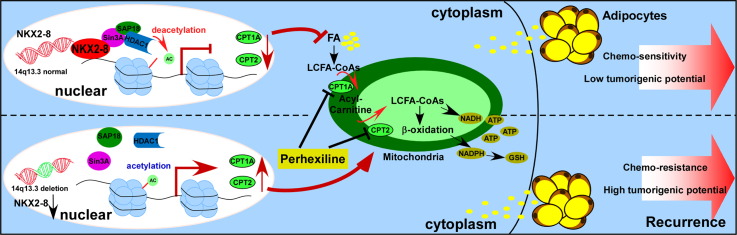
Abstract
Background
Aberrant fatty acid (FA) metabolism is a unique vulnerability of cancer cells and may present a promising target for cancer therapy. Our study aims to elucidate the molecular mechanisms by which NKX2–8 deletion reprogrammed FA metabolism-induced chemoresistance in epithelial ovarian cancer (EOC).
Methods
The deletion frequency and expression of NKX2–8 in 144 EOC specimens were assayed using Fluorescence in situ hybridization and immunochemical assays. The effects of NKX2–8 deletion and the fatty acid oxidation (FAO) antagonist Perhexiline on chemoresistance were examined by Annexin V and colony formation in vitro, and via an intraperitoneal tumor model in vivo. The mechanisms of NKX2–8 deletion in reprogrammed FA metabolism was determined using Chip-seq, metabolomic analysis, FAO assays and immunoprecipitation assays.
Findings
NKX2–8 deletion was correlated with the overall and relapse-free survival of EOC patients. NKX2–8 inhibited the FAO pathway by epigenetically suppressing multiple key components of the FAO cascade, including CPT1A and CPT2. Loss of NKX2–8 resulted in reprogramming of FA metabolism of EOC cells in an adipose microenvironment and leading to platinum resistance. Importantly, pharmacological inhibition of FAO pathway using Perhexiline significantly counteracted NKX2–8 deletion-induced chemoresistance and enhanced platinum's therapeutic efficacy in EOC.
Interpretation
Our results demonstrate that NKX2–8 deletion-reprogrammed FA metabolism contributes to chemoresistance and Perhexiline might serve as a potential tailored treatment for patients with NKX2–8-deleted EOC.
Fund
This work was supported by Natural Science Foundation of China; Guangzhou Science and Technology Plan Projects; Natural Science Foundation of Guangdong Province; The Fundamental Research Funds for the Central Universities.
Keywords: NKX2–8, Chemoresistance, Fatty acid oxidation, Metabolic reprogram, Epithelial ovarian cancer
Graphical abstract
Research in context.
Evidence before this study
Aberrant metabolism, especially fatty acid metabolism, plays vital roles in cancer progression and chemotherapy failure. we previously showed that silencing NKX2–8 promoted progression of bladder cancer and esophageal cancer. However, the molecular mechanisms of NKX2–8 downregulation and NKX2–8 deletion reprogrammed FA metabolism-induced chemoresistance in epithelial ovarian cancer (EOC) is yet unclear.
Added value of this study
In the present study, we reported that NK2 homeobox 8 (NKX2–8) inhibited the fatty acid oxidation (FAO) pathway by epigenetically suppressing multiple key components of the FAO cascade, including carnitine palmitoyltransferase CPT1A and CPT2, via interacting with Sin3A/HDAC1/SAP18 transcriptional repressor complex. Furthermore, genetic ablation of NKX2–8 resulted in reprogramming of FA metabolism of epithelial ovarian cancer (EOC) cells in the adipose microenvironment, leading to platinum resistance. Importantly, pharmacological inhibition of FAO pathway using Perhexiline, an FDA-approved drug approved for the treatment of angina and heart failure, significantly reduced NKX2–8 deletion-induced chemoresistance and enhanced the therapeutic efficacy of platinum in EOC.
Implications of all the available evidence
Our study uncovers a novel mechanism underlying FAO hyperactivation-mediated chemotherapy resistance in EOC and demonstrate that the combination of FAO inhibitor and chemotherapy is a potential tailored treatment for NKX2–8-deleted EOC.
Alt-text: Unlabelled Box
1. Introduction
Aberrant metabolism, an evolving hallmark of human cancer, plays vital roles in cancer progression and chemotherapy failure [[1], [2], [3]]. Various experimental and clinical studies demonstrated that metabolic reprogramming of cancer cells into dysregulated Warburg-like glucose metabolism, glutaminolysis and fatty acid metabolism may favor tumor cell growth and chemotherapy failure [[4], [5], [6]], suggesting that aberrant metabolism is a unique vulnerability of tumor cells and a promising target for cancer therapy.
Ovarian cancer metabolism is unique from that of other cancers because of its adipocyte-rich microenvironment, which relies markedly on fatty acid breakdown for the production of ATP and the NADPH biosynthetic intermediates that contribute to the growth and metastasis of ovarian cancer [[7], [8], [9]]. For instance, adipocytes promoted proliferation of metastatic ovarian cancer cells through transferring fatty acids into fatty acid oxidation (FAO)-hyperactivated tumor cells [9]. SIK2 coupled fatty acid metabolism with survival of ovarian cancer cells at the adipocyte-rich niche via activating both FAO and phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K)/AKT kinase (AKT) pathways [8]. Meanwhile, the polyunsaturated fatty acids secreted by platinum analogs-activated mesenchymal stem cells (MSCs) conferred the resistance of ovarian cancer to multiple types of chemotherapy via augmenting FAO activity [10]. Moreover, pharmacologic inhibition of FAO using carnitine palmitoyltransferase CPT1 and CPT2 inhibitors such as Perhexiline, an FDA-approved drug approved for the treatment of angina and heart failure, has been reported to show distinctive anti-tumor effects in multiple cancer types, such as chronic lymphocytic leukemia (CLL), neuroblastoma, leukemia and HK1−HK2+ liver cancer [[11], [12], [13], [14]]. Hence, all these studied yielded fundamental insights into activated FAO in ovarian cancer and suggested that targeting FAO might be a potential clinical strategy to treat epithelial ovarian cancer (EOC).
Human NKX2–8, a homeobox-containing developmental regulator, was reported to be downregulated in multiple tumors [[15], [16], [17], [18], [19], [20]]. Previously, we showed that silencing NKX2–8 promoted progression of bladder cancer and esophageal cancer [18,19]. Importantly, NKX2–8 knockout mice could spontaneously develop lung cancer at an early stage and exhibited multiple types of tumor in later stage, suggesting that loss of NKX2–8 contributes to tumorigenesis [21]. Herein, we demonstrated that heterogeneous deletion of NKX2–8 reprogrammed FA metabolism through epigenetic upregulation of multiple key components of the FAO cascade. Genetic ablation of NKX2–8 resulted in platinum resistance and a higher recurrence rate of epithelial ovarian cancer by activating the FAO pathway. FAO inhibitor Perhexiline was sufficient to sensitize NKX2–8-deleted EOC, but not NKX2–8-nondeleted EOC, to CDDP-based chemotherapy. These results highlight the vital role of NKX2–8 loss in reprogramming of FA metabolism–induced chemoresistance and may represent a novel therapeutic strategy to treat NKX2–8-deleted EOC.
2. Materials and methods
2.1. Cells
The OVCAR3, CAOV3 and OV90 EOC cell lines (ATCC) were cultured in the DMEM medium supplemented with 10% fetal bovine serum (FBS, Gibco, Grand Island, NY). Human Ovarian Surface Epithelial Cell (HOSEpiC) was obtained from ScienCell Research Laboratories and cultured in the specific medium according to the manufacturer's instructions. The above-mentioned cell lines were identified by performing short tandem repeat fingerprinting in Sun Yat-sen University (Guangzhou, China).
2.2. Tumor specimens
EOC specimens (n = 144) used in the current study were collected from Sun Yat-sen University Cancer Center (Guangdong, China) between 2005 and 2016. The clinical pathological characteristics of EOC patients has been shown in Supplementary Table S1–3. All the EOC patients received standard platinum-based chemotherapy in Sun Yat-sen University Cancer Center. Chemoresistance or chemosensitivity defined as relapse or progression within six months or after six months from the last chemotherapy, respectively. Prior donor consent was obtained from all EOC patients and the approval of using the specimens was obtained from the Institutional Research Ethics Committee.
2.3. DNA/RNA extraction and real-time quantitative PCR and genomic PCR
In the current study, total RNA and genomic DNA were extracted from the EOC cell lines and EOC tissue samples by using TRIzol (Life Technologies) and QIAamp DNA Mini Kit, respectively. cDNAs/DNAs were amplified via using FastStart Universal SYBR Green Master kit (ROX; Roche, Toronto, ON, Canada) and quantified by using the ABI Prism 7500 Sequence Detection System (Applied Biosystems, Foster City, CA). The expression data were normalized to housekeeping gene GAPDH or Line1 to control the variability in expression levels. Deletion of NKX2–8 was analyzed by genomic PCR amplification. The primer sequences were obtained from the Genome database was shown in Primers and Oligonucleotides table in Supplementary Table S4.
2.4. Vectors, retroviral infection and transfection
The retroviral vector pMSCV-neo was used to generate pMSCV-neo/NKX2–8 recombinant plasmid. Small interfering RNA of CPT1A, CPT2, Sin3A, SAP18 and HDAC1 were used to transient inhibit the gene expression. All the detailed sequences of siRNA oligonucleotides are shown in Supplementary Table S4. The promoter region of human CPT1A, including fragments covering nucleotides −853 to +475, and CPT2 promoter, including nucleotides covering −1450 to +483, were subcloned into the pGL3-Control vector luciferase reporter (Promega, USA), respectively. Recombinant plasmids or siRNA used in the current study were transfected by the Lipofectamine 3000 reagent (Invitrogen, Carlsbad, CA). Stable cell lines OV90/NKX2–8 was selected for 10 days with neomycin(1 mg/μl). The NKX2–8 protein level was detected by SDS–PAGE western blotting after ten-day selection to confirm stable expression of NKX2–8 in EOC cell lines. A list of primers used in the present study was shown in Supplementary Table S4.
2.5. Western blotting analysis
Western blotting analysis were performed in accordance with standard procedures. The antibodies used in this study were NKX2–8 (1:500, Abcam, ab125040), CPT1A (1:1000, Abcam, ab128568), CPT2 (1:1000, Abcam, ab181114), Sin3A (1:1000, Abcam, ab3479), SAP18 (1:1000, Abcam, ab31748), HDAC1 (1:2000, Abcam, ab7028), p-AKT1 (phospho S473) (1:500, Abcam, ab81283), p-mTOR (phospho S2448) (1:1000, Abcam, ab109268), p-S6K1 (phospho T389) (1:1000, Abcam, ab60948), p-Bad (phospho S112) (1:2000, Abcam, ab129192), activated caspase-3 (1:500, Abcam, ab2302) and Cytochrome C (5 μg/ml, Abcam, ab13575). Anti-α-Tubulin antibody (1:3000, Abcam, ab7291) was used as a loading control.
2.6. Immunohistochemistry (IHC) assay
IHC assay was performed to determine the protein expression of NKX2–8 (Abcam, ab125040), CPT1A (Abcam, ab128568) and CPT2 (Abcam, ab181114) in 144 EOC samples. The detailed processes of IHC assay were carried out according to previously published paper [22]. Axio Imager.Z2 system (Carl Zeiss Co. Ltd., Jena, Germany) was used to capture the immunohistochemistry images.
2.7. Fluorescence in situ hybridization (FISH)
Fluorescence in situ hybridization assay was conducted to analyze the DNA deletion level of NKX2–8 in human EOC tissues and EOC cell lines, and carried out according to previously published standard protocol [23] using an NKX2–8-specific probe (RiboBio, Guangzhou, China). To determine the NKX2–8 deletion status, the number of red and green signals per nuclei was scored in 200 nuclei from each specimen, which NKX2–8 was considered deleted in EOC when the red/green ratio was <0.85.
2.8. CRISPR/Cas9 system
CAOV3/NKX2–8 and OVCAR3/NKX2–8 knockout cells were conducted with CRISPR/Cas9 system as described previously [24]. GeneChem (Guangzhou, China) designed and cloned the corresponding single guide RNAs into GV392 plasmid (NKX2–8 sgRNA sequences showed in Table S4). Briefly, 3 × 105 CAOV3 and OVCAR3 cells were infected with lenti-CRISPR virus. After 24 h, infected cells were selected for 7 days using 0.5μg/ml puromycin. Afterwards, CAOV3/Cas9 and OVCAR3/Cas9 cells were re-infected with GV392-GFP-NKX2–8 gRNA lentivirus to ensure >95% cells were positive. The infected cells were sorted using flow cytometry and single-cell cloned, the CAOV3/NKX2–8+/− #1 and NKX2–8+/− #2; OVCAR3/NKX2–8+/− #1 and NKX2–8+/− #2 referred to single-cell clone 1 and 2, respectively. Genomic DNA was extracted from CAOV3/NKX2–8+/− #1 and NKX2–8+/− 2; OVCAR3/NKX2–8+/− #1 and NKX2–8+/− #2 cells using a QIAGEN genomic DNA Kit and the target sequence were amplified. Then, heterodimerization and digestion were performed using the Knockout and Mutation Detection Kit (GeneChem) according to the manufacturer's instructions. Mutagenesis was finally confirmed by nucleic acid sequencing. Primers used in the current study is shown in Supplementary Table S4.
2.9. Chromatin immunoprecipitation (ChIP) and ChIP-Immunobloting assay
ChIP assay was performed by using chromatin immunoprecipitation (ChIP) assay kit (Upstate/Millipore, Billerica, Maine, USA) according to the manufacturer's instructions. For ChIP-Immunobloting assay, the indicated EOC cells lysis were treated with or without formaldehyde (1%) for 10 min. Anti-NKX2–8 antibody (Abcam) and Sin3A (Abcam) was used for incubated the cell lysates overnight at 4 °C. The enriched DNA fragments was conducted PCR assay to detect the enrichment of NKX2–8 or Sin3A in the promoter of CPT1A and CPT2. Western blotting was performed to analyzed the protein fractions enriched by sucrose density gradient (1%–28% (W/V)). For ChIP-seq analysis, the enriched DNA fragments from cultured cell lines were subjected to sequence by using HiSeq 2000 (Illumina). The ChIP-seq data was available and downloaded from the NCBI's Sequence Read Archive (PRJNA487880).
2.10. In vivo experiments
Female BALB/c nude mice and NOD/Shi-SCID/IL-2Rγnull (NOG) were purchased from Vitalriver (Beijing, China) and housed under SPF level conditions. NOG mice were random assorted to different groups (n = 6/group) and embedded with fresh clinical EOC patient tissues (2–3 mm3 pieces) coated with a 1:1 ratio of Matrigel (BD Biosciences, San Jose, CA, USA). Nude mice were injected subcutaneously with indicated EOC cells (1 × 106) mixed with adipocytes (100 μl packed cell volume (PCV)) with growth factor-reduced Matrigel (50 μl). In the intraperitoneal tumor model, female nude mice were injected intraperitoneally with the indicated EOC cells (1 × 106). Tumor growth was monitored by living image software. The eq. (L*W2)/2 was used to determine the tumor volume of mice in the current study. The mice were treated with CDDP (5 mg/kg every 3 days) or vehicle (control) when the tumor became palpable (subcutaneous model) or the luminescence signal reached 1 × 107 p/s/cm2/sr (intraperitoneal model). Approved of the Institutional Animal Care and Use Committee of Sun Yat-sen University were obtained before all the in vivo experiment in this study.
2.11. Fatty acid oxidation (FAO) assay
To determine the FAO activity, the rate of carbon dioxide produced from the oxidation of [14C] palmitate (PerkinElmer) was measured in the Nuclear Medicine Department of Sun Yat-sen University. Briefly, tumor cells (2 × 104) were seeded in 24-well plates in the presence of 1 μCi [14C] palmitic acid-BSA complex and incubated for 3 h at 37 °C. The released [14C] carbon dioxide was trapped using filter paper soaked in 100 mM NaOH for 1 h at 37 °C. The rate of FAO activity was determined by calculating the amount of [14C] carbon dioxide in relative units produced per milligram of protein per hour.
2.12. Chemical reagents
Perhexiline was purchased from Sigma-Aldrich (Germany), and Trichostatin A (TSA,) purchased from Selleck Chemicals.
2.13. Metabolomic analysis
All the EOC samples (1 × 107cells/sample, n = 6 per group) were harvested firstly by centrifugation (3500 rpm, 10 min). The residue was suspended by adding equal volume of PBS and low temperature methanol, then centrifuged at 4 °C (3500 rpm, 10 min) to absorb the supernatant and immediately stored at −80 °C for analysis on the LC/MS platform, according to the manufacturer's instructions.
2.14. Adipocyte extraction and co-culture system
Adipocytes were extracted from omental tissues of ovarian cancer patient who undergo surgical procedures in the Sun Yat-sen University Cancer Center. Omental tissue was minced within DMEM/F12 medium which containing collagenase type I and bovine serum albumin. The above mixture was incubated and shake (80 g, 1 h) at 37 °C. Adipocytes cells was filtered by using a 250-μm mesh filter and removed the undigested tissue, and adipocytes were collected by centrifugation at 200 g and maintained in DMEM/F12 medium.
Adipocytes that extracted from omental tissues of ovarian cancer patient were co-cultured with ovarian cancer cell lines in vitro. Briefly, adipocytes (2 × 104) were cultured in DMEM/F12 medium containing collagenase type I, BSA and 50 μg/ml gentamicin reagent (Gibco) in the cell inserts (1.0 um-pores) coated with collagen type I. Meanwhile, EOC cell lines were seeded into the lower wells (2 × 104/well) coated with collagenase type I and the above-mentioned adipocytes cells were transferred to the 24 well culture insert companion plates.
2.15. Mean optical density (MOD) analysis
Mean optical density (MOD) was used to determine the immunostaining of EOC specimen. Briefly, the stained slides were evaluated by using the ZEISS Axio Scan.Z1 Digital Slide Scanner (Carl Zeiss, Germany) and the Image-Pro Plus 6.0 analysis software to measure the mean optical density (MOD). Ten random picked fields in each specimen were analyzed to determine the MOD of the specimen, based on which the mean MOD of a study group of samples was further generated for subsequent inter-group comparative analysis. The MOD data were statistically analyzed by using the Spearman's correlation analysis to compare the average MOD difference between different groups of tissues, and P < .05 was considered significant.
2.16. XF24 extracellular flux analyzer for measurement of cellular oxygen-consumption rate
Oxygen-consumption rate(OCR) was measured by using a Seahorse XF96 Extracellular Flux Analyzer according to the published protocol [25]. In brief, EOC cells (2 × 104) were plated on XF96 cell culture microplates (Seahorse Bioscience, Cat. No. 103010–100) and OCR assay was measured in the indicated cells treated with oligomycin (1.0 μM), FCCP (0.5 μM), 2-DG (10 mM) and Antimycin (2 μM) on a Seahorse XF96 bioanalyzer.
2.17. Annexin-V assay
For evaluation of apoptosis, Annexin-V assay was performed by using the PE Annexin V Apoptosis Detection Kit I (BD Pharmingen) according to the manufacturer's instructions. Briefly, the indicated treated cells (1 × 106) were collected and washed with PBS and the Annexin-V binding solution, and added 150 μl Annexin-V antibody,1.5 μl of PI at 1 mg/ml in binding buffer and incubated for 15 min at room temperature in the dark, subsequently analyzed with an EPICS XL flow cytometer (Beckman-Coulter).
2.18. ROS analysis
Intracellular ROS levels were determined by using the ROS detection kit (Beyotime, Jiangsu, China) according to the manufacturer's instructions. ROS levels was detected by using flow cytometry system (FACScan, Becton Dickenson, USA) and analyzed with FlowJo 7.6 software. Fluorescence values were normalized to protein load.
2.19. Streptavidin affinity purification of dCas9-captured DNA and proteins
Streptavidin affinity purification of dCas9-Captured DNA and proteins was performed following the published protocol [26]. Briefly, OVCAR3 cells (5 × 107)) which transfected with FB-dCas9 plasmid and CPT1A or CPT2 promoter sgRNAs were harvested when treated with 1% formaldehyde to cross-link the proteins to DNA. Anti- streptavidin antibody was used for incubated the cell lysates. The ChIP DNA was purified by QIA quick Spin columns (QIAGEN) and proteins were separated by SDS-PAGE gel, subsequencely analyzed by IP-MS analysis (Shanghai Applied Protein Technology Co.Ltd.).
2.20. Assessment of ATP, NADH and NADPH level
The indicated EOC cells were seeded in 96-well plates (2 × 104 cells) and treated with 10 μM Perhexiline for 24 h. The ATP, NADH and NADPH levels were determined after treatment with perhexiline (Sigma-Aldrich) by using the CellTiter-Glo Luminescent Cell Viability Assay (Promega), NAD/NADH Assay kit (Abcam, ab65348) and NADP/NADPH Quantitation Kit (Sigma-Aldrich, MAK038-1KT) according to the manufacturer's instructions, respectively.
2.21. Cell survival assay
The indicated cells were plated in 6-well plates (2 × 103 cells) and treated with CDDP (5 μM) and cultured for 10 days. The colonies were fixation with 10% formaldehyde for 5 min and washed three times, then stained with 1% crystal violet for 30 min. The number of colonies (defined as cell clusters consisting of at least 50 cells) was quantified by Analysis software (Olympus Biosystems).
2.22. Drug combination analysis
To evaluate the effects of CDDP and perhexiline combination treatment, NKX2–8+/− and NKX2–8+/+ cells were incubated with different ratios of CDDP and perhexiline and cell viability was analyzed using MTT assays. Briefly, 3000 cells/well were seeded into 96-well plates, grown for 24 h, and treated with CDDP and perhexiline alone, different weight ratios of CDDP and perhexiline combination (CDDP/perhexiline = 10:1, 5:1, 2:1, 1:1, 1:2, 1:5, and 1:10). After incubation for 24 h, 100 ml sterile MTT dye (0.5 mg/ml, Sigma) was added to each well for 4 h at 37 °C. Then, the media were removed and 150 μl dimethyl sulfoxide (DMSO, Sigma) was added. Finally, the absorbance of each well was measured at 570 nm using an EPICS XL flow cytometer (Beckman-Coulter).To evaluate the synergistic effects of CDDP and perhexiline combinations, Synergistic drug interactions were analyzed using the Chou and Talalay method [27] . The combination index (CI) for drug combinations is derived according to the equation below where n = number of drugs
According to the equation, CI <1 indicates synergy, CI = 1 indicates additivity, and CI >1 indicates antagonism. The above median effect equation and CI allow a plot of CI values at different effect levels (fas) to be determined using CalcuSyn software (Biosoft).
2.23. Statistical analysis
In the current study, all the data analysis was conducted by using SPSS 21.0 statistical software (IBM, Armonk, NY, USA). Student's t-test was performed to analysis differences between two variables. The relationship between NKX2–8 expression and the clinicopathological characteristics and prognosis of EOC patients were analyzed by using pearson's Chi-square test, log-rank analysis, Kaplan–Meier and Cox regression model. Data represented the mean ± SD. P values <.05 were considered statistically significant.
3. Results
3.1. Heterozygous deletion of NKX2–8 correlates with a worse prognosis in patients with cancer
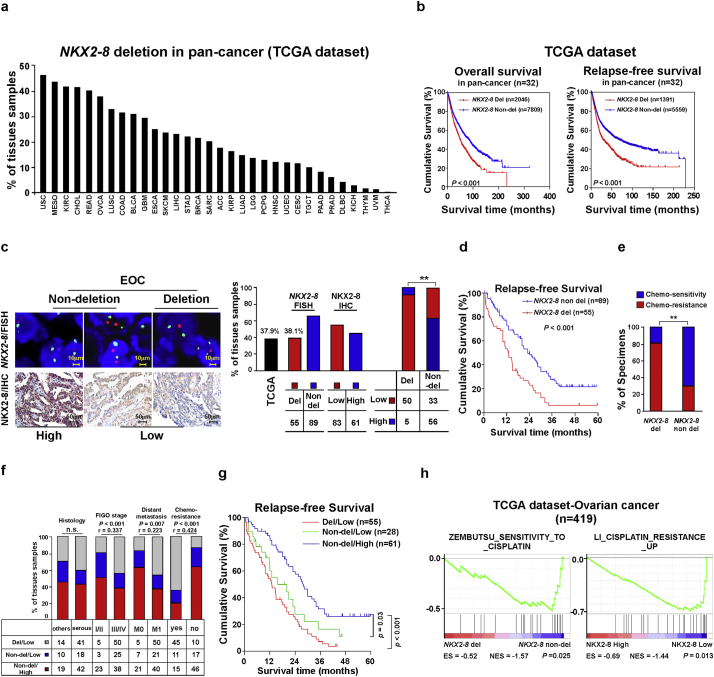
Previously, it has been shown that NKX2–8−/− or NKX2–8+/− mice spontaneously developed multiple tumors, suggesting a tumor suppressive role of NKX2–8 [21]. To decipher the molecular pathogenesis of NKX2–8 deletion in tumorigenesis, we examined the patterns of NKX2–8 deletion in 9855 tumors spanning 32 human tumor types (TCGA dataset). As shown in Fig. 1a, all 32 tumor types displayed heterozygous deletions to varying degrees of NKX2–8 and 16 types of tumor exhibited above 20% heterozygous deletion of NKX2–8. Importantly, patients with NKX2–8-deleted cancer had significant shorter overall and relapse-free survival compared with patients without NKX2–8 deletion (P < .001; P < .001; Fig. 1b), suggesting that loss of NKX2–8 results in a worse prognosis for patients with cancer.
Fig. 1.
NKX2–8 deletion correlates with worse prognosis in patients with EOC. a. GISTIC2.0 analysis of 32 human cancer types (The Cancer Genome Atlas, TCGA) showing varying degrees of heterozygous deletions of NKX2–8. b. Loss of NKX2–8 was correlated with overall and relapse-free survival in the TCGA pan-cancer cohort (P < .001; n = 9822; 32 cancer types). c. EOC was analyzed by FISH for NKX2–8 deletion (left, upper), IHC for NKX2–8 protein (left, lower), and the correlation between NKX2–8 deletion and protein expression (right) (P < .01). d. Loss of NKX2–8 was associated with shorter relapse-free survival in EOC (P < .001; n = 144). e. Significant correlation between NKX2–8 deletion and chemoresistance in EOC (P < .01; n = 144). f. Significant correlation between NKX2–8 protein expression and NKX2–8 deletion with the EOC clinical pathological parameters. NKX2–8 expression was stratified into three groups: (i) NKX2–8 del/low, (ii) NKX2–8 non-del/low and (iii) NKX2–8 non-del/high. g. Kaplan-Meier survival analysis of low protein expression/NKX2–8 deletion, low protein expression/NKX2–8 non-deletion, and high expression/NKX2–8 non-deletion. h. GSEA analysis showing that NKX2–8 deletion and NKX2–8 mRNA levels was correlated with CDDP resistance-related gene signature in the TCGA ovarian cancer dataset (P < .05; n = 419).
3.2. Loss of NKX2–8 is associated with chemoresistance in EOC
One of the major reasons for poor prognosis in cancer patients is chemotherapy resistance [28]. More than 75% of patients with EOC quickly become resistant to standard platinum-based treatment and suffer recurrence, therefore EOC was selected as our study model. Consistent with the data presented in the TCGA EOC profile (37.9%), fluorescence in situ hybridization (FISH) analysis of our EOC cohort (38.1%) revealed that the frequency of NKX2–8-deletion increased with increasing FIGO stage and shorter relapse-free survival (Fig. 1c and d; Supplementary Fig. S1a and b). We found that 81.8% of NKX2–8-deletion EOC cases exhibited chemoresistance whereas 70.8% of cases without NKX2–8 deletion were chemosensitive (Fig. 1e), suggesting that loss of NKX2–8 might be involved in chemotherapy failure.
Complementing NKX2–8 expression with NKX2–8 gene copy alterations revealed that 90.9% of NKX2–8-deleted EOC cases displayed low NKX2–8 expression whereas 62.9% of EOC cases without NKX2–8 deletion exhibited high NKX2–8 expression (Fig. 1c), suggesting that heterozygous deletions represent a major mechanism of NKX2–8 protein reduction. Next, 144 EOC specimens from patients who received CDDP-based chemotherapy were divided into three groups according to NKX2–8 deletion/protein expression patterns: (1) non-del/high, (2) non-del/low and (3) del/low. The NKX2–8-del/low group was characterized by a higher FIGO grade and failure of CDDP-based chemotherapy and had the shortest relapse-free survival time (Fig. 1f and g; Supplementary Table S1–3), which suggest that loss of NKX2–8 is involved in EOC chemoresistance and recurrence. This hypothesis was further confirmed by Gene Set Enrichment Analysis (GSEA) that NKX2–8-deletion and NKX2–8 mRNA levels were significantly associated with gene signature-related CDDP-resistance in EOC (Fig. 1h).
3.3. The adipose microenvironment enhances chemoresistance of NKX2–8-deleted EOC cells
To determine the effect of NKX2–8 deletion on EOC chemoresistance, two EOC cell lines CAOV3 and OVCAR3, which exhibited no NKX2–8 gene copy alterations, were selected to establish NKX2–8+/− cells via CRISPR/Cas9 genome editing technology (Supplementary Fig. S2a and b). Interestingly, genetic ablation of NKX2–8 only slightly enhanced the resistance capability of EOC cells to CDDP treatment, while co-cultured with adipocytes or treatment with adipocyte-derived conditioned medium (adipocyte-CM) resulted in increased tolerance of NKX2–8-deleted EOC cells to CDDP treatment, which was indicated by decreased numbers of apoptotic cells and an increased CDDP IC50 value (Supplementary Fig. S2c-e). On the other hand, co-cultured with adipocytes or treatment with adipocyte-CM significantly decreased the cytotoxic effect of CDDP in OV90 cells, which showed a heterozygous deletion of NKX2–8 (Supplementary Fig. S2a, g and h). However, overexpression of NKX2–8 significantly abrogated the effect of adipose microenvironment-induced CDDP resistance of OV90 cells (Supplementary Fig. S2f-h). These results suggest that adipose microenvironment is involved in the NKX2–8-deletion-induced chemoresistance.
3.4. NKX2–8 deletion contributes to EOC chemoresistance in the adipose microenvironment
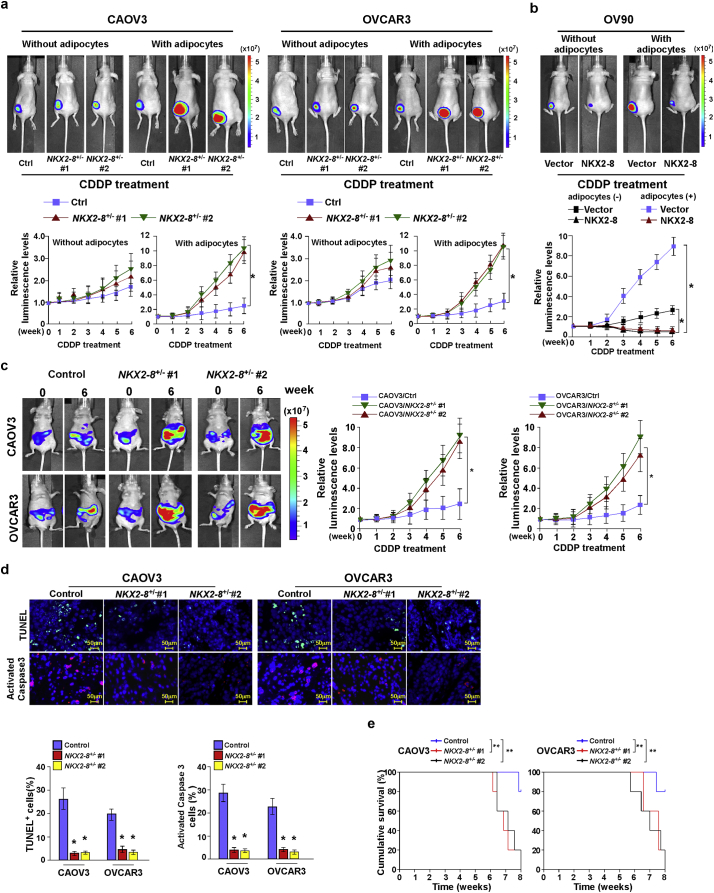
In agreement with the in vitro results, in vivo subcutaneous tumor model experiments showed that mixing CDDP with adipocytes significantly abolished its anti-tumor effect on subcutaneous tumor growth of NKX2–8-deleted EOC cells, as evidenced by rapid tumor progression, and lower proportions of TUNEL+- and activated caspase-3+-apoptotic cells. However, these effects were not observed using NKX2–8-nondeleted EOC cells (Fig. 2a and Supplementary Fig. S3a-c). Furthermore, adipose microenvironment-induced CDDP resistance of OV90 cells was dramatically reduced by restoring NKX2–8 expression (Fig. 2b and Supplementary Fig. S3d). These results indicate that the adipose microenvironment enhanced the chemoresistance of NKX2–8-deleted EOC cells.
Fig. 2.
NKX2–8 deletion contributes to EOC chemoresistance in adipose microenvironment. a-b. In vivo growth of subcutaneous tumors formed by the indicated EOC cells mixed with or without adipocytes injected in the dorsal flank of the mice. Representative images of tumor-bearing mice (left, upper) and quantification (right) of bioluminescence signal were measured on the indicated days (left, lower). (⁎P < .05, ANOVA, n = 6 mice/group). c. Representative images (left) and quantification (right) of intraperitoneal tumor-bearing nude mice in the indicated time (⁎P < .05, ANOVA, n = 6 mice/group). d. IHC staining for TUNEL and activated caspase-3 (left) and quantification of the apoptotic rate in the indicated xenograft tumors (right). ⁎P < .05. e. Kaplan-Meier survival of CDDP-treated intraperitoneal tumor-bearing nude mice. ⁎⁎P < .01.
Given that the omentum and mesentery are major repositories of visceral white adipose tissues and are frequently involved in the progression of ovarian cancer, an intraperitoneal xenograft model was then further employed (Fig. 2c). When the bioluminescence signal of the intraperitoneal tumors reached 1 × 107 p/s/cm2/sr, nude mice were treated with vehicle or CDDP for 6 weeks. Consistently, in the intraperitoneal adipose-enriched microenvironment, genetic deletion of NKX2–8 dramatically abolished the anti-tumor effect of CDDP on the intraperitoneal growth of EOC cells, as indicated by rapid tumor growth, lower proportion of TUNEL+- and caspase-3+-apoptotic cells and shorter survival time of tumor-bearing mice (Fig. 2c–e). These results demonstrate that NKX2–8+/−/EOC possesses a stronger capability of chemoresistance in the adipose microenvironment.
3.5. NKX2–8 deletion activates FAO signalling and reprograms fatty acid metabolism
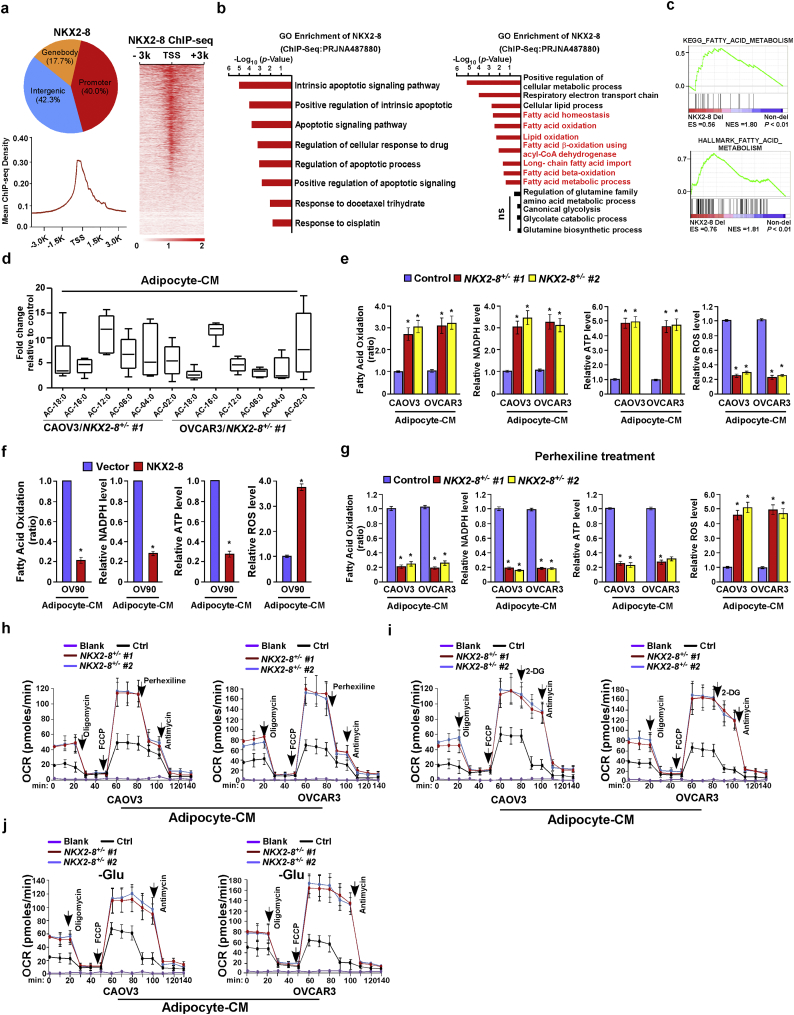
To further investigate the mechanism by which the adipose microenvironment enhances chemoresistance of NKX2–8-deleted cells, NKX2–8-associated gene promoters were profiled using ChIP-Seq (Fig. 3a). Gene ontology (GO) enrichment analysis revealed that genes with GO biological process terms “Intrinsic apoptotic signaling pathway”, “Apoptotic signaling pathway” and “Response to CDDP” were enriched (Fig. 3b). Strikingly, genes with GO terms such as “Long- chain fatty acid import”, “fatty acid beta-oxidation” and “fatty acid β-oxidation using acyl-CoA dehydrogenase” were also enriched. Similar enrichments were also observed in public dataset via GSEA and GO enrichment analysis (Fig. 3c and Supplementary Fig. S4a). The above results suggest an important role of NKX2–8 in regulation of FAO signalling. This hypothesis was further confirmed by multiple assays, in which in the presence of adipocyte-CM, metabolite analysis revealed a higher amount of long-chain acylcarnitine intermediates in NKX2–8-deleted EOC cells compared with that in control cells, and the FAO activity and production levels of NADPH and ATP were also significantly increased in NKX2–8-deleted EOC cells but decreased in NKX2–8-transduced cells compared with those in control cells (Fig. 3d–f). Conversely, the ROS level was significantly decreased in adipocyte-CM-treated NKX2–8+/−-cells but increased in adipocyte-CM-treated NKX2–8-transduced cells (Fig. 3e and f).
Fig. 3.
NKX2–8 deletion activates FAO signalling and reprograms fatty acid metabolism. a. Distribution of NKX2–8 binding sites in the promoter (−3 kb to +3 kb), gene body, and intergenic regions (top). NKX2–8 binding regions were enriched in the gene promoters (bottom). Heatmap of the ChIP enrichment signal in NKX2–8-binding regions (right). b. GO enrichment analysis of the NKX2–8 regulated-transcripts identified in ChIP-seq analysis in OVCAR3/NKX2–8 cells treatment with adipocyte-CM. c. The results of GSEA analysis associated with NKX2–8-deletion and fatty acid metabolism in the TCGA EOC dataset. d. Fold change in metabolite levels in NKX2–8 deleted EOC cells. The values are presented as minimum-to-maximum box plots for six experiments per group. e. Relative FAO activity and level of NADPH, ATP and ROS in adipocyte-CM-treated NKX2–8 deleted EOC cells compared with adipocyte-CM-treated control cells. f. Relative FAO activity and level of NADPH, ATP and ROS in the indicated adipocyte-CM-treated EOC cells. g. Relative FAO activity and level of NADPH, ATP and ROS in the indicated adipocyte-CM-treated EOC cells treatment with Perhexiline (10 μM) for 24 h. h–j. Effects of Perhexiline (H), 2-DG (I), or glutamine withdrawal (J) on the oxygen consumption rate (OCR) in the indicated adipocyte-CM-treated EOC cells. Each bar in Fig. 3e-j represents the mean ± SD of three independent experiments. ⁎P < .05.
Moreover, we observed that inhibition of FAO signalling with Perhexiline, a FAO inhibitor, in the adipocyte-CM-treated NKX2–8+/−-cells resulted in significantly decreasing of oxygen consumption rate (OCR), FAO activity, ATP and NADPH production and but led to more increased ROS level compared to control cells (Fig. 3g and h). However, treatment with glycolysis inhibitor 2-DG or withdrawing glutamine had no obvious effect on OCR in adipocyte-CM-treated NKX2–8+/−-cells (Fig. 3i and j). These results indicate that loss of NKX2–8 induces the reprogramming of fatty acid metabolism of EOC cells in the adipose microenvironment.
3.6. NKX2–8 transcriptionally represses multiple key components of the FAO cascade
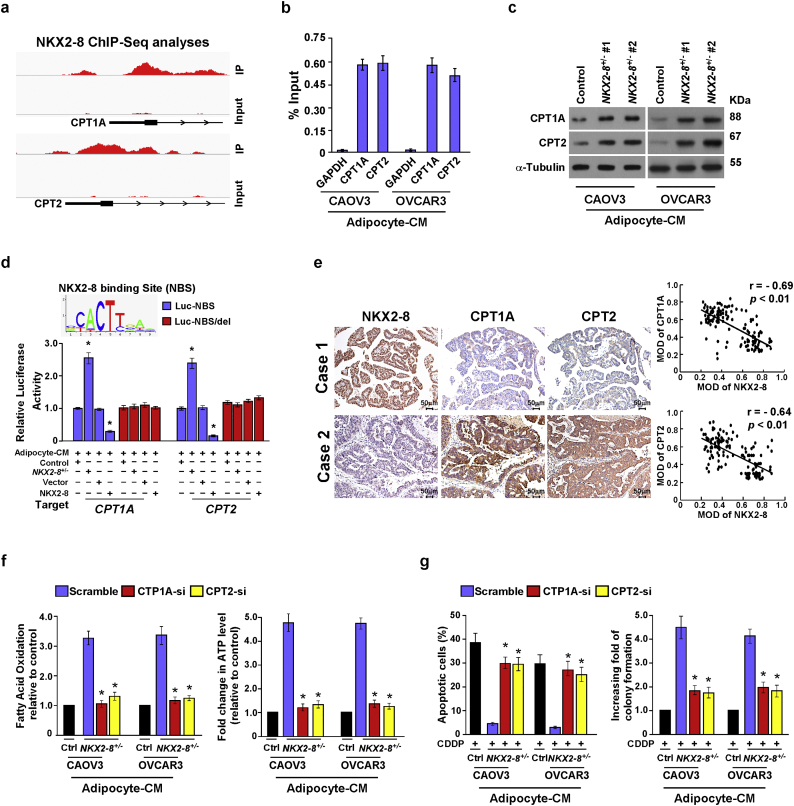
Analysis of the NKX2–8 ChIP-seq results suggested that the two key components (CPT1A and CPT2) in the FAO pathway might be downstream targets of NKX2–8 (Fig. 4a), which was further confirmed by a ChIP assay in which NKX2–8 was most significantly associated with the CPT1A and CPT2 promoters in adipocyte-CM-treated EOC cells (Fig. 4b). Moreover, we found that expression of CPT1A and CPT2, at both the mRNA and protein levels, and the luciferase activity driven by the promoters with a NKX2–8-specific binding site (NBS) of these genes was increased in NKX2–8-deleted cells but decreased in NKX2–8-transduced cells in the presence of adipocyte-CM (Supplementary Fig. S4b and c; Fig. 4c and d). However, no effect was shown on the luciferase activities of the promoter with a deleted NKX2–8-binding site (Fig. 4d). The adverse association of NKX2–8 with FAO activity was further confirmed via IHC analysis in EOC specimens, in which NKX2–8 levels correlated with the levels of CPT1A and CPT2 (Fig. 4e). Therefore, our results suggest that NKX2–8 regulates FAO-related genes via direct association with their promoters in the adipose microenvironment.
Fig. 4.
NKX2–8 transcriptionally represses multiple key components of the FAO cascade. a. NKX2–8 ChIP-Seq binding profiles on promoter of CPT1A and CPT2 genes. Images were created with the Integrative Genomic Viewer (IGV). b. ChIP analysis of the association between NKX2–8 and CPT1A and CPT2 genes transcripts in adipocyte-CM-treated EOC cells. c. Immunoblotting analysis of CPT1A and CPT2 protein expression in the indicated adipocyte-CM-treated EOC cells. α-Tubulin served as a loading control. d. Relative promoter luciferase activities of CPT1A and CPT2 in the indicated adipocyte-CM-treated EOC cells. e. IHC (left) and correlation analysis (right) of NKX2–8 and CPT1A or CPT2 in EOC tissues (n = 144), the ZEISS Axio Scan.Z1 Digital Slide Scanner (Carl Zeiss, Germany) and the Image-Pro Plus 6.0 analysis software were used to measure the mean optical density (MOD). f. Quantification of FAO activity (left) and ATP levels (right) in the indicated cells treatment with adipocyte-CM. g. The apoptotic index in the indicated adipocyte-CM-treated EOC cells, as analyzed using annexin V assay (left) and colony formation assay (right). Error bars in Fig. 4b, d, f, g represent the mean ± SD of three independent experiments. ⁎P < .05.
Additionally, individually silencing CPT1A and CPT2 not only significantly abolished the effects of NKX2–8-deletion on FAO activity and ATP production, but also drastically abrogated NKX2–8-deletion-induced chemoresistance in the adipose microenvironment (Supplementary Fig. S4d and Fig. 4f and g). Importantly, statistical analysis revealed that the expression levels of CPT1A and CPT2 correlated significantly with shorter relapse-free survival in patients with EOC (Supplementary Fig. S4e). Consistently, analysis of public accessed dataset showed that the mRNA expression levels (GSE 38666, GSE 18520) of CPT1A and CPT2 were also significantly increased in EOC and were positively correlated with shorter overall and relapse-free survival in EOC patients with platinum-based chemotherapy (Supplementary Fig. S4f-g). Taken together, our results further support the notion that fatty acid metabolism is involved in NKX2–8-deletion-induced chemoresistance in EOC cells in the adipose microenvironment.
3.7. The Sin3A/HDAC1/SAP18 repressor complex is involved in NKX2–8-inhibited FAO signalling
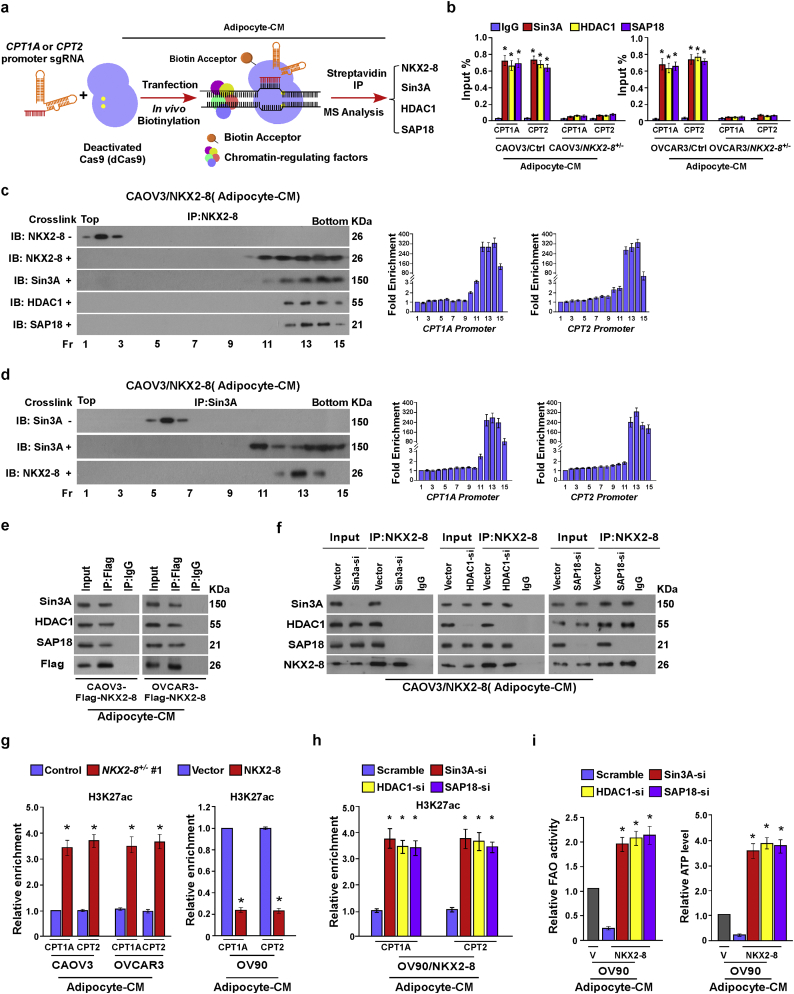
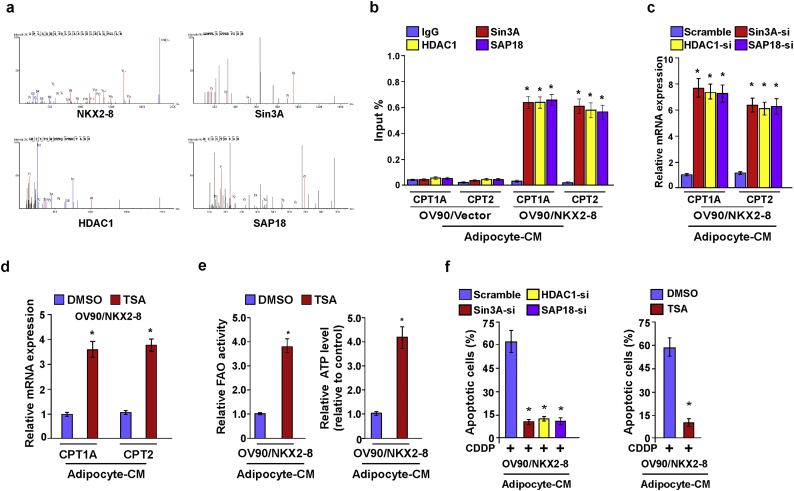
To further explore the mechanism by which NKX2–8 transcriptionally represses CPT1A and CPT2, biotinylated deactivated Cas9 (dCas9) capture analysis was performed. As shown in Fig. 5a and Supplementary Fig. S5a, NKX2–8 and Sin3A/HDAC1/SAP18 repressor complex were identified to be involved in the transcriptional regulation of CPT1A and CPT2 in EOC cells. Consistently, a ChIP assays showed that deletion of NKX2–8 significantly decreased, but overexpression of NKX2–8 increased, the level of Sin3A, HDAC1 and SAP18 on the promoter of CPT1A and CPT2 genes in adipocyte-CM-treated EOC cells (Fig. 5b and Supplementary Fig. S5b). ChIP-immunobloting assay showed that NKX2–8 interacted with Sin3A/HDAC1/SAP18 complex and at the promoters of CPT1A and CPT2 in adipocyte-CM-treated EOC cells (Fig. 5c and d). Moreover, we found that silencing of Sin3A dramatically decreased the interaction between NKX2–8 and HDAC1/SAP18, whereas silencing HDAC1 or SAP18 did not alter the association between NKX2–8 and Sin3A in adipocyte-CM-treated EOC cells (Fig. 5e and f), suggesting that Sin3A mediated the association of NKX2–8 with the Sin3A/HDAC1/SAP18 repressor complex in the adipose microenvironment.
Fig. 5.
The Sin3A/HDAC1/SAP18 repressor complex is involved in NKX2–8-inhibited FAO signalling. a. Schematic of dCas9-mediated capture of chromatin interactions beteween NKX2–8 and the Sin3A/HDAC1/SAP18 complex. b. ChIP analysis of the association of Sin3A, HDAC1 or SAP18 with the promoter of CPT1A and CPT2 gene in the indicated adipocyte-CM-treated EOC cells. c-d. The indicated adipocyte-CM-treated EOC cells were treated with 1% formaldehyde for crosslinking. Then anti-NKX2–8 (c) or anti-Sin3A antibody (d) was incubated with treated lysates for ChIP assays, followed by size fractionation with sucrose gradient ultracentrifugation. Eluate gradients were used for Western blot and PCR assays. e. immunoprecipitation assay showing that NKX2–8 specifically interacted with the Sin3a/HDAC1/SAP18 complex in the indicated adipocyte-CM-treated EOC cells. f. Depletion of Sin3A impaired the interaction between NKX2–8 and HDAC1/SAP18 (left). Depletion of HDAC1 did not affect the interaction between NKX2–8 and Sin3A/SAP18 (middle). Depletion of SAP18 did not alter the interaction between NKX2–8 and Sin3A/HDAC1 (right) in the indicated adipocyte-CM-treated EOC cells. g-h The binding between H3K27ac and CPT1A and CPT2 promoter was determined by ChIP analysis in the indicated adipocyte-CM-treated EOC cells. Total DNA and IgG were served as input and antibody control, respectively. i. Relative FAO activity (left) and ATP levels (right) in the indicated adipocyte-CM-treated EOC cells. Error bars in Fig. 5b, c, d, g, h, i represent the mean ± SD of three independent experiments. ⁎P < .05.
ChIP assays revealed that genetic deletion of NKX2–8 significantly increased the enrichment of H3K27ac at the promoters of CPT1A and CPT2 compared to the control cells, whereas but overexpression of NKX2–8 decreased the enrichment of H3K27ac at the promoters of CPT1A and CPT2 compared to the control cells in the presence of adipocyte-CM, which suggested that a histone epigenetic modification contributed to NKX2–8 deletion-mediated activation of FAO signalling in EOC cells in the adipose microenvironment (Fig. 5g). As expected, individually silencing Sin3A, HDAC1, or SAP18 dramatically increased the enrichment of H3K27ac in promoters of these genes and increased the transcriptional expression of CPT1A and CPT2, as well as FAO activity and ATP level, in NKX2–8-transduced EOC cells in the presence of adipocyte-CM (Fig. 5h and i and Supplementary Fig. S5c). Furthermore, treatment with Trichostatin A (TSA), a general histone deacetylase (HDAC) inhibitor, in NKX2–8-transduced cells not only increased CPT1A and CPT2 transcriptional levels but also elevated the FAO activity and ATP levels in the adipose microenvironment (Supplementary Fig. S5d and e). More importantly, the effect of NKX2–8-induced chemosensitivity in the adipose microenvironment was drastically abolished by individually silencing of Sin3A, HDAC1, SAP18, or TSA treatment (Supplementary Fig. S5f), which provide further evidence that the Sin3A/HDAC1/SAP18 repressor complex is essential for NKX2–8-mediated inhibition of FAO signalling and chemoresistance in the adipose microenvironment.
3.8. Blocking FAO signalling abrogated the adipose microenvironment-induced chemoresistance of NKX2–8-deleted EOC cells in vitro
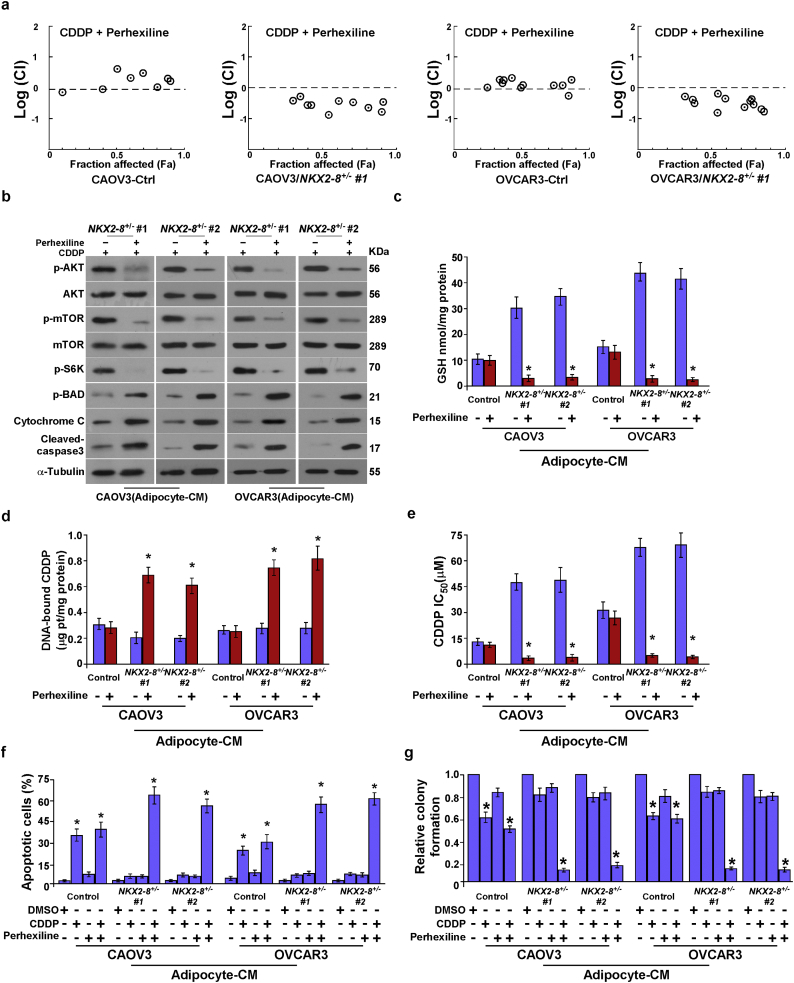
Furthermore, drug combination index analysis indicated a synergistic anti-tumor effect of FAO inhibition and CDDP in NKX2–8-deleted EOC cells but not in NKX2–8-nondeleted cells in an adipose microenvironment (Fig. 6a). We found that blocking FAO signalling using Perhexiline led to inhibition of Akt and mTOR/S6K pathways but increased levels of p-Bad expression in NKX2–8-deleted cells, which resulted in the release of more cytochrome c and increased activated caspase 3 (Fig. 6b). Interestingly, we found that the intracellular glutathione (GSH) level was much lower in NKX2–8+/− cells treated with Perhexiline, but the content of genomic DNA-bound CDDP was significantly increase in NKX2–8+/− cells when treated with Perhexiline (Fig. 6c and d), suggesting that Perhexiline treatment led to inhibit CDDP efflux in NKX2–8+/− cells. Consistently, Perhexiline treatment significantly decreased the IC50 values for CDDP in -treated NKX2–8+/− EOC cells under the adipose microenvironment (Fig. 6e), which further confirmed by colony formation and Annexin V assays that Perhexiline treatment significantly augmented the cellular toxic effects of CDDP in NKX2–8+/− cells but only had a minor effect in NKX2–8+/+ cells in the adipose microenvironment (Fig. 6f and g). Collectively, our results demonstrated that inhibition of the FAO pathway reversed the resistance capability of NKX2–8-deleted EOC cells to platinum treatment in the adipose microenvironment.
Fig. 6.
Blocking FAO signalling abrogates adipose microenvironment-induced chemoresistance of NKX2–8-deleted EOC cells in vitro. a. Logarithmic combination index plot for the combination of Perhexiline and CDDP in the indicated cells. b. Immunoblotting analysis of p-Akt, Akt, p-mTOR, mTOR, p-S6K, p-BAD, Cytochrome c and activated caspase 3 in the indicated adipocyte-CM-treated EOC cells. α-Tubulin served as a loading control. c. Intracellular GSH level in the indicated adipocyte-CM-treated EOC cells. d. Content of DNA-bound CDDP in the indicated adipocyte-CM-treated EOC cells. e. Relative IC50 value for CDDP in the indicated adipocyte-CM-treated cells using MTT cell viability assays. f. FACS analysis of annexin V staining in the indicated adipocyte-CM-treated EOC cells. g. Quantification of the number of colonies formed by the indicated adipocyte-CM-treated EOC cells. Error bars in Fig. 6c-g represent the mean ± SD of three independent experiments. ⁎P < .05.
3.9. Inhibiting FAO signalling sensitizes NKX-2-8-deleted EOC to CDDP therapy in vivo
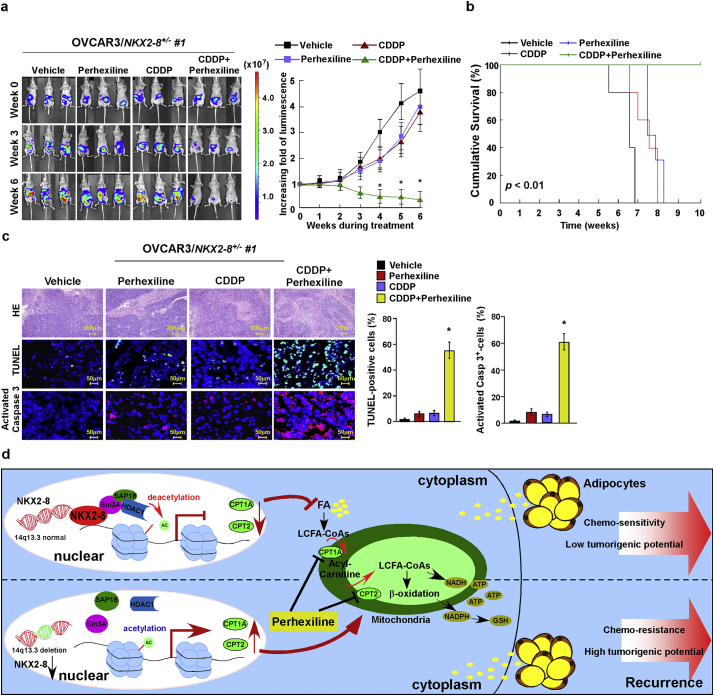
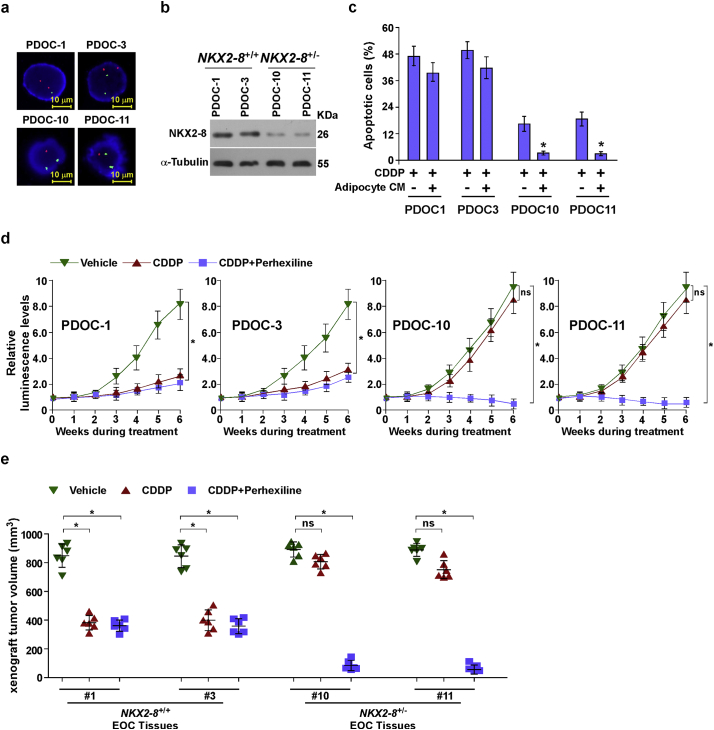
The therapeutic efficacy of inhibiting FAO with Perhexiline was further tested using intraperitoneal xenograft model. As shown in Fig. 7a–c, combined therapy with Perhexiline significantly enhanced the curative efficacy of CDDP on the growth of NKX2–8+/− tumors and prolonged the survival of the tumor-bearing mice of NKX2–8+/− tumors after CDDP therapy, providing further evidence that activation of FAO signalling plays a vital role in EOC recurrence. Furthermore, two NKX2–8+/+ and two NKX2–8+/−patient-derived EOC cells, isolated from fresh clinical EOC tissues, were tested (Supplementary Fig. S6a and b). As shown in Supplementary Fig. S6c, treatment with adipocyte-CM significantly decreased the cytotoxic effect of CDDP in NKX2–8+/−/ patient-derived ovarian cancer (PDOC) cells but not in NKX2–8+/+/PDOC cells. Consistently, the intraperitoneal xenograft model also showed that tumors formed by NKX2–8+/−/PDOC cells were more sensitive to co-treatment with Perhexiline and CDDP (Supplementary Fig. S6d and e). Taken together, our results demonstrate that NKX2–8-deletion contributes to chemoresistance via activation of FAO signalling in EOC cells in an adipose microenvironment (Fig. 7d).
Fig. 7.
Inhibiting FAO signalling sensitizes NKX-2-8-deleted EOC to CDDP therapy in vivo. a. Representative images (left) and quantification (right) of bioluminescence signal in intraperitoneal tumor-bearing nude mice treated with vehicle (control), or CDDP (5 mg/kg), or Perhexiline (3 mg/kg), or a combination of CDDP (5 mg/kg) and Perhexiline (3 mg/kg) treatment compared with pre-treatment tumors (week 0). (⁎P < .05, ANOVA, n = 6 mice/group). b. Kaplan-Meier survival of OVCAR3/NKX2–8+/−-mice treated with the indicated reagent(s). c. IHC staining for TUNEL and activated caspase-3 (left) and quantification (right) of the apoptotic rate in the indicated xenograft tumors (right). ⁎P < .05. d. Schematic diagram illustrating that NKX2–8-deleted reverses chemoresistance by activation of FAO signalling, consequently resulting in EOC recurrence.
4. Discussion
Metabolic energy changes have recently emerged as an important determinant of chemotherapy resistance [29,30]. Numerous studies linking aberrant energy metabolism to chemoresistance have revealed that elevated ATP and NADPH levels, which are produced from aerobic glycolysis in the cytoplasm and oxidative phosphorylation (OXPHOS) in the mitochondria, confer resistance of cancer cells to multiple types of chemotherapeutic drugs [11,[31], [32], [33]]. A series of recent reports suggested that mitochondrial FAO plays a major role in sustaining ATP levels and NADPH production [[34], [35], [36], [37], [38], [39]], suggesting that targeting FAO might be an alternative therapeutic approach for cancer. Herein, we reported that pharmacologically inhibiting FAO using Perhexiline, an anti-angina drug that inhibits carnitine palmitoyltransferases 1 and 2 (CPT-1 and CPT-2), considerably decreased energy metabolism to enhance the therapeutic efficacy of CDDP-chemotherapy in NKX2–8-deleted EOC. Therefore, these findings not only provide mechanistic and clinical insights into EOC chemoresistance, but also provide evidence that the FAO pathway is a druggable target to prevent chemoresistance.
The relationship between EOC progression and fatty acid metabolism has long been known. Previously, it was reported that an increased intake of foods with high levels of saturated fats or cholesterol leads to altered systemic lipid metabolism and elevated serum lipid levels, which are positively associated with the risk of ovarian cancer [6,40]. Recently, analysis of the ovarian metabolome showed dramatically increased FAO in primary and metastatic EOC [41]. In addition, a large-scale plasma metabolomics study indicated that the metabolic pathways in patients with EOC are mainly characterized by aggressive FAO and abnormal phospholipid metabolism [42]. Taken together, these studies provided fundamental insights into the activation of FAO in EOC. However, the precise mechanism underlying FAO hyperactivation in EOC is unknown. The current study provided several lines of evidence that loss of NKX2–8 induces FAO activity in EOC. First, NKX2–8-deletion was significantly associated with FAO activity and NADPH and ATP levels. Second, NKX2–8 transcriptionally repressed key components of the FAO cascade, such as CPT1A and CPT2, by recruiting the Sin3A/HDAC1/SAP18 transcriptional repression complex. Third, the PDX model indicated that inhibition of FAO was sufficient to sensitize NKX2–8-deleted EOC, but not NKX2–8-nondeleted EOC, to chemotherapy, resulting in longer overall and relapse-free survival. Therefore, our results uncover a novel molecular mechanism for FAO hyperactivation in chemoresistant EOC, and suggest that combined Perhexiline and chemotherapy could be a tailored treatment for patients with NKX2–8-deleted EOC.
Hyperactivation of the FAO pathway was recently reported to contribute to cancer metastasis and the maintenance of stem cells and cancer stem cells. For instance, activation of FAO via promyelocytic leukemia (PML) or antioxidants dramatically precluded the loss of matrix attachment-induced anoikis in breast cancer cells [43,44]. Metastatic EOC cancer cells exhibit elevated FAO activity [9,39]. Meanwhile, inhibiting FAO affected leukemia-initiating cells (LICs) and hematopoietic stem cells (HSCs), which resulted in the exhaustion of the stem cell pool [38,[45], [46], [47], [48]]. These studies provided fundamental insights into activated FAO in cancer progression and tumorigenesis. Interestingly, a previous study reported that heterozygous or homozygous NKX2–8-knockout mice had bronchial hyperplasia at birth, which developed to lung carcinoma and progressed to invasive lung cancer latterly. Meanwhile, the NKX2–8-knockout mice also exhibited increased numbers of putative tracheobronchial stem/progenitor cells throughout life [21]. However, the mechanism by which loss of NKX2–8 induced tumorigenesis, stem cell proliferation and cancer invasion/metastasis is remains unclear. In the current study, we demonstrated that loss of NKX2–8 induced FAO activity by directly regulating FAO components and inhibiting FAO remarkably enhanced the efficacy of chemotherapeutic drugs in NKX2–8-deleted EOC. Therefore, we speculate that FAO activation might also contribute to NKX2–8 loss-induced tumorigenesis and invasion/metastasis, which is under investigation in our laboratory.
Another key finding in the current study is the mechanism by which NKX2–8-mediates transcriptional repression. It has been reported that NKX2–8 inhibit the epithelial-mesenchymal transition phenotype of bladder urothelial carcinoma cells via transcriptionally repressed Twist1 expression via directly targeting to the Twist1 promoter region [49]. Our previous study also showed that NKX2–8 directly associated with the AKIP1 promoter and transcriptionally repressed the expression of AKIP1, which resulted in inhibition of NF-κB signalling pathway [19]. However, the molecular mechanism underlying NKX2–8-mediated transcriptional repression remains incompletely understood. Herein, we demonstrated that NKX2–8 transcriptionally repressed CPT1A and CPT2 via directly recruiting Sin3A/HDAC1/SAP18 transcriptional repressor complex to the promoters of CPT1A and CPT2. SIN3 complex mediated-repression of target genes was through recruiting the histone deacetylase 1/2 (HDAC1/2), which, resulted in deacetylation of histone H3 and H4 and blocking transcription [[50], [51], [52]]. For instance, Ji and colleagues also reported that the SIN3A-HDAC complex transcriptionally repressed CDKN1A by increasing levels of deacetylated H3 and H4. Consistently, we found that enrichment of H3K27ac on the promoters of CPT1A and CPT2 was decreased in NKX2–8+/− cells but increased in NKX2–8+/+ EOC cells, and silencing SIN3A-HDAC complex significantly reduced the level of H3K27ac in the promoters of CPT1A and CPT2 genes, suggesting the important role of Sin3A/HDAC1/SAP18 in NKX2–8-mediated transcriptional repression of CPT1A and CPT2. Therefore, the current study reveals a novel molecular mechanism of NKX2–8-mediated transcriptional repression of FAO signalling in EOC.
In conclusion, our findings reveal a precise mechanism by which loss of NKX2–8 activates the FAO signalling pathway and confers chemoresistance in EOC. A combination treatment using an FAO inhibitor and CDDP might serve as a targeted therapy option for NKX2–8-deleted EOC. Since NKX2–8 deletion was tightly associated with chemoresistance and shorter relapse-free survival of patients in multiple distinct cancer types via TCGA dataset analysis, which promoted us to investigate currently whether NKX2–8 deletion can also confer chemoresistance in other cancer types via activation of the FAO pathway. In this scenario, inhibition of the FAO pathway using a pharmacological inhibitor might serve as a novel and efficient approach to counterbalance NKX2–8 deletion-mediated drug resistance. Therefore, our results demonstrated that NKX2–8 might have the potential role to assist with prognosis evaluation and personalized treatment selection in multiple types of cancer.
The following are the supplementary data related to this article.
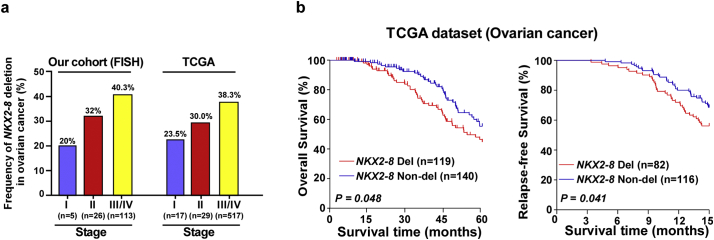
Supplementary Fig. S1.
NKX2-8 deletion is correlated with a worse prognosis of epithelial ovarian cancer (EOC). a. The frequency of NKX2-8 deletion in EOC with different FIGO, as analyzed by FISH in our collected EOC cohort (n = 144, left) and the TCGA EOC cohort (n = 563). b. Heterozygous deletion of NKX2-8 was correlated with overall survival and relapse-free survival in the TCGA EOC cohort (n = 563) (P = 0.048; P = 0.041).
Supplementary Fig. S2.
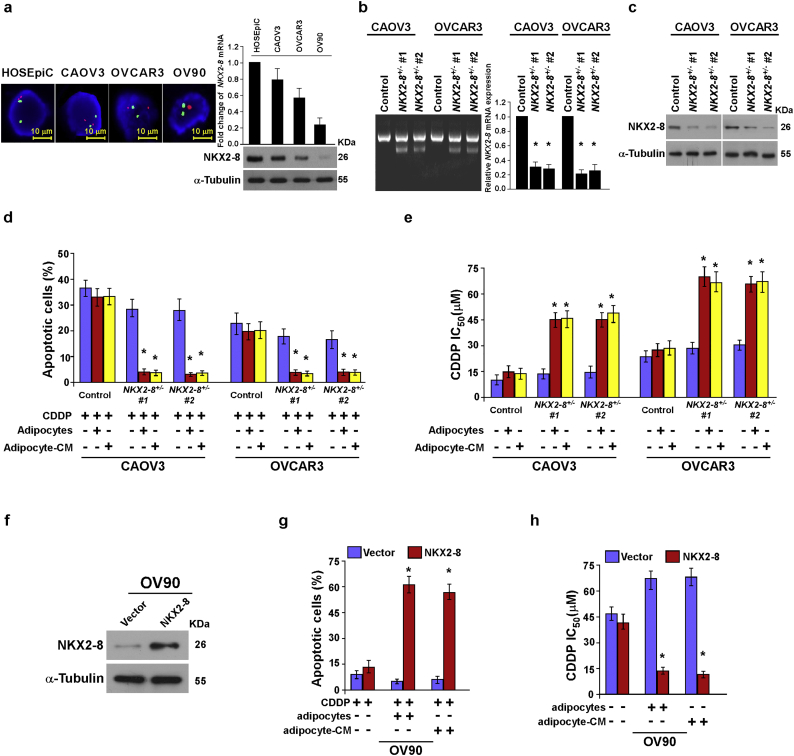
Adipose microenvironment enhances chemoresistance of NKX2-8-deleted EOC cells. a. Ovarian cancer cell lines were analyzed using FISH with green and red probes detecting NKX2-8 locus and the chromosome 14 centromere, respectively (left). The fold change in NKX2-8 mRNA and protein expression was normalized to human ovarian surface epithelial cells HOSEpiC using quantitative RT-PCR (upper) and immunoblotting (lower), respectively. GAPDH served as a loading control. α-Tubulin served as a loading control. b. NKX2-8 heterozygous knockout cells were established using a CRISPR/Cas9 system. T7 endonuclease I cleavage confirmed the efficiency of sgRNA(left). The mRNA expression of NKX2-8 in NKX2-8 heterozygous knockout cells was detected by Real time PCR assay(right). c. NKX2-8 KO efficiency was detected by Western blotting, α-Tubulin served as a loading control. d. The apoptotic index in the indicated adipocyte-CM-treated EOC cells, as analyzed using Annexin V assay. e. Relative IC50 value for CDDP in the indicated cells treated with vehicle or Perhexiline (10 μM) and then analyzed using MTT cell viability assays in the presence of adipocyte-CM. f. Immunoblotting analysis of NKX2-8 expression in the indicated cells. g. The apoptotic index in the indicated adipocyte-CM-treated EOC cells, as analyzed using annexin V assay. h. Relative IC50 value for CDDP in the indicated cells treated with vehicle or Perhexiline (10 μM) and then analyzed using MTT cell viability assays in the presence of adipocyte-CM. Error bars in Supplementary Fig. 2d, e, g, h represent the mean ± SD of three independent experiments. ⁎P < 0.05, ⁎⁎P < 0.01.
Supplementary Fig. S3.
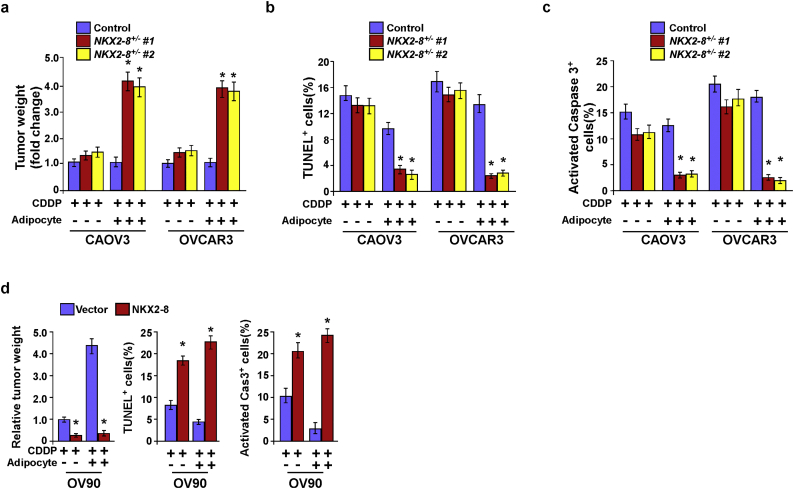
NKX2-8 deletion contributes to EOC chemoresistance in adipose microenvironment. a. Tumor weights were measured in each group. b-c. IHC staining for TUNEL(b) and activated caspase-3 (c) in the indicated xenograft tumors. d. Tumor weights were measured in each group (left). IHC staining for TUNEL (middle) and activated caspase-3 (right) in the indicated xenograft tumors. Error bars in Supplementary Fig. 3a-d represent the mean ± SD of three independent experiments. ⁎P < 0.05.
Supplementary Fig. S4.
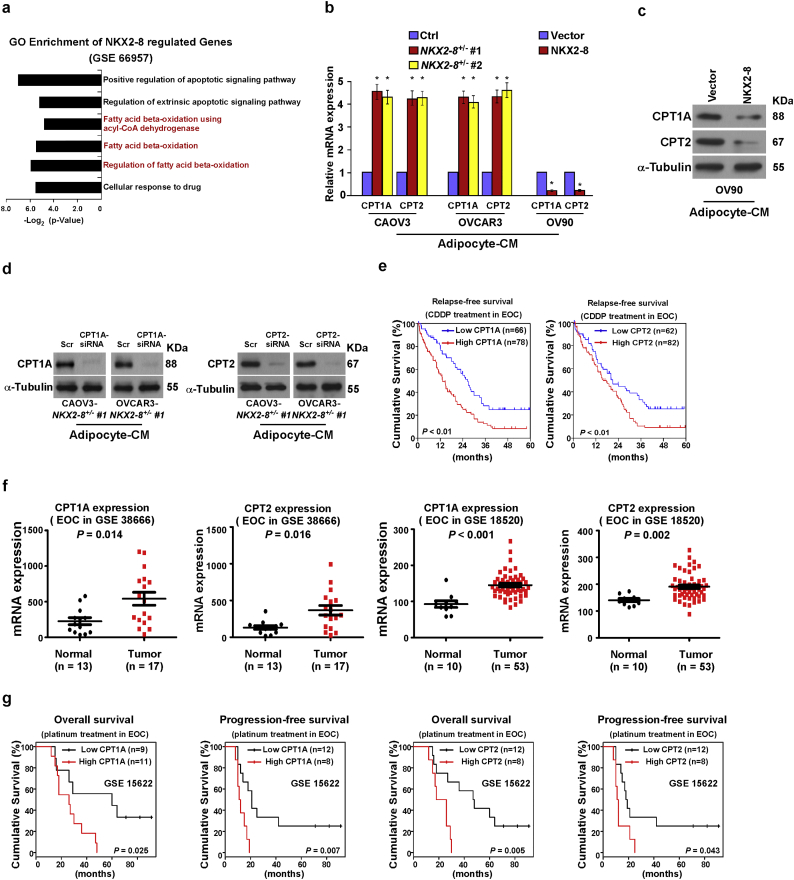
NKX2-8 transcriptionally represses multiple key components of the FAO cascade. a. GO enrichment analysis of the NKX2-8 regulated-transcripts identified in a published gene-set (GSE66957). b. Relative mRNA expression of CPT1A and CPT2 level in the indicated adipocyte-CM-treated EOC cells. Each bar represents the mean ± SD of three independent experiments, ⁎P < 0.05. c-d. Immunoblotting analysis of CPT1A and CPT2 in the indicated adipocyte-CM-treated EOC cells. α-Tubulin served as a loading control. e. Kaplan-Meier survival analysis of the association between CPT1A and CPT2 expression and shorter relapse-free survival in CDDP-treated EOC patients (P < 0.01; n = 144). f. Relative mRNA expression of CPT1A and CPT2 level in public accessed EOC dataset (GSE38666, n = 30; GSE18520, n = 63). g. Kaplan-Meier survival analysis of the association between CPT1A or CPT2 expression and overall survival, shorter relapse-free survival in EOC patients with platinum-based chemotherapy (GSE15622, n = 20).
Supplementary Fig. S5.
Sin3A/HDAC1/SAP18 repressor complex involves in NKX2-8-inhibited FAO signalling. a. Representative mass spectrometry plots and sequences of peptides from NKX2-8, Sin3A, HDAC1 and SAP18. b. ChIP analysis of the association of Sin3A, HDAC1 or SAP18 with the promoter of CPT1A and CPT2 transcripts in the indicated adipocyte-CM-treated EOC cells. c. CPT1A and CPT2 mRNA was determined by qRT-PCR in the indicated adipocyte-CM-treated EOC cells. d. CPT1A and CPT2 mRNA was determined by qRT-PCR in the indicated cells treat with TSA for 48hr in the presence of adipocyte-CM. e. Relative FAO activity (left) and ATP levels (right) in the indicated adipocyte-CM-treated EOC cells. f. The apoptotic index in the in the indicated adipocyte-CM-treated EOC cells, as analyzed using Annexin V assays. Error bars in Supplementary Fig. 5b-f represent the mean ± SD of three independent experiments. ⁎P < 0.05.
Supplementary Fig. S6.
Inhibiting FAO signalling sensitizes NKX-2-8-deleted EOC to CDDP therapy in vivo. a. A PDX model was established by inoculating NOG mice with 15 freshly collected clinical EOC tissues and treatment with CDDP (5 mg/kg). Two NKX2-8+/+- and two NKX2-8+/--EOC tissues were successfully engrafted in the NOG mice; Primary PDOC1, PDOC3, PDOC10 and PDOC11 EOC cells, which were isolated from EOC tissues #1, #3, #10 and #11, respectively, were analyzed using FISH to check NKX2-8 deletion. b. Immunoblotting analysis of NKX2-8 expression in PDOC1, PDOC3, PDOC10 and PDOC11 EOC cells, α-Tubulin served as a loading control. c. FACS analysis of annexin V staining in the indicated adipocyte-CM-treated EOC cells. Error bars represent the mean ± SD of three independent experiments. ⁎P < 0.05. d. Quantification of tumor-bearing mice treated with the indicated reagent(s); ns: no significance. (⁎P < 0.05, ANOVA, n = 6 mice/group). e. PDX model showing the relative volume change of PDX tumors (n = 6) treated with the indicated reagent(s) for 6 weeks compared with pretreated tumors (week 0). ⁎P < 0.05, ns: no significance.
Supplementary material
Funding sources
This work was supported by Natural Science Foundation of China (No. 81830082, 91740119, 91529301, 81621004, 91740118, 81773106 and 81530082); Guangzhou Science and Technology Plan Projects (201803010098); Natural Science Foundation of Guangdong Province (2018B030311009 and 2016A030308002); The Fundamental Research Funds for the Central Universities [No. 17ykjc02].
Declaration of interests
The authors declare no conflicts of interest.
Author contributions
Concept and design of study: Jinrong Zhu, Geyan Wu, Libing Song and Jun Li; Performed the xenograft tumor experiments: Jinrong Zhu, Geyan Wu, Lixue Cao; Performed the in vitro cell line studies: Jinrong Zhu, Geyan Wu, Lixue Cao, Zhanyao Tan; Designed and established the all plasmids: Jinrong Zhu, Ziwen Li; Performed immunohistochemical and pathological analysis: Miaoling Tang, Ziwen Li, Dongni Shi, Shuxia Zhang; Wrote the manuscript with input from the other authors: Jun Li.
References
- 1.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Cantor J.R., Sabatini D.M. Cancer cell metabolism: one hallmark, many faces. Cancer Discov. 2012;2:881–898. doi: 10.1158/2159-8290.CD-12-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pavlova N.N., Thompson C.B. The emerging hallmarks of cancer metabolism. Cell Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koppenol W.H., Bounds P.L., Dang C.V. Otto Warburg's contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 5.Yang L., Venneti S., Nagrath D. Glutaminolysis: a hallmark of cancer metabolism. Annu Rev Biomed Eng. 2017;19:163–194. doi: 10.1146/annurev-bioeng-071516-044546. [DOI] [PubMed] [Google Scholar]
- 6.Suh D.H., Kim M.K., No J.H. Metabolic approaches to overcoming chemoresistance in ovarian cancer. Ann N Y Acad Sci. 2011;1229:53–60. doi: 10.1111/j.1749-6632.2011.06095.x. [DOI] [PubMed] [Google Scholar]
- 7.Leinster D.A., Kulbe H., Everitt G. The peritoneal tumour microenvironment of high-grade serous ovarian cancer. J Pathol. 2012;227:136–145. doi: 10.1002/path.4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miranda F., Mannion D., Liu S. Salt-inducible kinase 2 couples ovarian cancer cell metabolism with survival at the adipocyte-rich metastatic niche. Cancer Cell. 2016;30:273–289. doi: 10.1016/j.ccell.2016.06.020. [DOI] [PubMed] [Google Scholar]
- 9.Nieman K.M., Kenny H.A., Penicka C.V. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat Med. 2011;17:1498–1503. doi: 10.1038/nm.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roodhart J.M., Daenen L.G., Stigter E.C. Mesenchymal stem cells induce resistance to chemotherapy through the release of platinum-induced fatty acids. Cancer Cell. 2011;20:370–383. doi: 10.1016/j.ccr.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 11.Carracedo A., Cantley L.C., Pandolfi P.P. Cancer metabolism: fatty acid oxidation in the limelight. Nat Rev Cancer. 2013;13:227–232. doi: 10.1038/nrc3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu P.P., Liu J., Jiang W.Q. Elimination of chronic lymphocytic leukemia cells in stromal microenvironment by targeting CPT with an antiangina drug perhexiline. Oncogene. 2016;35:5663–5673. doi: 10.1038/onc.2016.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vella S., Penna I., Longo L. Perhexiline maleate enhances antitumor efficacy of cisplatin in neuroblastoma by inducing over-expression of NDM29 ncRNA. Sci Rep. 2015;5 doi: 10.1038/srep18144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramu A., Fuks Z., Gatt S. Reversal of acquired resistance to doxorubicin in P388 murine leukemia cells by perhexiline maleate. Cancer Res. 1984;44:144–148. [PubMed] [Google Scholar]
- 15.Kajiyama Y., Tian J., Locker J. Regulation of alpha-fetoprotein expression by Nkx2.8. Mol Cell Biol. 2002;22:6122–6130. doi: 10.1128/MCB.22.17.6122-6130.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hagihara A., Miyamoto K., Furuta J. Identification of 27 5' CpG islands aberrantly methylated and 13 genes silenced in human pancreatic cancers. Oncogene. 2004;23:8705–8710. doi: 10.1038/sj.onc.1207783. [DOI] [PubMed] [Google Scholar]
- 17.Harris T., Pan Q., Sironi J. Both gene amplification and allelic loss occur at 14q13.3 in lung cancer. Clin Cancer Res. 2011;17:690–699. doi: 10.1158/1078-0432.CCR-10-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu C., Zhang Z., Liao W. The tumor-suppressor gene Nkx2.8 suppresses bladder cancer proliferation through upregulation of FOXO3a and inhibition of the MEK/ERK signaling pathway. Carcinogenesis. 2012;33:678–686. doi: 10.1093/carcin/bgr321. [DOI] [PubMed] [Google Scholar]
- 19.Lin C., Song L., Gong H. Nkx2-8 downregulation promotes angiogenesis and activates NF-kappaB in esophageal cancer. Cancer Res. 2013;73:3638–3648. doi: 10.1158/0008-5472.CAN-12-4028. [DOI] [PubMed] [Google Scholar]
- 20.Qu L., Deng B., Zeng Y. Decreased expression of the Nkx2.8 gene correlates with tumor progression and a poor prognosis in HCC cancer. Cancer Cell Int. 2014;14:28. doi: 10.1186/1475-2867-14-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tian J., Mahmood R., Hnasko R. Loss of Nkx2.8 deregulates progenitor cells in the large airways and leads to dysplasia. Cancer Res. 2006;66:10399–10407. doi: 10.1158/0008-5472.CAN-06-1564. [DOI] [PubMed] [Google Scholar]
- 22.Liu L., Lin C., Liang W. TBL1XR1 promotes lymphangiogenesis and lymphatic metastasis in esophageal squamous cell carcinoma. Gut. 2015;64:26–36. doi: 10.1136/gutjnl-2013-306388. [DOI] [PubMed] [Google Scholar]
- 23.Wu G., Cao L., Zhu J. Loss of RBMS3 confers platinum resistance in epithelial ovarian cancer via activation of miR-126-5p/beta-catenin/CBP signaling. Clin Cancer Res. 2019;25:1022–1035. doi: 10.1158/1078-0432.CCR-18-2554. [DOI] [PubMed] [Google Scholar]
- 24.Wu Y., Zhou L., Wang X. A genome-scale CRISPR-Cas9 screening method for protein stability reveals novel regulators of Cdc25A. Cell Discov. 2016;2 doi: 10.1038/celldisc.2016.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferrick D.A., Neilson A., Beeson C. Advances in measuring cellular bioenergetics using extracellular flux. Drug Discov Today. 2008;13:268–274. doi: 10.1016/j.drudis.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 26.Liu X., Zhang Y., Chen Y. In situ capture of chromatin interactions by biotinylated dCas9. Cell. 2017;170:1028–1043. doi: 10.1016/j.cell.2017.08.003. [e19] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chou T.C. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70:440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 28.Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer. 2007;7:573–584. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- 29.Moreno-Sanchez R., Rodriguez-Enriquez S., Marin-Hernandez A. Energy metabolism in tumor cells. FEBS J. 2007;274:1393–1418. doi: 10.1111/j.1742-4658.2007.05686.x. [DOI] [PubMed] [Google Scholar]
- 30.Amoedo N.D., Obre E., Rossignol R. Drug discovery strategies in the field of tumor energy metabolism: limitations by metabolic flexibility and metabolic resistance to chemotherapy. Biochim Biophys Acta Bioenerg. 2017;1858:674–685. doi: 10.1016/j.bbabio.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 31.Yu L., Lu M., Jia D. Modeling the genetic regulation of cancer metabolism: interplay between glycolysis and oxidative phosphorylation. Cancer Res. 2017;77:1564–1574. doi: 10.1158/0008-5472.CAN-16-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Solaini G., Sgarbi G., Baracca A. Oxidative phosphorylation in cancer cells. Biochim Biophys Acta. 2011;1807:534–542. doi: 10.1016/j.bbabio.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 33.Guerra F., Arbini A.A., Moro L. Mitochondria and cancer chemoresistance. Biochim Biophys Acta Bioenerg. 2017;1858:686–699. doi: 10.1016/j.bbabio.2017.01.012. [DOI] [PubMed] [Google Scholar]
- 34.Pacilli A., Calienni M., Margarucci S. Carnitine-acyltransferase system inhibition, cancer cell death, and prevention of myc-induced lymphomagenesis. J Natl Cancer Inst. 2013;105:489–498. doi: 10.1093/jnci/djt030. [DOI] [PubMed] [Google Scholar]
- 35.Chen C.L., Uthaya Kumar D.B., Punj V. NANOG metabolically reprograms tumor-initiating stem-like cells through tumorigenic changes in oxidative phosphorylation and fatty acid metabolism. Cell Metab. 2016;23:206–219. doi: 10.1016/j.cmet.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zaugg K., Yao Y., Reilly P.T. Carnitine palmitoyltransferase 1C promotes cell survival and tumor growth under conditions of metabolic stress. Genes Dev. 2011;25:1041–1051. doi: 10.1101/gad.1987211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.German N.J., Yoon H., Yusuf R.Z. PHD3 loss in Cancer enables metabolic reliance on fatty acid oxidation via deactivation of ACC2. Mol Cell. 2016;63:1006–1020. doi: 10.1016/j.molcel.2016.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Samudio I., Harmancey R., Fiegl M. Pharmacologic inhibition of fatty acid oxidation sensitizes human leukemia cells to apoptosis induction. J Clin Invest. 2010;120:142–156. doi: 10.1172/JCI38942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Camarda R., Zhou A.Y., Kohnz R.A. Inhibition of fatty acid oxidation as a therapy for MYC-overexpressing triple-negative breast cancer. Nat Med. 2016;22:427–432. doi: 10.1038/nm.4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tania M., Khan M.A., Song Y. Association of lipid metabolism with ovarian cancer. Curr Oncol. 2010;17:6–11. doi: 10.3747/co.v17i5.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fong M.Y., McDunn J., Kakar S.S. Identification of metabolites in the normal ovary and their transformation in primary and metastatic ovarian cancer. PLoS One. 2011;6 doi: 10.1371/journal.pone.0019963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ke C., Hou Y., Zhang H. Large-scale profiling of metabolic dysregulation in ovarian cancer. Int J Cancer. 2015;136:516–526. doi: 10.1002/ijc.29010. [DOI] [PubMed] [Google Scholar]
- 43.Carracedo A., Weiss D., Leliaert A.K. A metabolic prosurvival role for PML in breast cancer. J Clin Invest. 2012;122:3088–3100. doi: 10.1172/JCI62129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schafer Z.T., Grassian A.R., Song L. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature. 2009;461:109–113. doi: 10.1038/nature08268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ito K., Carracedo A., Weiss D. A PML-PPAR-delta pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. Nat Med. 2012;18:1350–1358. doi: 10.1038/nm.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gan B., Hu J., Jiang S. Lkb1 regulates quiescence and metabolic homeostasis of haematopoietic stem cells. Nature. 2010;468:701–704. doi: 10.1038/nature09595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gurumurthy S., Xie S.Z., Alagesan B. The Lkb1 metabolic sensor maintains haematopoietic stem cell survival. Nature. 2010;468:659–663. doi: 10.1038/nature09572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakada D., Saunders T.L., Morrison S.J. Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells. Nature. 2010;468:653–658. doi: 10.1038/nature09571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu C., Liu Z., Chen Q. Nkx2.8 inhibits epithelial-mesenchymal transition in bladder urothelial carcinoma via transcriptional repression of Twist1. Cancer Res. 2018;78:1241–1252. doi: 10.1158/0008-5472.CAN-17-1545. [DOI] [PubMed] [Google Scholar]
- 50.Laherty C.D., Yang W.M., Sun J.M. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 51.Hassig C.A., Fleischer T.C., Billin A.N. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 52.Heinzel T., Lavinsky R.M., Mullen T.M. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material