Summary
Maturity-onset diabetes of the young 1 (MODY1) is a monogenic diabetes condition caused by heterozygous HNF4A mutations. We investigate how HNF4A haploinsufficiency from a MODY1/HNF4A mutation influences the development of foregut-derived liver and pancreatic cells through differentiation of human induced pluripotent stem cells from a MODY1 family down the foregut lineage. In MODY1-derived hepatopancreatic progenitors, which expressed reduced HNF4A levels and mislocalized HNF4A, foregut genes were downregulated, whereas hindgut-specifying HOX genes were upregulated. MODY1-derived hepatocyte-like cells were found to exhibit altered morphology. Hepatic and β cell gene signatures were also perturbed in MODY1-derived hepatocyte-like and β-like cells, respectively. As mutant HNF4A (p.Ile271fs) did not undergo complete nonsense-mediated decay or exert dominant negativity, HNF4A-mediated loss of function is likely due to impaired transcriptional activation of target genes. Our results suggest that in MODY1, liver and pancreas development is perturbed early on, contributing to altered hepatic proteins and β cell defects in patients.
Subject Areas: Molecular Mechanism of Gene Regulation, Diabetology, Stem Cells Research, Developmental Biology
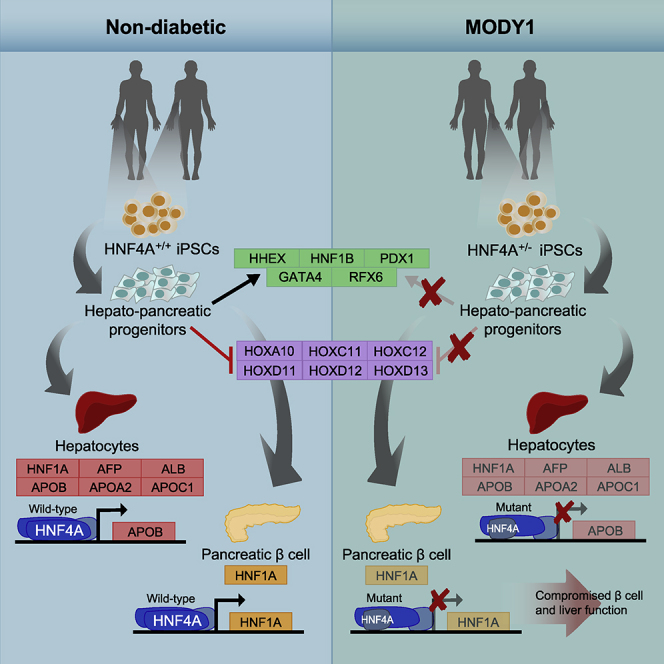
Graphical Abstract
Highlights
-
•
HNF4A is downregulated and predominantly mislocalized in the cytoplasm in MODY1
-
•
Foregut markers, pancreatic and hepatic genes, were downregulated in MODY1-HPPs
-
•
A reciprocal upregulation of hindgut HOX genes was observed in MODY1-HPPs
-
•
Mutant HNF4A resulted in loss of transcriptional activation of target genes
Molecular Mechanism of Gene Regulation; Diabetology; Stem Cells Research; Developmental Biology
Introduction
Maturity-onset diabetes of the young 1 (MODY1) is an autosomal dominant monogenic diabetes condition typically manifesting before the age of 25 years. This condition is caused by inactivating mutations in the hepatocyte nuclear factor 4A (HNF4A) gene (Yamagata et al., 1996) and is characterized by defects in glucose-stimulated insulin secretion (GSIS) from the pancreatic β cells (Byrne et al., 1995). Owing to the inaccessibility of human pancreatic tissue, rodent models have traditionally been used to study the molecular mechanisms underlying MODY1. Unfortunately, conditional knockout of Hnf4a in mouse pancreatic β cells did not result in a diabetic phenotype, although GSIS is impaired (Boj et al., 2010, Gupta et al., 2005, Miura et al., 2006). More importantly, Hnf4a+/- mice exhibit normal glucose tolerance (Stoffel and Duncan, 1997), indicating that rodent models do not accurately recapitulate the MODY1 phenotype in humans. Human induced pluripotent stem cell (hiPSC)-based disease modeling strategies (Teo et al., 2013a) therefore provide opportunities to investigate the impact of MODY1/HNF4A mutation on the development of the foregut lineage in humans. In particular, the ventral foregut endoderm gives rise to progenitors that subsequently form the liver, whose development and function is heavily dependent on regulation by HNF4A, or the pancreatic β cells, which are known to be implicated in MODY1 pathophysiology.
HNF4A is a member of the steroid hormone receptor superfamily and functions as a transcription factor upon homodimerization (Sladek et al., 1990). Its expression is regulated by either the P1 (proximal) or P2 (distal) promoter. The usage of alternate promoters and presence of alternative splicing results in up to 12 known HNF4A isoforms that are expressed in a developmental stage- and tissue-specific manner (Eeckhoute et al., 2003a, Harries et al., 2008, Huang et al., 2009, Jiang et al., 2003, Tanaka et al., 2006). Therefore HNF4A expression is dynamically regulated to ensure proper formation and function of multiple organs, in particular, the liver and pancreas (Lau et al., 2018), which are the tissues we focus on in our study.
Knockout of Hnf4a in mice is dispensable for early development of the liver, whereas it is required for driving hepatic specification at later stages and in maintaining proper liver function (Li et al., 2000). In an early human pluripotent stem cell differentiation study, HNF4A was found to be necessary for establishing the hepatic gene regulatory network and induction of hepatic cell fate (DeLaForest et al., 2011). This correlates with the observation that patients with an inactivating HNF4A mutation exhibit alterations in liver function (Gardner and Tai, 2012, Pearson et al., 2005, Shih et al., 2000). In addition to the liver, Hnf4a is also expressed in the maturing pancreas in mice and is largely confined to the developing islet and acinar cells (Nammo et al., 2008). A recent study showed that MODY1/HNF4A mutation does not prevent formation of INS+ cells from in vitro differentiations (Vethe et al., 2017). Nonetheless, the molecular and transcriptional impacts of heterozygous HNF4A mutation on early foregut endoderm, liver, and pancreas development leading to disease onset in humans remain largely unexplored.
We hypothesized that the MODY1/HNF4A mutation affects early human foregut development that can potentially lead to both liver and pancreas developmental defects. To circumvent the lack of access to human tissues during early development, we generated hiPSCs from members of a MODY1 family (with and without heterozygous HNF4A mutation) and differentiated them into hepatopancreatic foregut endoderm (henceforth termed hepatopancreatic progenitors [HPPs]), as well as hepatic and pancreatic β-like cells using independent, established protocols. Our data indicate that HNF4A haploinsufficiency, as a result of a loss-of-function MODY1 mutation, affects early human foregut development and that this deficiency is propagated to both hepatic and pancreatic cell fates. Our human disease model provides a platform for investigating why patients with MODY1 have specific hepatic and β cell developmental defects.
Results
Establishing a MODY1 Disease Model Using Patient-Derived iPSCs
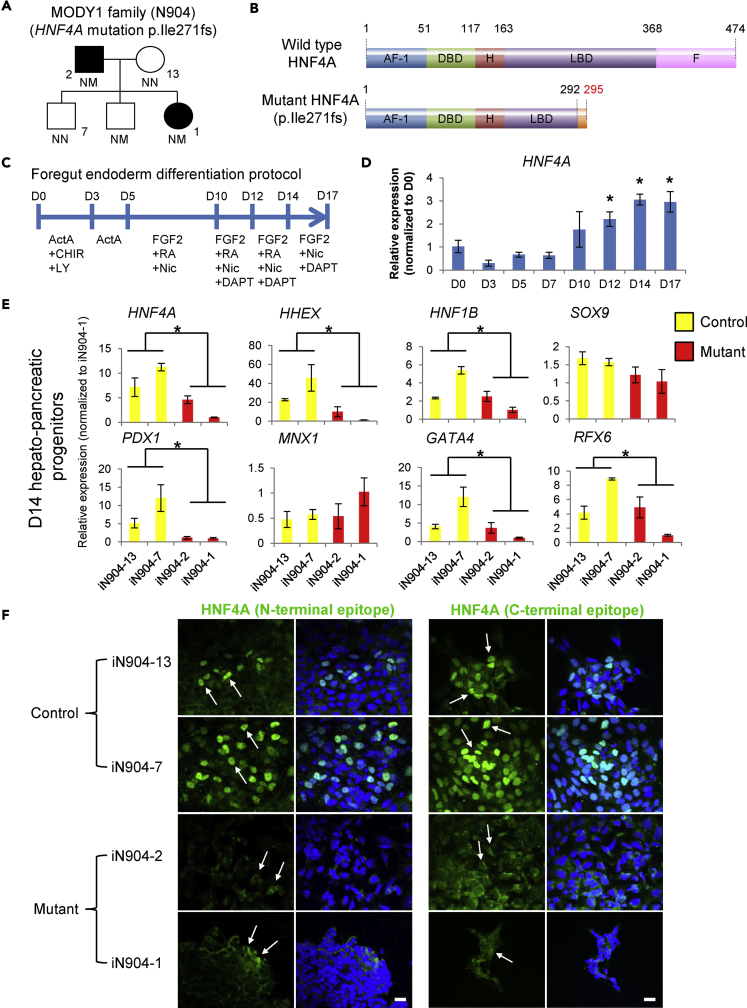
We previously reported the recruitment of two members of a MODY1 family harboring a heterozygous p.Ile271fs mutation in HNF4A (Figure 1A) (Teo et al., 2013b) resulting in premature truncation of the protein (Figure 1B). To facilitate rigorous and comprehensive hiPSC-based MODY1 disease modeling, we recruited more members of the same family and rederived a total of nine hiPSC lines composed of MODY1-hiPSCs from two patients (iN904-2 and iN904-1A/B/C) and control-hiPSCs from two non-diabetic family members (iN904-13A/B and iN904-7A/B/C) (Figures 1A and S1). Using a previously published 17-day foregut endoderm differentiation protocol, we observed that HNF4A expression peaked at day 14 (D14) (Figures 1C and 1D) (Teo et al., 2015b, Teo et al., 2016), and that ∼70% of D14 HPPs were HNF4A+ (Figure S2A), thereby providing a suitable model for studying HNF4A gene function and disease mechanisms underlying MODY1.
Figure 1.
HNF4A Mutation (p.Ile271fs) Causes Impaired Foregut/Early Hepatopancreatic Progenitor (HPP) Development
(A) MODY1 family node showing non-diabetic control-hiPSCs (iN904-13 and iN904-7) and MODY1-hiPSCs (iN904-2 and iN904-1).
(B) p.Ile271fs mutation results in C-terminally truncated HNF4A that lacks part of the ligand-binding domain (LBD) and the entire F repressor domain (not drawn to scale).
(C) The 17-day differentiation protocol for generating foregut endoderm and HPPs.
(D) qPCR analysis of HNF4A expression during HPP differentiation.
(E) qPCR analyses of HNF4A transcripts and foregut endoderm markers such as HHEX, HNF1B, PDX1, GATA4, and RFX6 in control and MODY1-HPPs.
(F) Immunofluorescent confocal images showing the localization of HNF4A protein in control and MODY1-HPPs, based on antibodies targeting the N- or C-terminal regions of HNF4A. Blue, DAPI; green, HNF4A; scale bars, 50 μm. White arrows point to the nuclear or cytoplasmic localization signal of HNF4A. Confocal images were acquired using similar scan settings across samples.
Data are represented as mean ± SD of n = 3; representative of three independent experiments. *p < 0.05 versus D0 or control samples by Student's t test. See also Figures S1 and S2.
HNF4A Mutation (p.Ile271fs) Causes Impaired Foregut/Early HPP Development
To elucidate the effects of the p.Ile271fs mutation, we simultaneously differentiated control- and MODY1-hiPSCs into HPPs. Both control- and MODY1-hiPSCs were able to differentiate into definitive endoderm cells at day 3 of differentiation (Figure S2B). At D14 of differentiation, although we observed no obvious morphological differences between control- and MODY1-HPPs (Figure S2C), the MODY1-HPPs expressed significantly lower levels of total HNF4A (Figure 1E). In fact, wild-type (WT) HNF4A protein was expressed at markedly lower levels in MODY1-HPPs based on protein expression data despite the presence of one copy of the WT allele at HNF4A (Figure S2D). To determine if P1- or P2-driven HNF4A transcripts were affected, we carried out isoform-specific qPCR analyses and showed that both P1- and P2-driven forms of HNF4A are potentially affected in the D14 HPPs (Figure S2E).
We further detected lower levels of foregut endoderm genes HHEX, HNF1B, PDX1, GATA4, and RFX6 in the MODY1-HPPs, whereas no differences were observed for other pancreas-related genes SOX9 or MNX1 (Figure 1E), reflecting a downregulation of specific gene targets of HNF4A affected by the p.Ile271fs mutation rather than a global downregulation of pancreatic developmental genes. Downregulation of PDX1 and GATA4 was confirmed at protein level by immunofluorescence staining (Figure S2F). Subsequent immunofluorescence analyses additionally revealed that HNF4A protein is largely sequestered in the cytoplasm of the MODY1-HPPs as opposed to the predominant nuclear localization observed in control-HPPs (Figure 1F). Mislocalization of the HNF4A protein could further account for the loss of its function as a transcription factor.
RNA Sequencing Analyses Reveal Downregulation of Pancreas- and Liver-Related Genes and Upregulation of Caudal HOX Genes
To thoroughly evaluate the genome-wide effects of the MODY1 mutation on foregut development, we performed RNA sequencing analyses on control and MODY1-hiPSC-derived D14 HPPs. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses revealed that genes involved in MODY and numerous liver functions were significantly downregulated (Figure 2A), consistent with the known functions of HNF4A target genes (Bolotin et al., 2010, Odom et al., 2004). The affected genes were involved in processes related to steroid metabolism and lipoprotein and sterol binding and transport (Figure S3A), providing clues to the role of HNF4A target genes. On the other hand, genes involved in DNA binding, transcription factor, and channel activity were upregulated, possibly due to compensatory regulatory mechanisms (Figure S3B).
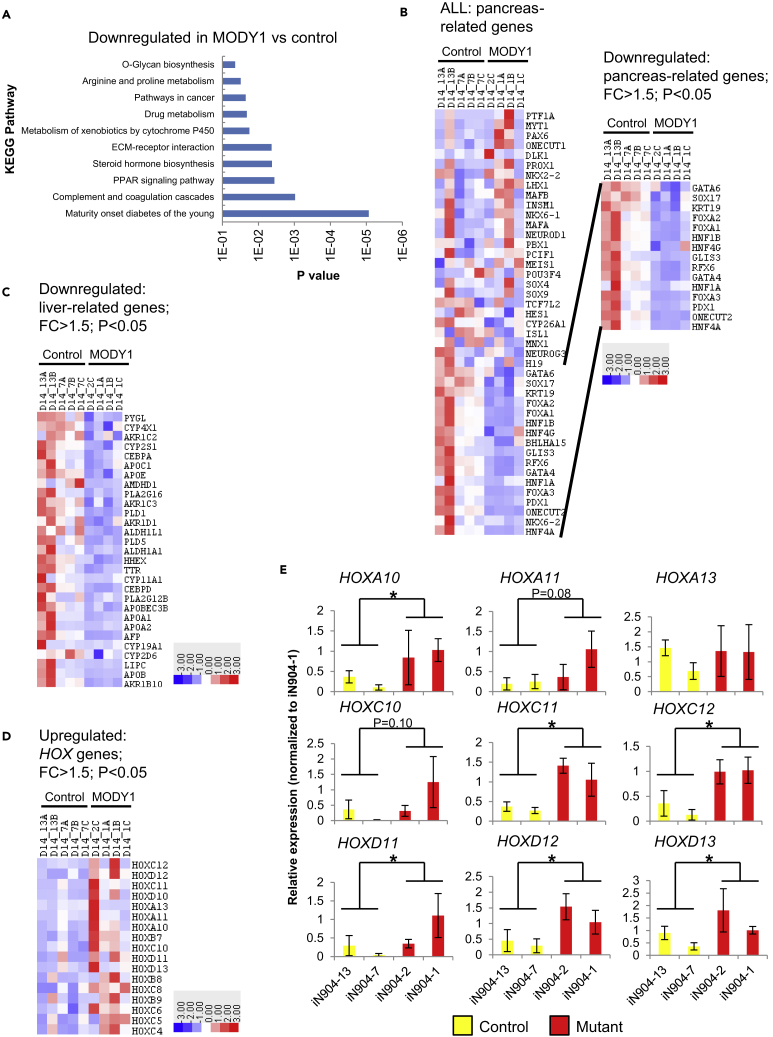
Figure 2.
RNA Sequencing Analyses Reveal Global Transcriptional Changes Induced by the HNF4A Mutation in MODY1-HPPs at D14
(A–C) (A) Analysis of downregulated genes via the KEGG pathway. Heatmap analyses of (B) pancreas-related and (C) liver-related genes that are downregulated with fold change (FC) > 1.5, p < 0.05.
(D) Heatmap analyses of numerous caudal HOX genes involved in hindgut specification that are upregulated with FC > 1.5, p < 0.05. Colors in the heatmap depict gene expression in units of SD from the mean across all samples (upregulation in red, downregulation in blue). Controls indicated as 13A, 13B, 7A, 7B, and 7C; MODY1 indicated as 2, 1A–1C.
(E) qPCR analyses of caudal HOX gene expression in control and MODY1-HPPs.
Data are represented as mean ± SD of n = 3, representative of four independent experiments. *p < 0.05 versus control samples by Student's t test. See also Figure S3 and Table S1.
Heatmap analyses revealed that a subset of pancreas-related genes was downregulated in MODY1-HPPs (fold change [FC] > 1.5; p < 0.05) (Figure 2B), including PDX1, the FOXA gene family, GATA4, RFX6, HNF1B, KRT19, and SOX17. In addition, numerous hepatic genes such as the apolipoprotein (APO) genes, AFP, TTR, and HHEX were also downregulated in the MODY1-HPPs (Figure 2C), consistent with findings from Hnf4a−/− mice (Li et al., 2000). In contrast, we were intrigued to observe an upregulation of numerous caudal HOX genes including HOXA10, HOXC11, HOXC12, HOXD11, HOXD12, and HOXD13 in the MODY1-HPPs (Figures 2D, 2E, and S3C). Although some of the changes were modest, likely due to the fact that the HPP protocol is suited for foregut, but not hindgut, differentiation, the trend toward increased levels of hindgut markers suggests a potential switch away from foregut specification. Indeed, these caudal HOX genes are typically upregulated only in differentiation conditions favorable for hindgut formation (high fibroblast growth factor 2 concentration) (Ameri et al., 2010) in which hindgut marker CDX2 is upregulated, but not foregut marker HNF4A (Figure S3D). The loss of the repressor domain in HNF4A owing to the p.Ile271fs-truncating mutation may account for this “derepression” phenomenon.
MODY1-Mediated Loss of HNF4A Transcriptional Function Affects Subsequent Hepatic and Pancreatic Development Signatures
Following the HNF4A loss-of-function observations in the HPPs, which are representative of a progenitor stage, we next investigated impacts on subsequent tissue development. We used established differentiation protocols that aimed to direct the differentiation of the hiPSCs into hepatocytes or pancreatic β-like cells (Hannan et al., 2013, Pagliuca et al., 2014), as these are more representative of the liver and β cell differentiation process. First, time course differentiation of control-hiPSCs into hepatocyte-like cells (Hannan et al., 2013) revealed that HNF4A expression peaked on day 8 (D8), when 70%–80% HNF4A+ cells may be obtained, whereas other hepatic genes displayed peak expression on days 16 (D16) or 24 (D24) (Figures 3A and S4A).
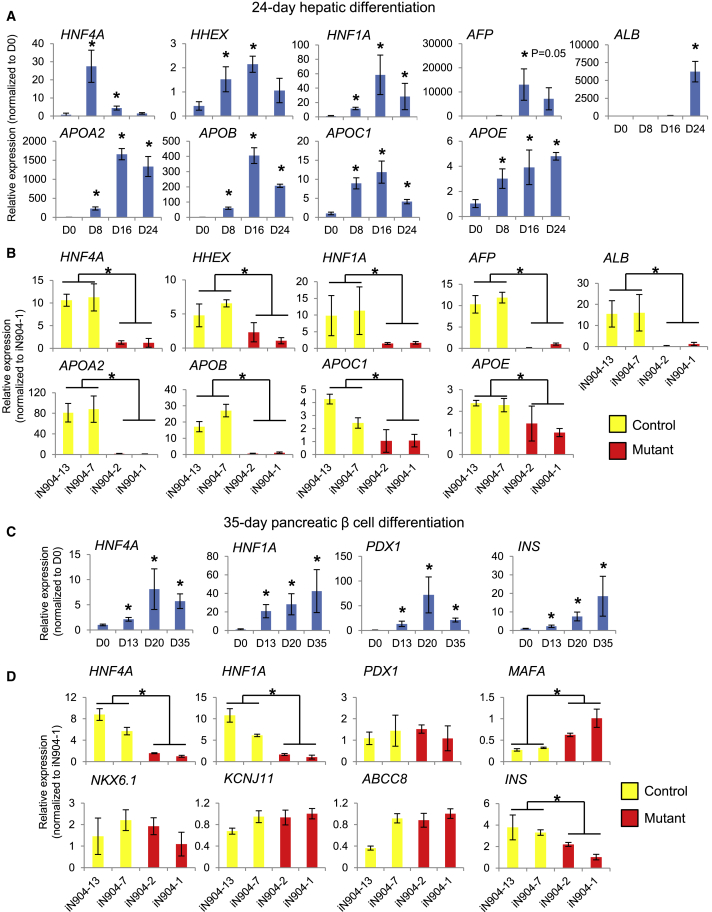
Figure 3.
MODY1-Mediated HNF4A Haploinsufficiency in Early Foregut Development Affects Subsequent Hepatic and Pancreatic Development Signatures
(A and B) (A) qPCR analyses showing pattern of hepatic gene expression over a 24-day differentiation in control-hiPSC-derived hepatic cells and (B) comparison of hepatic gene expression changes between control and MODY1 hepatic cells.
(C and D) (C) qPCR analyses showing pattern of pancreatic β cell gene expression over a 35-day differentiation in control-hPSC-derived pancreatic β-like cells and (D) comparison of β cell gene expression changes between control and MODY1 pancreatic β-like cells.
Data are represented as mean ± SD of n = 3, representative of three independent experiments. *p < 0.05 versus control samples by Student's t test. See also Figure S4.
During hepatic differentiation, we noted that control-hiPSCs formed polygonal hepatocyte-like cells, whereas MODY1-hiPSCs did not (Figure S4B). In addition, MODY1 hepatic progenitors expressed significantly lower levels of HNF4A and HHEX on D8, leading to reduced expression of HNF1A and hepatoblast marker AFP on D16, and finally reduced levels of key mature hepatocyte markers APOA2, APOB, APOC1, APOE, and ALB on D24 as opposed to control cells (Figure 3B). Residual HNF4A protein expressed in the MODY1 hepatic progenitors appeared to localize largely to the cytoplasm when compared with the controls (Figure S4), mirroring our earlier observations in the HPPs (Figure 1F). These data indicated that the early HNF4A loss-of-function effects (despite having a WT HNF4A allele) propagated into longer-term consequences that affected subsequent hepatic differentiation. This is consistent with the effects of shRNA-mediated knockdown of HNF4A in hPSCs that was reported previously (DeLaForest et al., 2011), although it is worth noting that a complete knockout of HNF4A in humans does not exist naturally.
We next investigated the impact of MODY1/HNF4A mutation on pancreatic β cell development using a published protocol for generating pancreatic β-like cells (Pagliuca et al., 2014). In differentiated WT β-like cells, HNF4A expression increased progressively over 35 days, together with other critical β cell transcripts such as HNF1A, PDX1, and INS (Figure 3C). Again, no difference was detected between the specification of control and MODY1 iPSCs into definitive endoderm cells, before the rise in HNF4A expression (Figure S4D). However, in MODY1 β-like cells both HNF4A and HNF1A were significantly downregulated, although this was not the case for a number of other β cell genes tested at D35 (Figure 3D). Despite some reduction in INS transcript levels in MODY1-derived β-like cells (Figure 3D), we could detect the expression of C-peptide in both control- and MODY1-derived β-like cells (Figure S4E). To corroborate the results observed in the MODY1-HPPs (Figure 1E), we also assessed gene expression changes in the pancreatic progenitors generated using the 35-day β cell differentiation protocol. We observed that the expression of PDX1 and other progenitor markers was indeed reduced in the D13 pancreatic progenitors (Figure S4F), although PDX1 reduction at protein level was not always consistently observed (Figure S4G). We postulate that the early perturbations may be less apparent later in the differentiation owing to the exogenous stimuli that drive the differentiation of PDX1- and INS-expressing cells during β cell differentiation. Nonetheless, it was clear from our multiple differentiated cell models that loss in both HNF4A and HNF1A function in early hepatic and β cell development may contribute to the impaired tissue function in MODY1.
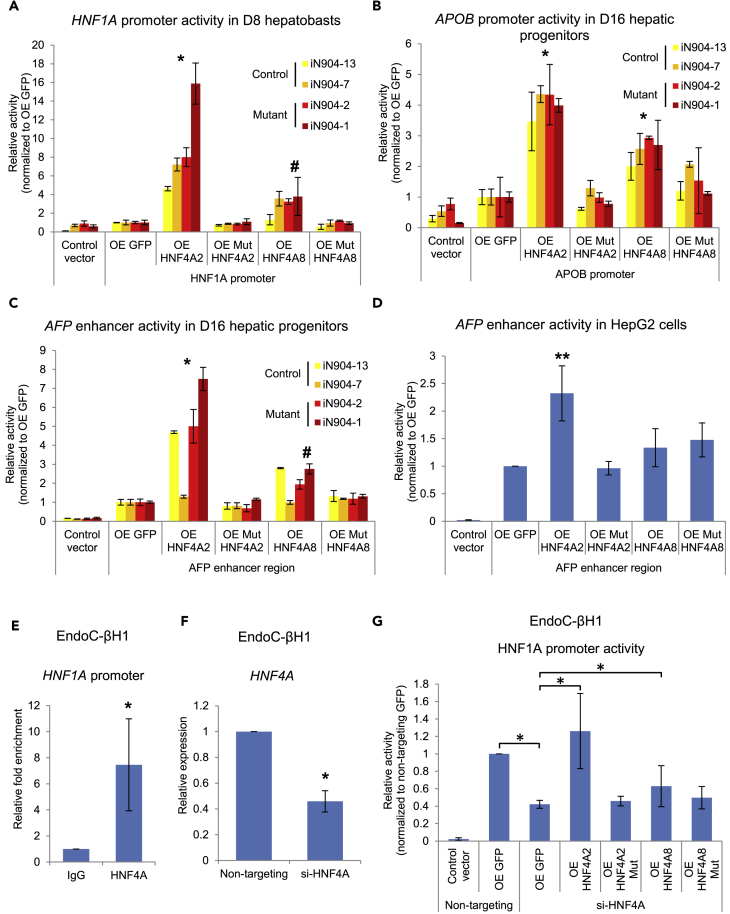
We then sought to define the molecular impact of HNF4A p.Ile271fs on its downstream targets including HNF1A, by evaluating the transcriptional potential of WT and mutant HNF4A in our hiPSC-derived hepatic differentiation models. As both P1- and P2-driven transcripts are present during foregut development (Figure S2E), the effects of both WT and mutant HNF4A2 and HNF4A8 (longest isoforms representative of P1- and P2-driven expression, respectively) were evaluated. When expressed in the D8 hepatic progenitors, WT HNF4A2 significantly activated HNF1A promoter activity, whereas mutant HNF4A did not elicit the same effect (Figure 4A). Similarly, in the D16 hepatic progenitors, WT but not mutant HNF4A2 resulted in activation of the APOB promoter and AFP enhancer (containing a HNF4A-binding motif) (Nakabayashi et al., 2004) (Figures 4B and 4C). The activation of the AFP enhancer by WT, but not mutant HNF4A2, was further replicated in HepG2 cells (Figure 4D), an AFP-producing human hepatoma cell line (Kawai et al., 2001). The lack of activation by mutant HNF4A may be explained in part by the reduced protein expression levels of mutant HNF4A compared with WT, although both HNF4A2/8 WT and mutants localized to the nuclei in the overexpression studies (Figures S5A and S5B). In all experiments, WT HNF4A8 exhibited a weaker transactivation potential when compared with HNF4A2, and in the case of the HNF1A promoter and AFP enhancer, the effect was only significant in MODY1-derived cells where endogenous HNF4A function is reduced (Figures 4A and 4C).
Figure 4.
MODY1/HNF4A Mutation Results in Loss of Ability to Activate Downstream Target Promoters in Hepatic and Pancreatic β Cells
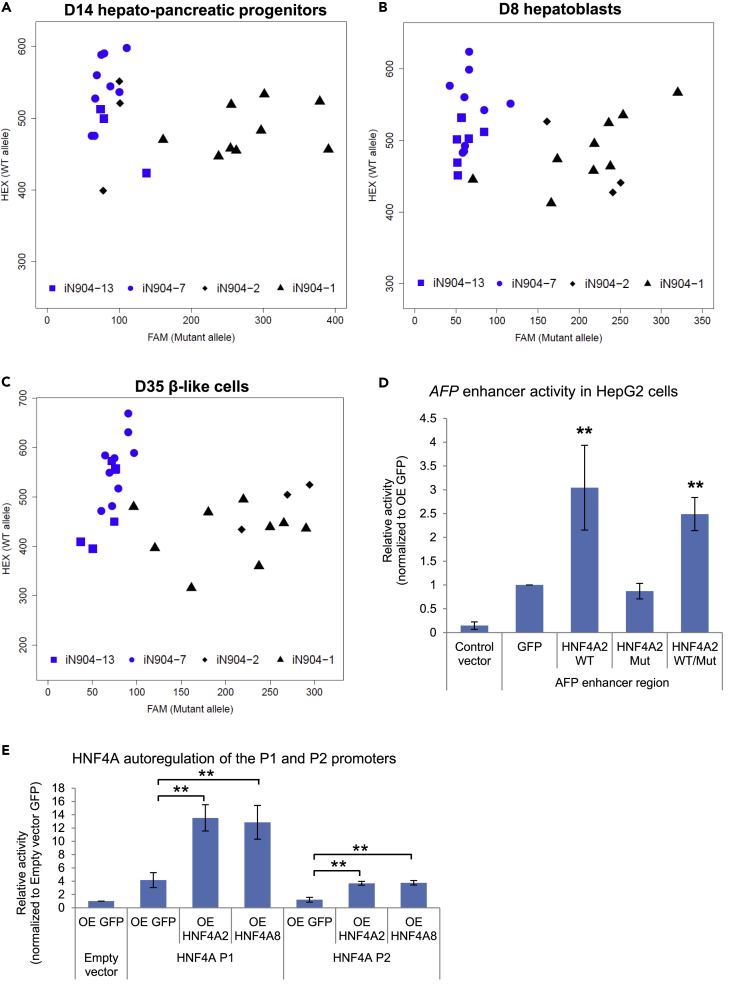
(A–D) Luciferase assays were performed to evaluate effects of WT or p.Ile271fs (Mut) HNF4A on the (A) HNF1A promoter, (B) APOB promoter, (C) AFP enhancer activity in hiPSC-derived hepatic cells, or (D) HepG2 cells. For (A–C), data are represented as mean ± SD n = 3, representative of two independent experiments. For (D), data are represented as mean ± SD of n = 12 from three independent experiments. *p < 0.05 versus GFP control in all hiPSC lines by two-way ANOVA; #p < 0.05 versus GFP control in mutant hiPSC lines only by two-way ANOVA. **p = 0.01 versus GFP control by Student's t test.
(E) Chromatin immunoprecipitation qPCR analysis of HNF4A binding onto HNF1A promoter in EndoC-βH1 cells.
(F) Small interfering RNA-mediated knockdown of HNF4A in EndoC-βH1 cells.
(G) Luciferase assay evaluating HNF1A promoter activity upon knockdown of HNF4A and rescue. For (E–G), data are represented as mean ± SD of n = 12 from three to four independent experiments. *p < 0.05 versus IgG/GFP control as indicated by Student's t test.
See also Figure S5.
We further set out to investigate the regulation of HNF1A by HNF4A WT or mutants in human pancreatic β cells. As the suspension cell clumps generated from the pancreatic β cell differentiation were less amenable for in vitro assays unlike monolayer differentiation cultures, we used the human β cell line EndoC-βH1. Chromatin immunoprecipitation analyses in EndoC-βH1 showed that HNF4A was bound to the HNF1A promoter (Figure 4E), and subsequent knockdown of HNF4A by ∼55% resulted in a corresponding reduction in HNF1A promoter activation (Figures 4F and 4G). This effect could be rescued by WT HNF4A overexpression but not mutant HNF4A (Figure 4G). Overall, we have shown that HNF4A directly regulates the transcription of key components of developing hepatic and pancreatic β cells, and that the MODY1/HNF4A mutation (p.Ile271fs) results in the inability to activate target promoters. Our patient-derived iPSC-based model thus provides an in vitro platform for the interrogation of the underlying disease mechanisms in the hepatic and β cells.
MODY1 hiPSC-Derived Cells Express Both WT and Mutant HNF4A Transcripts and Do Not Exhibit Dominant Negativity
Finally, we sought to address the question of why the decrease in total HNF4A levels in MODY1-derived cells is beyond the expected 2-fold change given the presence of a WT allele in heterozygote carriers. We first determined the expression of WT and mutant HNF4A (p.Ile271fs) transcripts using a custom-designed allele-specific assay (Figure S5C). As observed across multiple differentiated cell types, both WT and mutant HNF4A transcripts are expressed in the MODY1-derived cells, confirming heterozygosity at mRNA level (Figures 5A–5C). The detection of mutant transcripts indicated that there is an absence of complete nonsense-mediated decay (NMD) of the nonsense mutant transcripts (Zhang et al., 2009). Next, we checked if the MODY1 mutation could result in a dominant negative effect by co-expressing WT and mutant HNF4A to recapitulate a heterozygous condition. Gene regulatory assays showed that WT HNF4A was able to activate AFP enhancer activity normally in the presence of mutant protein, suggesting a lack of dominant negativity (Figure 5D). Given that HNF4A is also known to occupy its own promoter (Bolotin et al., 2010, Odom et al., 2004), we performed further gene regulatory assays involving both the HNF4A P1 and P2 promoters and demonstrated that HNF4A can activate both promoters and subsequently its own expression in a feedforward manner (Figure 5E). Therefore loss of HNF4A function or mislocalization may result in failure to undergo autoregulation, accounting for overall reduced HNF4A expression in MODY1.
Figure 5.
MODY1 hiPSC-Derived Cells Express Both WT and Mutant HNF4A Transcripts
(A–C) Allele-specific qPCR analyses in (A) D14 HPPs, (B) D8 hepatoblasts, and (C) D35 β-like cells evaluating both WT and mutant HNF4A transcripts in MODY1-derived cells. Axes show relative fluorescence units for each allele-specific TaqMan probe, for a representative differentiation experiment.
(D) Luciferase assays were performed to evaluate effects of WT and mutant HNF4A in combination with AFP enhancer activity in HepG2 cells.
(E) Luciferase assays were performed to evaluate effects of HNF4A2 and HNF4A8 on HNF4A P1 and P2 promoter activities in Ad293 cells.
Data are represented as mean ± SD of n = 4 independent experiments. **p < 0.01 versus GFP control unless otherwise indicated by Student's t test. See also Figure S5.
Discussion
Earlier studies have reported HNF4A nonsense or missense gene mutations leading to either a loss-of-function or a dominant negative effect (Laine et al., 2000, Lausen et al., 2000, Sladek et al., 1998). The p.Ile271fs mutation in our study introduces a frameshift and premature stop codon, which could lead to generation of unstable mRNA that may to some degree be degraded by NMD (Frischmeyer and Dietz, 1999), accounting for the overall lowered HNF4A levels in MODY1-hiPSC-derived cells. However, we did not observe complete NMD given that mutant transcripts were detectable in our MODY1-derived cells. At protein level, crystallographic studies have reported that several key residues of the ligand-binding domain are involved in charge-driven interactions that improve dimerization. Mutations in this region such as p.Ile271fs may therefore affect the formation of functional homodimers and impair DNA-binding activity, thereby abolishing transcriptional activity or coactivator recruitment (Eeckhoute et al., 2003b, Ek et al., 2005, Hani et al., 1998, Stoffel and Duncan, 1997) to affect downstream gene regulation. Our work conclusively showed that loss of HNF4A-mediated gene regulation due to the p.Ile271fs mutation in a heterozygous state in MODY1 affected foregut endoderm gene expression signatures.
Our observations on caudal HOX gene upregulation led us to hypothesize that HNF4A typically functions to suppress ectopic hindgut HOX gene expression to facilitate proper foregut endoderm development, whereas this suppressive effect is disrupted in cells carrying the p.Ile271fs mutation. Further studies are required to determine whether HOX gene derepression is indirect, or if HNF4A requires other co-factors for its repressor function. Future work should also determine whether p.Ile271fs affects the specific interaction of HNF4A with ligands or co-factors important for its function. We propose that the impact of HNF4A haploinsufficiency on the specification of the foregut versus hindgut lineage is cell autonomous given the well-established function of HNF4A as a transcription factor. Nonetheless, the possibility of non–cell autonomy cannot be ruled out as previous studies have provided evidence for non-cell-autonomous functions of homeobox genes and other early developmental genes (Balbinot et al., 2018, Becker et al., 2016). Future experiments that involve fluorescent labeling of the WT and mutant MODY1 hiPSCs followed by differentiation and fluorescence-activated cell sorting analyses may shed light on this.
HNF4A has been reported to be important for rodent hepatocyte development (Li et al., 2000) and is essential for specifying the early hepatic differentiation program (DeLaForest et al., 2011). However, the impact of MODY1/HNF4A mutation on hepatic development in humans has not been explored, given the intractability of human liver tissue. Here, we capitalize on our patient-derived iPSCs and ability to differentiate them into multiple relevant cell types to model cell-type-specific phenotypes and investigate underlying disease mechanisms. Our results indicated that the early loss of HNF4A expression in hepatoblasts propagated to long-term consequences on hepatic cell fate, as seen in the reduced expression of ALB and numerous APO genes. This is consistent with observations that patients with MODY1 with an inactivating HNF4A mutation exhibit reduced secretion of hepatocyte-specific proteins such as APOs (Lehto et al., 1999, Pearson et al., 2005, Shih et al., 2000). Nonetheless, these alterations in liver function may not be clinically significant given the lack of reports on liver deformities or severe liver dysfunction in these patients. Given the potential redundancy within the complex liver transcription factor network that HNF4A is involved in (Lau et al., 2018, Odom et al., 2004), there may be redundant mechanisms such as those involving HNF1A or ONECUT1 in the liver, or other compensatory mechanisms that enable largely normal liver development in vivo (Ober et al., 2006).
We also confirmed that although both P1- and P2-driven HNF4A are expressed during foregut differentiation, the primary isoform(s) that activates expression of key target genes such as HNF1A and AFP is likely encoded by the P1 promoter. These results are consistent with previous reports (Eeckhoute et al., 2003a) that P1-driven isoforms exhibit greater transcriptional potential than their P2-driven counterparts, and we show that this is the case in both human hepatic cells and β cells.
Besides a liver phenotype, patients with MODY1 are known to exhibit progressive β cell insulin secretory defects (Herman et al., 1997, Ryffel, 2001). Our studies provide valuable insights relating to the expression of early pancreatic genes affected by HNF4A haploinsufficiency such as HNF1B, PDX1, GATA4, and RFX6 in the pancreatic progenitors. Although key developmental genes may be perturbed at the progenitor stage, the terminally in vitro-differentiated β-like cells were still able to express select β cell markers and C-peptide. This observation is in line with a previous study also involving MODY1-derived cells (Vethe et al., 2017). However, the study did not provide data from any of the progenitor stages. In both our study and that by Vethe et al., the in vitro differentiations do not generate β-like cells that are functionally mature despite the presence of insulin, therefore the functional capacity of these cells cannot be appropriately elucidated. As HNF4A haploinsufficiency involves a heterozygous mutation, there could be compensatory effects that result in a reset of the regulatory network, therefore patients do not have pancreatic agenesis. Nonetheless, there is a distinctive decrease in HNF1A expression in our MODY1-derived β-like cells. These findings are also consistent with the prevailing notion that MODY1/HNF4A is clinically and genetically linked to MODY3/HNF1A considering that HNF4A directly regulates the expression of HNF1A (Ellard and Colclough, 2006, Lausen et al., 2000). Detailed assessment of how the HNF4A-HNF1A cross-regulatory circuit and downstream transcriptional network is perturbed in both MODY1 and MODY3 may shed further light on the convergent and divergent role of both genes in governing tissue function, especially in human β cells.
It is notable that mutations in HNF4A are not only relevant to MODY1 but also have been associated with the more commonly occurring type 2 diabetes (T2D). Specifically, single nucleotide polymorphisms in both the P2 and P1 promoter regions and those near or within the HNF4A gene have been linked to T2D susceptibility (Damcott et al., 2004, Ek et al., 2005, Hara et al., 2006, Kooner et al., 2011, Love-Gregory et al., 2004, Mahajan et al., 2018, Silander et al., 2004, Weedon et al., 2004). Pancreatic islets isolated from donors with T2D were also found to exhibit reduced HNF4A expression (Gunton et al., 2005). Collectively, we report the successful establishment of a MODY1 hiPSC model with HNF4A haploinsufficiency that arose from a naturally occurring heterozygous mutation. Our findings highlight MODY1-HNF4A as a developmental disease that begins in the foregut endoderm and extends to its derivatives—in particular the liver and the pancreas. Our approach and results will have important implications for the study and understanding of diabetes pathogenesis in the context of MODY and even T2D.
Limitations of the Study
In this study, we have validated our findings across multiple differentiation models that can generate known cell-type-specific markers, as well as established mechanisms in non-iPSC-based cell lines to substantiate our findings of HNF4A haploinsufficiency. However, there are well-recognized limitations of iPSC-based disease models that can affect the interpretation of results. First, the differentiation process is heterogeneous and therefore a bulk analysis approach may result in data with overall increased variability and reduced magnitude of effect. To circumvent this, single-cell studies may be used to interrogate cellular phenotypes at single-cell resolution (Petersen et al., 2017). Second, the use of isogenic controls generated using genome-editing tools may also help to reduce noise when compared with the use of family controls, which are still subject to differences in genetic background (Teo et al., 2015a). Next, directed differentiation protocols rely on the use of a cocktail of small molecules and growth factors to drive the differentiation process in vitro. This assumes that patient cells encounter these signals under an in vivo setting to drive tissue development. Therefore, an in vitro model may not accurately capture disease progression. On the contrary, currently available pancreatic β cell differentiation protocols are often unable to generate functional β cells in vitro and require transplantation into mice for in vivo maturation (Hrvatin et al., 2014, Loo et al., 2018). This hints at yet unknown molecular factors that are required to obtain β cells that can produce and secrete insulin in response to glucose stimuli. Therefore evaluation of the insulin secretory capacity of the MODY1-derived cells in the current differentiation model was not possible. Overcoming a number of these limitations will undoubtedly increase experimental robustness and reproducibility.
Methods
All methods and can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
The authors thank Andreas Alvin Purnomo Soetedjo and Chek Mei Bok for experimental assistance and also thank members of the Teo laboratory for the critical reading of this manuscript. N.H.J.N. is supported by the National Medical Research Council (NMRC) Open Fund-Young Individual Research Grant (OF-YIRG) OFYIRG18May. R.N.K. acknowledges support from National Institutes of Health Grant RO1 067536. H.R. is supported by the Bergen Forskningsstiftelse (BFS), the Western Norway Regional Health Authority, the Novo Nordisk Foundation, and Diabetesforbundet. L.V. is funded by the ERC advanced grant New-Chol, the Cambridge University Hospitals National Institute for Health Research Biomedical Research Centre and the core support grant from the Wellcome Trust and Medical Research Council to the Wellcome–Medical Research Council Cambridge Stem Cell Institute. A.K.K.T. is supported by the Institute of Molecular and Cell Biology (IMCB), A*STAR, A*STAR JCO Career Development Award (CDA) 15302FG148, NMRC OFYIRG16may014, A*STAR ETPL Gap Funding ETPL/18-GAP005-R20H, Lee Foundation Grant SHTX/LFG/002/2018, Skin Innovation Grant SIG18011, NMRC OF-LCG/DYNAMO, FY2019 SingHealth Duke-NUS Surgery Academic Clinical Program Research Support Program Grant, and the Precision Medicine and Personalised Therapeutics Joint Research Grant 2019.
Author Contributions
Conceptualization, A.K.K.T.; Methodology, N.H.J.N., J.B.J., and A.K.K.T.; Formal Analysis, N.H.J.N., J.B.J., and A.K.K.T.; Investigation, N.H.J.N., J.B.J., C.S.L., H.H.L., V.G.K., J.K., S.H., and A.K.K.T.; Resources, H.R., L.V., S.H., and A.K.K.T.; Writing – Original Draft, A.K.K.T.; Writing – Review & Editing, N.H.J.N., J.B.J., R.N.K., H.R., L.V., S.H., and A.K.K.T.; Visualization, N.H.J.N., J.B.J., and A.K.K.T.; Supervision, A.K.K.T.; Project Administration, A.K.K.T.; Funding Acquisition, A.K.K.T.
Declaration of Interests
The authors declare no competing interests.
Published: June 28, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2019.05.032.
Supplemental Information
The first two columns indicate the gene ID and official gene symbol for all protein-coding genes in the dataset. FPKM values are shown for control-hiPSC-derived cells (13A, 13B, 7A, 7B, and 7C) and MODY1-hiPSC-derived cells (2, 1A, 1B, and 1C).
References
- Ameri J., Stahlberg A., Pedersen J., Johansson J.K., Johannesson M.M., Artner I., Semb H. FGF2 specifies hESC-derived definitive endoderm into foregut/midgut cell lineages in a concentration-dependent manner. Stem Cells. 2010;28:45–56. doi: 10.1002/stem.249. [DOI] [PubMed] [Google Scholar]; Ameri, J., Stahlberg, A., Pedersen, J., Johansson, J.K., Johannesson, M.M., Artner, I., and Semb, H.. (2010). FGF2 specifies hESC-derived definitive endoderm into foregut/midgut cell lineages in a concentration-dependent manner. Stem Cells 28, 45-56. [DOI] [PubMed]
- Balbinot C., Armant O., Elarouci N., Marisa L., Martin E., De Clara E., Onea A., Deschamps J., Beck F., Freund J.N. The Cdx2 homeobox gene suppresses intestinal tumorigenesis through non-cell-autonomous mechanisms. J. Exp. Med. 2018;215:911–926. doi: 10.1084/jem.20170934. [DOI] [PMC free article] [PubMed] [Google Scholar]; Balbinot, C., Armant, O., Elarouci, N., Marisa, L., Martin, E., De Clara, E., Onea, A., Deschamps, J., Beck, F., Freund, J.N., et al. (2018). The Cdx2 homeobox gene suppresses intestinal tumorigenesis through non-cell-autonomous mechanisms. J. Exp. Med. 215, 911-926. [DOI] [PMC free article] [PubMed]
- Becker H., Renner S., Technau G.M., Berger C. Cell-autonomous and non-cell-autonomous function of hox genes specify segmental neuroblast identity in the gnathal region of the embryonic CNS in Drosophila. PLoS Genet. 2016;12:e1005961. doi: 10.1371/journal.pgen.1005961. [DOI] [PMC free article] [PubMed] [Google Scholar]; Becker, H., Renner, S., Technau, G.M., and Berger, C.. (2016). Cell-autonomous and non-cell-autonomous function of hox genes specify segmental neuroblast identity in the gnathal region of the embryonic CNS in Drosophila. PLoS Genet. 12, e1005961. [DOI] [PMC free article] [PubMed]
- Boj S.F., Petrov D., Ferrer J. Epistasis of transcriptomes reveals synergism between transcriptional activators Hnf1α and Hnf4α. PLoS Genet. 2010;6:e1000970. doi: 10.1371/journal.pgen.1000970. [DOI] [PMC free article] [PubMed] [Google Scholar]; Boj, S.F., Petrov, D., and Ferrer, J.. (2010). Epistasis of transcriptomes reveals synergism between transcriptional activators Hnf1α and Hnf4α. PLoS Genet. 6, e1000970. [DOI] [PMC free article] [PubMed]
- Bolotin E., Liao H., Chi Ta T., Yang C., Hwang-Verslues W., Evans J.R., Jiang T., Sladek F.M. Integrated approach for the identification of human hepatocyte nuclear factor 4α target genes using protein binding microarrays. Hepatology. 2010;51:642–653. doi: 10.1002/hep.23357. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bolotin, E., Liao, H., Chi Ta, T., Yang, C., Hwang-Verslues, W., Evans, J.R., Jiang, T., and Sladek, F.M.. (2010). Integrated approach for the identification of human hepatocyte nuclear factor 4α target genes using protein binding microarrays. Hepatology 51, 642-653. [DOI] [PMC free article] [PubMed]
- Byrne M.M., Sturis J., Fajans S.S., Ortiz F.J., Stoltz A., Stoffel M., Smith M.J., Bell G.I., Halter J.B., Polonsky K.S. Altered insulin secretory responses to glucose in subjects with a mutation in the mody1 gene on chromosome-20. Diabetes. 1995;44:699–704. doi: 10.2337/diab.44.6.699. [DOI] [PubMed] [Google Scholar]; Byrne, M.M., Sturis, J., Fajans, S.S., Ortiz, F.J., Stoltz, A., Stoffel, M., Smith, M.J., Bell, G.I., Halter, J.B., and Polonsky, K.S.. (1995). Altered insulin secretory responses to glucose in subjects with a mutation in the mody1 gene on chromosome-20. Diabetes 44, 699-704. [DOI] [PubMed]
- Damcott C.M., Hoppman N., Ott S.H., Reinhart L.J., Wang J., Pollin T.I., O'Connell J.R., Mitchell B.D., Shuldiner A.R. Polymorphisms in both promoters of hepatocyte nuclear factor 4-alpha are associated with type 2 diabetes in the Amish. Diabetes. 2004;53:3337–3341. doi: 10.2337/diabetes.53.12.3337. [DOI] [PubMed] [Google Scholar]; Damcott, C.M., Hoppman, N., Ott, S.H., Reinhart, L.J., Wang, J., Pollin, T.I., O'Connell, J.R., Mitchell, B.D., and Shuldiner, A.R.. (2004). Polymorphisms in both promoters of hepatocyte nuclear factor 4-alpha are associated with type 2 diabetes in the Amish. Diabetes 53, 3337-3341. [DOI] [PubMed]
- DeLaForest A., Nagaoka M., Si-Tayeb K., Noto F.K., Konopka G., Battle M.A., Duncan S.A. HNF4A is essential for specification of hepatic progenitors from human pluripotent stem cells. Development. 2011;138:4143–4153. doi: 10.1242/dev.062547. [DOI] [PMC free article] [PubMed] [Google Scholar]; DeLaForest, A., Nagaoka, M., Si-Tayeb, K., Noto, F.K., Konopka, G., Battle, M.A., and Duncan, S.A.. (2011). HNF4A is essential for specification of hepatic progenitors from human pluripotent stem cells. Development 138, 4143-4153. [DOI] [PMC free article] [PubMed]
- Eeckhoute J., Moerman E., Bouckenooghe T., Lukoviak B., Pattou F., Formstecher P., Kerr-Conte J., Vandewalle B., Laine B. Hepatocyte nuclear factor 4 alpha isoforms originated from the P1 promoter are expressed in human pancreatic beta-cells and exhibit stronger transcriptional potentials than P2 promoter-driven isoforms. Endocrinology. 2003;144:1686–1694. doi: 10.1210/en.2002-0024. [DOI] [PubMed] [Google Scholar]; Eeckhoute, J., Moerman, E., Bouckenooghe, T., Lukoviak, B., Pattou, F., Formstecher, P., Kerr-Conte, J., Vandewalle, B., and Laine, B.. (2003a). Hepatocyte nuclear factor 4 alpha isoforms originated from the P1 promoter are expressed in human pancreatic beta-cells and exhibit stronger transcriptional potentials than P2 promoter-driven isoforms. Endocrinology 144, 1686-1694. [DOI] [PubMed]
- Eeckhoute J., Oxombre B., Formstecher P., Lefebvre P., Laine B. Critical role of charged residues in helix 7 of the ligand binding domain in Hepatocyte Nuclear Factor 4α dimerisation and transcriptional activity. Nucleic Acids Res. 2003;31:6640–6650. doi: 10.1093/nar/gkg850. [DOI] [PMC free article] [PubMed] [Google Scholar]; Eeckhoute, J., Oxombre, B., Formstecher, P., Lefebvre, P., and Laine, B.. (2003b). Critical role of charged residues in helix 7 of the ligand binding domain in Hepatocyte Nuclear Factor 4α dimerisation and transcriptional activity. Nucleic Acids Res. 31, 6640-6650. [DOI] [PMC free article] [PubMed]
- Ek J., Rose C.S., Jensen D.P., Glumer C., Borch-Johnsen K., Jorgensen T., Pedersen O., Hansen T. The functional Thr130Ile and Val255Met polymorphisms of the hepatocyte nuclear factor-4 alpha (HNF4A): gene associations with type 2 diabetes or altered beta-cell function among Danes. J. Clin. Endocrinol. Metab. 2005;90:3054–3059. doi: 10.1210/jc.2004-2159. [DOI] [PubMed] [Google Scholar]; Ek, J., Rose, C.S., Jensen, D.P., Glumer, C., Borch-Johnsen, K., Jorgensen, T., Pedersen, O., and Hansen, T.. (2005). The functional Thr130Ile and Val255Met polymorphisms of the hepatocyte nuclear factor-4 alpha (HNF4A): gene associations with type 2 diabetes or altered beta-cell function among Danes. J. Clin. Endocrinol. Metab. 90, 3054-3059. [DOI] [PubMed]
- Ellard S., Colclough K. Mutations in the genes encoding the transcription factors hepatocyte nuclear factor 1 alpha (HNF1A) and 4 alpha (HNF4A) in maturity-onset diabetes of the young. Hum. Mutat. 2006;27:854–869. doi: 10.1002/humu.20357. [DOI] [PubMed] [Google Scholar]; Ellard, S., and Colclough, K.. (2006). Mutations in the genes encoding the transcription factors hepatocyte nuclear factor 1 alpha (HNF1A) and 4 alpha (HNF4A) in maturity-onset diabetes of the young. Hum. Mutat. 27, 854-869. [DOI] [PubMed]
- Frischmeyer P.A., Dietz H.C. Nonsense-mediated mRNA decay in health and disease. Hum. Mol. Genet. 1999;8:1893–1900. doi: 10.1093/hmg/8.10.1893. [DOI] [PubMed] [Google Scholar]; Frischmeyer, P.A., and Dietz, H.C.. (1999). Nonsense-mediated mRNA decay in health and disease. Hum. Mol. Genet. 8, 1893-1900. [DOI] [PubMed]
- Gardner D.S.L., Tai E.S. Clinical features and treatment of maturity onset diabetes of the young (MODY) Diabetes Metab. Syndr. Obes. 2012;5:101–108. doi: 10.2147/DMSO.S23353. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gardner, D.S.L., and Tai, E.S.. (2012). Clinical features and treatment of maturity onset diabetes of the young (MODY). Diabetes Metab. Syndr. Obes. 5, 101-108. [DOI] [PMC free article] [PubMed]
- Gunton J.E., Kulkarni R.N., Yim S., Okada T., Hawthorne W.J., Tseng Y.-H., Roberson R.S., Ricordi C., O’Connell P.J., Gonzalez F.J. Loss of ARNT/HIF1β mediates altered gene expression and pancreatic-islet dysfunction in human type 2 diabetes. Cell. 2005;122:337–349. doi: 10.1016/j.cell.2005.05.027. [DOI] [PubMed] [Google Scholar]; Gunton, J.E., Kulkarni, R.N., Yim, S., Okada, T., Hawthorne, W.J., Tseng, Y.-H., Roberson, R.S., Ricordi, C., O’Connell, P.J., Gonzalez, F.J., et al. (2005). Loss of ARNT/HIF1β mediates altered gene expression and pancreatic-islet dysfunction in human type 2 diabetes. Cell 122, 337-349. [DOI] [PubMed]
- Gupta R.K., Vatamaniuk M.Z., Lee C.S., Flaschen R.C., Fulmer J.T., Matschinsky F.M., Duncan S.A., Kaestner K.H. The MODY1 gene HNF-4α regulates selected genes involved in insulin secretion. J. Clin. Invest. 2005;115:1006–1015. doi: 10.1172/JCI200522365. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gupta, R.K., Vatamaniuk, M.Z., Lee, C.S., Flaschen, R.C., Fulmer, J.T., Matschinsky, F.M., Duncan, S.A., and Kaestner, K.H.. (2005). The MODY1 gene HNF-4α regulates selected genes involved in insulin secretion. J. Clin. Invest. 115, 1006-1015. [DOI] [PMC free article] [PubMed]
- Hani E., Suaud L., Boutin P., Chevre J.C., Durand E., Philippi A., Demenais F., Vionnet N., Furuta H., Velho G. A missense mutation in hepatocyte nuclear factor-4 alpha, resulting in a reduced transactivation activity, in human late-onset non-insulin-dependent diabetes mellitus. J. Clin. Invest. 1998;101:521–526. doi: 10.1172/JCI1403. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hani, E., Suaud, L., Boutin, P., Chevre, J.C., Durand, E., Philippi, A., Demenais, F., Vionnet, N., Furuta, H., Velho, G., et al. (1998). A missense mutation in hepatocyte nuclear factor-4 alpha, resulting in a reduced transactivation activity, in human late-onset non-insulin-dependent diabetes mellitus. J. Clin. Invest. 101, 521-526. [DOI] [PMC free article] [PubMed]
- Hannan N.R., Segeritz C.P., Touboul T., Vallier L. Production of hepatocyte-like cells from human pluripotent stem cells. Nat. Protoc. 2013;8:430–437. doi: 10.1038/nprot.2012.153. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hannan, N.R., Segeritz, C.P., Touboul, T., and Vallier, L.. (2013). Production of hepatocyte-like cells from human pluripotent stem cells. Nat. Protoc. 8, 430-437. [DOI] [PMC free article] [PubMed]
- Hara K., Horikoshi M., Kitazato H., Lto C., Noda M., Ohashi J., Froguel P., Tokunaga K., Tobe K., Nagai R. Hepatocyte nuclear factor-4 alpha P2 promoter haplotypes are associated with type 2 diabetes in the Japanese population. Diabetes. 2006;55:1260–1264. doi: 10.2337/db05-0620. [DOI] [PubMed] [Google Scholar]; Hara, K., Horikoshi, M., Kitazato, H., Lto, C., Noda, M., Ohashi, J., Froguel, P., Tokunaga, K., Tobe, K., Nagai, R., et al. (2006). Hepatocyte nuclear factor-4 alpha P2 promoter haplotypes are associated with type 2 diabetes in the Japanese population. Diabetes 55, 1260-1264. [DOI] [PubMed]
- Harries L.W., Locke J.M., Shields B., Hanley N.A., Hanley K.P., Steele A., Njolstad P.R., Ellard S., Hattersley A.T. The diabetic phenotype in HNF4A mutation carriers is moderated by the expression of HNF4A isoforms from the P1 promoter during fetal development. Diabetes. 2008;57:1745–1752. doi: 10.2337/db07-1742. [DOI] [PubMed] [Google Scholar]; Harries, L.W., Locke, J.M., Shields, B., Hanley, N.A., Hanley, K.P., Steele, A., Njolstad, P.R., Ellard, S., and Hattersley, A.T.. (2008). The diabetic phenotype in HNF4A mutation carriers is moderated by the expression of HNF4A isoforms from the P1 promoter during fetal development. Diabetes 57, 1745-1752. [DOI] [PubMed]
- Herman W.H., Fajans S.S., Smith M.J., Polonsky K.S., Bell G.I., Halter J.B. Diminished insulin and glucagon secretory responses to arginine in nondiabetic subjects with a mutation in the hepatocyte nuclear factor-4alpha/MODY1 gene. Diabetes. 1997;46:1749–1754. doi: 10.2337/diab.46.11.1749. [DOI] [PubMed] [Google Scholar]; Herman, W.H., Fajans, S.S., Smith, M.J., Polonsky, K.S., Bell, G.I., and Halter, J.B.. (1997). Diminished insulin and glucagon secretory responses to arginine in nondiabetic subjects with a mutation in the hepatocyte nuclear factor-4alpha/MODY1 gene. Diabetes 46, 1749-1754. [DOI] [PubMed]
- Hrvatin S., O'Donnell C.W., Deng F., Millman J.R., Pagliuca F.W., DiIorio P., Rezania A., Gifford D.K., Melton D.A. Differentiated human stem cells resemble fetal, not adult, beta cells. Proc. Natl. Acad. Sci. U S A. 2014;111:3038–3043. doi: 10.1073/pnas.1400709111. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hrvatin, S., O'Donnell, C.W., Deng, F., Millman, J.R., Pagliuca, F.W., DiIorio, P., Rezania, A., Gifford, D.K., and Melton, D.A.. (2014). Differentiated human stem cells resemble fetal, not adult, beta cells. Proc. Natl. Acad. Sci. U S A 111, 3038-3043. [DOI] [PMC free article] [PubMed]
- Huang J., Levitsky L.L., Rhoads D.B. Novel P2 promoter-derived HNF4α isoforms with different N-terminus generated by alternate exon insertion. Exp. Cell Res. 2009;315:1200–1211. doi: 10.1016/j.yexcr.2009.01.004. [DOI] [PubMed] [Google Scholar]; Huang, J., Levitsky, L.L., and Rhoads, D.B.. (2009). Novel P2 promoter-derived HNF4α isoforms with different N-terminus generated by alternate exon insertion. Exp. Cell Res. 315, 1200-1211. [DOI] [PubMed]
- Jiang S., Tanaka T., Iwanari H., Hotta H., Yamashita H., Kumakura J., Watanabe Y., Uchiyama Y., Aburatani H., Hamakubo T. Expression and localization of P1 promoter-driven hepatocyte nuclear factor-4α (HNF4α) isoforms in human and rats. Nucl. Recept. 2003;1:1–12. doi: 10.1186/1478-1336-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jiang, S., Tanaka, T., Iwanari, H., Hotta, H., Yamashita, H., Kumakura, J., Watanabe, Y., Uchiyama, Y., Aburatani, H., Hamakubo, T., et al. (2003). Expression and localization of P1 promoter-driven hepatocyte nuclear factor-4α (HNF4α) isoforms in human and rats. Nucl. Recept. 1, 1-12. [DOI] [PMC free article] [PubMed]
- Kawai H.F., Kaneko S., Honda M., Shirota Y., Kobayashi K. alpha-fetoprotein-producing hepatoma cell lines share common expression profiles of genes in various categories demonstrated by cDNA microarray analysis. Hepatology. 2001;33:676–691. doi: 10.1053/jhep.2001.22500. [DOI] [PubMed] [Google Scholar]; Kawai, H.F., Kaneko, S., Honda, M., Shirota, Y., and Kobayashi, K.. (2001). alpha-fetoprotein-producing hepatoma cell lines share common expression profiles of genes in various categories demonstrated by cDNA microarray analysis. Hepatology 33, 676-691. [DOI] [PubMed]
- Kooner J.S., Saleheen D., Sim X., Sehmi J., Zhang W., Frossard P., Been L.F., Chia K.S., Dimas A.S., Hassanali N. Genome-wide association study in individuals of South Asian ancestry identifies six new type 2 diabetes susceptibility loci. Nat. Genet. 2011;43:984–989. doi: 10.1038/ng.921. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kooner, J.S., Saleheen, D., Sim, X., Sehmi, J., Zhang, W., Frossard, P., Been, L.F., Chia, K.S., Dimas, A.S., Hassanali, N., et al. (2011). Genome-wide association study in individuals of South Asian ancestry identifies six new type 2 diabetes susceptibility loci. Nat. Genet. 43, 984-989. [DOI] [PMC free article] [PubMed]
- Laine B., Eeckhoute J., Suaud L., Briche I., Furuta H., Bell G.I., Formstecher P. Functional properties of the R154X HNF-4alpha protein generated by a mutation associated with maturity-onset diabetes of the young, type 1. FEBS Lett. 2000;479:41–45. doi: 10.1016/s0014-5793(00)01864-0. [DOI] [PubMed] [Google Scholar]; Laine, B., Eeckhoute, J., Suaud, L., Briche, I., Furuta, H., Bell, G.I., and Formstecher, P.. (2000). Functional properties of the R154X HNF-4alpha protein generated by a mutation associated with maturity-onset diabetes of the young, type 1. FEBS Lett. 479, 41-45. [DOI] [PubMed]
- Lau H.H., Ng N.H.J., Loo L.S.W., Jasmen J.B., Teo A.K.K. The molecular functions of hepatocyte nuclear factors - in and beyond the liver. J. Hepatol. 2018;68:1033–1048. doi: 10.1016/j.jhep.2017.11.026. [DOI] [PubMed] [Google Scholar]; Lau, H.H., Ng, N.H.J., Loo, L.S.W., Jasmen, J.B., and Teo, A.K.K.. (2018). The molecular functions of hepatocyte nuclear factors - in and beyond the liver. J. Hepatol. 68, 1033-1048. [DOI] [PubMed]
- Lausen J., Thomas H., Lemm I., Bulman M., Borgschulze M., Lingott A., Hattersley A.T., Ryffel G.U. Naturally occurring mutations in the human HNF4alpha gene impair the function of the transcription factor to a varying degree. Nucleic Acids Res. 2000;28:430–437. doi: 10.1093/nar/28.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lausen, J., Thomas, H., Lemm, I., Bulman, M., Borgschulze, M., Lingott, A., Hattersley, A.T., and Ryffel, G.U.. (2000). Naturally occurring mutations in the human HNF4alpha gene impair the function of the transcription factor to a varying degree. Nucleic Acids Res. 28, 430-437. [DOI] [PMC free article] [PubMed]
- Lehto M., Bitzen P.O., Isomaa B., Wipemo C., Wessman Y., Forsblom C., Tuomi T., Taskinen M.R., Groop L. Mutation in the HNF-4alpha gene affects insulin secretion and triglyceride metabolism. Diabetes. 1999;48:423–425. doi: 10.2337/diabetes.48.2.423. [DOI] [PubMed] [Google Scholar]; Lehto, M., Bitzen, P.O., Isomaa, B., Wipemo, C., Wessman, Y., Forsblom, C., Tuomi, T., Taskinen, M.R., and Groop, L.. (1999). Mutation in the HNF-4alpha gene affects insulin secretion and triglyceride metabolism. Diabetes 48, 423-425. [DOI] [PubMed]
- Li J., Ning G., Duncan S.A. Mammalian hepatocyte differentiation requires the transcription factor HNF-4α. Genes Dev. 2000;14:464–474. [PMC free article] [PubMed] [Google Scholar]; Li, J., Ning, G., and Duncan, S.A.. (2000). Mammalian hepatocyte differentiation requires the transcription factor HNF-4α. Genes Dev. 14, 464-474. [PMC free article] [PubMed]
- Loo L.S.W., Lau H.H., Jasmen J.B., Lim C.S., Teo A.K.K. An arduous journey from human pluripotent stem cells to functional pancreatic beta cells. Diabetes Obes. Metab. 2018;20:3–13. doi: 10.1111/dom.12996. [DOI] [PubMed] [Google Scholar]; Loo, L.S.W., Lau, H.H., Jasmen, J.B., Lim, C.S., and Teo, A.K.K.. (2018). An arduous journey from human pluripotent stem cells to functional pancreatic beta cells. Diabetes Obes. Metab. 20, 3-13. [DOI] [PubMed]
- Love-Gregory L.D., Wasson J., Ma J., Jin C.H., Glaser B., Suarez B.K., Permutt M.A. A common polymorphism in the upstream promoter region of the hepatocyte nuclear factor-4 alpha gene on chromosome 20q is associated with type 2 diabetes and appears to contribute to the evidence for linkage in an ashkenazi jewish population. Diabetes. 2004;53:1134–1140. doi: 10.2337/diabetes.53.4.1134. [DOI] [PubMed] [Google Scholar]; Love-Gregory, L.D., Wasson, J., Ma, J., Jin, C.H., Glaser, B., Suarez, B.K., and Permutt, M.A.. (2004). A common polymorphism in the upstream promoter region of the hepatocyte nuclear factor-4 alpha gene on chromosome 20q is associated with type 2 diabetes and appears to contribute to the evidence for linkage in an ashkenazi jewish population. Diabetes 53, 1134-1140. [DOI] [PubMed]
- Mahajan A., Wessel J., Willems S.M., Zhao W., Robertson N.R., Chu A.Y., Gan W., Kitajima H., Taliun D., Rayner N.W. Refining the accuracy of validated target identification through coding variant fine-mapping in type 2 diabetes. Nat. Genet. 2018;50:559–571. doi: 10.1038/s41588-018-0084-1. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mahajan, A., Wessel, J., Willems, S.M., Zhao, W., Robertson, N.R., Chu, A.Y., Gan, W., Kitajima, H., Taliun, D., Rayner, N.W., et al. (2018). Refining the accuracy of validated target identification through coding variant fine-mapping in type 2 diabetes. Nat. Genet. 50, 559-571. [DOI] [PMC free article] [PubMed]
- Miura A., Yamagata K., Kakei M., Hatakeyama H., Takahashi N., Fukui K., Nammo T., Yoneda K., Inoue Y., Sladek F.M. Hepatocyte nuclear factor-4alpha is essential for glucose-stimulated insulin secretion by pancreatic beta-cells. J. Biol. Chem. 2006;281:5246–5257. doi: 10.1074/jbc.M507496200. [DOI] [PubMed] [Google Scholar]; Miura, A., Yamagata, K., Kakei, M., Hatakeyama, H., Takahashi, N., Fukui, K., Nammo, T., Yoneda, K., Inoue, Y., Sladek, F.M., et al. (2006). Hepatocyte nuclear factor-4alpha is essential for glucose-stimulated insulin secretion by pancreatic beta-cells. J. Biol. Chem. 281, 5246-5257. [DOI] [PubMed]
- Nakabayashi H., Koyama Y., Suzuki H., Li H.M., Sakai M., Miura Y., Wong N.C., Nishi S. Functional mapping of tissue-specific elements of the human alpha-fetoprotein gene enhancer. Biochem. Biophys. Res. Commun. 2004;318:773–785. doi: 10.1016/j.bbrc.2004.04.096. [DOI] [PubMed] [Google Scholar]; Nakabayashi, H., Koyama, Y., Suzuki, H., Li, H.M., Sakai, M., Miura, Y., Wong, N.C., and Nishi, S.. (2004). Functional mapping of tissue-specific elements of the human alpha-fetoprotein gene enhancer. Biochem. Biophys. Res. Commun. 318, 773-785. [DOI] [PubMed]
- Nammo T., Yamagata K., Tanaka T., Kodama T., Sladek F.M., Fukui K., Katsube F., Sato Y., Miyagawa J., Shimomura I. Expression of HNF-4alpha (MODY1), HNF-1beta (MODY5), and HNF-1alpha (MODY3) proteins in the developing mouse pancreas. Gene Expr. Patterns. 2008;8:96–106. doi: 10.1016/j.modgep.2007.09.006. [DOI] [PubMed] [Google Scholar]; Nammo, T., Yamagata, K., Tanaka, T., Kodama, T., Sladek, F.M., Fukui, K., Katsube, F., Sato, Y., Miyagawa, J., and Shimomura, I.. (2008). Expression of HNF-4alpha (MODY1), HNF-1beta (MODY5), and HNF-1alpha (MODY3) proteins in the developing mouse pancreas. Gene Expr. Patterns 8, 96-106. [DOI] [PubMed]
- Ober E.A., Verkade H., Field H.A., Stainier D.Y. Mesodermal Wnt2b signalling positively regulates liver specification. Nature. 2006;442:688–691. doi: 10.1038/nature04888. [DOI] [PubMed] [Google Scholar]; Ober, E.A., Verkade, H., Field, H.A., and Stainier, D.Y.. (2006). Mesodermal Wnt2b signalling positively regulates liver specification. Nature 442, 688-691. [DOI] [PubMed]
- Odom D.T., Zizlsperger N., Gordon D.B., Bell G.W., Rinaldi N.J., Murray H.L., Volkert T.L., Schreiber J., Rolfe P.A., Gifford D.K. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]; Odom, D.T., Zizlsperger, N., Gordon, D.B., Bell, G.W., Rinaldi, N.J., Murray, H.L., Volkert, T.L., Schreiber, J., Rolfe, P.A., Gifford, D.K., et al. (2004). Control of pancreas and liver gene expression by HNF transcription factors. Science 303, 1378-1381. [DOI] [PMC free article] [PubMed]
- Pagliuca F.W., Millman J.R., Gurtler M., Segel M., Van Dervort A., Ryu J.H., Peterson Q.P., Greiner D., Melton D.A. Generation of functional human pancreatic beta cells in vitro. Cell. 2014;159:428–439. doi: 10.1016/j.cell.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]; Pagliuca, F.W., Millman, J.R., Gurtler, M., Segel, M., Van Dervort, A., Ryu, J.H., Peterson, Q.P., Greiner, D., and Melton, D.A.. (2014). Generation of functional human pancreatic beta cells in vitro. Cell 159, 428-439. [DOI] [PMC free article] [PubMed]
- Pearson E.R., Pruhova S., Tack C.J., Johansen A., Castleden H.A.J., Lumb P.J., Wierzbicki A.S., Clark P.M., Lebl J., Pedersen O. Molecular genetics and phenotypic characteristics of MODY caused by hepatocyte nuclear factor 4α mutations in a large European collection. Diabetologia. 2005;48:878–885. doi: 10.1007/s00125-005-1738-y. [DOI] [PubMed] [Google Scholar]; Pearson, E.R., Pruhova, S., Tack, C.J., Johansen, A., Castleden, H.A.J., Lumb, P.J., Wierzbicki, A.S., Clark, P.M., Lebl, J., Pedersen, O., et al. (2005). Molecular genetics and phenotypic characteristics of MODY caused by hepatocyte nuclear factor 4α mutations in a large European collection. Diabetologia 48, 878-885. [DOI] [PubMed]
- Petersen M.B.K., Azad A., Ingvorsen C., Hess K., Hansson M., Grapin-Botton A., Honore C. Single-cell gene expression analysis of a human ESC model of pancreatic endocrine development reveals different paths to beta-cell differentiation. Stem Cell Reports. 2017;9:1246–1261. doi: 10.1016/j.stemcr.2017.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; Petersen, M.B.K., Azad, A., Ingvorsen, C., Hess, K., Hansson, M., Grapin-Botton, A., and Honore, C.. (2017). Single-cell gene expression analysis of a human ESC model of pancreatic endocrine development reveals different paths to beta-cell differentiation. Stem Cell Reports 9, 1246-1261. [DOI] [PMC free article] [PubMed]
- Ryffel G.U. Mutations in the human genes encoding the transcription factors of the hepatocyte nuclear factor (HNF)1 and HNF4 families: functional and pathological consequences. J. Mol. Endocrinol. 2001;27:11–29. doi: 10.1677/jme.0.0270011. [DOI] [PubMed] [Google Scholar]; Ryffel, G.U.. (2001). Mutations in the human genes encoding the transcription factors of the hepatocyte nuclear factor (HNF)1 and HNF4 families: functional and pathological consequences. J. Mol. Endocrinol. 27, 11-29. [DOI] [PubMed]
- Shih D.Q., Dansky H.M., Fleisher M., Assmann G., Fajans S.S., Stoffel M. Genotype/phenotype relationships in HNF-4alpha/MODY1: haploinsufficiency is associated with reduced apolipoprotein (AII), apolipoprotein (CIII), lipoprotein(a), and triglyceride levels. Diabetes. 2000;49:832–837. doi: 10.2337/diabetes.49.5.832. [DOI] [PubMed] [Google Scholar]; Shih, D.Q., Dansky, H.M., Fleisher, M., Assmann, G., Fajans, S.S., and Stoffel, M.. (2000). Genotype/phenotype relationships in HNF-4alpha/MODY1: haploinsufficiency is associated with reduced apolipoprotein (AII), apolipoprotein (CIII), lipoprotein(a), and triglyceride levels. Diabetes 49, 832-837. [DOI] [PubMed]
- Silander K., Mohlke K.L., Scott L.J., Peck E.C., Hollstein P., Skol A.D., Jackson A.U., Deloukas P., Hunt S., Stavrides G. Genetic variation near the hepatocyte nuclear factor-4 alpha gene predicts susceptibility to type 2 diabetes. Diabetes. 2004;53:1141–1149. doi: 10.2337/diabetes.53.4.1141. [DOI] [PubMed] [Google Scholar]; Silander, K., Mohlke, K.L., Scott, L.J., Peck, E.C., Hollstein, P., Skol, A.D., Jackson, A.U., Deloukas, P., Hunt, S., Stavrides, G., et al. (2004). Genetic variation near the hepatocyte nuclear factor-4 alpha gene predicts susceptibility to type 2 diabetes. Diabetes 53, 1141-1149. [DOI] [PubMed]
- Sladek F.M., Dallas-Yang Q., Nepomuceno L. MODY1 mutation Q268X in hepatocyte nuclear factor 4alpha allows for dimerization in solution but causes abnormal subcellular localization. Diabetes. 1998;47:985–990. doi: 10.2337/diabetes.47.6.985. [DOI] [PubMed] [Google Scholar]; Sladek, F.M., Dallas-Yang, Q., and Nepomuceno, L.. (1998). MODY1 mutation Q268X in hepatocyte nuclear factor 4alpha allows for dimerization in solution but causes abnormal subcellular localization. Diabetes 47, 985-990. [DOI] [PubMed]
- Sladek F.M., Zhong W.M., Lai E., Darnell J.E., Jr. Liver-enriched transcription factor HNF-4 is a novel member of the steroid hormone receptor superfamily. Genes Dev. 1990;4:2353–2365. doi: 10.1101/gad.4.12b.2353. [DOI] [PubMed] [Google Scholar]; Sladek, F.M., Zhong, W.M., Lai, E., and Darnell, J.E., Jr. (1990). Liver-enriched transcription factor HNF-4 is a novel member of the steroid hormone receptor superfamily. Genes Dev. 4, 2353-2365. [DOI] [PubMed]
- Stoffel M., Duncan S.A. The maturity-onset diabetes of the young (MODY1) transcription factor HNF4 alpha regulates expression of genes required for glucose transport and metabolism. Proc. Natl. Acad. Sci. U S A. 1997;94:13209–13214. doi: 10.1073/pnas.94.24.13209. [DOI] [PMC free article] [PubMed] [Google Scholar]; Stoffel, M., and Duncan, S.A.. (1997). The maturity-onset diabetes of the young (MODY1) transcription factor HNF4 alpha regulates expression of genes required for glucose transport and metabolism. Proc. Natl. Acad. Sci. U S A 94, 13209-13214. [DOI] [PMC free article] [PubMed]
- Tanaka T., Jiang S., Hotta H., Takano K., Iwanari H., Sumi K., Daigo K., Ohashi R., Sugai M., Ikegame C. Dysregulated expression of P1 and P2 promoter-driven hepatocyte nuclear factor-4α in the pathogenesis of human cancer. J. Pathol. 2006;208:662–672. doi: 10.1002/path.1928. [DOI] [PubMed] [Google Scholar]; Tanaka, T., Jiang, S., Hotta, H., Takano, K., Iwanari, H., Sumi, K., Daigo, K., Ohashi, R., Sugai, M., Ikegame, C., et al. (2006). Dysregulated expression of P1 and P2 promoter-driven hepatocyte nuclear factor-4α in the pathogenesis of human cancer. J. Pathol. 208, 662-672. [DOI] [PubMed]
- Teo A.K., Gupta M.K., Doria A., Kulkarni R.N. Dissecting diabetes/metabolic disease mechanisms using pluripotent stem cells and genome editing tools. Mol. Metab. 2015;4:593–604. doi: 10.1016/j.molmet.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; Teo, A.K., Gupta, M.K., Doria, A., and Kulkarni, R.N.. (2015a). Dissecting diabetes/metabolic disease mechanisms using pluripotent stem cells and genome editing tools. Mol. Metab. 4, 593-604. [DOI] [PMC free article] [PubMed]
- Teo A.K., Lau H.H., Valdez I.A., Dirice E., Tjora E., Raeder H., Kulkarni R.N. Early developmental perturbations in a human stem cell model of MODY5/HNF1B pancreatic hypoplasia. Stem Cell Reports. 2016;6:357–367. doi: 10.1016/j.stemcr.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; Teo, A.K., Lau, H.H., Valdez, I.A., Dirice, E., Tjora, E., Raeder, H., and Kulkarni, R.N.. (2016). Early developmental perturbations in a human stem cell model of MODY5/HNF1B pancreatic hypoplasia. Stem Cell Reports 6, 357-367. [DOI] [PMC free article] [PubMed]
- Teo A.K., Tsuneyoshi N., Hoon S., Tan E.K., Stanton L.W., Wright C.V., Dunn N.R. PDX1 binds and represses hepatic genes to ensure robust pancreatic commitment in differentiating human embryonic stem cells. Stem Cell Reports. 2015;4:578–590. doi: 10.1016/j.stemcr.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]; Teo, A.K., Tsuneyoshi, N., Hoon, S., Tan, E.K., Stanton, L.W., Wright, C.V., and Dunn, N.R.. (2015b). PDX1 binds and represses hepatic genes to ensure robust pancreatic commitment in differentiating human embryonic stem cells. Stem Cell Reports.;4, 578-590. [DOI] [PMC free article] [PubMed]
- Teo A.K., Wagers A.J., Kulkarni R.N. New opportunities: harnessing induced pluripotency for discovery in diabetes and metabolism. Cell Metab. 2013;18:775–791. doi: 10.1016/j.cmet.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]; Teo, A.K., Wagers, A.J., and Kulkarni, R.N.. (2013a). New opportunities: harnessing induced pluripotency for discovery in diabetes and metabolism. Cell Metab. 18, 775-791. [DOI] [PMC free article] [PubMed]
- Teo A.K., Windmueller R., Johansson B.B., Dirice E., Njolstad P.R., Tjora E., Raeder H., Kulkarni R.N. Derivation of human induced pluripotent stem cells from patients with maturity onset diabetes of the young. J. Biol. Chem. 2013;288:5353–5356. doi: 10.1074/jbc.C112.428979. [DOI] [PMC free article] [PubMed] [Google Scholar]; Teo, A.K., Windmueller, R., Johansson, B.B., Dirice, E., Njolstad, P.R., Tjora, E., Raeder, H., and Kulkarni, R.N.. (2013b). Derivation of human induced pluripotent stem cells from patients with maturity onset diabetes of the young. J. Biol. Chem. 288, 5353-5356. [DOI] [PMC free article] [PubMed]
- Vethe H., Bjorlykke Y., Ghila L.M., Paulo J.A., Scholz H., Gygi S.P., Chera S., Raeder H. Probing the missing mature beta-cell proteomic landscape in differentiating patient iPSC-derived cells. Sci. Rep. 2017;7:4780. doi: 10.1038/s41598-017-04979-w. [DOI] [PMC free article] [PubMed] [Google Scholar]; Vethe, H., Bjorlykke, Y., Ghila, L.M., Paulo, J.A., Scholz, H., Gygi, S.P., Chera, S., and Raeder, H.. (2017). Probing the missing mature beta-cell proteomic landscape in differentiating patient iPSC-derived cells. Sci. Rep. 7, 4780. [DOI] [PMC free article] [PubMed]
- Weedon M.N., Owen K.R., Shields B., Hitman G., Walker M., McCarthy M.I., Love-Gregory L.D., Permutt M.A., Hattersley A.T., Frayling T.M. Common variants of the hepatocyte nuclear factor-4 alpha P2 promoter are associated with type 2 diabetes in the UK population. Diabetes. 2004;53:3002–3006. doi: 10.2337/diabetes.53.11.3002. [DOI] [PubMed] [Google Scholar]; Weedon, M.N., Owen, K.R., Shields, B., Hitman, G., Walker, M., McCarthy, M.I., Love-Gregory, L.D., Permutt, M.A., Hattersley, A.T., and Frayling, T.M.. (2004). Common variants of the hepatocyte nuclear factor-4 alpha P2 promoter are associated with type 2 diabetes in the UK population. Diabetes 53, 3002-3006. [DOI] [PubMed]
- Yamagata K., Furuta H., Oda N., Kaisaki P.J., Menzel S., Cox N.J., Fajans S.S., Signorini S., Stoffel M., Bell G.I. Mutations in the hepatocyte nuclear factor-4[alpha] gene in maturity-onset diabetes of the young (MODY1) Nature. 1996;384:458–460. doi: 10.1038/384458a0. [DOI] [PubMed] [Google Scholar]; Yamagata, K., Furuta, H., Oda, N., Kaisaki, P.J., Menzel, S., Cox, N.J., Fajans, S.S., Signorini, S., Stoffel, M., and Bell, G.I.. (1996). Mutations in the hepatocyte nuclear factor-4[alpha] gene in maturity-onset diabetes of the young (MODY1). Nature 384, 458-460. [DOI] [PubMed]
- Zhang Z., Xin D., Wang P., Zhou L., Hu L., Kong X., Hurst L.D. Noisy splicing, more than expression regulation, explains why some exons are subject to nonsense-mediated mRNA decay. BMC Biol. 2009;7:23. doi: 10.1186/1741-7007-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhang, Z., Xin, D., Wang, P., Zhou, L., Hu, L., Kong, X., and Hurst, L.D.. (2009). Noisy splicing, more than expression regulation, explains why some exons are subject to nonsense-mediated mRNA decay. BMC Biol. 7, 23. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The first two columns indicate the gene ID and official gene symbol for all protein-coding genes in the dataset. FPKM values are shown for control-hiPSC-derived cells (13A, 13B, 7A, 7B, and 7C) and MODY1-hiPSC-derived cells (2, 1A, 1B, and 1C).