Abstract
In the absence of DNA, a solution containing the four deoxynucleotidetriphosphates (dNTPs), a DNA polymerase, and a nicking enzyme generates a self-replicating mixture of DNA species called parasite. Parasites are problematic in template-based isothermal amplification schemes such as EXPAR as well as in related molecular programming approaches, such as the PEN DNA toolbox. Here we show that using a nicking enzyme with only three letters (C, G, T) in the top strand of its recognition site, such as Nb.BssSI, allows us to change the sequence design of EXPAR templates in a way that prevents the formation of parasites when dATP is removed from the solution. This method allows us to make the EXPAR reaction robust to parasite contamination, a common feature in the laboratory, while keeping it compatible with PEN programs, which we demonstrate by engineering a parasite-proof bistable reaction network.
Isothermal nucleic acid amplification methods1 are interesting alternatives to polymerase chain reaction (PCR) because they do not need thermocycling equipment and are thus suited for the detection of nucleic acids in resource-limited environments.2 Different molecular implementations exist, such as nucleic-acid sequence-based amplification (NASBA), strand displacement amplification (SDA), rolling circle amplification (RCA), loop-mediated isothermal amplification (LAMP), and exponential amplification reaction (EXPAR).3
Among the cited methods, EXPAR has two important advantages. First, it has a short detection time on the order of minutes.4 Second, it produces single-stranded DNA (ssDNA) as an output that can be used either to set up simple colorimetric detection methods based on the aggregation of DNA-decorated nanoparticles5 or to couple EXPAR to molecular programs capable of displaying complex spatiotemporal dynamics.6−9 EXPAR exponentially amplifies the concentration of a trigger ssDNA A in the presence of an ssDNA template T, a DNA polymerase, and a nicking endonuclease, called nickase, in the following (Figure 1a).
Figure 1.
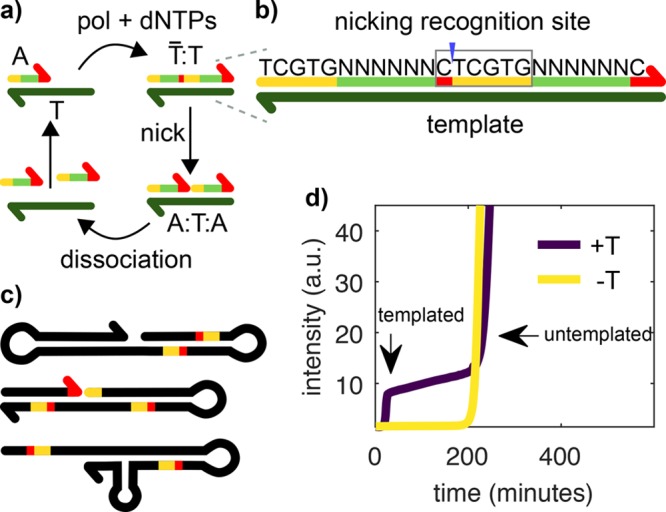
Templated and untemplated replication in the EXPAR reaction. (a) During templated replication, the trigger A is elongated on the template T by a polymerase (pol), consuming dNTPs. The double-stranded complex T̅:T is nicked by a nickase (nick), and two As are created, which can dissociate and replicate on other Ts. (b) Each trigger contains a split-up recognition site for the nickase (red and yellow), which is completed after elongation. The sequence of the six N nucleotides can be chosen freely. (c) In EXPAR experiments, an untemplated replicator, termed the parasite, emerges after some time. The parasite is not a single sequence but a pool of sequences. They are rich in secondary structures and bear recognition sites for the nickase. Unlike shown here, they can reach several kilobase pairs in length. (d) EvaGreen fluorescence versus time for an EXPAR reaction in the presence (+T, purple) and in the absence (−T, yellow) of template strand T1. EvaGreen is a DNA intercalator allowing us to track the concentration of dsDNA.
However, EXPAR has two important drawbacks induced by nonspecific reactions. The first one, usually known as early phase background amplification,4 or self-start, limits the detection of very low quantities of DNA. Several solutions have recently been proposed for this problem.10−12 The second problem, known as late-phase background amplification,4 or untemplated amplification, arises in systems where nucleic acids are exponentially amplified with the help of enzymes. Mutations lead to new sequences and, after some time, a parasitic sequence, or set of sequences, emerges, which is able to replicate more efficiently than the initial target sequence. One famous example is Sol Spiegelman’s monster, which is aroused during the in vitro replication of Qβ-RNA with Qβ-replicase and nucleotides.13 The EXPAR reaction produces a different kind of parasitic species containing repetitive and palindromic sequences where, typically, AT tracts are flanked by nickase recognition sites.4 Importantly, parasites appear by de novo, or untemplated, synthesis of DNA. Autocatalytic parasites have been observed in the presence of enzymes other than nickases, such as restriction enzymes,14 helicases,15 and possibly T7 RNA polymerase16 and also in PCR reactions either as a side effect17 or by design.18
Because parasite replication is as efficient as templated replication (Figure S5), parasites easily contaminate the whole laboratory and are difficult to eradicate, thus posing a problem in making EXPAR a robust analytical technique. Furthermore, when EXPAR is used to build more complex molecular programs, such as oscillators or bistable networks, the emergence of parasites limits the lifetime of these systems to typically 1 day, restricting the use of these powerful molecular programs19 for building nonequilibrium materials.9 In this work, we show that choosing a nickase with only three letters (C, G, T) in the top strand of its recognition site, such as Nb.BssSI, allows us to change the sequence design of EXPAR templates in a way that prevents the formation of parasites when dATP is removed from the solution. We further demonstrate that this approach permits the detection of DNA in the presence of contaminating parasites and that it is compatible with the design of bistable reaction networks.
Methods
Oligonucleotides were purchased from IDT, and their sequences displayed in Table 2 and Table S1. Template strands were HPLC-purified, and trigger strands were desalted. The enzymes we used were 8–40 U/mL Bst DNA Polymerase, Large Fragment (NEB), 20–500 U/mL Nb.BssSI (NEB), and 0 or 100 nM of in-house produced Thermus thermophilus RecJ exonuclease.20 We noticed a 3.4-fold batch-to-batch change in Nb.BssSI activity. The reaction buffer contained 20 mM Tris-HCl, 10 mM (NH4)2SO4, 50 mM NaCl, 1 mM KCl, 6 mM MgSO4, 1 g/L Synperonic F 108 (Sigma-Aldrich), 4 mM dithiothreitol, 0.1 g/L BSA (NEB), 1× EvaGreen Dye (Biotium), and 0.1× ROX (Invitrogen). Nucleotides (NEB or Invitrogen) were added in different concentrations and compositions. In some experiments, netropsin was added as indicated. Experiments were performed in a reaction volume of 20 μL at 44 °C, with 50 nM template concentration, when necessary, in a CFX96 Touch Real-Time PCR Detection System (Bio-Rad) or a Qiagen Rotor-Gene qPCR machine. The intensity of the green channel was recorded every minute. To avoid cross-contaminations, experiments involving pipetting of solutions containing parasites were performed in a different room using a different set of pipets.
Table 2. DNA Sequences for the Main Set of Species Used in This Work, All Working with Nb.BssSIa.
| species | sequence (5′-3′) |
|---|---|
| A1 | TCGTGTTCTGTC |
| T1 | G*A*C*AGAAC*ACGAGACAGAACACp |
| T1u | G*A*C*AGAACACGAGACAGAACACp |
| T13p | G*A*C*AGAA*C*A*CGAGACAGAACACp |
| T1fp | G*A*C*A*G*A*A*C*A*C*G*A*G*A*C*A*G*A*A*C*A*Cp |
| A1A1 | TCGTGTTCTGTCTCGTGTTCTGTC |
| R1 | A*A*A*AGACAGAACACGAp |
Asterisks indicate phosphorothioate bonds and p indicates phosphate modifications.
Results and Discussion
Suppressing Untemplated Replication
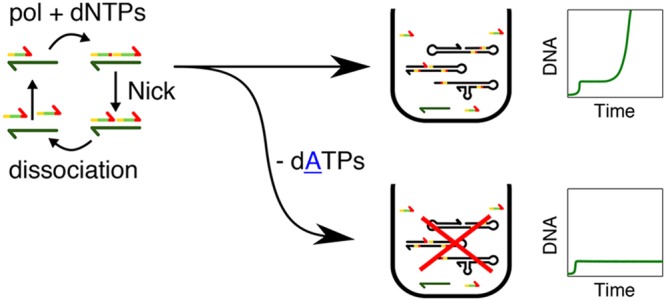
In EXPAR templated amplification, the trigger species A replicates on template T with the help of a polymerase and a nickase, noted pol and nick, respectively, in the following (Figure 1a). There is a series of sequence constraints for the reaction to happen. If we note a the sequence of A and a̅ its complementary, then T is a double repeat of a̅, noted a̅a̅. In addition, the double-stranded species T̅:T bears the recognition site of the nickase in such a way that the enzyme cuts T̅ to generate two species A that are bound to T (Figure 1b). As a result, when A binds to the 3′ end of T, it is extended by pol to make T̅:T, which is then cut by nick to form the complex A:T:A. The reaction is isothermal and set up close to the melting temperature of A:T, here 44 °C, to ensure that A can dehybridize and take part in reactions with other templates. To close the catalytic loop, either A:T:A dissociates to recycle species T, or it is extended again by pol, which is capable of strand displacement, regenerating T̅:T and producing an extra A.
Untemplated replication in the presence of pol, nick, and dNTPs is well documented,4,21 but its mechanism has not yet been elucidated.22 In the current working hypothesis, as soon as a sequence with a hairpin on the 3′ appears by de novo synthesis, it may be extended by pol. Subsequent rounds of hairpin formation and slippage, followed by polymerization, account for the synthesis of palindromic repetitive sequences. Nicking events, followed by de novo polymerization of a few bases that can self-hybridize, may explain the observation of the palindromic sequences flanked by nicking sites (Figure 1c).
Figure 1d displays a typical EXPAR experiment where the total concentration of double-stranded DNA (dsDNA) is followed by recording the fluorescence of the dsDNA intercalator EvaGreen. A solution containing pol, nick, dNTPs, and template strand T1 was incubated at constant temperature (purple line). In the beginning of the reaction, the signal coming from the dye is low because no dsDNA is present. When templated replication occurs, at 18 min, a rapid exponential phase leads to a signal increase until the signal saturates, indicating that all of the template is bound to the trigger. After 200 min, a second nonlinear signal increase takes place, which is due to untemplated replication. In the absence of template (yellow line), only the later signal increase is observed. Interestingly, the onset time of untemplated replication is similar in the presence or in the absence of template.
A strategy used so far to mitigate parasite replication is the addition of netropsin because the parasite sequence was found to be rich in AT repeats by Tan et al.4,19 Netropsin helps by binding such AT-rich sequences, but it cannot always prevent parasite formation, probably because other studies showed parasites without AT-rich stretches.21 Our solution to the problem consists of making it impossible to create secondary structures that can be nicked. Table 1 shows the sequences of the recognition sites for commercially available nickases. Most of the enzymes, including Nt.BstNBI, a common nickase used in EXPAR, contain all four bases (A, C, G, T) in each strand of their recognition site, and thus both templated and untemplated replication need the four dNTPs to proceed. In contrast, Nb.BssSI, Nt.BsmAI, and Nt.BspQI have recognition sites with only three bases (C, G, T) on their top strand. As a result, one can design a trigger with only C, G, and T and a template with only C, G, and A in their sequences. In the absence of dATP, such a system should be able to perform templated replication normally while being incapable of untemplated replication. Indeed, if mutations lead to the formation of unwanted products, then they might contain secondary structures but no recognition site for the nickase, which needs two adenines. This unwanted sequence will then only be able to grow by extending the 3′-end, which is not an exponential process and should not interfere too much with autocatalysis. In addition, the absence of dATP in the solution also precludes the formation of AT repeats that are rich in parasites, which might be another reason for reducing the untemplated replication. In the following, we fully characterize our strategy with Nb.BssSI, and we demonstrate that it also works with Nt.BsmAI and partially with Nt.BspQI.
Table 1. Top-Strand Sequences for the Recognition Site of Different Nicking Enzymes (from 5′ to 3′)a.
| enzyme | recognition site |
|---|---|
| Nb.BssSI | C′TCGTG |
| Nt.BsmAI | GTCTCN′N |
| Nt.BspQI | GCTCTTCN′ |
| Nt.BstNBI | GAGTCNNNN′N |
| Nb.BsmI | NG′CATTC |
| Nb.BsrDI | NN′CATTGC |
| Nb.BtsI | NN′CACTGC |
| Nt.BbvCI | CC′TCAGC |
| Nt.AlwI | GGATCNNNN′N |
Nicking sites are marked with ′. N means any base. In bold, the main enzyme used in this work. In italics, the three enzymes whose recognition sequences bear only three letters on the top strand tested in this study.
Figure 2 demonstrates that this approach works as designed. We incubated template T1 with pol and nick in the presence or in the absence of dATP. Because we will later use this system to design more complex molecular programs, template T1 lacked two bases on the 3′ side compared with the complementary of a double repeat of the trigger sequence A1; that is, A1 was a 12-mer, T1 was a 22-mer, and when A1 binds to T1 and is extended by the polymerase, the 24-mer A1A1 is formed (Table 2). In addition, ttRecJ, a single-stranded specific exonuclease with 5′ activity, noted exo, was added to the reaction to ensure that template replication was active throughout the duration of the experiment. T1 was protected from the exonuclease by three phosphorothioate (PTO) bonds at its 5′ end, and it bore a fourth one in the middle of the sequence to protect against background restriction activity (see below). Under these conditions, when dATP was present, both templated and untemplated replication were observed, whereas only templated amplification was observed in the absence of dATP (Figure 2a). The analysis of the reaction products on a denaturating polyacrylamide gel confirmed that at long times, large amounts of strands much longer than the trigger, which we identify with the parasite, form only when dATP is present (Figure 2b). Before the emergence of the parasite, the reaction products were indistinguishable both in the presence and in the absence of dATP: three bands at 12, 22, and 24-nt corresponding, respectively, to A1, T1, and A1A1. After 200 min, when the parasite has already appeared in the presence of dATP, no new bands appear in the −dATP sample. Finally, parasite suppression is compatible with different template sequences (Table S1), and the three nickases in Table 1 are suitable to our three-letter approach. Figure 2c shows three sets of sequences working with Nb.BssSI and one set working with Nt.BsmAI that displayed templated but not untemplated replication in the absence of dATP. Under our experimental conditions, parasite suppression worked with Nt.BspQI but not templated amplification, probably because this enzyme has low efficiency in the buffer used here (Figure S1). The experiments with Nb.BssSI in Figure 2c were performed in the absence of exo to demonstrate that degradation is not needed for parasite suppression. Because this approach allows us to perform EXPAR reactions that are robust against untemplated replication, we will call it rEXPAR in the following.
Figure 2.
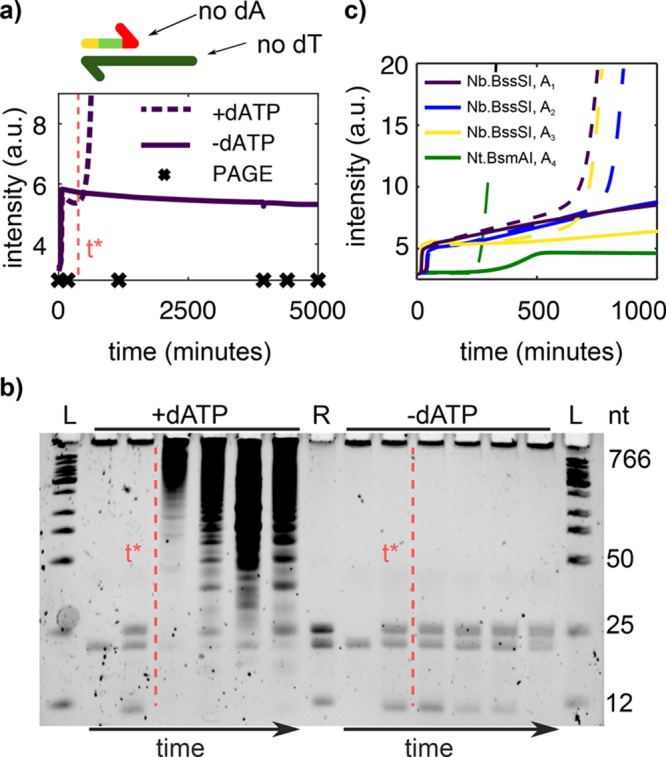
Suppressing dATP blocks untemplated replication without perturbing templated replication. (a) Top: We use Nb.BssSI, a nickase that allows us to design templates without thymine (dT) and triggers without adenine (dA). Bottom: EvaGreen fluorescence versus time for an EXPAR reaction of trigger A1 and template T1 in the presence (dashed) and in the absence (solid line) of dATP. Crosses indicate time points where aliquots were withdrawn for the gel in panel b. (b) Time evolution of the reactions in panel a on a PAGE denaturing gel. The red dashed line is a guide to the eye, indicating the onset of untemplated replication in the presence of dATP (t*). Lanes 4–7 have been diluted 20-fold for easier visualization of parasite bands. L is a ladder and R is a reference containing species A1, T1, and A1A1. (c) EXPAR reactions for three different sequences working with nickase Nb.BssSI and one sequence working with Nt.BsmAI in the presence (dashed) and in the absence (solid line) of dATP. Conditions: (a,b) 8 or (c) 4.8 U/mL pol, 20 U/mL nick, and (a,b) 1 or (c) 0.4 mM dNTPs. exo is 100 nM in panels a and b and in Nt.BsmAI reactions in panel c and 0 nM in Nb.BssSI reactions in panel c.
Nb.BssSI Nickase Displays a Background Restriction Activity That Can Be Easily Suppressed
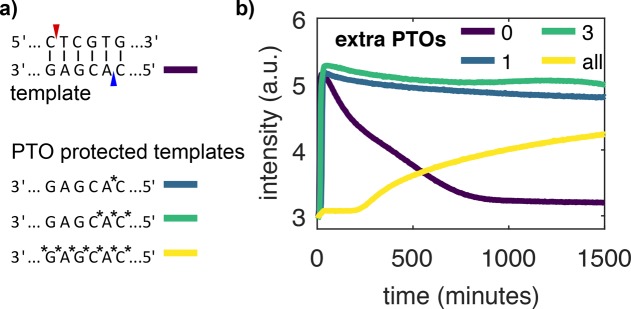
Nb.BssSI, the nickase used here, has been recently developed and has seldom been used in EXPAR experiments to our knowledge. In our preliminary experiments, we used template T1u, identical to T1 except for a missing PTO after the eighth nucleotide, and we observed a significant loss of fluorescence signal in many long-term experiments (Figure 3b, purple line). In EXPAR experiments with other nicking enzymes (Nb.BsmI, Nt.BstNBI), we did not observe this behavior. With T1, increasing Nb.BssSI concentrations promoted long-term signal loss, whereas the addition of T1u temporarily restored the signal (Figure S2). We hypothesized that because Nb.BssSI was derived from the restriction enzyme BssSI it may have a reminiscent restriction activity, which would cleave the template strand. To solve this problem, we tested different templates with additional PTO protection. (Note that all 5′-ends have three PTOs for protection from exonuclease.) Figure 3b shows EXPAR experiments with the protected templates. The unprotected template T1 shows degradation after only 20 min. The fully protected template T1fp has the PTOs not only in the recognition sequence but also between all 22 bases. This leads to a strong inhibition of templated replication that could arise from the higher melting temperature of PTO-modified strands or from the inhibition of pol or nick. In contrast, templates with 1, T1, or 3, T1, PTOs surrounding the putative second nicking site associated with the BssSI activity on the template strand show almost uninhibited replication and a stable signal in the steady state for at least 5000 min (Figure 2a).
Figure 3.
Nb.BssSI background restriction activity can be suppressed by adding a PTO bond to the template strand. (a) Nickase Nb.BssSI is supposed to cut at the site indicated in red. The restriction enzyme BssSI additionally cuts at the site indicated in blue. We tested four templates, T1u (purple), T1 (blue), T1 (green), and T1fp (yellow), with increasing number of PTOs around the BssSI site. (b) EvaGreen fluorescence versus time for rEXPAR experiments with templates in panel a. Conditions: 8 U/mL pol, 200 U/mL nick, 100 nM exo, and 0.4 mM dNTPs.
Standard Methods Delay Parasite Emergence but Do Not Suppress It
Standard methods to mitigate untemplated replication involve decreasing pol or increasing nick concentrations9 and adding netropsin.4,19Figure 4 shows that these approaches delay but do not suppress the onset of parasite emergence. In the absence of template, the parasite onset time, τu, is inversely proportional to the pol concentration, as expected from first-order kinetics (Figure 4a,b). In contrast, increasing nick concentration increases τu, especially at low pol concentration (Figure S3), suggesting that nick inhibits pol. Finally, adding up to 4 μM netropsin delays parasite emergence four-fold but also slows down templated replication by the same amount and reduces the fluorescence signal from the dsDNA intercalator dye (Figure 4c and Figure S4). In summary, all of these methods slow down untemplated replication at the cost of slowing down templated replication as well, which is an undesirable feature for rapid analysis.
Figure 4.
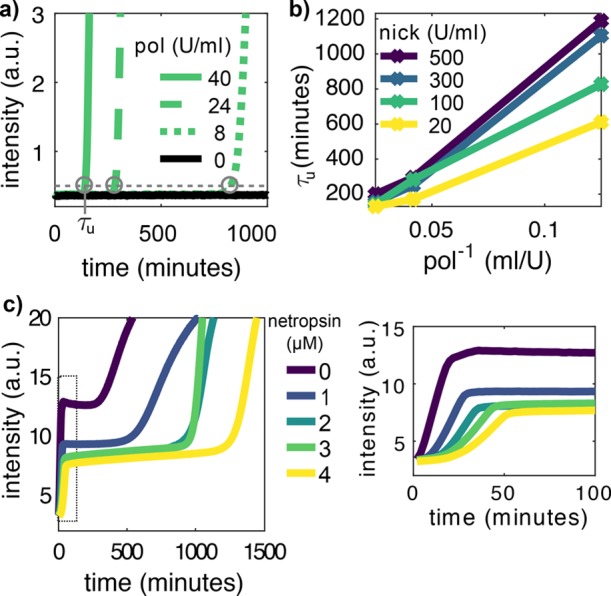
Standard approaches delay untemplated replication but do not suppress it. (a) EvaGreen fluorescence versus time in the absence of T1 for different polymerase (pol) concentrations at 100 U/mL nickase. Circles indicate the onset time of untemplated replication τu. (b) τu versus the inverse of pol concentration for different nick concentrations. (c) EvaGreen fluorescence versus time in the presence of T1 for different netropsin concentrations. The plot on the right is a zoom of the data on the left inside the dotted rectangle. Conditions: 0.4 mM dNTPs, 100 nM exo (for panels a and b) and 8 U/mL pol, 60 U/mL nick (for panel c).
rEXPAR Is Compatible with PEN Molecular Programs
If a chemical reaction is monostable in the sense of dynamical systems, then the involved species reach a single steady-state concentration independently of the initial conditions. Because the EXPAR autocatalytic network is intrinsically monostable, in the absence of a trigger strand (i.e., A = 0), an infinitesimally small addition of A will grow exponentially until the steady state is reached. This is problematic for detecting very low amounts of A because any unprimed synthesis of A will result in an undesired background, known as early-stage background amplification or self-start. One can make EXPAR robust to self-start by instead using a bistable autocatalytic network based on the polymerase, nickase, exonuclease dynamic network assembly toolbox (PEN DNA toolbox).10
The PEN DNA toolbox is an experimental framework based on the EXPAR reaction that allows the design of reaction networks that mimic the dynamics of gene regulatory networks in solution.6,19,23 This framework makes network design straightforward because network topology is defined by predictable interactions between short ssDNAs. In addition, the combination of its three core enzymes with large quantities of dNTPs provides a convenient way to keep the network out of equilibrium in a closed reactor for a very long time at steady state. These two unmatched properties have allowed the rational design of complex spatiotemporal behaviors such as oscillations,6,24 bistability,7,10 and reaction-diffusion patterns.8,25−27 Besides suppressing background amplification in EXPAR, such dynamic behaviors have important applications such as in nucleic acid detection,28 in material science,9 in the design of microrobots,29 and in protein directed evolution.30 However, untemplated amplification is an important obstacle for these applications because it precludes the use of PEN circuits for long periods of time.
PEN networks are usually built around one or more autocatalytic nodes based on EXPAR. An exonuclease enzyme is added such that nodes are not only dynamically produced but also degraded. In addition to the autocatalytic template strands intrinsic to EXPAR, that catalyze the reaction A → 2A, other templates can be used to catalyze other processes such as activation (A → A + B) and repression (A → ⌀). All of these template strands bear PTOs in 5′ to protect them from exonuclease degradation.
In this framework, to render an EXPAR reaction bistable and thus suppress self-start, one just needs to add a second template strand R that binds to trigger A, and, with the help of pol, it extends into the waste strand W, which can be degraded by exo but not recognized by nick (Figure 5a). In addition, the polymerization reaction of A on R needs to be faster than the one of A on its template T. To fulfill these two requirements, we chose to use a repressor strand R1 with a sequence of four adenines, followed by the reverse complementary sequence of A1 (Table 2) and a template strand that lacks on the 3′ end two bases to be fully complementary to A1, here T1u. R1 was protected against exonuclease degradation by three PTO bonds on the 5′ end. We incubated T1 in the presence of pol, nick, and exo and increasing concentrations of R1, noted R1, in the absence of trigger A1 and dATP. At the lowest R1 tested of 15 nM, untriggered templated amplification was observed within 30 min, indicating that the system is monostable with a single stable point at high A1 concentration. Increasing R1 resulted in a dramatic increase in the templated amplification time τt until it became undetectable (>1000 min, Figure S6) above R1 = 60 nM (Figure 5b,c). Above this threshold, the reaction network becomes bistable with a stable point at A1 = 0 and a second one at high A1. A plot of 1/τt as a function of the concentration of R1 indicates that R1 is a bifurcation parameter of the reaction network. Because dATP was absent, no untemplated amplification was observed in the system, demonstrating that rEXPAR is compatible with the construction of DNA-based reaction networks with complex dynamics, such as bistability.
Figure 5.
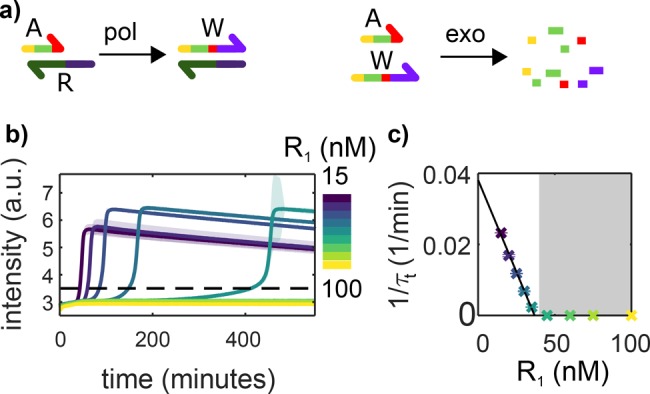
rEXPAR is compatible with PEN DNA bistable programs. (a) In addition to the autocatalytic network depicted in Figure 1a, a repressor template R is added that binds to trigger A and turns it into waste W, which can be degraded by exonuclease but cannot prime autocatalysis. (b) EvaGreen fluorescence versus time in the presence of T1u but in the absence of A1 for increasing concentrations of R1. The dashed line indicates the threshold corresponding to the onset time of templated amplification τt. (c) 1/τt versus R concentration from panel b. The black line corresponds to a linear fit of slope 1.0 × 10–3 nM–1 min–1. The gray area indicates where the system is bistable and thus robust to self-start. Experiments were performed in the absence of dATP. The shade and the error bars in panels b and c correspond to the standard deviation of a triplicate experiment. Conditions: 8 U/mL pol, 20 u/mL nick, 100 nM exo, and 0.4 mM dNTPs.
rEXPAR Is Robust to Parasite Contamination
Besides being an intriguing instance of molecular evolution, parasites can easily contaminate the laboratory31 and produce false-positives in EXPAR because they amplify as fast as target DNA and produce a higher fluorescent signal. Figure 6 shows EXPAR amplification experiments in the presence and in the absence of dATP (rEXPAR) for samples with or without parasite contamination. Contamination was performed by adding a 3000-fold diluted sample that had previously undergone untemplated amplification. This amount corresponds to the use of a pipet tip that has been filled and emptied with 1 μL of a solution containing parasite (Figure S5). We observed that templated and untemplated amplification occurred concomitantly in the sample containing both dATP and parasite. In contrast, rEXPAR samples without dATP were robust to untemplated amplification both in the absence and in the presence of contamination.
Figure 6.
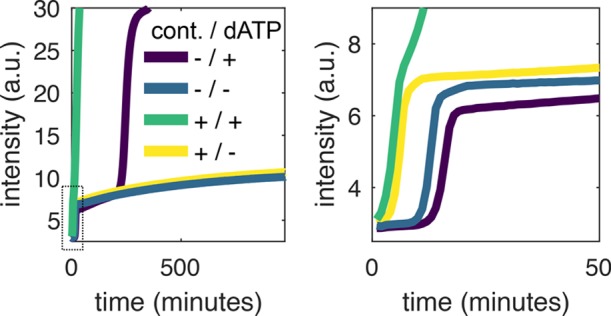
rEXPAR is robust to parasite contamination. EvaGreen fluorescence versus time for EXPAR experiments performed both with and without dATP and with and without contamination from a parasite solution diluted 3000-fold (cont.). The plot on the right is a zoom of the data on the left inside the dotted rectangle. Conditions: 40 U/mL pol, 500 U/mL nick, 0 nM exo, 60 nM R1, and 0.4 mM dNTPs.
In a second set of experiments (Figure 7 and Figure S9), we evaluated the performance of EXPAR and rEXPAR to detect trigger DNA in the presence of parasite contamination. To be more realistic about a fortuitous parasite contamination, we used a higher dilution of 106-fold. EXPAR and rEXPAR attained a similar limit of detection of 0.4 pM in the absence of contamination, although rEXPAR was 1.6 times faster (Figures S7 and S8). In contrast, in the presence of contamination, rEXPAR was able to detect the trigger without noticeable change, whereas EXPAR was unable to detect even the highest trigger concentration of 100 pM.
Figure 7.
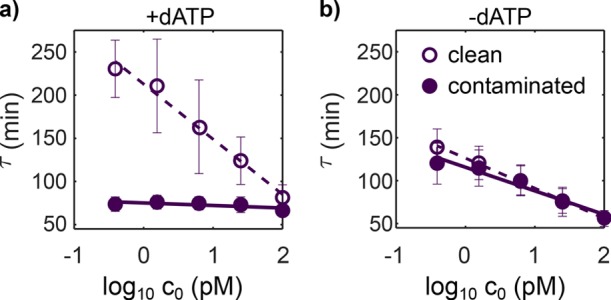
rEXPAR is able to detect trigger DNA in the presence of parasite contamination, in contrast with EXPAR. Amplification onset time, τ, versus the decimal logarithm of the initial trigger concentration, c0, for EXPAR reactions performed in the presence (a) and in the absence (b) of dATP, with (filled symbols) and without (empty symbols) contamination from a parasite solution diluted 106-fold. Error bars correspond to the standard deviation from three different experiments performed on different days with different stock solutions. Conditions: 8 U/mL pol, 20 U/mL nick, 100 nM exo, 15 nM R1, and 0.4 mM dNTPs.
Conclusions
We took advantage of a new nicking enzyme, Nb.BssSI, whose recognition site has only a three-letter code on the top strand (C′TCGTG), to perform EXPAR isothermal amplification experiments in the absence of one deoxynucleotide (dATP). Under these conditions, templated amplification proceeded normally and even slightly faster, whereas untemplated amplification, resulting in the production of an autocatalytic set of parasitic sequences, was completely suppressed. Our approach, called rEXPAR for robust EXPAR, contrasts with existing methods, such as the addition of netropsin, that mitigate but do not suppress untemplated amplification. rEXPAR is also compatible with other three-letter nicking enzymes, such as Nt.BsmAI. rEXPAR is compatible with EXPAR-based molecular programming languages such as the PEN DNA toolbox, which we demonstrated by implementing a bistable autocatalytic network that suppressed self-starting spurious reactions. In addition, rEXPAR, in contrast with EXPAR, allows the detection of trigger DNA in the presence of minute amounts of parasite contamination. As a result, we believe that rEXPAR will be useful, both for running out-of-equilibrium molecular programs over extended periods of time, which is essential for building “life-like” materials,9 and for making EXPAR more robust in analytical applications.
Acknowledgments
We thank Yannick Rondelez and Guillaume Ginés for insightful discussions.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.9b00063.
This work has been funded by the European Research Council (ERC) under the European’s Union Horizon 2020 program (grant no. 770940, A.E.-T.), by the Deutsche Forschungsgemeinschaft (grant no. 364775124, G.U.), by the Ville de Paris Emergences program (Morphoart, A.E.-T.), by a Marie Skłodowska-Curie fellowship (grant no. 795580, M.V.D.H.) from the European Union’s Horizon 2020 program, and by a PRESTIGE grant (grant no. 609102, M.V.D.H.) from the European Union’s Seventh Framework Programme.
The authors declare no competing financial interest.
Supplementary Material
References
- Zhao Y.; Chen F.; Li Q.; Wang L.; Fan C. (2015) Isothermal Amplification of Nucleic Acids. Chem. Rev. 115, 12491–12545. 10.1021/acs.chemrev.5b00428. [DOI] [PubMed] [Google Scholar]
- Reid M. S.; Le X. C.; Zhang H. (2018) Exponential isothermal amplification of nucleic acids and amplified assays for proteins, cells, and enzyme activities. Angew. Chem., Int. Ed. 57, 11856–11866. 10.1002/anie.201712217. [DOI] [PubMed] [Google Scholar]
- Van Ness J.; Van Ness L. K.; Galas D. J. (2003) Isothermal reactions for the amplification of oligonucleotides. Proc. Natl. Acad. Sci. U. S. A. 100, 4504–4509. 10.1073/pnas.0730811100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan E.; Erwin B.; Dames S.; Ferguson T.; Buechel M.; Irvine B.; Voelkerding K.; Niemz A. (2008) Specific versus nonspecific isothermal DNA amplification through thermophilic polymerase and nicking enzyme activities. Biochemistry 47, 9987–9999. 10.1021/bi800746p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Hu J.; Zhang C.-y. (2012) Sensitive Detection of Transcription Factors by Isothermal Exponential Amplification-Based Colorimetric Assay. Anal. Chem. 84, 9544–9549. 10.1021/ac3024087. [DOI] [PubMed] [Google Scholar]
- Montagne K.; Plasson R.; Sakai Y.; Fujii T.; Rondelez Y. (2011) Programming an in vitro DNA oscillator using a molecular networking strategy. Mol. Syst. Biol. 7, 466. 10.1038/msb.2010.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padirac A.; Fujii T.; Rondelez Y. (2012) Bottom-up construction of in vitro switchable memories. Proc. Natl. Acad. Sci. U. S. A. 109, E3212–E3220. 10.1073/pnas.1212069109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padirac A.; Fujii T.; Estévez-Torres A.; Rondelez Y. (2013) Spatial waves in synthetic biochemical networks. J. Am. Chem. Soc. 135, 14586–14592. 10.1021/ja403584p. [DOI] [PubMed] [Google Scholar]
- Zadorin A. S.; Rondelez Y.; Gines G.; Dilhas V.; Urtel G.; Zambrano A.; Galas J. C.; Estevez-Torres A. (2017) Synthesis and materialization of a reaction-diffusion French flag pattern. Nat. Chem. 9, 990–996. 10.1038/nchem.2770. [DOI] [PubMed] [Google Scholar]
- Montagne K.; Gines G.; Fujii T.; Rondelez Y. (2016) Boosting functionality of synthetic DNA circuits with tailored deactivation. Nat. Commun. 7, 13474. 10.1038/ncomms13474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.; Zhou X.; Ma Y.; Lin X.; Dai Z.; Zou X. (2016) Asymmetric exponential amplification reaction on a toehold/biotin featured template: an ultrasensitive and specific strategy for isothermal microRNAs analysis. Nucleic Acids Res. 44, gkw504 10.1093/nar/gkw504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid M. S.; Paliwoda R. E.; Zhang H.; Le X. C. (2018) Reduction of Background Generated from Template-Template Hybridizations in the Exponential Amplification Reaction. Anal. Chem. 90, 11033–11039. 10.1021/acs.analchem.8b02788. [DOI] [PubMed] [Google Scholar]
- Mills D. R.; Peterson R. L.; Spiegelman S. (1967) An extracellular Darwinian experiment with a self-duplicating nucleic acid molecule. Proc. Natl. Acad. Sci. U. S. A. 58, 217–224. 10.1073/pnas.58.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X.; Jensen K.; Frank-Kamenetskii M. D. (2004) Very Efficient Template/Primer-Independent DNA Synthesis by Thermophilic DNA Polymerase in the Presence of a Thermophilic Restriction Endonuclease. Biochemistry 43, 13459–13466. 10.1021/bi0489614. [DOI] [PubMed] [Google Scholar]
- Kaboev O. K.; Luchkina L. A. (2004) Template-Free Primer-Independent DNA Synthesis by Bacterial DNA Polymerases I Using the DnaB Protein from Escherichia coli. Dokl. Biochem. Biophys. 398, 265–267. 10.1023/B:DOBI.0000046633.66624.58. [DOI] [PubMed] [Google Scholar]
- Emery N. J.; Majumder S.; Liu A. P. (2018) Synergistic and non-specific nucleic acid production by T7 RNA polymerase and Bsu DNA polymerase catalyzed by single-stranded polynucleotides. Syst. Synth. Biol. 3, 130–134. 10.1016/j.synbio.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownie J.; Shawcross S.; Theaker J.; Whitcombe D.; Ferrie R.; Newton C.; Little S. (1997) The elimination of primer-dimer accumulation in PCR. Nucleic Acids Res. 25, 3235–3241. 10.1093/nar/25.16.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urtel G. C.; Rind T.; Braun D. (2017) Reversible Switching of Cooperating Replicators. Phys. Rev. Lett. 118, 1–5. 10.1103/PhysRevLett.118.078102. [DOI] [PubMed] [Google Scholar]
- Baccouche A.; Montagne K.; Padirac A.; Fujii T.; Rondelez Y. (2014) Dynamic DNA-toolbox reaction circuits: A walkthrough. Methods 67, 234–249. 10.1016/j.ymeth.2014.01.015. [DOI] [PubMed] [Google Scholar]
- Wakamatsu T.; Kitamura Y.; Kotera Y.; Nakagawa N.; Kuramitsu S.; Masui R. (2010) Structure of RecJ exonuclease defines its specificity for single-stranded DNA. J. Biol. Chem. 285, 9762–9. 10.1074/jbc.M109.096487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zyrina N. V.; Zheleznaya L. A.; Dvoretsky E. V.; Vasiliev V. D.; Chernov A.; Matvienko N. I. N. (2007) BspD6I DNA nickase strongly stimulates template-independent synthesis of non-palindromic repetitive DNA by Bst DNA polymerase. Biol. Chem. 388, 367–372. 10.1515/BC.2007.043. [DOI] [PubMed] [Google Scholar]
- Zyrina N. V.; Antipova V. N.; Zheleznaya L. A. (2014) Ab initio synthesis by DNA polymerases. FEMS Microbiol. Lett. 351, 1–1. 10.1111/1574-6968.12326. [DOI] [PubMed] [Google Scholar]
- Padirac A.; Fujii T.; Rondelez Y. (2013) Nucleic acids for the rational design of reaction circuits. Curr. Opin. Biotechnol. 24, 575–580. 10.1016/j.copbio.2012.11.011. [DOI] [PubMed] [Google Scholar]
- Fujii T.; Rondelez Y. (2013) Predator - Prey molecular ecosystems. ACS Nano 7, 27–34. 10.1021/nn3043572. [DOI] [PubMed] [Google Scholar]
- Zadorin A. S.; Rondelez Y.; Galas J.-C.; Estevez-Torres A. (2015) Synthesis of programmable reaction-diffusion fronts using DNA catalyzers. Phys. Rev. Lett. 114, 068301. 10.1103/PhysRevLett.114.068301. [DOI] [PubMed] [Google Scholar]
- Zambrano A.; Zadorin A. S.; Rondelez Y.; Estévez-Torres A.; Galas J. C. (2015) Pursuit-and-Evasion Reaction-Diffusion Waves in Microreactors with Tailored Geometry. J. Phys. Chem. B 119, 5349–5355. 10.1021/jp509474w. [DOI] [PubMed] [Google Scholar]
- Kurylo I.; Gines G.; Rondelez Y.; Coffinier Y.; Vlandas A. (2018) Spatiotemporal control of DNA-based chemical reaction network via electrochemical activation in microfluidics. Sci. Rep. 8, 6396. 10.1038/s41598-018-24659-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondelez Y., Gines G., Montagne K., and Fujii T. (2016) Method of Eliminating Background Amplification of Nucleic Acid Targets. Patent WO/2017/141067.
- Gines G.; Zadorin A. S.; Galas J.-C.; Fujii T.; Estevez-Torres A.; Rondelez Y. (2017) Microscopic agents programmed by DNA circuits. Nat. Nanotechnol. 12, 351–359. 10.1038/nnano.2016.299. [DOI] [PubMed] [Google Scholar]
- Drame-Maigné A. (2018) Compartmentalized Directed Evolution of Enzymes Using Molecular Programs as Self-Selection Function. Ph.D. Thesis, Université Paris Descartes. [Google Scholar]
- Marshall K. A.; Ellington A. D. (1999) Molecular Parasites That Evolve Longer Genomes. J. Mol. Evol. 49, 656–663. 10.1007/PL00006586. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.