Abstract
Simple Summary
In this article, we investigated the occurrence of Sarcocystis species in samples of sheep and goats obtained from slaughterhouses in Riyadh, Saudi Arabia. We searched for tongue, heart, esophagus, diaphragm and skeletal muscle tissues. Fragments of these tissues were investigated by macroscopic evaluations, direct optical microscopy of tissue fragments, optical microscopy of digested fragment sediment, transmission electron microscopy and PCR followed by nucleotide sequencing. From the set of information obtained, we searched 230 sheep, and 84 goats; 91 and 36 were found to be infected, respectively. Transmission electron microscopy (TEM) revealed Sarcocystis tenella (S. tenella) in sheep and Sarcocystis capracanis (S. capracanis) in goats. Sarcocystis species were confirmed in Saudi Arabian sheep and goats by molecular testing. S. capracanis was most closely related to S. tenella, with the mitochondrial cytochrome c oxidase subunit I gene (COX1) sequences sharing a (91.7%) identity.
Abstract
Sarcocystosis is induced by species of Sarcocystis, which is an intracellular protozoan parasite in the phylum Apicomplexa. The diversity and importance of Sarcocystis species in sheep and goats in Saudi Arabia are poorly understood. In this study, the tongue, esophagus, heart, diaphragm, and skeletal muscles were collected from 230 sheep and 84 goats, and the tissues were examined for the presence of Sarcocystis species by macroscopic examination and light microscopy. Microscopic Sarcocystis species cysts were found in both sheep and goats. Transmission electron microscopy (TEM) revealed S. tenella in sheep and S. capracanis in goats. Sarcocystis species were confirmed for the first time in Saudi Arabian sheep and goats by molecular testing. S. capracanis was most closely related to S. tenella, with the COX1 sequences sharing 91.7% identity. A phylogenetic analysis produced similar results and indicated that the Sarcocystis isolates were within a group of Sarcocystis species in which dogs were the final host. Finally, the Sarcocystis species cysts from sheep and goats could be grouped together, indicating that they were strongly related.
Keywords: Sarcocystis species, COX1, sheep and goats
1. Introduction
Sarcocystis species have an obligatory heteroxenous (prey-predator) life cycle [1]. Sarcocystis infection is common among various vertebrates, including people [2,3,4,5,6,7]. Sheep are intermediate hosts for at least six species, including S. tenella, S. arieticanis, S. gigantea, S. medusiformis, S. mihoensis, and S. microps, which are morphologically differentiated based on their sarcocyst wall ultrastructure. Sarcocystis tenella and S. arieticanis produce microscopic sarcocysts transmitted by canids, while S. gigantea and S. medusiformis produce macroscopic cysts transmitted by felids [2]. The remaining two species, S. mihoensis and S. microps, are transmitted by canids and are unusual or rare species of Sarcocystis. Sarcocystis mihoensis, reported only from Japan, and they produce macroscopic sarcocysts [8,9]. Goats can be infected by three different Sarcocystis species: S. capracanis and S. hircicanis, in which dogs are the final host, and S. moulei, in which cats are the final host [10,11]. In Saudi Arabia, sheep and goats are used for both meat and milk production. There are three main breeds of sheep. Niemy and Najdy are local sheep, while the Sawakny breed is imported from Sudan and is used for meat production only [12]. Previous studies have examined the prevalence of Sarcocystis in Najdy sheep and camels slaughtered in Riyadh [13,14], Sarcocystis in slaughtered camels in the Al-Ahssa region [15], and Sarcocystis parasites in the muscles of Arabian deer using optical and electron microscopy techniques [16]. In recent years, molecular techniques have been used as an epidemiological and diagnostic tool to determine which Sarcocystis species are involved in an infection [17,18]. Molecular characterization using appropriate markers has become an essential tool for accurate identification of Sarcocystis species and for research regarding the phylogenetic relationships of these species. As for Sarcocystis species in cervids, nucleotide sequences of the nuclear 18S ribosomal RNA gene (18S rRNA) and/or the mitochondrial cytochrome c oxidase subunit I gene (COX1) of approximately 30 species are currently available for comparative molecular studies [19,20]. Of these markers, COX1 has been found to be superior to 18S rRNA in resolving unclear species boundaries of closely related Sarcocystis species in various ruminant intermediate hosts [20,21,22]. The present study aimed to analyze the genes encoding COX1 in Sarcocystis species isolated from naturally infected sheep and goats in Riyadh, Saudi Arabia.
2. Materials and Methods
2.1. Sample Collection
The Institutional Committee of Postgraduate Studies and Research at King Saud University (Saudi Arabia) approved this study. Tissue samples were collected by veterinarians during postmortem inspections of slaughtered animals performed at the Al-Sada Abattoir in Riyadh, Saudi Arabia, from March 2016 to January 2017. Tissue samples were isolated from 230 sheep (including the Najdy, Niemy, and Sawakny breeds) and 84 goats. The entire tongue, heart, skeletal muscle, diaphragm, and esophagus were collected from each animal and individually stored in sealed plastic bags. The tissues were then transported to the laboratory in boxes containing ice packs.
2.2. Macroscopic Analysis
A macroscopic analysis was performed on the same day as tissue collection. Five transverse cuts were performed using a scalpel on the tongue and heart to reveal the macroscopic cysts. The entire esophagus was longitudinally sectioned to expose its lumen, and its internal and external walls were analyzed macroscopically [23].
2.3. Microscopic Analysis
The microscopic analysis for cysts using fresh tissues was performed using a squash preparation [24]. Separate fragments of each tissue measuring approximately 5 mm thick were firmly squashed between two slides and examined under an optical microscope at 40× and 100× magnifications. This procedure was performed in triplicate for each tissue. Sarcocystis species cysts were processed for light microscopy (LM), transmission electron microscopy (TEM), and DNA analysis.
2.4. Digestion Method
Approximately 20 g of each tissue was minced and then digested for 30 min at 37 °C in 100 mL of digestion medium containing 1.3 g of pepsin, 3.5 mL HCl, and 2.5 g NaCl in 500 mL of distilled water [25]. After digestion, the mixture was centrifuged for 3 min at 3500× g, and then the sediment was stained with Giemsa and examined by optical microscopy at 400× magnification [26].
2.5. Transmission Electron Microscopy (TEM)
Sarcocystis species cysts were collected, fixed in 2% glutaraldehyde solution in 0.1 M sodium cacodylate buffer (pH 7.4) for 2 h at 15–25 °C, and stored at 4 °C until processing. After fixation, the samples were washed in 0.1 M sodium cacodylate buffer (pH 7.4), fixed with 1% osmium tetroxide, dehydrated in different acetone solutions (30, 40, 50, 70, 90, and 100%), and treated in blocking buffer with 1% phosphotungstic acid and 1% uranyl acetate. Next, the 100% acetone solution was replaced with Polybed resin, followed by paraffin embedding and polymerization in an oven at 60°C. Semithin cuts were made to observe the Sarcocystis species cysts by light microscopy. Ultrathin sections were stained with uranyl acetate and lead citrate and then examined using a JEM100-CX transmission electron microscope at 80 kV. [23].
2.6. Molecular Analysis
2.6.1. DNA Extraction and PCR Amplification
Small pieces from each individual microscopic Sarcocystis isolate were washed five times with sterile distilled water, and genomic DNA (gDNA) was extracted using the QIAamp DNA mini kit for tissue and blood according to the manufacturer’s instructions (Qiagen GmbH, Hilden, Germany Cat. No. 51304). The mitochondrial COX1 gene was amplified using specific primer pairs (Table 1) [21,22]. Amplification was carried out in a thermocycler (Veriti®96-well thermal cycler, model 9902, Biosystem) in a 20-μL reaction mixture containing 4 μL of master mix (5×), 12.8 μL of RNase-free water, 1.2 μL of both forward and reverse primers, and 2 μL of DNA template. The PCR program consisted of an initial denaturation step at 94 °C for 5 min, followed by 40 cycles of denaturation at 94 °C for 45 seconds, annealing at 50 °C for 45 seconds, and a final extension step at 72 °C for 10 min. The PCR products were analyzed by 1% agarose gel electrophoresis.
Table 1.
List of primers used for the amplification of genes encoding c oxidase subunit I gene (COX1) in Sarcocystis species.
2.6.2. DNA Sequencing and Phylogenetic Analysis
The PCR products derived from the target COX1 gene were purified and subjected to sequencing with both forward and reverse complements on the Genetic Analyzer at the Central Lab of King Saud University. The sequences were analyzed using Geneious software version 11.1.4. All sequences were slightly truncated using an error probability method with a limit of 0.05 at both ends. A BLAST search was performed for each sequence to find related sequences.
The neighbor-joining method was used with the Tamura Nei model in order to generate a phylogenetic tree [27]. The bootstrap method was used for resampling with the number of replicates set to 1000.
2.7. Statistical Analysis
Statistical analysis was performed using the Statistical Package for Social Sciences (SPSS) software (version 17, SPSS, Inc., Chicago, IL, USA). Continuous and categorical variables were displayed as the means ± standard deviation (SD) and percentages, respectively. Differences in the prevalence between males and females and different age groups were analyzed by a Chi-square test (χ2). p Values < 0.05 were considered statistically significant. The lengths and widths of at least ten cysts from each organ (heart, tongue, and esophagus) were determined by light microscopy and expressed as the mean sizes and amplitudes of variation.
3. Results
3.1. Prevalence of Natural Infection
An investigation of random muscles samples obtained from 314 slaughtered sheep and goats for the presence of Sarcocystis species revealed that the prevalence of Sarcocystis in naturally infected Niemy sheep, Najdy sheep, Sawakny sheep, and goats was 35.18%, 43.33%, 43.75%, and 42.85%, respectively (Table 2).
Table 2.
Prevalence of Sarcocystis species in sheep and goats.
| Animals | No. Examined (%) | No. Infected (%) |
|---|---|---|
| Sheep | Niemy 108 | 38 (35.18%) |
| Najdy 90 | 39 (43.33%) | |
| Sawakny 32 | 14 (43.75%) | |
| 230 | 91 (39.56%) | |
| Goats | 84 | 36 (42.85%) |
| Total | 314 | 127 (40.44%) |
The prevalence of Sarcocystis species in young and adult male sheep was 43.53% and 35.84%, respectively. The prevalence of Sarcocystis species was 42.85% and 33.33%, respectively, in young and adult female sheep. In young and adult male goats, the prevalence was 47.45% and 40%, respectively. Finally, the prevalence in young female goats was 35.29%, whereas the examined adult female goats were not infected (Table 3).
Table 3.
Prevalence of Sarcocystis species among different categories of sheep and goats.
| Categories | Sheep | Goats | ||
|---|---|---|---|---|
| No. Examined | No. Infected (%) | No. Examined | No. Infected (%) | |
| Young males (under 1 year) | 147 | 64 (43.53%) | 59 | 28 (47.45%) |
| Adult males (above 1 year) | 53 | 19 (35.84%) | 5 | 2 (40%) |
| Young females (under 1 year) | 21 | 9 (42.85%) | 17 | 6 (35.29%) |
| Adult females (above 1 year) | 9 | 3 (33.33%) | 3 | 0 (0%) |
| Total | 230 | 95 (41.30%) | 84 | 36 (42.85%) |
The distribution of Sarcocystis cysts varied from one sheep organ to another, and from one sheep breed to another. The distribution was different than that in goats, but in general, the diaphragm and skeletal muscle showed the highest infection levels (Niemy sheep, 84.21%; Najdy sheep, 61.53%; Sawakny sheep, 85.71%; goats, 61.11%) of all the organs examined. The tongue was the organ with the lowest infection rate (Najdy sheep, 12.82%; Sawakny sheep, 21.42%; goats, 19.44%), with the exception of Niemy sheep, in which the esophagus had the lowest infection rate (28.94%) (Table 4).
Table 4.
Organ distribution of Sarcocystis species in sheep and goats.
| Animals | No. Infected | Esophagus | Tongue | Diaphragm | Heart | Skeletal Muscles |
|---|---|---|---|---|---|---|
| No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | ||
| Niemy | 38 | 11 | 14 | 26 | 23 | 32 |
| (28.94%) | (36.84%) | (68.42%) | (60.52%) | (84.21%) | ||
| Najdy | 39 | 8 | 5 | 24 | 14 | 17 |
| (20.51%) | (12.82%) | (61.53%) | (35.89%) | (43.58%) | ||
| Sawakny | 14 | 5 | 3 | 12 | 8 | 11 |
| (35.71%) | (21.42%) | (85.71%) | (57.14%) | (78.57%) | ||
| Goats | 36 | 9 | 7 | 21 | 17 | 22 |
| (25%) | (19.44%) | (58.33%) | (47.22%) | (61.11%) | ||
| Total | 127 | 33 | 29 | 83 | 62 | 82 |
| (25.98%) | (22.83%) | (65.35%) | (48.81%) | (64.56%) |
3.2. Morphological Characteristics of the Cysts
An analysis of the esophagus, tongue, diaphragm, skeletal muscle, and heart revealed microscopic S. tenella and S. capracanis cysts in sheep and goats, respectively. Macroscopic cysts were not recorded in the present study.
3.2.1. S. tenella Cysts
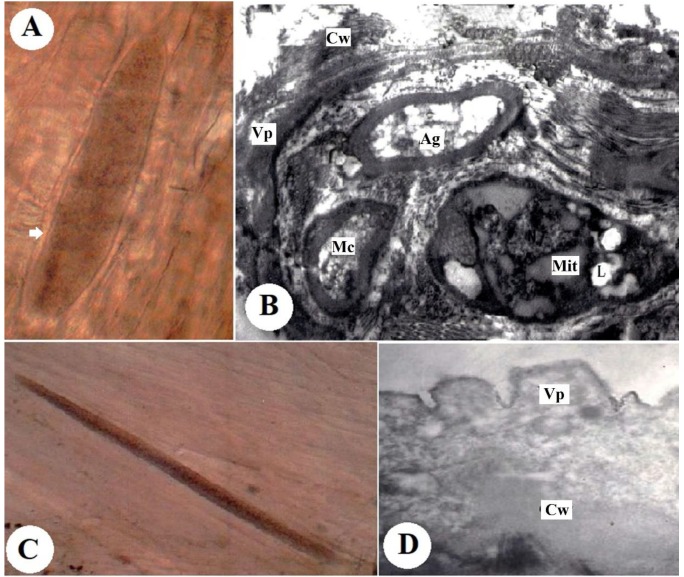
The fresh microscopic cysts were spindle-shaped with fully formed walls. They measured 851 µm × 75 µm to 185 µm × 31 µm (Figure 1A). An ultrastructure analysis of the cyst wall revealed that it was 0.74-µm thick and consisted of finger-like villar projections (Vp). The rim of the Vp consisted of minute undulations over the basal third. The microtubules were often arranged in bundles (Figure 1B).
Figure 1.
(A) Fresh cyst of S. tenella in the muscle of naturally infected sheep (digital camera). (B) Ultrastructure of S. tenella with a fully formed cyst wall (Cw) showing finger-like villar projections (Vp) (×16,000). (C). Fresh cyst of S. capracanis in the muscle of naturally infected goats (digital camera). (D) Ultrastructure of S. capracanis with the fully formed cyst wall (Cw) showing finger-like villar projections (Vp), metrocytes (Mc), lipid (L), amylopectin granules (Ag) and mitochondria (Mit) (×6300).
3.2.2. S. capracanis Cysts
The fresh microscopic cysts had a spindle shape with a thin wall and measured 869 µm × 648 µm to 370 µm × 70 µm (Figure 1C). The ultrastructural analysis showed that the cyst wall had palisade-like Vp with long constrictions at the base (Figure 1D).
3.3. Molecular Analysis
Partial amplification of the COX1 gene (1085 bp) is shown in all DNA samples of microscopic Sarcocystis species.
3.4. Molecular Characterization of the COX1 Gene
A total of 23 samples, including seven Sawakny samples, nine Najdy samples, two Naimy samples, and five goat samples, were examined. The samples were 1034-1106 nucleotides long and shared a 91.7% pairwise identity.
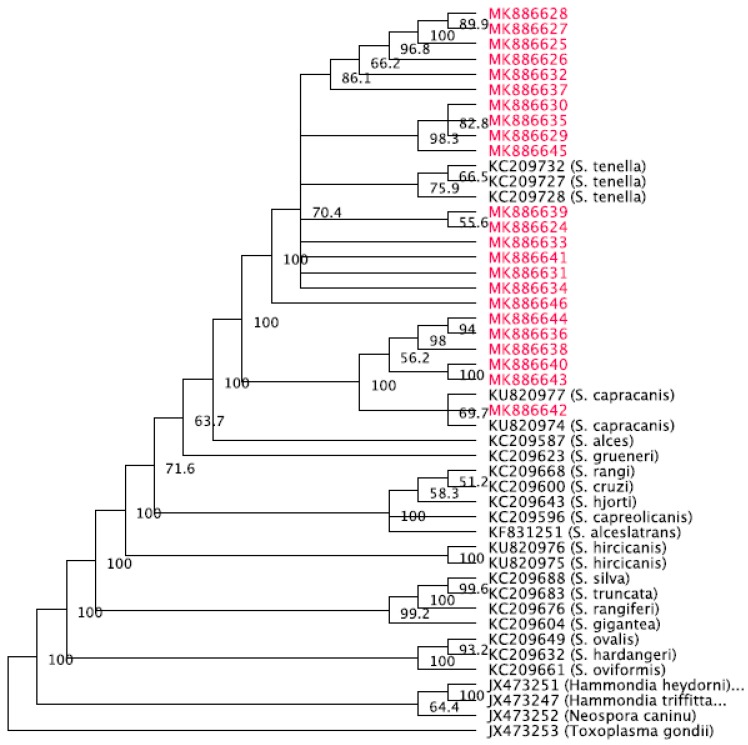
The phylogenetic tree was generated with Toxoplasma gondii, accession number JX473253, as the out-group. The phylogenetic tree with consensus support values is shown in Figure 2. The samples form a clade with S. tenella sequences given by accession numbers KC209727, KC209728, and KC209732 from Norwegian sheep. In addition, the clade contains S. capracanis sequences with accession numbers KU820974 and KU820977 from Chinese goats.
Figure 2.
Phylogenetic tree of the 23 samples along with several sequences published in GenBank.
4. Discussion
To determine the presence of Sarcosystis spp., the present study examined the esophagus, tongue, diaphragm, skeletal muscle, and heart tissue from 230 sheep and 84 goat specimens. It was concluded that the overall prevalence for the Sarcosystis spp. were as follows: Sheep specimens, 39.5%, and goat specimens, 42.8%. It was unexpected that adult animals had a lower prevalence Sarcosystis; this could be due to low number of adult animal samples. Microscopic Sarcocystis species cysts were detected in the current work by means of tissue squash, pepsin-hydrochloric acid digestion, and Hematoxylin and eosin stain. The tissue squash method was a fast and practical diagnostic tool and detected a higher number of positive animals than the pepsin-hydrochloric acid digestion technique, although these methods did not permit differentiation among the Sarcocystis species. There was great variation in the lengths and widths of the observed cysts, which may be related to the age of the cysts.
As mentioned previously, the only documented evidence of Sarcosystis species in Saudi Arabia was based on TEM [12,28]. The present study included the separation of S. arieticanis cysts. In all previous studies, the absence of septa in S. arieticanis cysts [29] was probably due to the low number of cysts examined (n = 3). Previous reports throughout the world have recorded S. tenella infection in sheep from Brazil [18], Tunisia [30], and Iraq [31]. The cysts examined by TEM in the goat tissues in the present study had wall morphological characteristics of S. capracanis, with finger-like or palisade Vp [32]. The morphology of S. capracanis observed in the present study conforms to the TEM classification by [33] and was similar to the morphology of S. capracanis in domestic goats observed in Japan [34], the Philippines [35], Egypt [36], and China [37]. In addition, similar sarcocyst morphology has been described from other domestic ruminant animals, such as S. tenella in domestic sheep [38]. In the present study, certain DNA samples from the microscopic Sarcocystis species yielded no bands during the partial PCR amplification of the COX1 gene. This may be due to the low number of COX1 gene copies. Therefore, it is essential to utilize additional PCR cycles in these studies. When these sequences were queried via Basic Local Alignment Search Tool (BLAST), the sequence of COX1 from S. tenella (18 sheep) shared high identity (91.7%) with that from S. capracanis (5 goats). The high identity between the COX1 sequences from S. tenella and S. capracanis suggests that sheep and goats can harbor the same Sarcocystis species [39].
Since this study demonstrated a high prevalence of Sarcocystis infection by microscopic cyst forming species, sheep and goats could be considered alternative intermediate hosts for S. tenella and S. capracanis. The complete sequences of the gene that encoded COX1, or the sequence analysis of other genetic loci, are essential in order to understand the genetic diversity found across the world among Sarcocystis species. Our data collected in this study highlight the need to carry a more in-depth phylogenetic analysis and study of Sarcocystis species with more taxa and different molecular markers. Because of its high variance, studies with the internal transcribed spacer gene 1 (ITS-1) region should be performed to further distinguish among the closely related Sarcocystis species.
Author Contributions
Conceptualization: D.M.M. Data curation: I.M.A.-T. Formal analysis: M.A.A.-D. Funding acquisition: M.F.E.-K. Investigation: D.M.M. Methodology: M.A.A.-D. Project administration: D.M.M. Resources: M.A.A.-D. and Software: I.M.A.-T. Supervision: D.M.M. Validation: D.M.M. Visualization: M.F.E.-K. Writing: original draft: D.M.M. Writing, review and editing: M.F.E.-K., D.M.M., and I.M.A.-T.
Funding
The authors would like to extend their sincere appreciation to the Deanship of Scientific Research at King Saud University for funding this research through the Research Group project no. RG-1439-034.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Abdel-Ghaffar F., Mehlhorn H., Bashtar A.R., Al-Rasheid K., Sakran T., El-Fayoumi H. Life cycle of Sarcocystis camelicanis infecting the camel (Camelus dromedarius) and the dog (Canis familiaris), light and electron microscopic study. Parasitol. Res. 2009;106:189–195. doi: 10.1007/s00436-009-1648-x. [DOI] [PubMed] [Google Scholar]
- 2.Mehlhorn H., Heydorn A.O. Advances in Parasitology. Academic Press; Cambridge, MA, USA: 1978. The Sarcosporidia (Protozoa, Sporozoa): Life cycle and fine structure; pp. 43–91. [DOI] [PubMed] [Google Scholar]
- 3.Ghaffar F.A., Hilali M., Scholtyseck E. Ultrastructural study of Sarcocystis fusiformis (Railliet, 1897) infecting the Indian water buffalo (Bubalus bubalis) of Egypt. Trop. Parasitol. 1978;29:289–294. [PubMed] [Google Scholar]
- 4.Dubey J.P., Kistner T.P., Callis G. Development of Sarcocystis in mule deer transmitted through dogs and coyotes. Can. J. Zool. 1983;61:2904–2912. doi: 10.1139/z83-378. [DOI] [Google Scholar]
- 5.Entzroth R. Invasion and early development of Sarcocystis muris (Apicomplexa, Sarcocystidae) in tissue cultures. J. Protozol. 1985;32:446–453. doi: 10.1111/j.1550-7408.1985.tb04042.x. [DOI] [PubMed] [Google Scholar]
- 6.Abdel-Ghaffar F., Bashtar A.R., El-Sayed M. Electron microscopic studies on Sarcocystis infection in sheep in Upper Egypt. Bull. Fac. Sci. Cairo Univ. 1990;58:33–49. [Google Scholar]
- 7.Abdel-Ghaffar F., Al-Johany A. A light and electron microscope study of Sarcocystis mitrani (sp. nov.) infecting the skink Scincus mitranus in the central region of Saudi Arabia. Parasitol. Res. 2002;88:102–106. doi: 10.1007/s004360100506. [DOI] [PubMed] [Google Scholar]
- 8.Collins G.H., Atkinson E., Charleston W.A. Studies on Sarcocystis species III: The macrocystic species of sheep. N. Z. Vet. J. 1979;27:204–206. doi: 10.1080/00480169.1979.34651. [DOI] [PubMed] [Google Scholar]
- 9.Heydorn A.O., Mehlhorn H. Fine structure of Sarcocystis arieticanis Heydorn, 1985 in its intermediate and final hosts (sheep and dog) Zentralbl. Bakteriol. Mikrobiol. Hyg. Ser. A. 1987;264:353–362. doi: 10.1016/S0176-6724(87)80056-1. [DOI] [PubMed] [Google Scholar]
- 10.Heydorn A.O., Gestrich R., Mehlhorn H., Rommel M. Proposal for a new nomenclature of the Sarcosporidia. Parasitol. Res. 1975;48:73–82. doi: 10.1007/BF00389639. [DOI] [PubMed] [Google Scholar]
- 11.Heydorn A.O., Matuschka F.R. Zur Endwirtspezifität der vom Hund übertragenen Sarkosporidienarten, Final host specificity of Sarcocystis species transmitted by dogs. Zeitschrift für Parasitenkunde. 1981;66:231–234. doi: 10.1007/BF00925730. [DOI] [PubMed] [Google Scholar]
- 12.Al Quraishy S., Morsy K., Bashtar A.R., Ghaffar F.A., Mehlhorn H. Sarcocystis arieticanis (Apicomplexa: Sarcocystidae) infecting the heart muscles of the domestic sheep, Ovis aries (Artiodactyla: Bovidae), from KSA on the basis of light and electron microscopic data. Parasitol. Res. 2014;113:3823–3831. doi: 10.1007/s00436-014-4050-2. [DOI] [PubMed] [Google Scholar]
- 13.Hussein H.S. The prevalence of Sarcocystis infection in Saudi Arabia Najdi sheep and camels. Biol. Sci. 1991;1:43–56. [Google Scholar]
- 14.Hilali M., Fatani A., Al-Atiya S. Isolation of tissue cysts of Toxoplasma, Isospora, Hammondia and Sarcocystis from camel (Camelus dromedarius) meat in Saudi Arabia. Vet. Parasitol. 1995;58:353–356. doi: 10.1016/0304-4017(94)00727-T. [DOI] [PubMed] [Google Scholar]
- 15.Fatani A., Hilali M., Al-Atiya S., Al-Shami S. Prevalence of Sarcocystis in camels (Camelus dromedarius) from Al-Ahsa, Saudi Arabia. Vet. Parasitol. 1996;62:241–245. doi: 10.1016/0304-4017(95)00843-8. [DOI] [PubMed] [Google Scholar]
- 16.Shazly M.A. Light and electron microscopic studies on Sarcocystis infecting the Dromedaries in Saudi Arabia. Egypt. J. Zool. 2000;35:273–285. [Google Scholar]
- 17.Mohammed O.B. Ph.D. Thesis. Kingston University; London, UK: 2000. Prevalence, Identity and Phylogeny of Sarcocystis Parasites from Gazelles in Saudi Arabia. [Google Scholar]
- 18.da Silva R.C., Su C., Langoni H. First identification of Sarcocystis tenella (Railliet, 1886) Moule, 1886 (Protozoa: Apicomplexa) by PCR in naturally infected sheep from Brazil. Vet. Parasitol. 2009;165:332–336. doi: 10.1016/j.vetpar.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 19.Gjerde B., Luzón M., Alunda J.M., de la Fuente C. Morphological and molecular characteristics of six Sarcocystis species from red deer (Cervus elaphus) in Spain, including Sarcocystis cervicanis and three new species. Parasitol. Res. 2017;116:2795–2811. doi: 10.1007/s00436-017-5590-z. [DOI] [PubMed] [Google Scholar]
- 20.Rudaitytė-Lukošienė E., Prakas P., Butkauskas D., Kutkienė L., Vepštaitė-Monstavičė I., Servienė E. Morphological and molecular identification of Sarcocystis species from the sika deer (Cervus nippon), including two new species Sarcocystis frondea and Sarcocystis nipponi. Parasitol. Res. 2018;117:1305–1315. doi: 10.1007/s00436-018-5816-8. [DOI] [PubMed] [Google Scholar]
- 21.Gjerde B. Phylogenetic relationships among Sarcocystis species in cervids, cattle and sheep inferred from the mitochondrial cytochrome c oxidase subunit I gene. Int. J. Parasitol. 2013;43:579–591. doi: 10.1016/j.ijpara.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 22.GJERDE B. Sarcocystis species in red deer revisited: With a re-description of two known species as Sarcocystis elongata n. sp. and Sarcocystis truncata n. sp. based on mitochondrial COX1 sequences. Parasitology. 2014;141:441–452. doi: 10.1017/S0031182013001819. [DOI] [PubMed] [Google Scholar]
- 23.Bittencourt M.V., Meneses I.D., Ribeiro-Andrade M., de Jesus R.F., de Araújo F.R., Gondim L.F. Sarcocystis species in sheep and goats: Frequency of infection and species identification by morphological, ultrastructural, and molecular tests in Bahia, Brazil. Parasitol. Res. 2016;115:1683–1689. doi: 10.1007/s00436-016-4909-5. [DOI] [PubMed] [Google Scholar]
- 24.Odening K., Stolte M., Bockhardt I. On the diagnostics of Sarcocystis in cattle: Sarcocysts of a species unusual for Bos taurus in a dwarf zebu. Vet. Parasitol. 1996;66:19–24. doi: 10.1016/S0304-4017(96)00993-4. [DOI] [PubMed] [Google Scholar]
- 25.Dubey J.P., Speer C.A., Charleston W.A. Ultrastructural differentiation between sarcocysts of Sarcocystis hirsuta and Sarcocystis hominis. Vet. Parasitol. 1989;34:153–157. doi: 10.1016/0304-4017(89)90177-5. [DOI] [PubMed] [Google Scholar]
- 26.Hamidinejat H., Moetamedi H., Alborzi A., Hatami A. Molecular detection of Sarcocystis species in slaughtered sheep by PCR–RFLP from south-western of Iran. J. Parasit. Dis. 2014;38:233–237. doi: 10.1007/s12639-012-0231-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saitou N., Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 28.Al-Qureishy S.A. Ph.D. Thesis. King Saud University; Riyadh, Saudi Arabia: 2005. Sarcocystis Parasites (Coccidia, Apicomplexa) Infecting Some Slaughtered Animals in Riyadh City, Saudi Arabia. [Google Scholar]
- 29.Dubey J.P., Lindsay D.S., Speer C.A., Fayer R., Livingston C.W., Jr. Sarcocystis arieticanis and other Sarcocystis species in sheep in the United States. J. Parasitol. 1988;74:1033–1038. doi: 10.2307/3282228. [DOI] [PubMed] [Google Scholar]
- 30.Amairia S., Amdouni Y., Rouatbi M., Rjeibi M.R., Awadi S., Gharbi M. First detection and molecular identification of Sarcocystis species in small ruminants in North-West Tunisia. Transbound. Emerg. Dis. 2018;65:441–446. doi: 10.1111/tbed.12722. [DOI] [PubMed] [Google Scholar]
- 31.Whaeeb S.T., Khalaf A.A.F.I.A. Molecular Study of Four Species of Sarcocystis Isolated from Sheeps Esophagus and Inter Skeleton Muscle in Baghdad. WJPR. 2016;5:158–165. [Google Scholar]
- 32.Heydorn A.O., Haralambidis S. Development of Sarcocystis capracanis Fischer, 1979. Berl. Munch. Tierarztl. Wochenschr. 1982;95:265–271. [PubMed] [Google Scholar]
- 33.Dubey J.P., Calero-Bernal R., Rosenthal B.M., Speer C.A., Fayer R. Sarcocystosis of Animals and Humans. CRC Press; Boca Raton, FL, USA: 2015. [Google Scholar]
- 34.Saito M., Shibata Y., Kobayashi T., Kobayashi M., Kubo M., Itagaki H. Ultrastructure of the cyst wall of Sarcocystis species with canine final host in Japan. J. Vet. Med. Sci. 1996;58:861–867. doi: 10.1292/jvms.58.861. [DOI] [PubMed] [Google Scholar]
- 35.Claveria F.G., Pedro-Lim M.R.S., Tan J.E., Flores-Cruz M.J. Sarcocystis capracanis infection in Philippine domestic goats (Capra hircus): Ultrastructural studies. Philipp. J. Sci. 2004;133:33–37. [Google Scholar]
- 36.Morsy K., Saleh A., Al-Ghamdi A., Abdel-Ghaffara F., Al-Rasheid K., Bashtar A.R., Al Quraishy S., Mehlhorn H. Prevalence pattern and biology of Sarcocystis capracanis infection in the Egyptian goats: A light and ultrastructural study. Vet. Parasitol. 2011;181:75–82. doi: 10.1016/j.vetpar.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 37.Hu J.J., Liu T.T., Liu Q., Esch G.W., Chen J.Q., Huang S., Wen T. Prevalence, morphology, and molecular characteristics of Sarcocystis species in domestic goats (Capra hircus) from Kunming, China. Parasitol. Res. 2016;115:3973–3981. doi: 10.1007/s00436-016-5163-6. [DOI] [PubMed] [Google Scholar]
- 38.Vlemmas I., Kanakoudis G., Tsangaris T.H., Theodorides I., Kaldrymidou E. Ultrastructure of Sarcocystis tenella (Sarcocystis ovicanis) Vet. Parasitol. 1989;33:207–217. doi: 10.1016/0304-4017(89)90130-1. [DOI] [PubMed] [Google Scholar]
- 39.Formisano P., Aldridge B., Alony Y., Beekhuis L., Davies E., Del Pozo J., Dunn K., English K., Morrison L., Sargison N., et al. Identification of Sarcocystis capracanis in cerebrospinal fluid from sheep with neurological disease. Vet. Parasitol. 2013;193:252–255. doi: 10.1016/j.vetpar.2012.12.016. [DOI] [PubMed] [Google Scholar]