Abstract
Owls are nocturnal raptors that are prevalently infected with haemosporidian parasites wordwide. These birds were commonly submitted to the Kasetsart University Raptor Rehabilitation Unit, Kasetsart University, Thailand and were examined using PCR-based methods for the presence of haemosporidian infections of by the genera Plasmodium and Haemoproteus. Blood samples from 167 individual owls belonging to 12 species common in Thailand were collected between September 2012 and February 2018. The overall prevalence of haemosporidians was 34.1%, with Haemoproteus infections (25.1%) being more prevalent than Plasmodium infections (9.0%). The prevalence of both Haemoproteus and Plasmodium parasites was similar in all seasons of the year. Molecular characterization revealed 17 new haemosporidian parasite lineages (11 Haemoproteus and six Plasmodium), with genetic variation among partial cytochrome b sequences ranging from 0.0% to 3.6% in Haemoproteus lineages and 0.2%–8.8% in Plasmodium lineages. Phylogenetic analysis showed that all Haemoproteus lineages detected in owls appeared in one well-supported clade together with other parasites belonging to the Parahaemoproteus subgenus, indicating their close evolutionary relationship and common transmission modality by Culicoides biting midges. This study showes the existence of prominent non-described haemosporidian parasite diversity in Thai owls and provides baseline molecular information for further research on the genetic diversity of owl haemosporidian parasites. New DNA sequence information can be used for the diagnosis of owl infections, which have been often reported during rehabilitation planning.
Keywords: Avian malaria, Haemosporidian parasites, Haemoproteus, PCR, Plasmodium, Strigiformes
Graphical abstract
Highlights
-
•
Molecular prevalence of haemosporidian parasites in owls from Thailand was high.
-
•
Haemoproteus and Plasmodium infections in owls were found in all seasons of Thailand.
-
•
Seventeen new haemosporidian parasite lineages were isolated from Thai owls.
-
•
Cytochrome b sequences showed genetic variable of Haemoproteus and Plasmodium in owls.
1. Introduction
Kasetsart University Raptor Rehabilitation Unit (KURRU) was established in 2007 for the rehabilitation of both migratory and resident raptors (Accipitriformes, Falconiformes and Strigiformes) in Thailand (Salakij et al., 2015b). KURRU admits birds brought by local authorities and citizens throughout Thailand. The rehabilitation procedures include a health assessment and subsequent treatment, behaviour monitoring, and then release back into the wild. Furthermore, KURRU has made efforts to raise public awareness of raptor conservation through the mainstream media (Kidsin et al., 2012).
Owls (Strigiformes) are nocturnal or crepuscular predators that hunt small vertebrates, especially small rodents (Kunsorn et al., 2015). There are 19 species of resident owls and two species of non-breeding migratory owls in Thailand (BCST, 2018), including the near threatened reddish scops-owl, Otus rufescens, and the vulnerable white-fronted scops-owl, Otus sagittatus, (IUCN, 2019). Most of the owls are legally protected by the Wild Animal Reservation and Protection Act, B.E. 2535 (1992) of Thailand, which prohibits trading, hunting and keeping these birds as pets. The owls are considered ‘umbrella’ species due to their role as top predators in the food chain which contributes to ecological balance in their environments (Krone et al., 2008). In agricultural areas, owls are used as an alternative biological control agent of pests (Puan et al., 2011). In southern Thailand, artificial nests have been used to increase the number of owls in an attempt to eliminate small rodents in oil palm plantations (Tavitchasri et al., 2016).
Between 2008 and 2011, owls were the most frequently admitted birds at KURRU (Kidsin et al., 2012). The most common reasons for admission were trauma in adults and malnutrition and parental death or abandonment in chicks. Blood parasite infections (Salakij et al., 2012a, 2012b, 2018, 2015a), especially Haemoproteus spp. and Plasmodium spp. that cause haemoproteosis and avian malaria, respectively (Valkiūnas et al., 2005), were often found in admitted raptors. Parasites belonging to these two genera are evolutionary closely related and are responsible for vector-borne infections transmitted by blood-sucking insects of the Hippoboscidae for Haemoproteus, Ceratopogonidae for Parahaemoproteus, and Ceratopogonidae, Culicidae, and Phlebotomidae for Plasmodium (Valkiūnas, 2005).
Haemoproteus spp. and Plasmodium spp. might be harmful and even cause lethal diseases in some non-adapted avian species (Atkinson and van Riper, 1991; Valkiūnas, 2005). Although Plasmodium spp. are generally considered more pathogenic, Haemoproteus spp. infections can also be occasionally lethal, especially when non-adapted hosts are exposed to these parasites (Baker et al., 2018; Kelly et al., 2018; Lee et al., 2018; Remple, 2004; Valkiūnas and Iezhova, 2017). With regards specifically to owls, Evans and Otter (1998) reported the case of a snowy owl (Bubo scandiacus) that died due to a co-infection by Haemoproteus noctuae and Leucocytozoon danilewskyi.
Molecular markers are valuable for haemosporidian infection diagnostics (Chagas et al., 2015; Dimitrov et al., 2016), but sequencing data for lineages that infect owls are still limited. During an ongoing project (Salakij et al., 2018) that focused on the haematology and cytochemistry of blood cells and molecular characteristics of blood parasites in raptors, preliminary data on Haemoproteus infections in barn owls and collared scops-owls were collected, but limited data were obtained on the prevalence and diversity of these parasites. This study fills this gap and focuses on the prevalence and genetic diversity of Haemoproteus spp. and Plasmodium spp. in owls in Thailand. The infection data from a previous report (Salakij et al., 2018) were re-analysed, allowing us to access complete information about the prevalence of parasite lineages found in owls.
In addition, this study aimed to gain new knowledge about the genetic diversity of Haemoproteus spp. and Plasmodium spp. in all sampled owls based on a 6-year survey at the KURRU, Faculty of Veterinary Medicine, Kasetsart University, Thailand. The prevalence data were based on molecular detection using a polymerase chain reaction (PCR) protocol (Hellgren et al., 2004), which has been widely used in wildlife haemosporidian research (Bensch et al., 2009; Bukauskaitė et al., 2015; Chagas et al., 2015; Dimitrov et al., 2016; Hellgren et al., 2004; Valkiūnas et al., 2008).
2. Materials and methods
2.1. Sample collection
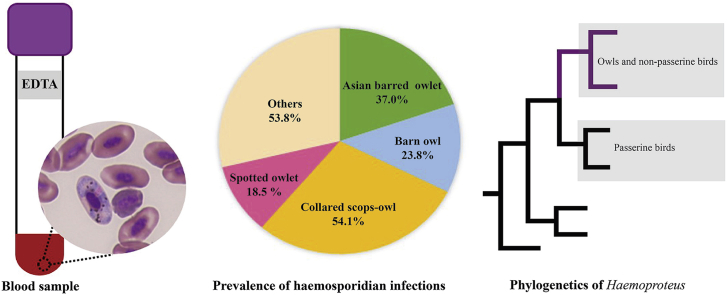
From September 2012 to February 2018, 1 ml of EDTA-blood was opportunistically collected from owls admitted to the KURRU (14°1′ N, 99°58′ E). In total, samples from 167 owls were collected (Table 1) and maintained under refrigeration (4 °C) until DNA extraction.
Table 1.
Prevalence of Haemoproteus and Plasmodium infections of owls in Thailand during September 2012–February 2018.
| Bird species | No. examined | Haemoproteus | Plasmodium | H–Pa | Total |
|---|---|---|---|---|---|
| Asian barred Owlet (ABO, Glaucidium cuculoides) | 27 | 2 (7.4)b | 8 (29.6) | 0 | 10 (37.0) |
| Barn owl (BO, Tyto alba) | 63 | 13 (20.6) | 2 (3.2) | 0 | 15 (23.8) |
| Collared scops-owl (CSO, Otus lettia) | 37 | 19 (51.4) | 1 (2.7) | 0 | 20 (54.1) |
| Spotted owlet (SO, Athene brama) | 27 | 4 (14.8) | 1 (3.7) | 0 | 5 (18.5) |
| Barred eagle-owl (BEO, Bubo sumatranus) | 1 | 0 | 1 | 0 | 1 |
| Buffy fish owl (BFO, Ketupa ketupu) | 2 | 0 | 1 | 0 | 1 |
| Brown hawk owl (BHO, Ninox scutulata) | 4 | 2 | 1 | 0 | 3 |
| Brown wood owl (BWO, Strix leptogrammica) | 1 | 0 | 0 | 0 | 0 |
| Oriental bay-owl (OBO, Phodilus badius) | 1 | 1 | 0 | 0 | 1 |
| Oriental scops-owl (OSO, Otus sunia) | 2 | 0 | 0 | 1 | 1 |
| Short-eared owl (SEO, Asio flammeus) | 1 | 0 | 0 | 0 | 0 |
| Spot-bellied eagle-owl (SBEO, Bubo nipalensis) |
1 |
0 |
0 |
0 |
0 |
| Grand total | 167 | 41 (24.6) | 15 (9.0) | 1 (0.6) | 57 (34.1) |
Haemoproteus-Plasmodium co-infection.
Number of infected individuals are given, followed by prevalence data (in parentheses, percentage).
2.2. Microscopic examinations
Parasite morphology was not described due to the limitation of the sampling procedure, resulting in a lack of fresh blood smears (Salakij et al., 2018). Because intracellular haemosporidian parasites morphologically change and undergo exflagellation within several minutes after exposure to air (Valkiūnas, 2005), it was impossible to use our EDTA-blood films for morphological examinations. However, some parasitaemia detections using microscopy were performed. At least two blood smears were obtained from each owl and stained with Wright's stain (Salakij et al., 2018). Parasitaemia observation was performed at a high power field (400 × ) for 100 fields and oil-immersion lenses (1000 × ) for at least 100 fields (Valkiūnas et al., 2008).
2.3. DNA extraction, PCR amplification and DNA sequencing
Total DNA was extracted from 50 μL of EDTA-blood using a Blood Genomic DNA Extraction Mini Kit (FavorPrep, Pingtung, Taiwan). Nested PCR for cytochrome b (cyt b) gene amplification was performed according to Hellgren et al. (2004). In brief, HaemNFI and HaemNR3 primers were used to amplify the cyt b gene of both Haemoproteus and Plasmodium in the first reaction. In the second reaction, HaemF and HaemR2 primers (Bensch et al., 2000) were used to amplify a 479 base pair (bp) region of those parasites. All PCRs were performed in 20 μL total volumes, which contained 2 μL of template DNA, 10 μL of DreamTaq Green PCR Master Mix (2 × ) (ThermoScientific, Waltham, Massachusetts, USA), 1 μL of each primer at 10 μM concentration and 6 μL of water. A positive control and non-template control were applied in all reactions. The amplification started with 3 min of pre-denaturation at 94 °C, followed by 20 cycles (first reaction) or 35 cycles (second reaction) at 94 °C for 30 s, 50 °C for 45 s, and 72 °C for 30 s. Then, the reaction ended with a final extension at 72 °C for 10 min. Amplicons were electrophoresed on a 1.5% agarose gel at 132 V for 20 min to detect parasite-specific bands. Amplicons were purified using the GEL/PCR Purification Mini Kit (FavorPrep, Pingtung, Taiwan). Purified DNA was submitted to Apical Scientific (Selangor, Malaysia) for nucleotide sequencing. The quality of the sequences was screened and contigs were assembled by using BioEdit (Hall, 1999). Double peaks in the sequence electropherograms were considered to be co-infections. The sequences showing double peak in chromatogram were considered co-infections and were excluded from the sequence and phylogenetic analyses.
2.4. Sequence and phylogenetic analysis
Only one sequence amplified from an oriental scops owl was excluded from the analysis because it showed double peaks in the chromatogram. Thirty-seven sequences were aligned with Haemoproteus and Plasmodium parasite sequences deposited in the MalAvi database (Bensch et al., 2009) with a consensus length of 479 bp. Nineteen sequences from a previous report (Salakij et al., 2018) were re-analysed. All 56 sequences detected were BLAST searched in the MalAvi database to identify the similarities between existing sequences and to determine if they were new lineages. The sequences with at least 1 base pair difference from existing sequences were considered new lineages (Chagas et al., 2017; Ivanova et al., 2015). These sequences were named according to the MalAvi nomenclature (Bensch et al., 2009) and deposited on GenBank and MalAvi.
Bayesian phylogenetic analysis was performed using haemosporidian parasite cyt b sequences detected during this study and other sequences from haemosporidians of owls deposited in the MalAvi database (Bensch et al., 2009). Additionally, sequences of morphologically identified species of Haemoproteus and Plasmodium parasites were included to better understand the possible species identity of owl parasites detected in Thailand. Some missing data (gap position) in each lineage were coded as “N” (Xue et al., 2015). The best-fit model, which was general time reversible model selected including the invariant site and gamma distribution (GTR + I + G) for both Haemoproteus and Plasmodium, was selected by the software jModelTest2 (Darriba et al., 2012). The phylogenetic tree was created by MrBayes version 3.2.6 (Ronquist and Huelsenbeck, 2003). Markov chain Monte Carlo (MCMC) mofel was run for a total of 3 million generations for Haemoproteus and Plasmodium, with sampling every 1000 generations. The first 25% of trees were discarded as “burn in”. The remaining trees were used to construct a consensus tree. The sequence divergence between the different lineages was calculated using the Jukes-Canter model of substitution, with all substitutions weighted equally, implemented by MEGA7 (Kumar et al., 2016). 2.5 Statistical analysis.
Prevalence was calculated from combined molecular results from September 2012 and February 2018. Confidence intervals (CIs, 95%), were calculated by the function bionom.approx in R software (R development core team, 2016). According to the Thai Meteorological Department (TMD), the climate of Thailand is influenced by monsoon winds (i.e., southwest and northeast monsoon) and the climate can be divided into three seasons: summer, the hottest period (late-February to early-May); rainy, rainfall period (late-May to early-October) and cold-dry, coldest period (late-October to early-February). The seasonal prevalence analysis in this report was performed according to the TMD classification. Pearson's Chi-square (χ2, α = 0.05) was used to assess differences in the prevalence of parasite infection in different seasons and different species of owls. Due to the available sample size, the group of species were divided into 5 groups, including Asian barred-owlet (Glaucidium cuculoides), barn owl (Tyto alba), collared scops-owl (Otus lettia), spotted owlet (Athene brama) and other owls (8 species, Table 1).
3. Results
3.1. Parasite detection and molecular prevalence
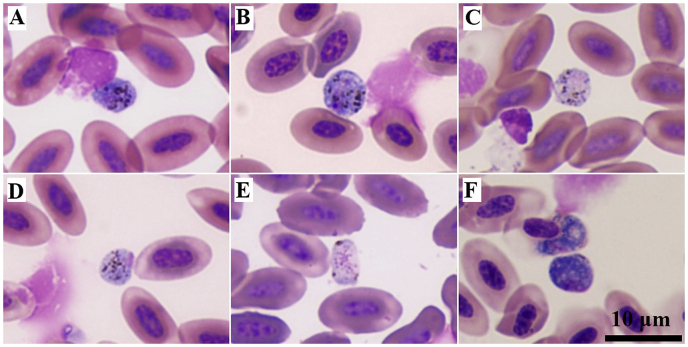
Microscopic examinations showed extracellular gametocytes in some samples (Fig. 1). The number of parasite infections identified by microscopic examination was 42, which was lower than that identified by molecular examinations (57 samples). In all 57 PCR-positive samples, one sample from an Oriental scops owl (OSO) showed a double peak in the chromatogram, indicating Haemoproteus-Plasmodium co-infection (Table 1). This one sample was included in the overall prevalence calculation but was excluded form the phylogenetic analysis.
Fig. 1.
Extracellular gametocytes of Haemoproteus spp. in an Asian barred owlet (A), barn owl (B), brown hawk owl (C), collared scops-owl (D), Oriental scops-owl (E), and spotted owlet (F). Wright's stain. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Based on molecular examination, the most commonly submitted owls were the Asian barred owlet (ABO), barn owl (BO), collared scops owl (CSO) and spotted owlet (SO). The overall prevalence of parasite infections between the different species of owl was different. Parasite prevalence in the CSO was significantly higher than in the other bird species (χ2 = 14.79, p = 0.005). The prevalence of parasites in cold-dry (43.9%, 95% CI: 28.7–59.1), rainy (28.0%, 95% CI: 15.6–40.4), and summer (32.9%; 95% CI 22.3–43.5) seasons were not significantly different (χ2 = 2.63, p = 0.269). This indicates active haemosporidian transmission throughout the year.
The prevalence of Haemoproteus species (24.6%, 95% CI: 18.0–31.1) infections was higher than that of Plasmodium spp. (9.0%, 95% CI: 4.6–13.3). Haemoproteus spp. were found during the entire year (Fig. 2). The highest prevalence of Haemoproteus spp. were recorded in August and October, at 50.0% (95% CI: 10.0–90.0) for both months. The highest prevalence of Plasmodium spp. infection was recorded in September, at 27.3% (95% CI: 1.0–53.6). Plasmodium infections were not detected during January, May, June, July, and August.
Fig. 2.
Monthly molecular prevalence of Haemoproteus species and Plasmodium spp. in owls in Thailand during 2012–2018. Vertical lines are 95% confidence intervals. The average temperature and rainfall in Kamphaeng Saen were reported by the Nakhon Pathom meteorological station, Thai Meteorological Department.
3.2. Sequence and phylogenetic analysis
Among the 56 detected cyt b sequences, 23 distinct lineages of haemosporidian parasites were reported (Table 2): 12 belonged to Haemoproteus and 11 belonged to Plasmodium. Seventeen of the reported lineages were new, including 11 Haemoproteus lineages and six Plasmodium lineages. Six other lineages were identical to previously deposited lineages in the MalAvi database, including Haemoproteus lineage NINOX07 and Plasmodium lineages ACCBAD01, FANTAIL01, MILANS06, ORW1, and TEPON02 (Table 2).
Table 2.
Haemoproteus and Plasmodium lineages isolated from 56 owls in Thailand during September 2012–February 2018.
| Lineagesa | Parasite genus | Host species |
Host family | Locality | Age | GenBank | ||
|---|---|---|---|---|---|---|---|---|
| Scientific name | Common name | IDb | ||||||
| ATHBRA01 | Haemoproteus | Athena brama | Spotted owlet | KU210 | Stringidae | Bangkok | Adult | MK390801 |
| Athena brama | Spotted owlet | KU382 | Stringidae | Bangkok | Adult | MK390802 | ||
| Athena brama | Spotted owlet | KU494 | Stringidae | Chaiyaphum | Adult | MK390803 | ||
| Athena brama | Spotted owlet | KU598 | Stringidae | Bangkok | Juvenile | MK390804 | ||
| GLACUC03 | Haemoproteus | Glaucidium cuculoides | Asian barred owlet | KU278 | Stringidae | Bangkok | Adult | MK390805 |
| GLACUC04 | Haemoproteus | Glaucidium cuculoides | Asian barred owlet | KU597 | Stringidae | Bangkok | Juvenile | MK390806 |
| NINOX07 | Haemoproteus | Ninox scutulata | Brown hawk owl | KU320 | Stringidae | Bangkok | Adult | MK390822 |
| Ninox scutulata | Brown hawk owl | KU608 | Stringidae | Bangkok | Adult | MK390823 | ||
| OTULET01 | Haemoproteus | Otus lettia | Collared scops-owl | KU127 | Stringidae | Rayong | Adult | KJ561457c |
| OTULET02 | Haemoproteus | Otus lettia | Collared scops-owl | KU234 | Stringidae | Bangkok | Adult | KU528645c |
| PHOBAD01 | Haemoproteus | Phodilus badius | Oriental bay owl | KU571 | Tytonidae | Bangkok | Adult | MK390808 |
| TYTAL3 | Haemoproteus | Tyto alba | Barn owl | KU421 | Tytonidae | Bangkok | Adult | MK390809 |
| TYTAL4 | Haemoproteus | Tyto alba | Barn owl | KU481 | Tytonidae | Pathum Thani | Juvenile | MK390810 |
| Tyto alba | Barn owl | KU532 | Tytonidae | Bangkok | Adult | MK390811 | ||
| Tyto alba | Barn owl | KU537 | Tytonidae | Bangkok | Adult | MK390812 | ||
| Otus lettia | Collared scops-owl | KU340 | Stringidae | Bangkok | Adult | MK390813 | ||
| Otus lettia | Collared scops-owl | KU442 | Stringidae | Bangkok | Adult | MK390814 | ||
| Otus lettia | Collared scops-owl | KU465 | Stringidae | Bangkok | Adult | MK390815 | ||
| Otus lettia | Collared scops-owl | KU482 | Stringidae | Pathum Thani | Adult | MK390816 | ||
| Otus lettia | Collared scops-owl | KU545 | Stringidae | Bangkok | Adult | MK390817 | ||
| Otus lettia | Collared scops-owl | KU568 | Stringidae | Samut Sakhon | Adult | MK390818 | ||
| Otus lettia | Collared scops-owl | KU582 | Stringidae | Kanchanaburi | Adult | MK390819 | ||
| Otus lettia | Collared scops-owl | KU588 | Stringidae | Ratchaburi | Adult | MK390820 | ||
| Otus lettia | Collared scops-owl | KU181 | Stringidae | Nakhon Ratchasima | Juvenile | KJ561458c | ||
| Otus lettia | Collared scops-owl | KU222 | Stringidae | Bangkok | Adult | KU528643c | ||
| Otus lettia | Collared scops-owl | KU226 | Stringidae | Bangkok | Adult | KU528644c | ||
| Otus lettia | Collared scops-owl | KU259 | Stringidae | Bangkok | Adult | KU528648c | ||
| Otus lettia | Collared scops-owl | KU318 | Stringidae | Bangkok | Adult | KU528650c | ||
| TYTAL5 | Haemoproteus | Tyto alba | Barn owl | KU516 | Tytonidae | Bangkok | Adult | MK390821 |
| TYTAL6 | Haemoproteus | Tyto alba | Barn owl | KU71 | Tytonidae | Pathum Thani | Adult | KU528634c |
| TYTAL6 | Haemoproteus | Tyto alba | Barn owl | KU86 | Tytonidae | Prachuap Khiri Khan | Adult | KU528635c |
| Tyto alba | Barn owl | KU112 | Tytonidae | Bangkok | Adult | KU528636c | ||
| Tyto alba | Barn owl | KU120 | Tytonidae | Ratchaburi | Adult | KU528637c | ||
| Tyto alba | Barn owl | KU132 | Tytonidae | Bangkok | Adult | KU528638c | ||
| Tyto alba | Barn owl | KU200 | Tytonidae | Ayutthaya | Adult | KU528639c | ||
| Tyto alba | Barn owl | KU202 | Tytonidae | Ayutthaya | Adult | KU528640c | ||
| Otus lettia | Collared scops-owl | KU163 | Stringidae | Bangkok | Juvenile | KU528642c | ||
| Otus lettia | Collared scops-owl | KU248 | Stringidae | Bangkok | Adult | KU528646c | ||
| Otus lettia | Collared scops-owl | KU250 | Stringidae | Bangkok | Juvenile | KU528647c | ||
| Otus lettia | Collared scops-owl | KU284 | Stringidae | Samut Prakan | Adult | KU528649c | ||
| TYTAL7 | Haemoproteus | Tyto alba | Barn owl | KU203 | Tytonidae | Sing Buri | Juvenile | KU528641c |
| ACCBAD01 | Plasmodium | Tyto alba | Barn owl | KU236 | Tytonidae | Ayutthaya | Adult | MK390829 |
| Tyto alba | Barn owl | KU323 | Tytonidae | Nakhon Ratchasima | Nestling | MK390830 | ||
| Bubo sumatranus | Barred eagle owl | KU485 | Stringidae | Bangkok | Adult | MK390831 | ||
| Athena brama | Spotted owlet | KU270 | Stringidae | Samut Prakan | Adult | MK390832 | ||
| FANTAIL01 | Plasmodium | Glaucidium cuculoides | Asian barred owlet | KU240 | Stringidae | Pathum Thani | Adult | MK390834 |
| GLACUC05 | Plasmodium | Glaucidium cuculoides | Asian barred owlet | KU213 | Stringidae | Nakhon Ratchasima | Adult | MK390824 |
| GLACUC06 | Plasmodium | Glaucidium cuculoides | Asian barred owlet | KU328 | Stringidae | Nakhon Pathom | Juvenile | MK390825 |
| GLACUC07 | Plasmodium | Glaucidium cuculoides | Asian barred owlet | KU406 | Stringidae | Chiang Mai | Juvenile | MK390826 |
| GLACUC08 | Plasmodium | Glaucidium cuculoides | Asian barred owlet | KU408 | Stringidae | Bangkok | Adult | MK390827 |
| MILANS06 | Plasmodium | Glaucidium cuculoides | Asian barred owlet | KU454 | Stringidae | Prachin Buri | Juvenile | MK390835 |
| NISCU2 | Plasmodium | Ninox scutulata | Brown hawk owl | KU523 | Stringidae | Bangkok | Adult | MK390833 |
| ORW1 | Plasmodium | Glaucidium cuculoides | Asian barred owlet | KU281 | Stringidae | Bangkok | Juvenile | MK390836 |
| OTULET03 | Plasmodium | Otus lettia | Collared scops-owl | KU576 | Stringidae | Bangkok | Adult | MK390828 |
| TEPON02 | Plasmodium | Glaucidium cuculoides | Asian barred owlet | KU220 | Stringidae | Bangkok | Adult | MK390837 |
| Ketupa ketupu | Buffy fish owl | KU244 | Stringidae | Bangkok | Adult | MK390838 | ||
New lineages are given in Bold.
ID of owls providing by KURRU.
Lineages had been reported in barn owls and collared scops-owls [Salakij et al., 2018], used for analysis of prevalence and re-analysed of lineage nomenclature.
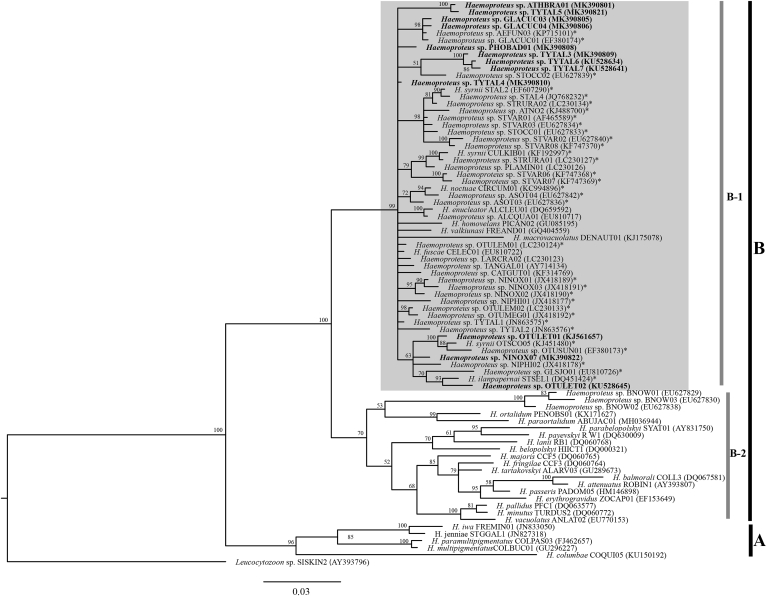
Phylogenetic inference (Fig. 3) revealed that Haemoproteus species could be separated into two well-supported clades, Haemoproteus clade (clade A) and Parahaemoproteus clade (clade B). Based on phylogenetic branches, clade B could also be subdivided into two well-supported clades. Most of the Haemoproteus lineages isolated from strigiform owls clustered together (Fig. 3, clade B-1). However, there were three lineages of Haemoproteus spp. isolated from Strigiformes owls (barn owls: lineages BNOW01, BNOW02, and BNOW03) that grouped in another clade (Fig. 3, clade B-2).
Fig. 3.
Bayesian phylogeny based on partial cytochrome b gene (479 base pairs) of Haemoproteus species lineages. The lineages reported in this study are given in bold. MalAvi lineage codes and GenBank accession numbers are given after species names. Node values (in percentages) indicate posterior clade probabilities. Vertical bars indicate clades of Haemoproteus subgenus (A), Parahaemoproteus (B). Almost all of the Parahaemoproteus lineages recovered from owls were grouped together (clade B-1, grey box). * indicates lineages infecting Strigiformes.
In addition, clade B-1 contained Haemoproteus lineages detected in birds belonging non-Strigiformes orders: Anseriformes (Haemoproteus macrovacuolatus lineage DENAUT01), Charadriiformes (Haemoproteus sp. lineage LARCRA02), Coraciiformes (Haemoproteus enucleator lineage ALCLEU01, Haemoproteus fuscae lineage CELEC01, Haemoproteus sp. lineage TANGAL01, and Haemoproteus sp. lineage ALCQUA01), Pelecaniformes (Haemoproteus sp. lineage PLAMIN01), Piciformes (Haemoproteus homovelans lineage PICAN02), Suliformes (Haemoproteus valkiunasi lineage FREAND01), and Passeriformes (Haemoproteus sp. lineage CATGUT01).
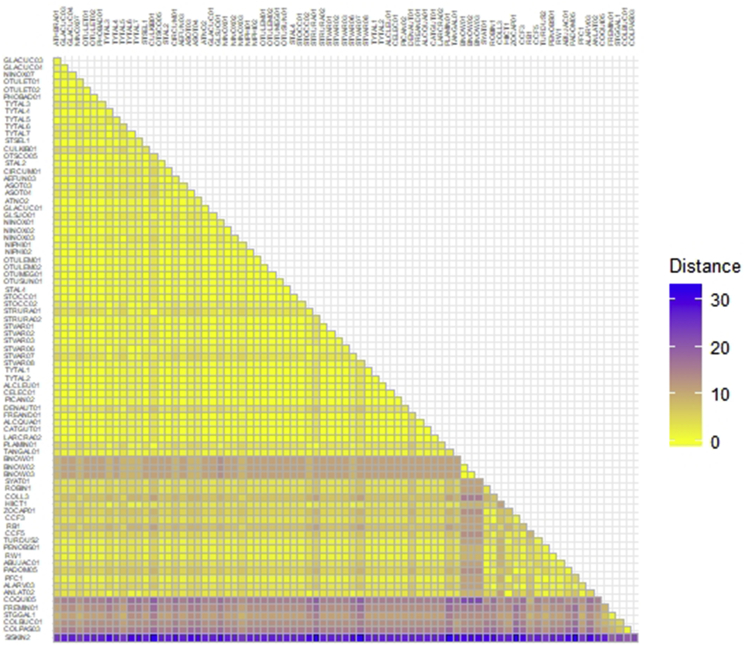
The genetic distances between the lineages in clade B-1 were 0.0%–7.4% (Fig. 4). The genetic distances between the lineages infecting strigiform owls in Africa, Asia, Europe and North America were 0.0%–5.5%. The genetic distance between the 11 new lineages identified in the present study ranged from 0.0% to 3.6%. Regarding other owl parasites, these new lineages had between a 0.0% and 1.8% divergence from H. ilanpapernai and between a 1.8%–3.6% divergence from H. noctuae, while they had between a 0.0% and 5.5% divergence from H. syrnii. The Haemoproteus spp. lineages GLACUC03, GLACUC04, OTULET01, OTULET02, PHOBAD01 and TYTAL4 were identical to H. ilanpapernai (STSEL1, DQ451424), H. syrnii (OTSCO05, KJ451480), and H. syrnii (STAL2, EF607290). Additionally, these 6 lineages were closely related to H. noctuae (CIRCUM01, KC994896), with a 1.8% genetic distance. In addition, the Haemoproteus spp. lineages TYTAL3, TYTAL6 and TYTAL7 were close to H. syrnii (CULKIB01, KF192997), H. syrnii (OTSCO05, KJ451480) and H. syrnii (STAL2, EF607290), with 1.8% genetic distance.
Fig. 4.
Colour heatmap of pairwise genetic distances estimated from nucleotide sequences of the cytochrome b gene (479 bp) of Haemoproteus spp. based on the Jukes-Canter model. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
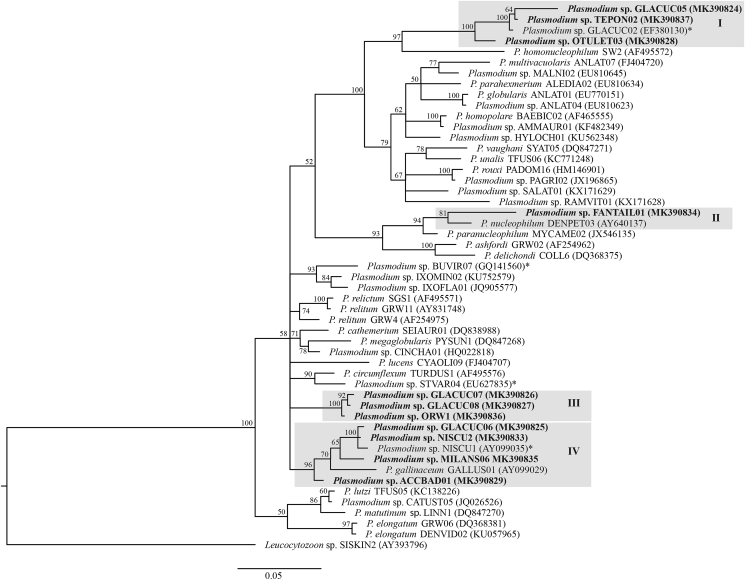
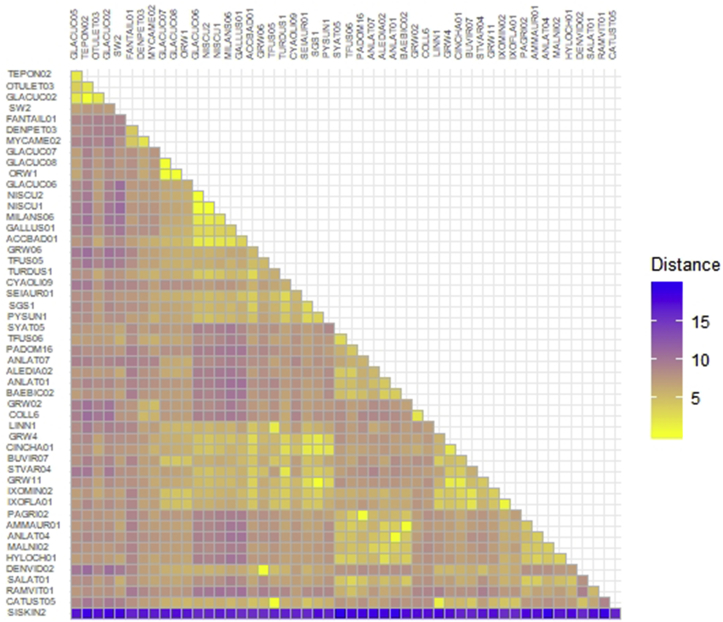
Plasmodium phylogenetic inference revealed that 11 reported lineages in this study, which were classified as group I–IV, were randomly distributed in the tree (Fig. 5). The 11 reported lineages showed genetic distances ranging from 0.2% to 9.5% (Fig. 6). The genetic distance among the six newly reported lineages was 0.2%–8.8%. Regarding the described species Plasmodium, our reported Plasmodium in group I (GLACUC02, TEPON02 and OTULET03) was close to P. homonucleophilum (SW2, AF495572), with a 6.9%–7.6% genetic distance. Plasmodium in group II was close to P. nucleophilum, with a 4.3% genetic distance. Plasmodium in group IV was close to P. gallinaceum, with a 2.4%–3.8% genetic distance. However, Plasmodium group III was unclear.
Fig. 5.
Bayesian phylogeny based on the partial cytochrome b gene (479 base pairs) of Plasmodium species lineages. Lineages reported in this study are given in bold. MalAvi lineages codes and GenBank accession numbers are given after species names. Node values (in percentages) indicate posterior clade probabilities. Four groups (I-IV) of closely related reported lineages are highlighted.
Fig. 6.
Colour heatmap of pairwise genetic distances estimated from nucleotide sequences of the cytochrome b gene (479 bp) of Plasmodium spp. based on the Jukes-Canter model. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
Species of Plasmodium and Haemoproteus are prevalent in owls in eastern and southern Asia (McClure, 1974; Salakij et al., 2012b, 2015a); however, molecular data of these parasites were insufficient (Salakij et al., 2018). This study provided new information on this subject by testing 12 out of 21 species of reported owls in Thailand (BCST, 2018) that were submitted to KURRU for rehabilitation. We report the first data on the molecular prevalence of haemosporidian parasites in 12 species of owls in Thailand. In total, 23 haemosporidian parasite lineages were detected, and 17 of them were new lineages.
The EDTA-blood smears showed extracellular gametocytes. This may have been affected by storage time or anticoagulant addition that induced exflagellation of the haemosporidian parasite (Palinauskas et al., 2013; Valkiūnas, 2005). The number of PCR-positive samples were higher than the number of microscopically positive samples, which supports the suggestion that PCR is a more sensitive method than microscopy (Valkiūnas et al., 2009).
Air temperatures above 20 °C and 12 °C are suitable for completing the sporogony development of Haemoproteus spp. and Plasmodium spp. in their vectors (Bukauskaitė et al., 2015; LaPointe et al., 2010). This is in accord with our findings, which showed Haemoproteus infections during the entire year, and Plasmodium infections were reported almost throughout the year. This was likely due to the average temperatures in Thailand, which are above 20 °C (Fig. 2). Plasmodium infection fluctuated throughout the year, with a peak in September (the end of the rainy season), but infection was absent from May to August (the end of the summer to the rainy season) and January (the cold-dry season). This may be associated with the abundance and activity of their dipteran vector. Thus, further studies addressing vector diversity and abundance during the different seasons are required for better understand the seasonal dynamics of haemosporidian infections in Thailand.
The detection of Haemoproteus spp. and Plasmodium spp. when the birds are admitted to the KURRU is important because it helps to develop preventive measures to minimize possible transmission of haemosporidian parasites within this and other rehabilitation centres. The PCR-based protocol often detects numerous haemosporidian infections (Hellgren et al., 2004; Valkiūnas et al., 2008). The reported prevalence of Haemoproteus and Plasmodium infection, especially Haemoproteus (24.6%), was high in Strigiformes owls in Thailand (34.1%). This results was similar to the prevalence of Haemoprteus in Strigiformes owls in Japan, 56.7% (Murata, 2002) and 57.1% (Inumaru et al., 2017) and Korea 62.8% (Rhim et al., 2018). These results indicate extensive transmission of Haemoproteus species and Plasmodium spp. in Strigiformes owls in Asia.
It is important to note that the PCR protocol used in this study is sensitive to detect mono-infection but insufficiently sensitive to identify Haemoproteus-Plasmodium co-infections, which are common in wildlife (Bernotienė et al., 2016). This can explain why only one co-infection was reported in this study. During PCR-based research, these co-infections can be detected by screening the chromatograms (Dimitrov et al., 2016). However, since preferable amplification of one parasite DNA during co-infections is common, application of one set of general primers provides only partial information about the true parasite diversity and must be carefully analysed in biodiversity and epidemiology research. This study calls for the application of specific primers and nested multiplex (single tube) PCR protocols for the detection of haemosporidian co-infections by Haemoproteus and Plasmodium spp. (Pacheco et al., 2018). In addition, the preferential amplification of primers or intensity of parasitaemia (Bernotienė et al., 2016; Martínez et al., 2009) can be used to explain an absence of Plasmodium-positive samples during Jan, May, Jun, Jul, and Aug, when Haemoproteus-positive samples were found.
To date, approximately 150 species of Haemoproteus and 55 species of Plasmodium have been identified and described in birds (Valkiūnas, 2005; Valkiūnas and Iezhova, 2018). Sequences of three described species of Haemoproteus, including H. noctuae, H. syrnii, and H. ilanpapernai, in owls were deposited on MalAvi database (Bensch et al., 2009). The reported new Haemoproteus lineages ATHBRA01, TYTAL3, TYTAL5, TYTAL6, and TYTAL7 were different from reported species lineages (by at least 1.8%). A genetic distance greater than 5% for the cyt b gene likely indicates different morphospecies of haemosporidian parasites (Hellgren et al., 2007), and even smaller genetic divergences (less than 1%) may be a result of DNA amplification of different parasite species (Križanauskienė et al., 2010). Our data likely indicate the existence of non-described Haemoproteus species diversity in Thai owls.
The Plasmodium lineages, formed four separate groups (I-IV) that were closely related to parasites of the Haemamoeba and Novyella subgenera in our phylogenetic analysis (Valkiūnas and Iezhova, 2018), except the lineages in group III, whose relationship with parasites of other subgenera of Plasmodium remain unclear. There are five Plasmodium species reported in owls: Plasmodium elongatum, P. fallax, P. gundersi, P. hexamerium and P. subpraecox (Valkiūnas, 2005). Of these parasites, only cyt b sequences of P. elongatum were determined and are available in the MalAvi database (Bensch et al., 2009). None of the lineages reported in this study are related to P. elongatum. Available data indicate that owls in Thailand might be infected with non-described species of Haemoproteus and Plasmodium, and this warrants further investigation, by a combination of microscopic and molecular techniques.
Vectors of Haemoproteus parasites of owls and other birds remain unidentified in Thailand and other South Asian countries. This study shows that all lineages of owl Haemoproteus parasites appeared in the clade of Parahaemoproteus parasites (Fig. 3, clade B), which are transmitted by Culicoides biting midges. Owl parasites are absent in clade A, which contains haemosporidians transmitted by louse flies of the family Hippoboscidae. Because phylogenies based on partial cyt b sequences indicate parasite-vector relationships (Bukauskaitė et al., 2018), it is likely that all lineages of owl Haemoproteus parasites reported in this study are transmitted by biting midges of the family Ceratopogonidae. When planning vector studies of owl haemosporidians, attention should be paid mainly to Culicoides biting midges.
Notably, our phylogenetic analysis showed that almost all available Haemoproteus sequences from owls from Africa, Asia, Europe, and North America (Bensch et al., 2009) are phylogenetically related to each other. On the basis of this information, it can be theorized that Haemoproteus that infect owls worldwide possibly share the same ancestor even though they are geography distant. However, three lineages isolated from barn owls (BNOW01, BNOW02, and BNOW03) in California, USA (Ishak et al., 2008) are distinguished from others owl Haemoproteus spp., especially the seven barn owls lineages TYTAL1, TYTAL2, TYTAL3, TYTAL4, TYTAL5, TYTAL6, and TYTAL7. This may suggest that some owl Haemoproteus lineages are unique in different geographical locations.
5. Conclusion
This study reports new haemosporidian parasite lineages, contributes new knowledge for a better understanding of the genetic diversity of owl parasites and analyses the phylogenetic relationships of detected infections with other owl parasite lineages from a global perspective. The data from this study can be used as baseline information for further studies, and particularly for haemosporidian infection diagnostics, which are essential during rehabilitation procedures to prevent the transmission of haemosporidians in admitted raptors. The molecular prevalence of avian haemosporidian parasites in owls in Thailand is high, suggesting the existence of non-described parasite diversity. This raises a question about the influence of these infections on bird health. In total, 17 new lineages of these parasites were detected and deposited in GenBank and MalAvi. This provides opportunities to diagnose the diseases caused by haemosporidians, especially the blood stages, in owls. The further investigation of parasite diversity, sporogonic development, and pathological changes caused by haemosporidian parasites in owls and other avian hosts is important. Due to the high prevalence of haemosporidian parasites throughout the year in Thailand, these infections may represent a problem in owls that should be considered in rehabilitation planning.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethics approval
This study was approved by the Institutional Laboratory Animal Care and Use Committee of Kasetsart University, Thailand under protocol number ACKU 01560.
Acknowledgements
This work was supported by the Faculty of Veterinary Medicine, the Center for Advanced Studies in Agriculture and Food (CASAF), the Kasetsart University Research and Development Institute (grant number 55.61), Thailand, and the Research Council of Lithuania (nr. DOTSUT-137-09.3.3-LMT-K-712-02-0004), Lithuania.
References
- Atkinson C.T., van Riper C., III. Pathogenicity and epizootiology of avian haematozoa: Plasmodium, Leucocytozoon, and Haemoproteus. In: Loye J.E., Zuk M., editors. Bird–Parasite Interactions. Ecology, Evolution, and Behavior. Oxford University Press; New York: 1991. pp. 19–48. [Google Scholar]; Atkinson, C.T., van Riper, C. III. 1991. Pathogenicity and epizootiology of avian haematozoa: Plasmodium, Leucocytozoon, and Haemoproteus. in: Loye, J.E., Zuk, M. (Eds.), Bird-Parasite Interactions. Ecology, Evolution, and Behavior. Oxford University Press, New York, pp. 19-48.
- Baker K.C., Rettenmund C.L., Sander S.J., Rivas A.E., Green K.C., Mangus L., Bronson E. Clinical effect of haemoparasite infections in Snowy owls (Bubo scandiacus) J. Zoo Wildl. Med. 2018;49:143–152. doi: 10.1638/2017-0042R.1. [DOI] [PubMed] [Google Scholar]; Baker, K.C., Rettenmund, C.L., Sander, S.J., Rivas, A.E., Green, K.C., Mangus, L., Bronson, E. 2018. Clinical effect of haemoparasite infections in Snowy owls (Bubo scandiacus) J. Zoo. Wildl. Med. 49, 143-152. https://doi.org/10.1638/2017-0042R.1. [DOI] [PubMed]
- BCST . The Bird Conservation Society of Thailand; Nonthaburi, Thailand: 2018. Checklist Thai Birds 2018.https://www.bcst.or.th/report-archives [Google Scholar]; BCST. 2018. Checklist Thai Birds 2018. The Bird Conservation Society of Thailand, Nonthaburi, Thailand. https://www.bcst.or.th/report-archives. Accessed by 28 Febuary 2019.
- Bensch S., Hellgren O., Pérez-Tris J. MalAvi: a public database of malaria parasites and related haemosporidians in avian hosts based on mitochondrial cytochrome b lineages. Mol. Ecol. Resour. 2009;9:1353–1358. doi: 10.1111/j.1755-0998.2009.02692.x. [DOI] [PubMed] [Google Scholar]; Bensch, S., Hellgren, O., Perez-Tris, J. 2009. MalAvi: a public database of malaria parasites and related haemosporidians in avian hosts based on mitochondrial cytochrome b lineages. Mol. Ecol. Resour. 9, 1353-1358. https://doi.org/10.1111/j.1755-0998.2009.02692.x [DOI] [PubMed]
- Bensch S., Stjerman M., Hasselquist D., Östman Ö., Hansson B., Westerdahl H., Pinheiro R.T. Host specificity in avian blood parasites: a study of Plasmodium and Haemoproteus mitochondrial DNA amplified from birds. Proc. Roy. Soc. Lond. B. 2000;267:1583–1589. doi: 10.1098/rspb.2000.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bensch, S., Stjerman, M., Hasselquist, D., Ostman, O., Hansson, B., Westerdahl, H., Pinheiro, R.T. 2000. Host specificity in avian blood parasites: a study of Plasmodium and Haemoproteus mitochondrial DNA amplified from birds. Proc. R. Soc. Lond. B. 267, 1583-1589. https://doi.org/10.1098/rspb.2000.1181. [DOI] [PMC free article] [PubMed]
- Bernotienė R., Palinauskas V., Iezhova A., Murauskait D., Valkiunas G. Avian haemosporidian parasites (Haemosporida): a comparative analysis of different polymerase chain reaction assays in detection of mixed infections. Exp. Parasitol. 2016;163:31–37. doi: 10.1016/j.exppara.2016.01.009. [DOI] [PubMed] [Google Scholar]; Bernotienė, R., Palinauskas, V., Iezhova, A., Murauskait, D., Valkiunas, G. 2016. Avian haemosporidian parasites (Haemosporida): A comparative analysis of different polymerase chain reaction assays in detection of mixed infections. Exp. Parasitol. 163, 31-37. http://dx.doi.org/10.1016/j.exppara.2016.01.009. [DOI] [PubMed]
- Bukauskaitė D., Iezhova T.A., Ilgūnas M., Valkiūnas G. High susceptibility of the laboratory-reared biting midges Culicoides nubeculosus to Haemoproteus infections, with review on Culicoides species that transmit avian haemoproteids. Parasitology. 2018:1–9. doi: 10.1017/S0031182018001373. [DOI] [PubMed] [Google Scholar]; Bukauskaitė, D., Iezhova, T.A., Ilgūnas, M., Valkiūnas, G. 2018. High susceptibility of the laboratory-reared biting midges Culicoides nubeculosus to Haemoproteus infections, with review on Culicoides species that transmit avian haemoproteids. Parasitology, 1-9. [DOI] [PubMed]
- Bukauskaitė D., Ziegyte R., Palinauskas V., Iezhova T.A., Dimitrov D., Ilgunas M., Bernotiene R., Markovets M.Y., Valkiunas G. Biting midges (Culicoides, Diptera) transmit Haemoproteus parasites of owls: evidence from sporogony and molecular phylogeny. Parasites Vectors. 2015;8:303. doi: 10.1186/s13071-015-0910-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bukauskaitė, D., Ziegyte, R., Palinauskas, V., Iezhova, T. A., Dimitrov, D., Ilgunas, M., Bernotiene, R., Markovets, M. Y., Valkiunas, G. 2015. Biting midges (Culicoides, Diptera) transmit Haemoproteus parasites of owls: evidence from sporogony and molecular phylogeny. Parasit. Vectors. 8, 303. https://doi.org/10.1186/s13071-015-0910-6. [DOI] [PMC free article] [PubMed]
- Chagas C.R.F., Guimarães L.O., Monteiro E.F., Valkiūnas G., Katayama M.V., Santos S.V., Guida F.J.V., Simões R.F., Kirchgatter K. Hemosporidian parasites of free-living birds in the São Paulo Zoo, Brazil. Parasitol. Res. 2015;115:1443–1452. doi: 10.1007/s00436-015-4878-0. [DOI] [PubMed] [Google Scholar]; Chagas, C.R.F., Guimaraes, L.O., Monteiro, E.F., Valkiūnas, G., Katayama, M.V., Santos, S.V., Guida, F.J.V., Simoes, R.F., Kirchgatter, K. 2015. Hemosporidian parasites of free-living birds in the Sao Paulo Zoo, Brazil. Parasitol. Res. 115, 1443-1452. https://doi.org/10.1007/s00436-015-4878-0. [DOI] [PubMed]
- Chagas C.R.F., Valkiūnas G., Guimarães L.O., Monteiro E.F., Guida F.J.V., Simões R.F., Rodrigues P.T., Luna E.J.A., Kirchgatter K. Diversity and distribution of avian malaria and related haemosporidian parasitesin captive birds from a Brazilian megalopolis. Malar. J. 2017;16:83. doi: 10.1186/s12936-017-1729-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chagas, C.R.F., Valkiūnas, G., Guimaraes, L.O., Monteiro, E.F., Guida, F.J.V., Simoes, R.F., Rodrigues, P.T., Luna, E.J.A., Kirchgatter, K. 2017. Diversity and distribution of avian malaria and related haemosporidian parasitesin captive birds from a Brazilian megalopolis. Malar. J. 16, 83. https://doi.org/10.1186/s12936-017-1729-8. [DOI] [PMC free article] [PubMed]
- Darriba D., Taboada G.L., Doallo R., Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]; Darriba, D., Taboada, G.L., Doallo, R., Posada, D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods. 9, 772. https://doi.org/10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed]
- Dimitrov D., Iezhova T.A., Zehtindjiev P., Bobeva A., Ilieva M., Kirilova M., Bedev K., Sjöholm C., Valkiūnas G. Molecular characterisation of three avian haemoproteids (Haemosporida, Haemoproteidae), with the description of Haemoproteus (Parahaemoproteus) palloris n. sp. Syst. Parasitol. 2016;93:431–449. doi: 10.1007/s11230-016-9638-8. [DOI] [PubMed] [Google Scholar]; Dimitrov, D., Iezhova, T.A., Zehtindjiev, P., Bobeva, A., Ilieva, M., Kirilova, M., Bedev, K., Sjoholm, C., Valkiūnas, G. 2016. Molecular characterisation of three avian haemoproteids (Haemosporida, Haemoproteidae), with the description of Haemoproteus (Parahaemoproteus) palloris n. sp. Syst. Parasitol. 93, 431-449. https://doi.org/10.1007/s11230-016-9638-8. [DOI] [PubMed]
- Evans M., Otter A. Fatal combined infection with Haemoproteus noctuae and Leucocytozoon ziemanni in juvenile snowy owls (Nyctea scandiaca) Vet. Rec. 1998;143:72–76. doi: 10.1136/vr.143.3.72. [DOI] [PubMed] [Google Scholar]; Evans, M., Otter, A. 1998. Fatal combined infection with Haemoproteus noctuae and Leucocytozoon ziemanni in juvenile snowy owls (Nyctea scandiaca). Vet. Rec. 143, 72-76. https://dx.doi.org/10.1136/vr.143.3.72. [DOI] [PubMed]
- Hall T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]; Hall, T.A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic. Acids. Symp. Ser. 41, 95-98.
- Hellgren O., Križanauskiene A., Valkiūnas G., Bensch S. Diversity and phylogeny of mitochondrial cytochome b lineages from six morphospecies of avian Haemoproteus (Haemosporida: haemoproteidae) J. Parasitol. 2007;93:889–896. doi: 10.1645/GE-1051R1.1. [DOI] [PubMed] [Google Scholar]; Hellgren, O., Križanauskiene, A., Valkiūnas, G., Bensch, S. 2007. Diversity and phylogeny of mitochondrial cytochome b lineages from six morphospecies of avian Haemoproteus (Haemosporida: Haemoproteidae). J. Parasitol. 93, 889-896. https://doi.org/10.1645/GE-1051R1.1. [DOI] [PubMed]
- Hellgren O., Waldenström J., Bensch S. A new PCR assay for simultaneous studies of Leucocytozoon, Plasmodium, and Haemoproteus from avain blood. J. Parasitol. 2004;90:797–802. doi: 10.1645/GE-184R1. [DOI] [PubMed] [Google Scholar]; Hellgren, O., Waldenstrom, J., Bensch, S. 2004. A new PCR assay for simultaneous studies of Leucocytozoon, Plasmodium, and Haemoproteus from avain blood. J. Parasitol. 90, 797-802. https://doi.org/10.1645/GE-184R1. [DOI] [PubMed]
- Inumaru M., Murata K., Sato Y. Prevalence of avian haemosporidia among injured wild birds in Tokyo and environs, Japan. Int. J. Parasitol. Parasites. Wild. 2017;6:299–309. doi: 10.1016/j.ijppaw.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; Inumaru, M., Murata, K., Sato, Y. 2017. Prevalence of avian haemosporidia among injured wild birds in Tokyo and environs, Japan. Int. J. Parasitol. Parasites. Wild. 6, 299-309. http://doi.org/10.1016/j.ijppaw.2017.09.007. [DOI] [PMC free article] [PubMed]
- Ishak H.D., Dumbacher J.P., Anderson N.L., Keane J.J., Valkiūnas G., Haig S.M., Tell L.A., Sehgal R.N.M. Blood parasites in owls with conservation implications for the spotted owl (Strix occidentalis) PLoS One. 2008;3 doi: 10.1371/journal.pone.0002304. e2304. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ishak, H.D., Dumbacher, J.P., Anderson, N.L., Keane, J.J., Valkiūnas, G., Haig, S.M., Tell, L.A., Sehgal, R.N.M. 2008. Blood Parasites in Owls with Conservation Implications for the Spotted Owl (Strix occidentalis). PLOS ONE 3, e2304. https://doi.org/10.1371/journal.pone.0002304. [DOI] [PMC free article] [PubMed]
- IUCN . International Union for Conservation of Nature and Natural Resources; Gland, Switzerland: 2019. The IUCN Red List of Threatend Speceis. Version 2018-2.https://www.iucnredlist.org [Google Scholar]; IUCN. 2019. The IUCN Red List of Threatend Speceis. Version 2018-2. International Union for Conservation of Nature and Natural Resources, Gland, Switzerland. https://www.iucnredlist.org. Accessed by 28 Febuary 2019.
- Ivanova K., Zehtindjiev P., Mariaux J., Georgiev B.B. Genetic diversity of avian haemosporidians in Malaysia: cytochrome b lineages of the genera Plasmodium and Haemoproteus (Haemosporida) from Selangor. Infect. Genet. Evol. 2015;31:33–39. doi: 10.1016/j.meegid.2015.01.004. [DOI] [PubMed] [Google Scholar]; Ivanova, K., Zehtindjiev, P., Mariaux, J., Georgiev, B.B. 2015. Genetic diversity of avian haemosporidians in Malaysia: Cytochrome b lineages of the genera Plasmodium and Haemoproteus (Haemosporida) from Selangor. Infect. Genet. Evol. 31, 33-39. https://doi.org/10.1016/j.meegid.2015.01.004. [DOI] [PubMed]
- Kelly E.J., Baldwin T.J., Frame D.D., Childress A.L., Wellehan J.F.X. Haemoproteus (Parahaemoproteus) spp. in captive-bred bobwhite quail (Colinus virginianus) in sounthern utha, USA. J. Wildl. Dis. 2018;54:726–733. doi: 10.7589/2017-01-014. [DOI] [PubMed] [Google Scholar]; Kelly, E.J., Baldwin, T.J., Frame, D.D., Childress, A.L., Wellehan, J.F.X. 2018. Haemoproteus (Parahaemoproteus) spp. in captive-bred Bobwhite quail (Colinus virginianus) in sounthern Utha, USA. J. Wildl. Dis. 54, 726-733. https://doi.org/10.7589/2017-01-014. [DOI] [PubMed]
- Kidsin K., Sanythitiseree P., Pothieng D., Wajjwalku W., Kasorndorkbua C. A restrospective study of morbidity and mortality of raptors in Kasetsart University Raptor Rehabilitation Unit, 2008-2011. Kor. J. Orni. 2012;19:87–92. [Google Scholar]; Kidsin, K., Sanythitiseree, P., Pothieng, D., Wajjwalku, W., Kasorndorkbua, C. 2012. A restrospective study of morbidity and mortality of raptors in Kasetsart University Raptor Rehabilitation Unit, 2008-2011. Kor. J. Orni. 19, 87-92.
- Križanauskienė A., Palinauskas V., Hellgren O., Bensch S., Valkinas G. Molecular phylogenetic and morphological analysis of haemosporidian parasites (Haemosporida) in a naturally infected European songbird, the Blackcap Sylvia atricapilla, with description of Haemoproteus pallidulus sp. nov. Parasitology. 2010;137:217–227. doi: 10.1017/S003118200999123579. [DOI] [PubMed] [Google Scholar]; Križanauskienė, A., Palinauskas, V., Hellgren, O., Bensch, S, Valkinas, G. 2010. Molecular phylogenetic and morphological analysis of haemosporidian parasites (Haemosporida) in a naturally infected European songbird, the Blackcap Sylvia atricapilla, with description of Haemoproteus pallidulus sp. nov. Parasitology 137, 217-227. https://doi.org/10.1017/S003118200999123579. [DOI] [PubMed]
- Krone O., Waldenström J., Valkiūnas G., Lessow O., Müller K., Iezhova T.A., Fickel J., Bensch S. Haemosporidian blood parasites in European birds of prey and owls. J. Parasitol. 2008;94:709–715. doi: 10.1645/GE-1357.1. [DOI] [PubMed] [Google Scholar]; Krone, O., Waldenstrom, J., Valkiūnas, G., Lessow, O., Muller, K., Iezhova, T. A., Fickel, J., Bensch, S. 2008. Haemosporidian blood parasites in European birds of prey and owls. J. Parasitol. 94, 709-715. https://doi.org/10.1645/GE-1357.1. [DOI] [PubMed]
- Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kumar, S., Stecher, G., Tamura, K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870-1874. https://doi.org/10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed]
- Kunsorn A., Chomdej S., Sitasuwan N., Wangpakapattawong P., Suwannapoom C., Sandercock B.K. First investigation on the diet of the eastern grass owl during the nesting period in Thailand. Raffles Bull. Zool. 2015;63:27–32. [Google Scholar]; Kunsorn, A., Chomdej, S., Sitasuwan, N., Wangpakapattawong, P., Suwannapoom, C., Sandercock, B.K. 2015. First investigation on the diet of the eastern grass owl during the nesting period in Thailand. Raffles. Bull. Zool. 63, 27-32.
- LaPointe D.A., Goff M.L., Atkinson C.T. Thermal constraints to the sporogonic development and altitudinal aistribution of avian malaria Plasmodium relictum in Hawai'i. J. Parasitol. 2010;96:318–324. doi: 10.1645/GE-2290.1. [DOI] [PubMed] [Google Scholar]; LaPointe, D.A., Goff, M.L., Atkinson, C.T. 2010. Thermal constraints to the sporogonic development and altitudinal aistribution of avian malaria Plasmodium relictum in Hawai'i. J. Parasitol. 96, 318-324. https://doi.org/10.1645/GE-2290.1. [DOI] [PubMed]
- Lee S.-H., Kwak D., Kim K.-T. The first clinical cases of Haemoproteus infection in a Snowy owl (Bubo scandiacus) and a Goshawk (Accipiter gentilis) at a zoo in the Republic of Korea. J. Vet. Med. Sci. 2018;80:1255–1258. doi: 10.1292/jvms.18-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lee, S-H., Kwak, D., Kim, K-T. 2018. The first clinical cases of Haemoproteus infection in a Snowy owl (Bubo scandiacus) and a Goshawk (Accipiter gentilis) at a zoo in the Republic of Korea. J. Vet. Med. Sci. 80, 1255-1258. https://doi.org/10.1292/jvms.18-0072. [DOI] [PMC free article] [PubMed]
- Martínez J., Martínez-De La Puente J., Herrero J., Del Cerro S., Lobato E., Rivero-De Aguilar J., Vásquez R.A., Merino S. A restriction site to differentiate Plasmodium and Haemoproteus infections in birds: on the inefficiency of general primers for detection of mixed infections. Parasitology. 2009;136:713–722. doi: 10.1017/S0031182009006118. [DOI] [PubMed] [Google Scholar]; Martinez, J., Martinez-De La Puente, J., Herrero, J., Del Cerro, S., Lobato, E., Rivero-De Aguilar, J., Vasquez, R.A., Merino, S. 2009. A restriction site to differentiate Plasmodium and Haemoproteus infections in birds: on the inefficiency of general primers for detection of mixed infections. Parasitology 136, 713-722. http://doi.org/doi:10.1017/S0031182009006118. [DOI] [PubMed]
- McClure H.E. U.S. Army Component, SEATO Medical Research Laboratory; Bangkok: 1974. Migration and Survival of the Birds of Asia. [Google Scholar]; McClure, H.E. 1974. Migration and Survival of the Birds of Asia. U.S. Army Component, SEATO Medical Research Laboratory, Bangkok.
- Murata K. Prevalence of blood parasites in Japanese wild birds. Parasitology. 2002;64:785–790. doi: 10.1292/jvms.64.785. [DOI] [PubMed] [Google Scholar]; Murata, K. 2002. Prevalence of blood parasites in Japanese wild birds. Parasitology 64, 785-790. https://doi.org/10.1292/jvms.64.785. [DOI] [PubMed]
- Pacheco M.A., Cepedab A.S., Bernotienėc R., Lottab I.A., Mattab N.E., Valkiūnasc G., Escalantea A.A. Primers targeting mitochondrial genes of avian haemosporidians: PCR detection and differential DNA amplification of parasites belonging to different genera. Int. J. Parasitol. 2018;48:657–670. doi: 10.1016/j.ijpara.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; Pacheco, M.A., Cepedab, A.S, Bernotienėc, R., Lottab, I.A., Mattab, N.E., Valkiūnasc, G., Escalantea, A.A. 2018. Primers targeting mitochondrial genes of avian haemosporidians: PCR detection and differential DNA amplification of parasites belonging to different genera. Int. J. Parasitol. 48, 657-670. https://doi.org/10.1016/j.ijpara.2018.02.003. [DOI] [PMC free article] [PubMed]
- Palinauskas V., Križanauskiene˙ A., Iezhova T.A., Bolshakov C.V., Jönsson J., Bensch S., Valkinas G. A new method for isolation of purified genomic DNA from haemosporidian parasites inhabiting nucleated red blood cells. Exp. Parasitol. 2013;133:275–280. doi: 10.1016/j.exppara.2012.12.003. [DOI] [PubMed] [Google Scholar]; Palinauskas, V., Križanauskiene˙, A., Iezhova, T.A., Bolshakov, C.V., Jonsson, J., Bensch, S., Valkinas, G. 2013. A new method for isolation of purified genomic DNA from haemosporidian parasites inhabiting nucleated red blood cells. Exp. Parasitol. 133, 275-280. https://doi.org/10.1016/j.exppara.2012.12.003. [DOI] [PubMed]
- Puan C.L., Goldizen A.W., Zakaria M., Hafidzi M.N., Baxter G.S. Absence of differential predation on rats by Malaysian barn owls in oil palm plantations. J. Raptor Res. 2011;45:71–78. doi: 10.3356/JRR-10-18.1. [DOI] [Google Scholar]; Puan, C.L., Goldizen, A.W., Zakaria, M., Hafidzi, M.N., Baxter, G.S. 2011. Absence of differential predation on rats by Malaysian barn owls in oil palm plantations. J. Raptor. Res. 45, 71-78. https://doi.org/10.3356/JRR-10-18.1.
- R development core team . R Foundation for Statistical computing; Vienna, Austria: 2016. R: A Language and Environment for Statisticcal Computing. [Google Scholar]; R development core team. 2016. R: A language and environment for statisticcal computing. R Foundation for Statistical computing, Vienna, Austria.
- Remple J.D. Intracellular hematozoa of raptors: a review and update. J. Avian Med. Surg. 2004;18:75–88. doi: 10.1647/2003-008. [DOI] [Google Scholar]; Remple, J.D. 2004. Intracellular hematozoa of raptors: A review and update. J. Avian. Med. Surg. 18, 75-88. https://doi.org/10.1647/2003-008.
- Rhim H., Bae J., Kim H., Han J.-I. Prevalence and phylogenetic analysis of avian aaemosporidia in wild birds in the Republic of Korea. J. Wildl. Dis. 2018;54 doi: 10.7589/2018-01-009. [DOI] [PubMed] [Google Scholar]; Rhim, H., Bae, J., Kim, H., Han, J-I. 2018. Prevalence and phylogenetic analysis of avian aaemosporidia in wild birds in the Republic of Korea. J. Wildl. Dis. 54. http://doi.org/10.7589/2018-01-009. [DOI] [PubMed]
- Ronquist F., Huelsenbeck J.P. MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]; Ronquist, F., Huelsenbeck, J.P. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19, 1572-1574. https://doi.org/10.1093/bioinformatics/btg180. [DOI] [PubMed]
- Salakij C., Kasorndorkbua C., Lertwatcharasarakul P., Salakij J. Hematology, molecular phylogeny and ultra-structure of Trypanosoma corvi in a Shikra. Comp. Clin. Pathol. 2012;21:1757–1761. doi: 10.1007/s00580-012-1612-5. [DOI] [Google Scholar]; Salakij, C., Kasorndorkbua, C., Lertwatcharasarakul, P., Salakij, J. 2012a. Hematology, molecular phylogeny and ultra-structure of Trypanosoma corvi in a Shikra. Comp. Clin. Pathol. 21, 1757-1761. https://doi.org/10.1007/s00580-012-1612-5.
- Salakij C., Kasorndorkbua C., Lertwatcharasarakul P., Salakij J. Ultra-structure of blood cells and molecular characteristics of Haemoproteus sp. in Blyth's hawk eagle. Comp. Clin. Pathol. 2015;24:1293–1299. doi: 10.1007/s00580-015-2161-5. [DOI] [Google Scholar]; Salakij, C., Kasorndorkbua, C., Lertwatcharasarakul, P., Salakij, J. 2015a. Ultra-structure of blood cells and molecular characteristics of Haemoproteus sp. in Blyth’s hawk eagle. Comp. Clin. Pathol. 24, 1293-1299. http://doi.org/10.1007/s00580-015-2161-5.
- Salakij C., Kasorndorkbua C., Salakij J., Suwannasaeng P., Jakthong P. Quantitative and qualitative morphologic, cytochemical and ultrastructural characteristics of blood cells in the Crested Serpent eagle and Shikra. Jpn. J. Vet. Res. 2015;63:95–105. doi: 10.14943/jjvr.63.3.95. [DOI] [PubMed] [Google Scholar]; Salakij, C., Kasorndorkbua, C., Salakij, J., Suwannasaeng, P., Jakthong, P. 2015b. Quantitative and qualitative morphologic, cytochemical and ultrastructural characteristics of blood cells in the Crested Serpent eagle and Shikra. Jpn. J. Vet. Res. 63, 95-105. https://doi.org/10.14943/jjvr.63.3.95. [PubMed]
- Salakij C., Pornpanom P., Lertwatcharasarakul P., Kasorndorkbua C., Salakij J. Haemoproteus in barn and collared scops owls from Thailand. J. Vet. Sci. 2018;19:280–289. doi: 10.4142/jvs.2018.19.2.280. [DOI] [PMC free article] [PubMed] [Google Scholar]; Salakij, C., Pornpanom, P., Lertwatcharasarakul, P., Kasorndorkbua, C., Salakij, J. 2018. Haemoproteus in barn and collared scops owls from Thailand. J. Vet. Sci. 19, 280-289. https://doi.org/10.4142/jvs.2018.19.2.280. [DOI] [PMC free article] [PubMed]
- Salakij J., Lertwatcharasarakul P., Kasorndorkbua C., Salakij C. Plasmodium circumflexum in a Shikra (Accipiter badius): phylogeny and ultra-structure of the haematozoa. Jpn. J. Vet. Res. 2012;60:105–109. doi: 10.14943/jjvr.60.2-3.105. [DOI] [PubMed] [Google Scholar]; Salakij, J., Lertwatcharasarakul, P., Kasorndorkbua, C., Salakij, C. 2012b. Plasmodium circumflexum in a Shikra (Accipiter badius): Phylogeny and ultra-structure of the haematozoa. Jpn. J. Vet. Res. 60, 105-109. https://doi.org/10.14943/jjvr.60.2-3.105. [PubMed]
- Tavitchasri P., Klaudkaew K., Kanloung T., Hamarit K., Wajjwalku W. A survey of barn owl and owl species in Chumkho sub-district, Pathiu district, Chumphon province. Khon. Kaen. Agr. J. 2016;44:389–394. (in Thai, with English abstract) [Google Scholar]; Tavitchasri, P., Klaudkaew, K., Kanloung, T., Hamarit, K., Wajjwalku, W. 2016. A survey of barn owl and owl species in Chumkho sub-district, Pathiu district, Chumphon province. Khon. Kaen. Agr. J. 44, 389-394 (in Thai, with English abstract).
- Valkiūnas G. CRC Press; Boca Raton: 2005. Avian Malaria Parasites and Other Haemosporidia. [Google Scholar]; Valkiūnas, G. 2005. Avian Malaria Parasites and Other Haemosporidia. CRC Press, Boca Raton.
- Valkiūnas G., Anwar A.M., Atkinson C.T., Greiner E.C., Paperna I., Peirce M.A. What distinguishes malaria parasites from other pigmented haemosporidians? Trends Parasitol. 2005;21:357–358. doi: 10.1016/j.pt.2005.06.005. [DOI] [PubMed] [Google Scholar]; Valkiūnas, G., Anwar, A.M., Atkinson, C.T, Greiner, E.C., Paperna, I., Peirce, M.A. 2005. What distinguishes malaria parasites from other pigmented haemosporidians? Trends. Parasitol. 21, 357-358. https://doi.org/10.1016/j.pt.2005.06.005. [DOI] [PubMed]
- Valkiūnas G., Iezhova T.A. Exo-erythrocytic development of avian malaria and related haemosporidian parasites. Malar. J. 2017;16:101. doi: 10.1186/s12936-017-1746-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; Valkiūnas, G., Iezhova, T.A. 2017. Exo-erythrocytic development of avian malaria and related haemosporidian parasites. Malar. J. 16, 101. https://doi.org/10.1186/s12936-017-1746-7. [DOI] [PMC free article] [PubMed]
- Valkiūnas G., Iezhova T.A. Keys to the avian malaria parasites. Malar. J. 2018;17:212. doi: 10.1186/s12936-018-2359-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; Valkiūnas, G., Iezhova, T.A. 2018. Keys to the avian malaria parasites. Malar. J. 17, 212. https://doi.org/10.1186/s12936-018-2359-5. [DOI] [PMC free article] [PubMed]
- Valkiūnas G., Iezhova T.A., Križanauskiene˙ A., Palinauskas V., Sehgal R.N.M., Bensch S. A comparative analysis of microscopy and PCR-based detection methods for blood parasites. J. Parasitol. 2008;94:1395–1401. doi: 10.1645/GE-1570.1. [DOI] [PubMed] [Google Scholar]; Valkiūnas, G., Iezhova, T.A., Križanauskiene˙, A., Palinauskas, V., Sehgal, R.N.M., Bensch, S. 2008. A comparative analysis of microscopy and PCR-based detection methods for blood parasites. J. Parasitol. 94, 1395-1401. https://doi.org/10.1645/GE-1570.1. [DOI] [PubMed]
- Valkiūnas G., Iezhova T.A., Loiseau C., Sehgal R.N.M. Nested cytochrome b polymerase chain reaction diagnostics detected sporozoites of hemosporidian parasites in peripheral blood of naturally infected birds. J. Parasitol. 2009;95:1512–1515. doi: 10.1645/GE-2105.1. [DOI] [PubMed] [Google Scholar]; Valkiūnas, G., Iezhova, T.A., Loiseau, C., Sehgal, R.N.M. 2009. Nested cytochrome b polymerase chain reaction diagnostics detected sporozoites of hemosporidian parasites in peripheral blood of naturally infected birds. J. Parasitol. 95, 1512-1515. [DOI] [PubMed]
- Xue H.R., Yamaguchi N., Driscoll C.A., Han Y., Bar-Gal G.K., Zhuang Y., Mazak J.H., Macdonald D.W., O'Brien S.J., Luo S.J. Genetic ancestry of the extinct Javan and Bali tigers. J. Hered. 2015;106:247–257. doi: 10.1093/jhered/esv002. [DOI] [PMC free article] [PubMed] [Google Scholar]; Xue, H.R., Yamaguchi, N., Driscoll, C.A., Han, Y., Bar-Gal, G.K., Zhuang, Y., Mazak, J.H., Macdonald, D.W., O'Brien, S.J., Luo, S.J. 2015. Genetic ancestry of the extinct Javan and Bali tigers. J. Hered. 106, 247-257. http://doi.org/10.1093/jhered/esv002. [DOI] [PMC free article] [PubMed]