Abstract
Objective
We aim to evaluate the effectiveness of advanced deep learning models (eg, capsule network [CapNet], adversarial training [ADV]) for single-domain and multidomain relation extraction from electronic health record (EHR) notes.
Materials and Methods
We built multiple deep learning models with increased complexity, namely a multilayer perceptron (MLP) model and a CapNet model for single-domain relation extraction and fully shared (FS), shared-private (SP), and adversarial training (ADV) modes for multidomain relation extraction. Our models were evaluated in 2 ways: first, we compared our models using our expert-annotated cancer (the MADE1.0 corpus) and cardio corpora; second, we compared our models with the systems in the MADE1.0 and i2b2 challenges.
Results
Multidomain models outperform single-domain models by 0.7%-1.4% in F1 (t test P < .05), but the results of FS, SP, and ADV modes are mixed. Our results show that the MLP model generally outperforms the CapNet model by 0.1%-1.0% in F1. In the comparisons with other systems, the CapNet model achieves the state-of-the-art result (87.2% in F1) in the cancer corpus and the MLP model generally outperforms MedEx in the cancer, cardiovascular diseases, and i2b2 corpora.
Conclusions
Our MLP or CapNet model generally outperforms other state-of-the-art systems in medication and adverse drug event relation extraction. Multidomain models perform better than single-domain models. However, neither the SP nor the ADV mode can always outperform the FS mode significantly. Moreover, the CapNet model is not superior to the MLP model for our corpora.
Keywords: natural language processing, relation extraction, deep learning, electronic health record note, single and multidomain
INTRODUCTION
Electronic health record (EHR) notes are important resources for clinical knowledge, epidemiological study, drug safety research, and pharmacovigilance.1,2 Relations between medication and adverse drug events (ADEs) are important knowledge that can be extracted from EHR notes. In general, the task of relation extraction is to identify predefined relations between entity pairs. For example, given a sentence, “Due to the renal failure, I stopped antibiotics,” a causal relation can be extracted between the entity pair renal failure and antibiotics. Because of the importance of relation extraction, the biomedical natural language processing (NLP) community has published a number of corpora and shared tasks.3–10 These corpora and shared tasks may focus on different objects (eg, chemical-disease,4 bacteria location6) and derive from different text sources (eg, clinical text,3 knowledge bases,5 biomedical literature9). In this work, we focused on extracting medication–attribute, medication–indication, and medication–ADE relations from EHR notes.
Related work
First, our work is related to relation extraction in the biomedical domain3 and closely related to ADE extraction or detection.4,10 A typical method is to cast relation extraction as a classification problem and use machine learning models to solve it. For example, Xu et al11 used support vector machines (SVMs) to determine the relations between drug–disease pairs, while Henriksson et al2 used the random forest model. However, there are also other methods to enrich this research area. For example, Liu et al12 aimed to not only predict ADEs, but also identify the factors that contribute significantly to ADEs. They fulfilled this goal by using causality analysis to select important features. Kilicoglu et al13 enhanced relation extraction from another aspect. They used coreference resolution to link the entities in different sentences and thus extended relation extraction from sentence level to discourse level.
Our work is methodologically different from the aforementioned approaches. Our work is centered on deep learning. Munkhdalai et al14 used the recurrent neural network to extract relations from clinical notes, but they found that the performance of the recurrent neural network was inferior to that of SVM. Luo et al15 employed the convolutional neural network (CNN) to extract relations from clinical notes and found that their model was better than non–deep learning models. Therefore, it is important to conduct a comprehensive evaluation to compare deep learning approaches with the traditional machine learning approaches, as we have done in this study.
Deep learning–based approaches have advanced rapidly in the open domain. Recently, Sabour et al16 proposed a novel capsule network (CapNet) with excellent performance for hierarchical relation recognition such as segmenting highly overlapped digits. The CapNet may be applicable to NLP because natural languages are tree-structured. Therefore, in this study, we evaluated whether the CapNet is also effective for clinical relation extraction. Our work is also motivated by Chen and Cardie17 and Rios et al,18 who explored domain adaptation and adversarial training (ADV) techniques to improve information extraction in the open domain or in different tasks from ours.
Last, our work is also related to other variations of relation extraction using deep learning. For example, Verga et al19 proposed a self-attention model to extract the relations across all the sentences in a document. Although our models can also extract intersentence relations, they are limited in a fixed sentence window. Recently, end-to-end relation extraction is receiving increasing attention in both the open domain and biomedical domain.20–22 Unlike our study, such a method does not depend on an entity recognition model23 to provide entities. However, we explored multidomain adaptation and CapNets that were not explored in these studies.
Objective
Our objectives are 2-fold. First, we aimed to investigate the effectiveness of an up-to-date model (ie, the CapNet model) for the task of clinical relation extraction from EHR notes. The CapNet model receives much attention in the computer vision community,16,24 but it has not been well studied in biomedical NLP tasks. In this study, we adapted the CapNet model and compared it with a multilevel perceptron (MLP) network, a widely used classifier for various classification tasks.
Second, we built multidomain learning models and evaluated them for multidomain relation extraction from EHR notes. Specifically, we first built a popular shared-private (SP) network.17,25 The shared part learns domain-invariant knowledge and the private part learns domain-specific knowledge. However, the shared part may also learn some domain-specific knowledge because it is trained using the data of multiple domains. To solve this problem, we built an ADV network that was integrated with the state-of-the-art technique for multidomain adaptation.17,18,25
Contributions
The main contributions of this study are the following:
MATERIALS AND METHODS
Corpora
We employed 3 corpora to evaluate our models for both single-domain and multidomain relation extraction. First, we evaluated our models with 2 expert-annotated EHR corpora from 2 clinical domains: cancer and cardiovascular diseases (cardio). Our models extract the relations among medications, indications, ADEs, and their attributes from EHR notes. The relations were annotated based on linguistic expressions in the EHR notes. We took both intrasentence and intersentence relations into account, which means that there may be a long distance between the argument entities of a relation.
The cancer corpus consists of 1089 EHRs of patients with cancer. This corpus is released as a part of the MADE1.0 challenge.27 The corpus is separated into 876 EHRs for training and 213 EHRs for testing. Therefore, we kept such setting to compare with the systems in the challenge. The cardio corpus comprises of 485 EHRs of patients with cardiovascular diseases. The cardio corpus is divided into 390 and 85 EHRs for training and testing, respectively, similar to the proportion of the cancer corpus. In both corpora, we predefined 9 entity types, namely medication, indication, frequency, severity, dosage, duration, route, ADE, and SSLIF (any sign, symptom, and disease that is not an ADE or indication). The 7 types of relations among these entity types are shown in Figure 1.
Figure 1.
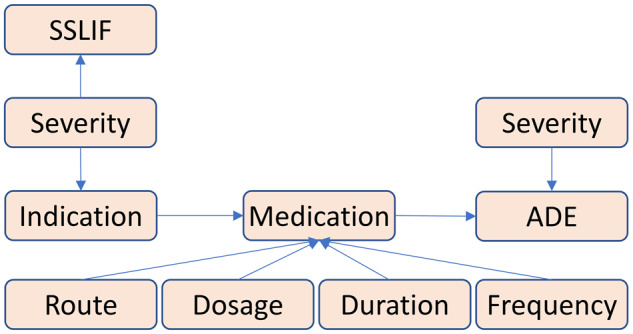
Relations in our corpora. Relations are identified among medications, indications, adverse drug events (ADEs), and attributes (ie, severity, route, dosage, duration, and frequency). SSLIF: any sign, symptom, and disease that is not an adverse drug event or indication.
Second, we compared our models with other state-of-the-art systems for relation extraction in medications, indications, ADEs and their attributes. We compared the performance of our models with the performance of NLP systems in the MADE1.0 challenges (cancer corpus). We also compared our models with MedEx,26 using the cancer and cardio corpora as well as the corpus in the i2b2 2009 Medication Challenge.28 The i2b2 corpus focuses on the identification of medications and their dosages, routes, frequencies, and durations. It consists of 1243 clinical notes, but only 251 with gold standard annotations are available for us. Therefore, we used these notes to compare with MedEx.
Deep learning models
Single-domain relation extraction using the MLP network
To compare with other models in this study, we built a strong baseline model as shown in Figure 2. There are 3 main parts in this model and we describe them in the following sections.
Figure 2.
Architecture of the single-domain relation extraction model using the mutlilayer perceptron network. Given an instance, that is, 2 target entities , and a word sequence that incorporates and , 2 kinds of features are extracted, namely sequence-level features and instance-level features. A sequence-level feature corresponds to a word (eg, word embedding) while an instance-level feature corresponds to an instance (eg, entity type). After feature extraction, all features are fed into the classifier to determine which kind of relations and have. ADE: adverse drug event; CNN: convolutional neural network; POS: part of speech.
Sequence-level feature extraction
The word sequence of an instance can be represented as , where denotes the first word in , denotes the part of speech (POS) tag of this word, and denote the relative positions29 from this word to the entity and . The concatenation of these features is used to represent the th word, namely . Therefore, the word sequence corresponds to a feature sequence .
As the length of the feature sequence is variable, a popular architecture30 of CNNs is utilized to transform the feature sequence into a fixed-length representation. This architecture has 3 CNNs with the window sizes as 3, 4 and 5 respectively. For example, if the window size is 3, the input of the CNN can be , , …, . After each CNN transforms the feature sequence into a vector, these vectors are concatenated and their dimensions are decreased by a nonlinear transformation. The final representation of the feature sequence can be formalized as
| (1) |
where indicates the rectified-linear activation function and is the parameter matrix. , , and indicate the CNNs with the window sizes as 3, 4, and 5, respectively.
Instance-level feature extraction
We extracted 4 types of instance-level features, including
Words of 2 target entities: ,
Types of 2 target entities: ,
Distance (word-token) between 2 target entities:
Number of the entities between 2 target entities:
Take the instance in Figure 2 as an example, its instance-level features can be , , , , and .
For , , , and , each of them is directly represented as a fixed-length vector, which is similar to the method for generating word embeddings. For and , the attention method31 is employed to encode multiple entity words into a fixed-length vector, as there may be more than 1 word in an entity. As shown in Figure 2, for each entity word , a weight is computed by
| (2) |
where is the parameter matrix. Then the feature can be generated by the weighted-sum computation, given by
| (3) |
Classifier
We employed an MLP model as the classifier to determine the relation of an instance. Both the sequence-level and instance-level features are fed in to the MLP model, formalized as:
| (4) |
| (5) |
where and are the parameter matrices, denotes the output of the hidden layer, and denotes relation types. A softmax layer calculates the probabilities of all relation types and the relation type with the maximum probability is selected as the prediction result. During training, we minimized the cross-entropy loss, given by
| (6) |
where denotes all the parameters in the classifier and Σ denotes the loss of each instance is accumulated.
Single-domain relation extraction using the CapNet
To investigate the effectiveness of CapNet in our task, we built a capsule classifier as shown in Figure 3. In the original work of the CapNet, a capsule is a group of neurons that denotes an object (eg, a face or nose).16 The CapNet can learn the hierarchical relationships of objects (eg, a nose is in the middle of a face). For our task, such relationships also exist in the entities, which can be considered as patterns (eg, aspirin 81 mg/d). Therefore, we also aimed to model such patterns via the CapNet.
Figure 3.
Architecture of the capsule network classifier. The architecture of the single-domain relation extraction model using the capsule network is identical to the model using the multilayer perceptron classifier (Figure 2), except that the capsule network classifier replaces the multilayer perceptron classifier.
In this study, a vector is used to denote a capsule, so all the features from the feature extractors need to be reshaped to match the vector dimension of capsules. For example, if we have a 256-dimension feature and the vector dimension of capsules is 8, we will get 32 capsules after reshaping. This procedure can be formalized as
| (7) |
where denotes the number of capsules. Then the dynamic routing algorithm16 is leveraged to transfer information between the lower and higher capsule layers. After that, we got the capsules whose number is the same as the number () of relation types, formalized as
| (8) |
During inference, the capsule whose vector has the maximal norm is selected, so its corresponding relation is also selected as the prediction result. During training, we minimized the margin loss, given by
| (9) |
where indicates a relation type, is 1 if the gold answer is , and is the norm of the capsule. The training objective is to make the norm of the capsule corresponding to the gold answer be much longer than the norms of other capsules corresponding to nongold answers.
Multidomain relation extraction using the fully shared or shared-private modes
To utilize the data from different domains for mutual benefit, we proposed a multidomain relation extraction model using the fully shared (FS) or shared-private (SP) modes.17,25 Because the FS mode is a simple case of the SP mode, the SP mode is introduced first. If we have domains denoted as (eg, denotes cancer, denotes cardio), the training and test sets will become and . As shown in Figure 4, each domain corresponds to a private feature extractor and all domains share a shared feature extractor . Both and can be considered as the feature extraction layer in Figure 2.
Figure 4.

Architecture of the multidomain relation extraction model using the shared-private mode. A feature extractor denotes both sequence-level and instance-level feature extraction in Figure 2. A classifier can be either the multilayer perceptron model or capsule network. The blue or green rectangle denotes the private feature extractor. The white rectangle denotes the shared feature extractor.
During training for the domain , is fed into and is fed into . Therefore, learns domain-specific knowledge and learns shared knowledge for all domains. During evaluation for the domain , is fed into and to generate shared features and domain-specific features. For simplicity, here we also define the generated features as or . Then both shared features and domain-specific features are input into the classifier, formalized by
| (10) |
where denotes the classifier that can be either the MLP network or CapNet, and denotes relation types. For the FS mode, all the domains share 1 feature extractor and they have no private feature extractor. Therefore, Equation 10 can be changed as .
Multidomain relation extraction using the ADV mode
Because the shared feature extractor is trained using the data of all domains, it may also learn some domain-specific knowledge. To make the shared feature extractor learn less domain-specific knowledge but more shared knowledge, we employed the ADV mode17 as shown in Figure 5.
Figure 5.

Architecture of the multidomain relation extraction model using the adversarial training. The yellow arrow line denotes the adversarial training.
The ADV mode can be roughly divided into 2 steps. First, a domain classifier is added upon the shared feature extractor. It is trained to determine which domain an instance belongs to, based on the knowledge provided by the shared feature extractor:
| (11) |
where is actually an MLP and its loss function is defined as:
| (12) |
where denotes domain types and denotes the parameters of the domain classifier. The first step is to train the domain classifier to predict the domain type accurately.
Second, the gradient reverse (ie, ) is used to train the shared feature extractor to confuse the domain classifier to make inaccurate predictions. Therefore, the domain classifier and share feature extractor compete with each other in the 2 steps of the ADV mode.
Intuitively, the shared feature extractor uses domain-specific knowledge to help the domain classifier discriminate the domain type. If the shared feature extractor is trained using the reversed gradient, it is not able to help the domain classifier very well. Therefore, we can consider that the shared feature extractor contains less domain-specific knowledge.
RESULTS
To get the experimental settings related to the results, please refer to Supplementary Appendix 1.
Comparisons of our models
Comparing MLP and CapNet
From Table 1, we can see that for single-domain relation extraction, the F1 of the MLP model is 0.1% lower than that of the CapNet in the cancer corpus but 0.3% higher in the cardio corpus. For multidomain relation extraction using the SP mode, the F1s of the MLP model are 0.1% and 0.8% higher than those of the CapNet in the cancer and cardio corpora, respectively. For multidomain relation extraction using the ADV mode, the MLP model also outperforms the CapNet by 0.1% and 0.8% in F1. Therefore, the MLP model is generally superior to the CapNet in our task.
Table 1.
Result comparisons of our models
|
Cancer
(%)
|
Cardio (%) |
||||||
|---|---|---|---|---|---|---|---|
| P | R | F1 | P | R | F1 | ||
| Single domain | MLP | 88.3 | 85.8 | 87.1 | 91.0 | 82.0 | 86.3 |
| CapNet | 88.7 | 85.6 | 87.2 | 91.6 | 81.4 | 86.0 | |
| Multidomain (S) | MLP | 88.9 | 86.7 | 87.8 | 90.9 | 84.5 | 87.6 |
| CapNet | 88.3 | 86.5 | 87.4 | 88.5 | 84.8 | 86.6 | |
| Multidomain (SP) | MLP | 88.8 | 86.8a | 87.8 | 90.7 | 84.9a | 87.7a |
| CapNet | 91.1a | 84.4 | 87.7 | 92.1 | 82.3 | 86.9 | |
| Multidomain (ADV) | MLP | 89.9 | 86.3 | 88.1a | 92.3a | 83.3 | 87.6 |
| CapNet | 90.6 | 85.5 | 88.0 | 89.2 | 84.5 | 86.8 | |
Statistical significance is provided in Supplementary Appendix 2.
ADV: adversarial training; CapNet: capsule network; MLP: multilayer perceptron; S: fully shared; SP: shared-private.
Highest score in the column.
Comparing single-domain and multidomain models
As shown in Table 1, when multidomain relation extraction models use the SP mode, the F1s of the MLP model increase by 0.7% and 1.4% in 2 corpora, compared with the F1s of single-domain models. Similarly, the F1s of the CapNet also increase by 0.5% and 0.9% in 2 corpora. When multidomain relation extraction models use the ADV mode, the F1s of the MLP model rise by 1.0% and 1.3% in 2 corpora, compared with the F1s of single-domain models. Likewise, the F1s of the CapNet also rise by 0.8% in 2 corpora. Therefore, multidomain relation extraction, either using the SP mode or ADV mode, is capable of improving the performances.
Comparing the FS, SP, and ADV modes
As shown in Table 1, the CapNet is more sensitive to the mode than the MLP model. Overall, the performance of the SP mode is slightly better than that of the FS mode, but the difference is not always significant as shown in Supplementary Appendix 2. This result was also demonstrated in prior work.17,25 In terms of the ADV mode, Table 1 shows that in the cancer corpus, the F1 of the MLP model using the ADV mode is 0.3% higher than that of the MLP model using the SP mode. The F1 of the CapNet is also improved by 0.3% when the ADV mode is used. By contrast, in the cardio corpus, both the F1s of the MLP model and CapNet decrease by 0.1% when the ADV mode is used. Therefore, we cannot get a conclusion that the ADV mode is always effective for our corpora.
Comparisons with the systems in the MADE1.0 challenge
Because the cancer corpus is also used in the MADE1.0 challenge, the top 3 systems are compared with our best model trained only in this corpus (ie, the CapNet for single-domain relation extraction). As each system has only 1 or 2 submissions, we reported their best result. By contrast, we reported our result using the mean ± 95% confidence interval. As shown in Table 2, the best and third-best system employed traditional machine learning methods such as the random forest algorithm and SVM. Our model outperforms their systems by 0.4% and 4.0%, respectively. The second-best system used deep learning methods and our model also outperforms it by 3.2%.
Table 2.
Results of comparisons with the systems in the MADE1.0 challenge
RNN: recurrent neural network; SVM: support vector machine.
Comparisons with MedEx
MedEx26 is a competitive system to recognize medications and their attributes from free-text clinical records. In this section, we compared the single-domain MLP model with MedEx in the cancer, cardio, and i2b2 corpora.28 Please note that we used the entities that were recognized by MedEx as the inputs of our model and our model was trained without using any data in the i2b2 corpus. As shown in Table 3, the F1 scores of our model are better than those of MedEx in most categories in the cancer and i2b2 corpus. In the cardio corpus, however, MedEx outperforms the single-domain MLP model in medication–route and medication–frequency.
Table 3.
Results of comparisons with MedEx
| Corpus | Relation Type | MedEx |
Single-Domain MLP |
||||
|---|---|---|---|---|---|---|---|
| P | R | F1 | P | R | F1 | ||
| Cancer | Medication–route | 71.9 | 47.9 | 57.5 | 88.8 | 54.3 | 67.4a,b |
| Medication–dosage | 29.7 | 3.5 | 6.2 | 57.4 | 6.2 | 11.3a,b | |
| Medication–duration | 25.5 | 15.6 | 19.4 | 34.7 | 17.7 | 23.4a,b | |
| Medication–frequency | 52.5 | 36.2 | 42.8 | 57.7 | 37.4 | 45.4a,b | |
| Cardio | Medication–route | 89.0 | 46.5 | 61.1a,b | 87.2 | 43.5 | 58.0 |
| Medication–dosage | 83.8 | 4.4 | 8.3 | 83.3 | 4.9 | 9.3a,b | |
| Medication–duration | 42.1 | 21.6 | 28.6 | 50 | 21.6 | 30.2a,b | |
| Medication–frequency | 77.6 | 44.2 | 56.3a,b | 70.6 | 42.1 | 52.8 | |
| i2b2 | Medication–route | 84.2 | 74.8 | 79.2 | 89.2 | 75.6 | 81.9a,b |
| Medication–dosage | 82.1 | 69.8 | 75.5 | 84.1 | 69.9 | 76.3a,b | |
| Medication–duration | 15.3 | 6.5 | 9.1 | 18.8 | 6.5 | 9.6a | |
| Medication–frequency | 75.2 | 65.8 | 70.2 | 79.2 | 66.1 | 72.1a,b | |
Higher F1 scores between 2 systems. bF1 of one system is significantly higher than the F1 of another system at P = .05.
MLP: multilayer perceptron.
DISCUSSION
Method analysis
For the CapNet, it does not show advantages compared with the MLP model in our task. One possible reason is that the instance-level features are more effective to be used directly. In the MLP model, these features are directly sent to the output layer. In the CapNet, these features are regrouped and transformed so their effects may be influenced. To demonstrate this, we removed instance-level features and only used sequence-level features. Experimental results show that the CapNet performs better than the MLP model (F1 in the cancer corpus: 86.3% vs 85.7%; F1 in the cardio corpus: 80.4% vs 80.2%). In addition, as the size of the cardio corpus is smaller and the CapNet achieves worse results in the cardio corpus than in the cancer corpus, we also studied whether the data size is a factor to influence the CapNet. First, the performances of small-sized relations are analyzed and then the CapNet is trained using half of the training data in the cancer corpus. However, we have not found any evidence to demonstrate that the data size impacts the results.
For multidomain relation extraction, either using the SP mode or ADV mode, it is an effective method to improve the performances. Single-domain relation extraction models can only learn from the corpus of 1 domain, while multidomain relation extraction models can learn from the corpora of all domains. Therefore, common knowledge from different domains can be learned by the models for mutual benefit. Such a technique shows effectiveness in many prior studies such as text classification17 and relation extraction.18
For the ADV mode, the performances of both MLP and CapNet go up in our experiments of the cancer corpus, but the performances both go down in the experiments of the cardio corpus. Therefore, the ADV mode is not effective for all domains in our task. We found similar phenomena in previous work25 and they also got worse results for some domains. The effect of the ADV mode may be influenced by many factors such as the data distribution, domain relevancy, and optimization techniques.35
Last, compared with other systems in the MADE1.0 challenge, our models perform better. One possible reason is that they only used words and POS tags between and neighboring 2 target entities, while we used all the words and POS tags in the sentences. Moreover, they only encoded the word information into their models, while we did not only use such information but also manually designed features. Therefore, the performance improvement of our models is due to the usage of various methods such as feature engineering, deep learning, multidomain learning, and ADV mode.
Error analysis
As shown in Supplementary Appendix 3, Table 1, our models perform well in medication–route and medication–dosage but do poorly in medication–ADE and medication–indication relation extraction. Therefore, the error analysis is performed on these relations. We manually reviewed the false positive and false negative instances of medication–ADE and medication–indication relations. We found that the errors of our models are due to 3 main reasons.
First of all, more than 80% false positive errors are because relations exist in the instance, but the relations are not related to the target entities. Take the first instance in Table 4 as an example: “peripheral neuropathy” and “thalidomide” have no medication–ADE relation, but the model incorrectly predicts their relation due to the words secondary to related to Velcade and peripheral neuropathy. This phenomenon indicates that it is difficult for our models to distinguish which context is related to which entity pair, when several entity pairs occur in the same instance.
Table 4.
Main error reasons for extracting medication–ADE and medication-indication relations
| Error Type | Error Reason | Example | |
|---|---|---|---|
| False positive | Relations exist but not related to target entities | His current therapy includes [thalidomide]e1 50 mg a day for 2 weeks of the month. He had been on Velcade, which was stopped secondary to increasing [peripheral neuropathy]e2 | Medication–ADE |
| According to the patient, she is also taking [Flovent]e1, ProAir and Spiriva. She is using Chantix to help her [quit smoking]e2 | Medication–indication | ||
| False negative | No obvious patterns | I do want to continue to hold the [Velcade]e1 as his [peripheral neuropathy]e2 continues to improve | Medication–ADE |
| She usually takes a baby [aspirin]e1 and she feels the [chest pain]e2 gets better after a while | Medication–indication | ||
| Long context | She had contacted the physician complaining of [chest discomfort]e1 and gurgling in chest. She had increased edema and shortness of breath. She was advised to go to the emergency room, but refused. She took an additional dose of [Lasix]e2 40 mg and symptoms improved. | Medication–indication | |
ADE: adverse drug event.
The second reason leading to many errors is that there are no obvious patterns such as trigger words in the instance. Most NLP systems depend on pattern features to extract relations. For example, medication–ADE relations may often be accompanied by the trigger words such as secondary to, develop, associated with, and medication–indication relations may often co-occur with treat, therapy, and prescription, etc. When these patterns do not exist, it is harder to predict relations correctly. Natural language understanding in the semantic level is still very challenging for most NLP systems due to many limitations such as small corpus size and lack of background knowledge.
The third error reason is that the context of the instance is long. This situation frequently happens in the instances of medication–indication relations since the treatment procedure is usually expressed via several sentences in EHRs. Extracting such relations is challenging because there may be more noise as the context becomes long. Moreover, it requires the models to be stronger to understand document-level natural languages.
Limitations and future work
One limitation of our work is that our approaches are single note–centric and do not take into consideration the longitudinality,36,37 which we will explore in our future work. Other limitations include limited clinical domains (cancer and cardiovascular diseases) and limited annotated corpora. On the other hand, cancer and cardiovascular diseases are common in the United States. Due to comorbidity, our corpora include a wide range of diseases (Supplementary Appendix 4). Annotation is expensive and in the future work we may explore the additions of other existing annotated corpora.3,4,10
CONCLUSION
In this study, we investigated the effectiveness of the CapNet in the task of relation extraction from EHRs. Results show that the CapNet does not achieve better performance compared with the MLP model. We also investigated multidomain relation extraction. Results show that although the performance can be improved by adding the data from a different domain, neither the SP nor ADV mode consistently outperforms the FS mode significantly. Furthermore, we found that the ADV mode for multidomain relation extraction is not always effective. In the future, we will explore how to effectively use the CapNet and evaluate our models on more domains of EHR relation extraction.
FUNDING
This work was supported by National Institutes of Health grant 5R01HL125089 and Health Services Research & Development Program of the U.S. Department of Veterans Affairs Investigator-Initiated Research grant 1I01HX001457-01.
AUTHOR CONTRIBUTIONS
FL and HY conceptualized and designed this study. FL implemented the tools. HY processed the data. FL wrote the manuscript. HY reviewed and contributed to formatting the manuscript.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Supplementary Material
ACKNOWLEDGMENTS
The authors gratefully acknowledge Abhyuday N Jagannatha and Bhanu Pratap Singh for technical support on implementing the tools and Weisong Liu for technical support on building the development environment.
Conflict of interest statement
None declared.
REFERENCES
- 1. Turchin A, Shubina M, Breydo E, et al. Comparison of information content of structured and narrative text data sources on the example of medication intensification. J Am Med Inform Assoc 2009; 163: 362–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Henriksson A, Kvist M, Dalianis H, et al. Identifying adverse drug event information in clinical notes with distributional semantic representations of context. J Biomed Inform 2015; 57: 333–49. [DOI] [PubMed] [Google Scholar]
- 3. Uzuner Ö, South BR, Shen S, et al. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc 2011; 185: 552–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wei C-H, Peng Y, Leaman R, et al. Assessing the state of the art in biomedical relation extraction: overview of the BioCreative V chemical-disease relation (CDR) task. Database 2016; 2016: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wei W-Q, Cronin RM, Xu H, et al. Development and evaluation of an ensemble resource linking medications to their indications. J Am Med Inform Assoc 2013; 205: 954–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Deléger L, Bossy R, Chaix E, et al. Overview of the bacteria biotope task at BioNLP Shared Task 2016 In: Proceedings of the 4th BioNLP Shared Task Workshop, 2016: 12–22. [Google Scholar]
- 7. Segura-Bedmar I, Martınez P, Herrero-Zazo M.. Semeval-2013 task 9: Extraction of drug-drug interactions from biomedical texts (DDIExtraction 2013) In: Seventh International Workshop on Semantic Evaluation (SemEval 2013), 2013: 341–50. Association for Computational Linguistics. [Google Scholar]
- 8. Kim J-D, Ohta T, Tateisi Y, et al. GENIA corpus—a semantically annotated corpus for bio-textmining. Bioinformatics 2003; 19 (Suppl 1): i180–2. [DOI] [PubMed] [Google Scholar]
- 9. Krallinger M, Rabal O, Akhondi SA, et al. Overview of the BioCreative VI chemical-protein interaction track In: Proceedings of the BioCreative VI Workshop, 2017: 141–6. BioCreative. [Google Scholar]
- 10. Gurulingappa H, Rajput AM, Roberts A, et al. Development of a benchmark corpus to support the automatic extraction of drug-related adverse effects from medical case reports. J Biomed Inform 2012; 455: 885–92. [DOI] [PubMed] [Google Scholar]
- 11. Xu J, Wu Y, Zhang Y, et al. CD-REST: a system for extracting chemical-induced disease relation in literature. Database 2016; 2016: 1–9. doi:10.1093/database/baw036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu M, Cai R, Hu Y, et al. Determining molecular predictors of adverse drug reactions with causality analysis based on structure learning. J Am Med Inform Assoc 2014; 212: 245–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kilicoglu H, Rosemblat G, Fiszman M, et al. Sortal anaphora resolution to enhance relation extraction from biomedical literature. BMC Bioinformatics 2016; 17: 1–16. doi:10.1186/s12859-016-1009-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Munkhdalai T, Liu F, Yu H.. Clinical relation extraction toward drug safety surveillance using electronic health record narratives: classical learning versus deep learning. JMIR Public Health Surveill 2018; 42: e29.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Luo Y, Cheng Y, Uzuner Ö, et al. Segment convolutional neural networks (Seg-CNNs) for classifying relations in clinical notes. J Am Med Inform Assoc 2018; 251: 93–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sabour S, Frosst N, Hinton GE.. Dynamic routing between capsules In: Guyon I, Luxburg UV, Bengio S, et al. , eds. Advances in Neural Information Processing Systems. Long Beach, CA: Curran Associates; 2017: 3856–66. [Google Scholar]
- 17. Chen X, Cardie C.. Multinomial adversarial networks for multi-domain text classification In: Proceedings of the 2018 Conference of the North American Chapter of the Association for Computational Linguistics, 2018: 1226–40; New Orleans, Louisiana, USA. [Google Scholar]
- 18. Rios A, Kavuluru R, Lu Z.. Generalizing biomedical relation classification with neural adversarial domain adaptation. Bioinformatics 2018; 34 (17): 2973–81. doi:10.1093/bioinformatics/bty190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Verga P, Strubell E, McCallum A.. Simultaneously self-attending to all mentions for full-abstract biological relation extraction In: Proceedings of the 2018 Conference of the North American Chapter of the Association for Computational Linguistics, 2018: 872–84; New Orleans, Louisiana, USA. [Google Scholar]
- 20. Mehryary F, Hakala K, Kaewphan S, et al. End-to-end system for bacteria habitat extraction In: BioNLP 2017, 2017: 80–90; Vancouver, Canada. [Google Scholar]
- 21. Miwa M, Bansal M.. End-to-end relation extraction using LSTMs on sequences and tree structures In: Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics, 2016: 1105–16; Berlin, Germany. doi:10.18653/v1/P16-1105. [Google Scholar]
- 22. Li F, Zhang M, Fu G, et al. A neural joint model for entity and relation extraction from biomedical text. BMC Bioinformatics 2017; 181: 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jagannatha AN, Yu H.. Structured prediction models for RNN based sequence labeling in clinical text. Proc Conf Empir Methods Nat Lang Process 2016; 2016: 856–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hinton G, Sabour S, Frosst N.. Matrix capsules with EM routing In: International Conference on Learning Representations, Vancouver, Canada; 2018: 1–15. https://openreview.net/forum? id=HJWLfGWRb. [Google Scholar]
- 25. Liu P, Qiu X, Huang X.. Adversarial multi-task learning for text classification In: Proceedings of the 55th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Articles), 2017: 1–10. [Google Scholar]
- 26. Xu H, Stenner SP, Doan S, et al. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc 2010; 171: 19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jagannatha A, Liu F, Liu W, et al. Overview of the first natural language processing challenge for extracting medication, indication, and adverse drug events from electronic health record notes (MADE 1.0). Drug Saf 2019; 421: 99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Uzuner O, Solti I, Cadag E.. Extracting medication information from clinical text. J Am Med Inform Assoc 2010; 175: 514–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zeng D, Liu K, Lai S, et al. Relation classification via convolutional deep neural network In: Proceedings of COLING 2014, 2014: 2335–44; Dublin, Ireland. [Google Scholar]
- 30. Kim Y. Convolutional neural networks for sentence classification In: Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP), 2014: 1746–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Luong T, Pham H, Manning CD.. Effective approaches to attention-based neural machine translation In: Proceedings of the 2015 Conference on Empirical Methods in Natural Language Processing, 2015: 1412–21; Lisbon, Portugal. [Google Scholar]
- 32. Chapman AB, Peterson KS, Alba PR, et al. Hybrid system for adverse drug event detection In: International Workshop on Medication and Adverse Drug Event Detection, PMLR;2018: 16–24. [Google Scholar]
- 33. Dandala B, Joopudi V, Devarakonda M.. IBM research system at MADE 2018: detecting adverse drug events from electronic health records In: International Workshop on Medication and Adverse Drug Event Detection, PMLR; 2018: 39–47. [Google Scholar]
- 34. Xu D, Yadav V, Bethard S.. UArizona at the MADE1.0 NLP challenge In: International Workshop on Medication and Adverse Drug Event Detection, PMLR; 2018: 57–65. [PMC free article] [PubMed] [Google Scholar]
- 35. Salimans T, Goodfellow I, Zaremba W, et al. Improved techniques for training GANs In: Lee DD, Sugiyama M, Luxburg UV, Guyon I, Garnett R. eds, Advances in Neural Information Processing Systems, Barcelona Spain: Curran Associates, Inc; 2016: 1–9. [Google Scholar]
- 36. Sun W, Rumshisky A, Uzuner O.. Evaluating temporal relations in clinical text: 2012 i2b2 challenge. J Am Med Inform Assoc 2013; 205: 806–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Eriksson R, Werge T, Jensen LJ, et al. Dose-specific adverse drug reaction identification in electronic patient records: temporal data mining in an inpatient psychiatric population. Drug Saf 2014; 374: 237–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.




