901834 (+) | Siderophore biosynthesis non‐ribosomal peptide synthetase modules |
| D1030_04520 | 901841–903292 (+) | Enterobactin exporter EntS |
| D1030_04525 | 903318–904544 (+) | Isochorismate synthase |
| D1030_04530 | 904535–906157 (+) | 2,3‐hydroxybenzoate‐AMP ligase |
| D1030_04535 | 906177–907064 (+) | Isochorismatase |
| D1030_04540 | 907082–907828 (+) | 2,3‐dihydro‐2,3‐dihydroxybenzoate dehydrogenase |
Figure 2.
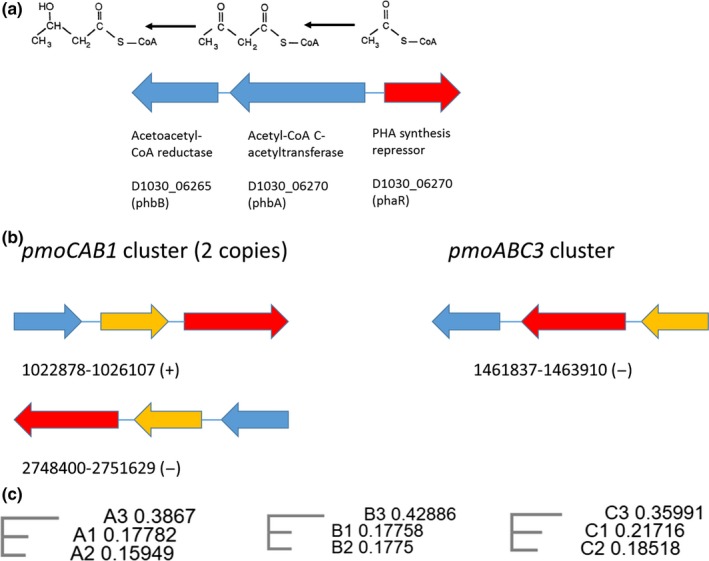
PHB synthesis and methane oxidation cluster. (a) PHB synthesis cluster. (b) Two clusters coding pMMO enzymes. The pmoCAB1 cluster is duplicated and is identical to the cluster found in Methylocystis sp. SC2. The pmoABC3 cluster differs in the order in which the subunits are arranged. (c) Evolutionary distance between proteins in the pmoCAB1, pmoABC3, and pmoCAB2 clusters. pmoCAB2 is present in Methylocystis sp. SC2 but is absent in Methylocystis hirsuta CSC1
Methylocystis hirsuta CSC1 contained two different gene clusters coding the three subunits (A, B, and C) of particulate methane monooxygenase (pMMO) enzyme, one of them was present in two copies (Figure 2b) and was identical to the pmoCAB1 cluster present in Methylocystis sp. SC2 (Dam et al., 2012). One of the copies in M. hirsuta CSC1 was oriented in the reverse sense compared to Methylocystis sp. SC2 as a result of a chromosomal inversion. The pMMO coded by the pmoCAB1 cluster has been reported to oxidize methane at gas concentrations higher than 600 ppmv (Baani & Liesack, 2008). Methylocystis sp. SC2 contains a second pMMO cluster, pmoCAB2, which codes an enzyme with higher affinity for methane at low concentrations. This cluster was absent in M. hirsuta CSC1, which contained instead a second cluster (Figure 2b) with the pMMO subunits arranged in the order ABC and that is designated as pmoABC3 cluster. In order to identify the evolutionary relations among these three gene clusters, a multiple alignment of each of their proteins was performed using the software MUSCLE (Edgard, 2004). Figure 2c shows that the proteins in the pmoABC3 cluster are more evolutionary distant than those in the two other clusters (pmoCAB1 and 2). A BLAST search revealed that the pmoABC3 cluster can be also found in M. hirsuta SB2 (Vorobev et al., 2014) with a 96% nucleotide identity. M. hirsuta SB2 actually contains all the 3 pMMO clusters discussed previously. Finally, the catalytic sites of all the pMMO subunits in both clusters are well conserved, which suggests the existence of two active pMMOs that could be tailored to work under different environmental conditions.
The genome sequence contained two malyl‐CoA lyases (D1030_00725 and D1030_16225), which suggests that M. hirsuta CSC1 uses the serine cycle to assimilate C1 compounds, similarly to most type II methanotrophs (Hanson & Hanson, 1996).
A complete nif operon (involved in nitrogen fixation) was found (with genomic coordinates 818892–827543). The operon contained the nifH, D, K, E, N, and X genes. This suggests that M. hirsuta CSC1 was able to fix atmospheric nitrogen similarly to Methylocystis sp. SC2 (Dam, Dam, Blom, & Liesack, 2013). M. hirsuta CSC1 contained also genes involved in denitrification. However, no nitrous oxide reductases were annotated (in contrast to Methylocystis sp. SC2 that contains an operon with six genes involved in N2O reduction). Therefore, M. hirsuta CSC1 possesses the potential to perform only a partial denitrification, with an associated production of N2O under anaerobic conditions.
CONFLICT OF INTEREST
The authors declare not to have any conflict of interest.
AUTHORS CONTRIBUTION
SB performed the bioinformatics analysis and wrote the manuscript. ER carried out the microbial cultures and sample preparation. RM conceived and supervised the work. All the authors edited the manuscript.
ETHICS STATEMENT
None required.
ACKNOWLEDGEMENTS
This work was performed with the support of the Marie Curie grant H2020‐MSCA‐IF‐2016 CH4BioVal (GA no. 750126). The financial support of the Regional Government of Castilla y León (UIC 71), the FEDER Program and the Ministry of Science, Innovation and Universities (CTM2015‐70442‐R) are gratefully acknowledged.
Bordel S, Rodríguez E, Muñoz R. Genome sequence of Methylocystis hirsuta CSC1, a polyhydroxyalkanoate producing methanotroph. MicrobiologyOpen. 2019;8:e771 10.1002/mbo3.771
DATA ACCESSIBILITY
This whole genome shotgun project has been deposited at DDBJ/ENA/GenBank under the accession number QWDD00000000 (BioProject: PRJNA487728).
REFERENCES
- Baani, M. , & Liesack, W. (2008). Two isozymes of particulate methane monooxygenase with different methane oxidation kinetics are found in Methylocystis sp. strain SC2. Proceedings of the National Academy of Sciences USA, 105, 10203–10208 10.1073/pnas.0702643105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin, K. , Wolf, T. , Chevrette, M. G. , Lu, X. , Schwalen, C. J. , Kautsar, S. A. , … Medema, M. H. (2017). antiSMASH 4.0‐improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Research, 45, W36–W41. 10.1093/nar/gkx319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin, C. S. , Alexander, D. H. , Marks, P. , Klammer, A. A. , Drake, J. , Heiner, C. , … Korlach, J. (2013). Nonhybrid, finished microbial genome assemblies from long‐read SMRT sequencing data. Nature Methods, 10, 563–569. 10.1038/nmeth.2474 [DOI] [PubMed] [Google Scholar]
- Dam, B. , Dam, S. , Blom, J. , & Liesack, W. (2013). Genome analysis coupled with physiological studies reveals a diverse nitrogen metabolism in Methylocystis sp. strain SC2. PLoS One, 8, e74767 10.1371/journal.pone.0074767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dam, B. , Dam, S. , Kube, M. , Reinhardt, R. , & Liesack, W. (2012). Complete genome sequence of Methylocystis sp. strain SC2, an aerobic methanotroph with high affinity methane oxidation potential. Journal of Bacteriology, 194, 6008–6009. 10.1128/JB.01446-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai, M. , & Harvey, R. P. (2017). Inventory of U.S. Greenhouse gas emissions and sinks: 1990–2015. Federal Register, 82, 10767 10.1002/yd.370 [DOI] [Google Scholar]
- Edgard, R. C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 32, 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García‐Pérez, T. , López, J. C. , Passos, F. , Lebrero, R. , Revah, S. , & Muñoz, R. (2018). Simultaneous methane abatement and PHB production by Methylocystis hirsuta in a novel gas‐recycling bubble column bioreactor. Chemical Engineering Journal, 334, 691–697. 10.1016/j.cej.2017.10.106 [DOI] [Google Scholar]
- Hanson, R. S. , & Hanson, T. E. (1996). Methanotrophic bacteria. Microbiological Reviews, 60, 439–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebrero, R. , Lopez, J. C. , Lehtinen, I. , Perez, R. , Quijano, G. , & Muñoz, R. (2016). Exploring the potential of fungi for methane abatement: Performance evaluation of a fungal‐bacterial biofilter. Chemosphere, 144, 97–106. 10.1016/j.chemosphere.2015.08.017 [DOI] [PubMed] [Google Scholar]
- Lidner, A. S. , Pacheco, A. , Aldrich, H. C. , Costello Staniec, A. , Uz, I. , & Ogram, A. V. (2007). Methylocystis hirsuta sp.nov., a novel methanotroph isolated from a groundwater aquifer. International Journal of Systematic and Evolutionary Microbiology, 57, 1891–1900. 10.1099/ijs.0.64541-0 [DOI] [PubMed] [Google Scholar]
- Mokhtari‐Hosseini, Z. B. , Vasheghani‐Farahani, E. , Heidarzadeh‐Vazifekhoran, A. , Shojaosadati, S. A. , Karimzadeh, R. , & Darani, K. K. (2009). Statistical media optimization for growth and PHB production from methanol by a methylotrophic bacterium. Bioresource Technology, 8, 2436–2443. 10.1016/j.biortech.2008.11.024 [DOI] [PubMed] [Google Scholar]
- Overbeek, R. , Olson, R. , Pusch, G. D. , Olsen, G. J. , Davis, J. J. , Disz, T. , … Stevens, R. (2014). The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Research, 42, D206–D214. 10.1093/nar/gkt1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieja, A. J. , Morse, M. C. , & Cal, A. J. (2017). Methane to bioproducts: The future of the bioeconomy? Current Opinion in Chemical Biology, 41, 123–131. 10.1016/j.cbpa.2017.10.024 [DOI] [PubMed] [Google Scholar]
- Pieja, A. J. , Rostowski, K. H. , & Criddle, C. S. (2011). Distribution and selection of poly‐3‐hydroxybutyrate production capacity in methanotrophic proteobacteria. Microbial Ecology, 62, 564 10.1007/s00248-011-9873-0 [DOI] [PubMed] [Google Scholar]
- Richter, M. , & Rosselló‐Móra, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proceedings of the National Academy of Sciences USA, 106, 19126–19131. 10.1073/pnas.0906412106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter, M. , Rosselló‐Móra, R. , Glöckner, F. O. , & Peplies, J. (2015). JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics, 32, 929–931. 10.1093/bioinformatics/btv681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine, M. , Tanikawa, S. , Omata, S. , Saito, M. , Fujisawa, T. , Tsukatani, N. , … Harayama, S. (2006). Sequence analysis of three plasmids harboured in Rhodococcus erythropolis strain PR4. Environmental Microbiology, 8, 334–346. 10.1111/j.1462-2920.2005.00899.x [DOI] [PubMed] [Google Scholar]
- Tatusova, T. , Dicuccio, M. , Badretdin, A. , Chetvernin, V. , Nawrocki, E. P. , Zalavsky, L. , … Ostell, J. (2016). NCBI procaryotic annotation pipeline. Nucleic Acids Research, 44, 6614–6624. 10.1093/nar/gwk569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobev, A. , Jagadevan, S. , Jain, S. , Anantharaman, K. , Dick, G. J. , Vuilleumier, S. , & Semrau, J. D. (2014). Genomic and transcriptomic analyses of the facultative methanotroph Methylocystis sp. strain SB2 grown on methane or ethanol. Applied and Environment Microbiology, 80, 3044–3052. 10.1128/AEM.00218-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This whole genome shotgun project has been deposited at DDBJ/ENA/GenBank under the accession number QWDD00000000 (BioProject: PRJNA487728).


