Abstract
Background/purpose
Helicobacter pylori (H. pylori) infection is the most common in the world and is associated with various gastrointestinal pathologies, including chronic gastritis, peptic ulcers, and gastric cancer. The prevalence is associated with socioeconomic conditions, with this infection being more common in developing countries than in developed countries. The presence and permanence of H. pylori in the oral cavity has been reported, but its role is controversial. The aim of this study was to determine the prevalence of H. pylori in dental plaque of patients with periodontitis.
Materials and methods
A cross-sectional study was carried out and Periodontal Screening and Recording (PSR) index was determined. 38 dental plaque samples were taken and total DNA was extracted and qPCR was performed.
Results
60.5% of the samples (n = 23) were positive for the presence of H. pylori by the amplification of the 16S rRNA and vacA genes. In addition, cagA gene was detected in 21.7% (n = 5) of H. pylori-positive. A significant relationship between periodontal status and H. pylori oral infection was found (P ≤ 0.05); patients with initial and moderate periodontitis were the most affected with 39.1% and 30.4%, respectively.
Conclusion
Our results suggest that the prevalence of H. pylori in the oral cavity could be related to the progression of periodontal disease. Therefore, oral hygiene and treatment for the elimination of oral H. pylori could stop the progression of periodontal disease.
Keywords: Helicobacter pylori, cagA, vacA, Oral cavity
Introduction
Helicobacter pylori (H. pylori) belongs to the subdivision of Proteobacteria, order Campylobacter, of the family Helicobacter and 20 species are recognized. This microorganism has the property of being microaerophilic and of being catalase, oxidase, and urease positive in most cases.1 The lineages in which we can find this species can be subdivided into gastric and hepatic enterococci Helicobacter; these show a high prevalence of being organ-specific, since gastric Helicobacter do not have the capacity to colonize intestine and liver.1,2 H. pylori presents, a genome of 1.66 × 107 bp included in a single circular chromosome and a G + C content of 39%, within the genome has five regions that have a different composition of G + C, in the which two possess one or more copies of IS605 insertion sequences, 5S ribosomal RNA sequences at the ends and a 521 bp duplication close to each other.3
H. pylori infection causes mainly gastritis and one of the main risk factors for developing gastric adenocarcinoma and MALT lymphoma, the development of these clinical pathologies is influenced by various aspects, genetic susceptibility of the host, the environment, and the factors of virulence of the infecting strain; this being one of the most important. Virulence factors are bacterial products or strategies that confer pathogenicity. H. pylori several virulence factors have been proposed, such as cagA, vacA, among others, none of them implies the development of a specific disease. Risk of developing some clinical pathology increases the more virulence factors the bacteria accumulate.4,5
CagA is a protein with an approximate weight of 140 kDa, is highly immunogenic and is encoded by the cagA gene which is present in western countries in 60–70% of the isolated bacteria. H. pylori strains associated with cag A are a genomic marker of the pathogenicity island (PAI).6
The most studied molecular target for cagA is SHP-2 phosphatase (tyrosine phosphatase protein). Mutations and polymorphisms related to gastric carcinogenesis have been found into coding gene.7 Also, changes that occur in gene expression after H. pylori infection in epithelial cells are usually dependent on the secretion system encoded by cag PAI.8
On the other hand, in several studies showed that some strains of H. pylori produced a protein in the culture supernatant said proteins induced vacuolization in epithelial cells, cell death and the destruction of epithelial integrity by such was designated to the protein as cytotoxin vacuolizing (VacA).9 VacA protein is encoded by the vacA gene and is present in all strains of H. pylori; however, only 50% of the strains show detectable cytotoxic activity. This forms part of a multimeric complex of approximately 100 kD and presents at least two variable parts.10 However, these present variations in their vacuolizing activity due to heterogeneity within the vacA gene.11
The VacA protein could induce programmed death, independently of vacuolization, since it induces the release of cytochrome C from mitochondria through proapoptotic Bax and Bak proteins. This participates in the process of apoptosis by the activation of the Fas/CD95 receptor, through caspase 3 and the breakdown of the mitochondrial membrane that alters the cell cycle by disturbing the cellular ATP concentration.12
Geographic location, ethnic group, socioeconomic status, and age are variables that directly influence the prevalence of H. pylori. Several studies have revealed that there are several ways in which this bacterium could be acquired, within these forms have been considered a route of gastro-oral, oral-fecal transmission that is believed to be the main form of transmission especially in underdeveloped countries.13
Several studies have evaluated dental plaque and saliva as possible samples for the diagnosis of H. pylori using various techniques. The culture of the bacteria from the oral cavity has seldom been successfully isolated. With the different molecular techniques, better results have been reported when using dental plaque and saliva as samples for the detection of H. pylori.14 The oral cavity has been considered a reservoir of infection of this microorganism, in different studies have shown results has been assumed as a reservoir of H. pylori infection.
Most studies have shown the presence of H. pylori in the oral cavity from patients who show to have some periodontal complication, so the American Dental Association developed the Periodontal Screening and Recording (PSR) index to improve the detection of periodontal disease.15,16 Presence of this bacterium in the oral cavity has been postulated as an important potential source of reinfection after it has been eradicated from stomach.17 Bürgers et al.18 in their research showed that the presence of H. pylori can occur in the oral cavity independently of colonization of the stomach.
Mexico has a high prevalence (70%) of the microorganism and severe pathologies associated with it, it is relevant the genotypic diagnosis of H. pylori, using the qPCR for the detection of virulence factors and thereby determine the circulating variants. In addition, it is important that it be done with a reduced invasiveness to the patient, since usually, gastric biopsies are used for this purpose.
The aim of this study was to determine the prevalence of H. pylori in dental plaque of patients with periodontitis.
Materials and methods
Study and patients
A cross-sectional study was carried out. This study was submitted and approved by the Ethics Committee in Biological Science College at Universidad Autónoma de Nuevo León (FCB-UANL) and Centro de investigation y Desarrollo en Ciencias de la Salud (CIDICS-UANL).
Patients were attended at dental clinic in Esquipulas, Chiapas, Mexico, during April 2016 and selected according to the following exclusion criteria: 1) current history of antibiotic usage or during the previous six months, 2) had a systemic condition that required prophylactic antibiotics, 3) dyspeptic symptoms, 4) history of gastrointestinal diseases and 5) were pregnant.
The patients who accepted their participation in this study signed the informed consent form. After, the dental clinical history, PSR score by sextant and dental plaque samples were taken. All experiments on human subjects were conducted in accordance with the Declaration of Helsinki and all procedures were carried out with the adequate understanding and written consent of the subjects.
Periodontal examination
PSR is a quick, reliable, and reproducible method for identifying patients that may require a more complete evaluation of their periodontal health status.19 The plastic PSR Periodontal Examination Probe was used throughout this study. The probe has a 0.5 mm diameter ball tip and a color-coded band extending 3.5 mm–5.5 mm from the tip. The PSR probe was gently inserted into the gingival sulcus of each tooth until the light resistance was reached and then the probe was moved around the circumference of the tooth. The depth of the probe in each sextant of the mouth was determined and recorded. Intraoral sextants were designated S1—S6, beginning in the maxillary right sextant (S1), proceeding in a clockwise manner, and finishing in the mandibular right sextant (S6). Each sextant was assigned a code based on the highest probing value obtained on any tooth in that sextant.
PSR codes were based on the following system: 0) Colored area of probe remains completely visible in the deepest crevice in the sextant. No calculus or defective margins are detected. Gingival tissues are healthy with no bleeding after gentle probing. 1) Colored area of probe remains completely visible in the deepest probing depth in the sextant. No calculus or margins are detected. There is bleeding after gentle probing. 2) Colored area of probe remains completely visible in the deepest probing depth in the sextant. Supra- or subgingival calculus and/or defective margins are detected. 3) Colored area of probe remains partly visible in the deepest probing depth in the sextant. 4) Colored area of probe completely disappears, indicating probing depth of greater than 5.5 mm.19,20
Collection of dental plaque samples
Subgingival plaque samples were collected with the help of sterile curette after careful removal of supragingival plaque. In each subject, samples were collected from all posterior teeth. Subgingival plaque samples were suspended in 500 μL of 30 g/L TS broth (Becton Dickinson) supplemented with 30% (w/v) glycerol solution for the maintenance of bacterial viability. All samples were stored at −80 °C, until use.
DNA isolation
For total DNA extraction, TRIzol reagent (Thermo Fisher Scientific, Waltham, MA) was used according to manufacturer's instructions. The concentration and purity of DNA samples, was analyzed in a NanoDrop 8000 UV–Vis spectrophotometer (Thermo Fisher Scientific). The 230/260 absorbance ratio of DNA samples was between 1.9 and 2.1. In addition, DNA samples were loaded in a 1% (w/v) agarose gel then stained with 0.5 μg/mL ethidium bromide solution and visualized under UV light.
H. pylori vacA and cagA genes status and detection
H. pylori detection was performed by analysis of real-time PCR amplification (qPCR) using oligonucleotides targeting against to 16S rRNA gene. In addition, vacA and cagA genes status was analyzed by qPCR. The oligonucleotide sequence were previously designed in our laboratory by Urrutia-Baca et al.21 as shown in Table 1. The qPCR reactions were performed in 96-well plates, as follows: SYBR qPCR master mix 2X (Thermo Fisher Scientific), 0.1 μM each primer, 40 ng of DNA sample and nuclease-free water to a final volume of 25 μL qPCR reactions was amplified in a LighCycler 480 thermal cycler (Roche, Basel, Switzerland), programmed with the Sybr Green I detection system, in four stages: i) pre-incubation: one denaturing cycle of 95 °C per 10 min; ii) Amplification: forty of amplify cycle (95 °C per 10 s and with a decrease of 4 °C/s, alignment at 55 °C for 10 s and decrease of 2 °C/s), one extension cycle at 72 °C per 10 s, with a decrease of 4 °C/s; iii) Dissociation curve: one cycle of 95 °C per 5 s with a decrease of 4 °C/s, at 65 °C for 1 min with a decrease of 2 °C/s and 97 °C with a decrease of 5 °C/s; and iv) Cooling: one cycle of 40 °C per 30 s and 2 s at 1.5 °C/s.
Table 1.
qPCR Primers used for identification and genotyping of H. pylori.
| Gene | Forward | Reverse | Tm (°C) | Reference |
|---|---|---|---|---|
| 16s rRNA | ggagtacggtcgcaagattaaa | ctagcggattctccaatgtcaa | 55 | Urrutia-Baca et al., 2018 |
| cagA | gaccgactcgatcaaatagca | ttagctgaaagccctaccttac | 55 | |
| vacA | cctactgagaatggtggcaata | gttcttcacgagagcgtagtt | 55 |
Statistical analysis
The results were collected in a database using SPSS software v22.0 and the descriptive statistical analysis was performed. In order to determine the association between each nominal variable, Fisher's exact tests was applied; one-way ANOVA and post-hoc Tukey tests were used for numeric variables. The P < 0.05 value was considered statistically significant.
Results
Description of patients
A total of 38 patients with periodontal disease, 4 males and 34 females ranging in age from 34 to 60 years with an average age of 47.1 ± 12.48, were included. In the periodontal examination of patients based on PSR index, showed that 39.5% of patients were diagnosed with moderate periodontitis, 34.2% with periodontitis initial, 10.5% with gingivitis and 15.8% with severe periodontitis (Table 2). In addition, 100% of male were diagnosed with moderate periodontitis.
Table 2.
Distribution of patients by gender, age and periodontal status.
| Variable | Patients N = 38 |
|---|---|
| Gender n (%) | |
| Male | 4 (10.6) |
| Female | 34 (89.4) |
| Age | |
| Mean (±SD) | 47.1 ± 12.48 |
| PSR classification n (%) | |
| 1 | 4 (10.5) |
| 2 | 13 (34.2) |
| 3 | 15 (39.5) |
| 4 | 6 (15.8) |
H. pylori cagA and vacA genes status and detection in dental plaque
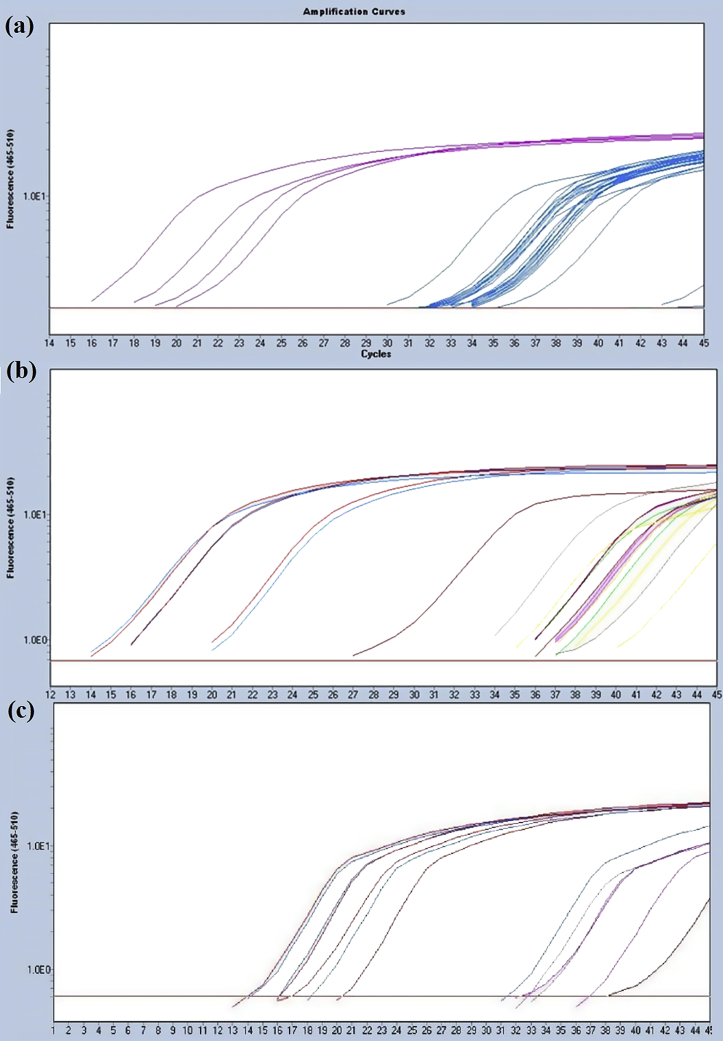
H. pylori detection in the dental plaque samples collected from the patients was performed by qPCR assay. The qPCR amplification and melting curves obtained from each DNA sample were compared to DNA H. pylori J99 strain as control positive (Fig. 1). The results showed that 60.5% of the patients (23/38) were positive for the amplification of H. pylori16S rRNA gene. Likewise, the H. pylori vacA and cagA genes were detected in 100% (23/23) and 21.7% (5/23) of H. pylori-positive patients. The mean values of cycle threshold (Ct) obtained from each positive sample shown in Table 3.
Figure 1.
Amplification curves for detection of H. pylori genes by qPCR. (a) 16S rRNA, (b) vacA and (c) cagA genes.
Table 3.
Cycle threshold values for 16S rRNA, vacA and cagA genes by qPCR assay.
| Patients | Cp 16S rRNA | Cp vacA | Cp cagA |
|---|---|---|---|
| P1 | 32.68 | 34.68 | |
| P3 | 33.36 | 36.44 | |
| P5 | 33.6 | 35.66 | |
| P8 | 33.52 | 35.65 | |
| P9 | 31.69 | 35.55 | |
| P10 | 33.36 | 33.69 | 33.51 |
| P11 | 35.03 | 34.65 | 36.91 |
| P12 | 42.37 | 32.57 | |
| P14 | 32.23 | 37.07 | 32.84 |
| P17 | 31.41 | 35.52 | 38.03 |
| P18 | 41.09 | 37.05 | |
| P19 | 29.41 | 25.63 | |
| P21 | 31.89 | 35.5 | |
| P22 | 31.52 | 37.03 | |
| P25 | 30.85 | 33.55 | |
| P26 | 31.82 | 34.67 | |
| P27 | 32.89 | 34.47 | |
| P28 | 31.12 | 36.01 | 32.63 |
| P30 | 32.89 | 34.52 | |
| P32 | 33.65 | 32.5 | |
| P33 | 32.67 | 35.5 | |
| P35 | 33.5 | 32.67 | |
| P36 | 31.67 | 35.35 |
Relationship between periodontal status and H. pylori oral infection
A significant relationship was found between the periodontal status calculated by PSR and oral H. pylori infection using 16s rRNA and vacA genes (P < 0.05). The 39.1%, 30.4% and 26.0% of the infected patients were found in the 2nd, 3rd and 4th classification of the PSR index, respectively. The PSR index describes 2nd, 3rd and 4th classification as initial, moderate and severe periodontitis, respectively (Table 4). On the other hand, cagA gene was detected in 21.7% 5/23 of H. pylori-positive patients and 40% of cagA-positive patients were within 3rd and 4th classification, both cases; moderate and severe periodontitis according to PSR examination, respectively (Table 5).
Table 4.
Relationship between the PSR classification and H. pylori detection.
| PSR classification n (%) |
H. pylori 16s rRNA status |
P value | |
|---|---|---|---|
| Positive N = 23 |
Negative N = 15 |
||
| 1 | 1 (4.3) | 3 (20.0) | 0.044a |
| 2 | 9 (39.1) | 4 (26.7) | |
| 3 | 7 (30.4) | 8 (53.3) | |
| 4 | 6 (26.0) | 0 | |
Exact Fisher test.
Table 5.
The cagA gene status in H. pylori-positive patients by PSR classification.
| PSR classification n (%) |
cagA gene status in H. pylori-positive patients |
P value | |
|---|---|---|---|
| Positive N = 5 |
Negative N = 18 |
||
| 1 | 0 | 1 (5.6) | 0.648a |
| 2 | 1 (20.0) | 8 (44.4) | |
| 3 | 2 (40.0) | 5 (27.8) | |
| 4 | 2 (40.0) | 4 (22.3) | |
Exact Fisher test.
Discussion
In Mexico, the seroprevalence of H. pylori has been reported to around 20% in children at one year of age, 50% at 10 years of age and the highest prevalence has been reported between 25 and 30 years of age where gastrointestinal pathologies related to this infection are also frequent. Almost always infected people may develop some clinical gastrointestinal pathology such as gastritis, peptic ulcer or gastric cancer.22 Therefore, the study of the oral cavity is of great importance since it has been proposed as a native reservoir for H. pylori where reinfections after the eradication treatment against this bacterium.23
Kignel et al.14 have suggested that the colonization of H. pylori in dental plaque does not go beyond some kind of local disease. However, in recent years a high prevalence of bacteria in this reservoir has been reported in patients with periodontal diseases. Several studies based on molecular techniques have been carried out to detect H. pylori, which show high sensitivity and specificity.24 In the present study, we use the real-time PCR method that allows quantification, shorter diagnostic times and greater sensitivity for the diagnosis of H. pylori; the 16S RNAr gene was used for the identification of H. pylori and the virulence genes vacA and cagA for genotyping.
In our study we observed a high prevalence (60.5%) of oral H. pylori infection in patients with periodontal clinical manifestations. Our data were higher than those obtained by De la Garza-Ramos et al.23 they reported 13% of oral infection per H. pylori in dental plaque samples from thirty Mexican patients using conventional PCR. Other studies have reported a prevalence of oral H. pylori around 38%, however those studies were carried out in asymptomatic Mexican children.24, 25, 26
Several studies have detected H. pylori in the oral cavity in both dental plaque and saliva.27 It was demonstrated that the presence of oral H. pylori is related to the reappearance of gastric infection by H. pylori causing the failure of the therapies, and that the periodontal treatment and good oral hygiene in infected patients significantly increases the eradication of H. pylori in the stomach.28 However, there is a lot of disagreement among researchers about the role of dental plaque as an extragastric reservoir in the transmission of H. pylori.29 Also, the oral H. pylori infection has been related to periodontal problems.30,31 Reports including clinical and experimental studies have reported that people with H. pylori infection tend to periodontal disease.27,32 However, some studies showed that there was no correlation between H. pylori infection and periodontal status.33 In our study, PSR analysis showed that the majority of patients have moderate periodontitis (39.5%), followed by periodontitis in the initial phase (34.2%).
It is known that some H. pylori strains directly influence the severity of gastrointestinal disease in infected patients. It is reported that individuals infected with cagA-positive H. pylori strains have a higher risk of developing peptic ulcer and gastric cancer.34 It is possible that the accumulation of periodontopathogenic bacteria, the permanence of pathogenic H. pylori strains and increased concentration of bacterial metabolites in the periodontal tissues could contribute to a chronic inflammatory process. Hu et al.35 evaluated the correlation between H. pylori infection with periodontal parameters, periodontal pathogens and inflammation in 14 patients. They obtained frequencies of Porphyromonas gingivalis, Prevotella intermedia, Fusobacterium nucleatum and Treponema denticola significantly higher with H. pylori infection than those without H. pylori infection. In that same study, they observed an increase in the expression of periodontitis-related molecules Wnt5a, interleukin 8 (IL-8), interleukin 6 (IL-6) and interferon gamma (IFN-γ) in human acute monocytic leukemia cells (THP-1) stimulated with cagA-positive strain (H. pylori 26695).
In our study, a 21.7% (5/23) positivity was found for the cagA gene among the infected patients, however, a relationship of this gene with the periodontal state of the patients was not observed.
In this way we agree with what was published by Kignel et al.,14 when it is stated that in previous studies the use of dental plaque has been achieved as a possible non-invasive sample for the diagnosis of H. pylori. Likewise, with the results obtained in this research. We agree with published studies since they imply that H. pylori is found as a reservoir in the oral cavity, especially in dental plaque, and that the real-time PCR technique (qPCR) is the most appropriate due to its high sensitivity and specificity in order to detect a low number of H. pylori bacteria and their virulence genes in non-invasive samples.15,23,24
Our results allow us to confirm that it is possible to detect the presence and circulating genotype of H. pylori by amplifying the 16s rRNA gene and their cagA and vacA virulence genes using qPCR in dental plaque in patients from Esquipulas, Mexico, since this has been a technique that shows satisfactory results in terms of sensitivity, specificity, low cost and easy processing, although of course, complementary studies with a greater number of samples would provide data with greater forcefulness.
qPCR has great advantages for those epidemiological studies in which we intend to study the prevalence of infection in the population and its relationship with different gastrointestinal pathologies. These procedures will allow the diagnosis to be extended in those patients who are asymptomatic or who have symptoms of some gastrointestinal pathology.
Conflicts of interest
None declared.
Acknowledgments
We thank Maria Teresa Gutierrez Lopez of the Community Clinic in Esquipulas, Chiapas, Mexico and the patients for their participation in the study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jds.2019.01.010.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Marshall B.J., Goodwin C.S. Notes: revised nomenclature of Campylobacter pyloridis. Int J Syst Evol Microbiol. 1987;37:8–68. [Google Scholar]
- 2.Goodwin C.S., Armstrong J.A., Chilvers T. Transfer of Campylobacter pylori and Campylobacter mustelae to Helicobacter gen. nov. as Helicobacter pylori comb. nov. and Helicobacter mustelae comb. nov., Respectively. Int J Syst Bacteriol. 1989;39:397–405. [Google Scholar]
- 3.Tomb J.F., White O., Kerlavage A.R. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 4.Kao C.Y., Sheu B.S., Wu J.J. Helicobacter pylori infection: an overview of bacterial virulence factors and pathogenesis. Biomed J. 2016;39:14–23. doi: 10.1016/j.bj.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wen S., Moss S.F. Helicobacter pylori virulence factors in gastric carcinogenesis. Cancer Lett. 2009;282:1–8. doi: 10.1016/j.canlet.2008.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hatakeyama M. Oncogenic mechanisms of the Helicobacter pylori CagA protein. Nat Rev Cancer. 2004;4:688–694. doi: 10.1038/nrc1433. [DOI] [PubMed] [Google Scholar]
- 7.Saadat I., Higashi H., Obuse C. Helicobacter pylori CagA targets PAR1/MARK kinase to disrupt epithelial cell polarity. Nature. 2007;447:330–333. doi: 10.1038/nature05765. [DOI] [PubMed] [Google Scholar]
- 8.Montecucco C., Rappuoli R. Living dangerously: how Helicobacter pylori survives in the human stomach. Nat Rev Mol Cell Biol. 2001;2:457–466. doi: 10.1038/35073084. [DOI] [PubMed] [Google Scholar]
- 9.Leunk R.D., Johnson P.T., David B.C., Kraft W.G., Morgan D.R. Cytotoxic activity in broth-culture filtrates of Campylobacter pylori. J Med Microbiol. 1988;26:93–99. doi: 10.1099/00222615-26-2-93. [DOI] [PubMed] [Google Scholar]
- 10.Phadnis S.H., Ilver D., Janzon L., Normark S., Westblom T.U. Pathological significance and molecular characterization of the vacuolating toxin gene of Helicobacter pylori. Infect Immun. 1994;62:1557–1565. doi: 10.1128/iai.62.5.1557-1565.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cover T.L., Tummuru M.K., Cao P., Thompson S.A., Blaser M.J. Divergence of genetic sequences for the vacuolating cytotoxin among Helicobacter pylori strains. J Biol Chem. 1994;269:10566–10573. [PubMed] [Google Scholar]
- 12.Reyrat J.M., Pelicic V., Papini E., Montecucco C., Rappuoli R., Telford J.L. Towards deciphering the Helicobacter pylori cytotoxin. Mol Microbiol. 1999;34:197–204. doi: 10.1046/j.1365-2958.1999.01592.x. [DOI] [PubMed] [Google Scholar]
- 13.Fennerty M.B. Helicobacter pylori: why it still matters in 2005. Cleve Clin J Med. 2005;72:S14–S21. doi: 10.3949/ccjm.72.suppl_2.s1. [DOI] [PubMed] [Google Scholar]
- 14.Kignel S., de Almeida Pina F., Andre E.A., Alves Mayer M.P., Birman E.G. Occurrence of Helicobacter pylori in dental plaque and saliva of dyspeptic patients. Oral Dis. 2005;11:17–21. doi: 10.1111/j.1601-0825.2004.01043.x. [DOI] [PubMed] [Google Scholar]
- 15.Gebara E.C., Pannuti C., Faria C.M., Chehter L., Mayer M.P., Lima L.A. Prevalence of Helicobacter pylori detected by polymerase chain reaction in the oral cavity of periodontitis patients. Oral Microbiol Immunol. 2004;19:277–280. doi: 10.1111/j.1399-302X.2004.00153.x. [DOI] [PubMed] [Google Scholar]
- 16.Landry R.G., Jean M. Periodontal Screening and Recording (PSR) Index: precursors, utility and limitations in a clinical setting. Int Dent J. 2002;52:35–40. doi: 10.1111/j.1875-595x.2002.tb00595.x. [DOI] [PubMed] [Google Scholar]
- 17.Mapstone N.P., Lynch D.A., Lewis F.A. Identification of Helicobacter pylori DNA in the mouths and stomachs of patients with gastritis using PCR. J Clin Pathol. 1993;46:540–543. doi: 10.1136/jcp.46.6.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brgers R., Schneider-Brachert W., Reischl U. Helicobacter pyloriin human oral cavity and stomach. Eur J Oral Sci. 2008;116:297–304. doi: 10.1111/j.1600-0722.2008.00543.x. [DOI] [PubMed] [Google Scholar]
- 19.Khocht A., Zohn H., Deasy M., Chang K.M. Screening for periodontal disease: radiographs vs. PSR. J Am Dent Assoc. 1996;127:749–756. doi: 10.14219/jada.archive.1996.0310. [DOI] [PubMed] [Google Scholar]
- 20.Covington L.L., Breault L.G., Hokett S.D. The application of periodontal screening and recording™(PSR) on amilitary population. J Contemp Dent Pract. 2003;4:21–30. [PubMed] [Google Scholar]
- 21.Urrutia-Baca V.H., Escamilla-Garcia E., de la Garza-Ramos M.A., Tamez-Guerra P., Gomez-Flores R., Urbina-Rios C.S. In vitro antimicrobial activity and downregulation of virulence gene expression on Helicobacter pylori by reuterin. Probiot Antimicrob Proteins. 2018;10:168–175. doi: 10.1007/s12602-017-9342-2. [DOI] [PubMed] [Google Scholar]
- 22.Calva-Rodriguez R., Luna-Alcantara J.J., Lagunes-Yannelli B., Rivera-Dominguez M.E., Calva-Cerqueira D., Santos-Marcial E. Prevalence and risk faactors of Helicobacter pylori infection in three populations of children in Puebla, Mexico. Rev Gastroenterol Mex. 2006;71:440–445. [PubMed] [Google Scholar]
- 23.De la Garza-Ramos M.A., Jesuacute s AVG., Rogelio A.E.P., Benito PAr, Rauacute l GC., Francisco GlS. Prevalence of Helicobacer pylori in saliva and dental plaque related to periodontal disease and gastritis. Afr J Microbiol Res. 2013;7:2505–2509. [Google Scholar]
- 24.Valdez-Gonzalez J.A., Mares-Moreno P.C., Kowolik M.J., Vargas-Villlarreal J., Gonzalez-Salazar F., De la Garza-Ramos M.A. Detection of Helicobacter pylori in dental plaque of mexican children by real-time PCR. Health. 2014;6:231–235. [Google Scholar]
- 25.Castro-Munoz L.J., Gonzalez-Diaz C.A., Munoz-Escobar A. Prevalence of Helicobacter pylori from the oral cavity of Mexican asymptomatic children under 5 years of age through PCR. Arch Oral Biol. 2017;73:55–59. doi: 10.1016/j.archoralbio.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 26.Mendoza-Cantu A., Urrutia-Baca V.H., Urbina-Rios C.S., De la Garza-Ramos M.A., Garcia-Martinez M.E., Torre-Martinez H.H.H. Prevalence of Helicobacter pylori vacA genotypes and cagA gene in dental plaque of asymptomatic Mexican children. BioMed Res Int. 2017;2017:4923640. doi: 10.1155/2017/4923640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anand P.S., Kamath K.P., Anil S. Role of dental plaque, saliva and periodontal disease in Helicobacter pylori infection. World J Gastroenterol. 2014;20:5639–5653. doi: 10.3748/wjg.v20.i19.5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X.M., Yee K.C., Hazeki-Taylor N. Oral Helicobacter pylori, its relationship to successful eradication of gastric H. pylori and saliva culture confirmation. J Physiol Pharmacol. 2014;65:559–566. [PubMed] [Google Scholar]
- 29.Al Sayed A., Anand P.S., Kamath K.P., Patil S., Preethanath R., Anil S. Oral cavity as an extragastric reservoir of Helicobacter pylori. ISRN Gastroenterol. 2014;2014 doi: 10.1155/2014/261369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng P., Zhou W. Relation between periodontitis and Helicobacter pylori infection. Int J Clin Exp Med. 2015;8:16741. [PMC free article] [PubMed] [Google Scholar]
- 31.Sujatha S., Jalihal U.M., Sharma S. Association between periodontal disease and oral and gastric Helicobacter pylori infection. Indian J Gastroenterol. 2015;34:343–344. doi: 10.1007/s12664-015-0569-0. [DOI] [PubMed] [Google Scholar]
- 32.Dye B.A., Kruszon-Moran D., McQuillan G. The relationship between periodontal disease attributes and Helicobacter pylori infection among adults in the United States. Am J Publ Health. 2002;92:1809–1815. doi: 10.2105/ajph.92.11.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salehi M.R., Shah Aboei M., Naghsh N., Hajisadeghi S., Ajami E. A comparison in prevalence of Helicobacter pylori in the gingival crevicular fluid from subjects with periodontitis and healthy individuals using polymerase chain reaction. J Dent Res Dent Clin Dent Prospects. 2013;7:238–243. doi: 10.5681/joddd.2013.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chiarini A., Calà C., Bonura C. Prevalence of virulence-associated genotypes of Helicobacter pylori and correlation with severity of gastric pathology in patients from western Sicily, Italy. Eur J Clin Microbiol Infect Dis. 2008;28:437–446. doi: 10.1007/s10096-008-0644-x. [DOI] [PubMed] [Google Scholar]
- 35.Hu Z., Zhang Y., Li Z. Effect of Helicobacter pylori infection on chronic periodontitis by the change of microecology and inflammation. Oncotarget. 2016;7:1. doi: 10.18632/oncotarget.11449. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.