Abstract
Aim:
First, evaluate if patients carrying putatively diminished activity CYP2C8 genotype have longer paclitaxel exposure (e.g., time above threshold concentration of 0.05 μM [Tc >0.05]). Second, screen additional pharmacogenes for associations with Tc >0.05.
Methods:
Pharmacogene panel genotypes were translated into genetic phenotypes for associations with Tc >0.05 (n = 58).
Results:
Patients with predicted low-activity CYP2C8 had shorter Tc >0.05 after adjustment for age, body surface area and race (9.65 vs 11.03 hrs, β = 5.47, p = 0.02). This association was attributed to CYP2C8*3 (p = 0.006), not CYP2C8*4 (p = 0.58). Patients with predicted low-activity SLCO1B1 had longer Tc >0.05 (12.12 vs 10.15 hrs, β = 0.85, p = 0.012).
Conclusion:
Contrary to previous publications, CYP2C8*3 may confer increased paclitaxel metabolic activity. SLCO1B1 and CYP2C8 genotype may explain some paclitaxel pharmacokinetic variability.
Keywords: : CYP2C8, OATP1B1, paclitaxel, pharmacogenomics, SLCO1B1
Paclitaxel is used in the treatment of several solid tumors and has two major dose limiting toxicities; neutropenia and peripheral neuropathy (PN)[1]. Paclitaxel systemic drug concentrations, or exposure, has been associated with these toxicities [2,3] and to efficacy [4]. The exposure parameter most commonly associated with treatment outcomes is ‘time above threshold’ which is the number of h a patient's systemic plasma concentration remains above 0.05 μM (Tc >0.05), which has been shown to be predictive of PN. We and other groups have found that another pharmacokinetic (PK) parameter, maximum concentration (Cmax), was similarly predictive of PN [3,5] and is more conveniently collected, which is a critical advantage for clinical translation.
There is substantial interpatient variability in paclitaxel exposure when receiving standard body surface area (BSA) based dosing, and accurate prediction of exposure could enable dose individualization to optimize efficacy and reduce toxicity [6]. Paclitaxel exposure is likely affected by activity of drug metabolizing enzymes and drug transporters, for which phenotypic activity (i.e., poor [PM], intermediate [IM], normal [NM] or ultra-rapid [UM] metabolizer) can be predicted based on the presence of functional polymorphisms using the standard approach from the Clinical Pharmacogenetics Implementation Consortium [7]. Paclitaxel is primarily metabolized by CYP2C8 through the conversion to 6-hydroxypaclitaxel [8]. The CYP2C8 genetic polymorphisms, CYP2C8*3 and CYP2C8*4, have been associated with modest decreases in paclitaxel clearance in clinical studies [9,10] and in in vitro studies [8,11–16]. CYP2C8*3 has also been associated with increased paclitaxel-induced PN [17,18]. Based on the available literature, it is expected that CYP2C8*3 confers decreased activity for paclitaxel metabolism, though this polymorphism confers increased activity for other substrates such as the thiazolidinediones [19,20]. Polymorphisms in the P-gp efflux transporter (ABCB1) have also been associated with paclitaxel PK [9,10].
Further research is needed to elucidate the impact of genetic polymorphisms on paclitaxel PK to determine whether pharmacogenetics can be used to inform individualized dosing to optimize therapeutic outcomes. We conducted a prospective clinical trial to discover PK, genetic, and other predictive biomarkers of paclitaxel-induced PN in patients with early stage breast cancer. The overall objective of this secondary pharmacogenetic analysis is to identify pharmacogenetic and clinical factors associated with paclitaxel exposure. The primary objective was to determine whether patients with genotype-predicted low-activity CYP2C8 phenotype (i.e., carriers of CYP2C8*3 or CYP2C8*4) have greater paclitaxel exposure, as estimated by a longer Tc >0.05. A secondary objective of the study was to screen for associations between genotype-predicted phenotypic activity of other genes relevant to paclitaxel metabolism or transport with paclitaxel exposure estimated by Tc >0.05 or Cmax.
Methods
Patient cohort
Patients with breast cancer receiving weekly paclitaxel 80 mg/m2 1-h infusions were enrolled in an observational clinical registry (UMCCC 2014.002, NCT02338115) assessing paclitaxel exposure during the first dose and paclitaxel-induced PN. Detailed information about these patients, treatment, sampling time points and the primary analyses have been previously reported [5]. Briefly, eligible women were >18 years old, had a diagnosis of invasive breast cancer, and were scheduled to receive Cremophor EL-based paclitaxel 80 mg/m2 1-h infusions weekly for 12 weeks with a curative intent. The study was approved by the University of Michigan IRBMed and conducted in accordance with recognized ethical guidelines including the Declaration of Helsinki and Belmont report. All enrolled subjects signed written informed consent.
Sample & data collection
Patient demographic and treatment information was collected at baseline including age, race, ethnicity and limited medical history. Blood samples were collected at the first paclitaxel infusion for a complete blood count, comprehensive metabolic profile, and extraction of genomic DNA. Additional blood samples were collected within 10 min prior to the end of the first paclitaxel infusion and 16–26 h after infusion for PK analyses. The end of infusion sample was collected from the peripheral vein contralateral to the infusion and the 16–26 h level was collected from a peripheral vein or a properly flushed port. Samples were collected in Na-Heparin tubes, immediately placed on ice, and centrifuged within 10 min of collection. Plasma was transferred to a secondary cryotube and stored at -20°C until analysis.
Pharmacokinetic methodology
Total paclitaxel concentration was measured in plasma samples by liquid chromatography-mass spectroscopy by the University of Michigan College of Pharmacy Pharmacokinetics Core. Tc >0.05 was calculated using the MyCare™ Dose Exposure Calculator (Saladax Biomedical, Inc, PA, USA). MyCare Dose Exposure Calculator is a PC-based software that uses Phoenix WinNonlin modeling software (Certara USA, Inc., NJ, USA) to determine exposure to therapeutic agents based on a previously published population-PK model [21]. The following parameters were used to calculate Tc >0.05: absolute dose of paclitaxel (in mg), infusion start and end time, time of sample collection and sample concentration. The concentration determined from the plasma sample collected at the end of infusion was used as Cmax.
Genotyping & activity phenotype prediction
DNA samples were genotyped for 266 variants in 36 genes relevant to drug metabolism and transport on the iPLEX ADME PGx ProPanel by Agena Bioscience (CA, USA). All genetic information was subjected to appropriate quality control including assessment of sample call rate. Similar to our previous publication [22], each allele was translated into a predicted activity (low, normal and high) based on published data (Supplementary Table 1). Then, each patient's diplotype was translated into a predicted activity phenotype: PM, IM, NM or UM, for analysis (Supplementary Table 2). Hardy Weinberg equilibrium was confirmed for all variants included in significant findings from the primary and secondary analyses using a χ2 test with simulated p-value based on 10,000 replicates, which was performed using R.
Statistical analysis
All patients with available genetic and exposure (Tc >0.05 and Cmax) data were included in the analysis. For the primary analysis, genotype-predicted CYP2C8 activity was tested in a dominant model (PM/IM vs NM) for an association with Tc >0.05. The secondary analysis was conducted using an additive model for associations between Tc >0.05 and Cmax with the following genes with known relevance to paclitaxel metabolism or transport: CYP3A4, CYP3A5, ABCB1, ABCC2, ABCG2, SLCO1B1 and SLCO1B3. Finally, all remaining genes analyzed by the iPLEX ADME chip with no known relevance to paclitaxel metabolism or transport: COMT, CYP1A1, CYP1A2, CYP2A6, CYP2B6, CYP2C19, CYP2C9, CYP2D6, CYP2E1, DPYD, GSTM1, GSTP1, GSTT1, GSTT2b, NAT1, NAT2, SLC15A2, SLC22A1, SLC22A2, SLC22A6, SLCO2B1, SULT1A1, TPMT, UGT1A1, UGT2B15, UGT2B17, UGT2B7, VKORC1, were included in a hypothesis generating screen for associations with Tc >0.05 or Cmax. For the secondary analysis and the hypothesis generating screen, if the number of patients in an extreme genetic phenotype (PM or UM) was less than three, that phenotype was collapsed into the next phenotype level (i.e., PM/IM or NM/UM). Additional post-hoc analyses were conducted to investigate associations for individual CYP2C8 genotypes (CYP2C8*3 or CYP2C8*4) with Tc >0.05. For the analysis of each variant, patients who were heterozygous (i.e., CYP2C8*1/*3) or homozygous (i.e., CYP2C8*3/*3) for that variant were compared only to patients homozygous for the wild-type genotype (i.e., CYP2C8*1/*1) by excluding all carriers of the alternative variant. Clinical variables used in the analysis included age and BSA as continuous variables, race (Caucasian or other), alkaline phosphatase (elevated >116 IU/l), aspartate aminotransferase or alanine aminotransferase (elevated >40 IU/l), albumin (low ≤3.0 g/dl), prior or concurrent pertuzumab/trastuzumab therapy and prior or concurrent chemotherapy as categorical variables. The associations between genetic phenotype or clinical variables and the Cmax and logarithm of Tc >0.05 were analyzed using univariate linear regression with an unadjusted α = 0.05 (two-sided). None of the clinical covariates was significant in the univariate association, therefore, genetic associations were adjusted for the putatively relevant covariates age, race and BSA. Statistical analysis was performed using SAS v9.4.
Results
Patient demographic, PK, & genetic data
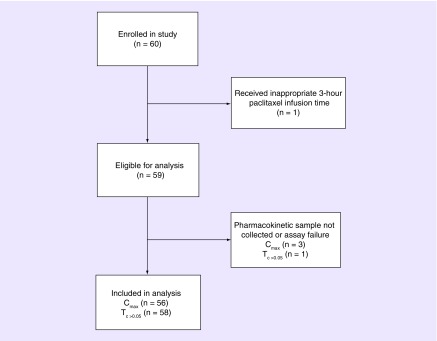
After excluding one patient that received a 3-h paclitaxel infusion, 56 and 58 patients were included in the Tc >0.05 and Cmax analyses, respectively (Figure 1). Patient demographic data and relevant baseline laboratory levels are included in Table 1. The mean age of the patients was 51.4 years (standard deviation [SD]: 12.55), mean BSA was 1.82 m2 (SD 0.21), and 93.2% of patients self-reported as Caucasian (Table 1). The mean Tc >0.05 and Cmax was 10.72 h (n = 58, SD 2.73) and 2390.18 ng/ml (n = 56, SD 640.85), respectively.
Figure 1. . CONSORT Diagram of patient inclusion in the secondary pharmacogenomic analysis.
This diagram shows the reasons for exclusion from the parent clinical registry into the secondary pharmacogenomics analysis.
Table 1. . Patient demographics.
| Clinical variables | Level | n = 59; mean (SD)/n (%) |
|---|---|---|
| Age | Years | 51.41 (12.55) |
| BSA | m2 | 1.82 (0.21) |
| Race | Caucasian | 55 (93.2%) |
| Other | 4 (6.8%) | |
| Alkaline phosphatase | Low/normal | 51 (86.4%) |
| Elevated | 8 (13.6%) | |
| ALT or AST | Normal | 53 (89.8%) |
| Elevated | 6 (10.2%) | |
| Albumin | Low | 1 (1.7%) |
| Normal | 58 (98.3%) | |
| Prior or concurrent chemotherapy | Yes | 55 (93.2%) |
| No | 4 (6.8%) | |
| Prior or concurrent HER2 therapy | Yes | 28 (47.5%) |
| No | 31 (52.5%) | |
BSA: Body surface area; SD: Standard deviation.
CYP2C8 diplotype was determined for all patients included in the analysis. For the 57 patients included in the primary analysis, the distribution of CYP2C8 diplotypes and corresponding genetic phenotypes was: 40 CYP2C8*1/*1 (NM), 11 CYP2C8*1/*3 (IM), 3 CYP2C8*1/*4 (IM), 1 CYP2C8*3/*3 (PM), 1 CYP2C8*3/*4 (PM) and 1 CYP2C8*4/*4 (PM), resulting in a phenotype distribution of 3 PMs (5%), 14 IMs (25%) and 40 NMs (70%). No patients were carriers of CYP2C8*2, *5, *7 or *8. The CYP2C8 and SLCO1B1 variants were confirmed to be in Hardy–Weinberg equilibrium and the minor allele frequency was similar to that expected for a primarily Caucasian population.
Associations of clinical & genetic variables with Tc >0.05
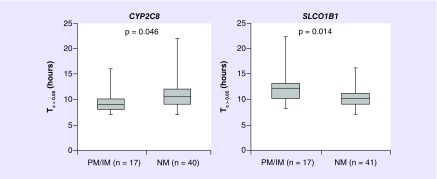
In the primary analysis, patients with low-activity CYP2C8 genetic phenotype (PM or IM) had shorter Tc >0.05 (PM/IM mean = 9.65 h, NM mean = 11.03 h, linear regression β-coefficient = 1.13, 95% CI: 1.01–1.28, p = 0.046, Table 2 & Figure 2 left). None of the clinical variables were associated with Tc >0.05; however, associations for increasing BSA (β = 1.27, 95% CI: 0.96–1.68, p = 0.10) and Caucasian race (β = 1.22, 95% CI: 0.97–1.53, p = 0.09, Table 3) with longer Tc >0.05 approached significance. The association with CYP2C8 maintained significance after adjustment for age, BSA and race (β = 5.47, 95% CI: 2.99–9.99, p = 0.020).
Table 2. . Paclitaxel Tc >0.05 and Cmax by phenotype predicted metabolizer status.
| Gene | Poor | Intermediate | Normal | Ultra-rapid | Univariate association |
|---|---|---|---|---|---|
| Primary analysis | Mean Tc >0.05 (h ) (SD) stratified by phenotype | β (95% CI), p-value | |||
| CYP2C8 | 9.65 (2.06) n = 17 |
11.03 (2.74) n = 40 |
† | β = 1.13 (1.01–1.28) p = 0.046* |
|
| Secondary analysis | Mean Tc >0.05 (h ) (SD) stratified by phenotype or genotype | β (95% CI), p-value | |||
| CYP2C8 *1/*3 and *3/*3 | 8.92 (1.00) n = 12 |
11.03 (2.74) n = 40 |
† | β = 0.82 (0.72–0.94) p = 0.006* |
|
| CYP2C8 *1/*4 and *4/*4 | 10.25 (1.71) n = 4 |
11.03 (2.74) n = 40 |
† | β = 0.94 (0.76–1.17) p = 0.58 |
|
| CYP3A4 | 12.00 (2.83) n = 8 |
10.52 (2.68) n = 50 |
† | β = 1.14 (0.97–1.35) p = 0.12 |
|
| CYP3A5 | 10.64 (2.72) n = 48 |
10.83 (2.04) n = 6 |
– n = 0 |
† | β = 1.03 (0.85–1.24) p = 0.76 |
| ABCB1 | 10.91 (3.48) n = 22 |
10.55 (2.33) n = 20 |
10.29 (1.33) n = 14 |
† | β = 0.99 (0.92–1.06) p = 0.73 |
| ABCC2 | 10.67 (1.53) n = 3 |
10.83 (3.05) n = 30 |
10.67 (2.28) n = 18 |
9.33 (1.37) n = 6 |
β = 0.96 (0.89–1.04) p = 0.33 |
| ABCG2 | – n = 0 |
9.67 (2.50) n = 9 |
10.92 (2.74) n = 49 |
† | β = 0.88 (0.75–1.04) p = 0.14 |
| SLCO1B1 | 12.12 (3.60) n = 17 |
10.15 (2.06) n = 41 |
† | β = 1.17 (1.03–1.33) p = 0.014* |
|
| SLCO1B3 | 10.88 (2.92) n = 40 |
10.79 (2.39) n = 14 |
8.67 (1.15) n = 3 |
† | β = 0.95 (0.86–1.05) p = 0.32 |
| Secondary analysis | Mean Cmax (ng/ml ) (SD) stratified by phenotype | β (95% CI), p-value | |||
| CYP2C8 | 2187.33 (656.90) n = 15 |
2455.75 (632.08) n = 40 |
† | β = 268.42 (-119.46–656.30) p = 0.17 |
|
| CYP2C8 *1/*3 and *3/*3 | 1985.00 (594.76) n = 12 |
2455.75 (632.08) n = 40 |
† | β = -320.75 (-767.56–126.06) p = 0.16 |
|
| CYP2C8 *1/*4 and *4/*4 | 2550.00 (693.83) n = 4 |
2455.75 (632.08) n = 40 |
† | β = 94.25 (-568.47–756.97) p = 0.78 |
|
| CYP3A4 | 2250.00 (458.11) n = 7 |
2410.20 (664.14) n = 49 |
† | β = -160.20 (-682.31–361.91) p = 0.54 |
|
| CYP3A5 | 2413.26 (679.75) n = 46 |
2226.67 (500.63) n = 6 |
– n = 0 |
† | β = -186.59 (-765.50–392.31) p = 0.52 |
| ABCB1 | 2219.05 (636.73) n = 21 |
2405.79 (782.05) n = 19 |
2585.00 (403.94) n = 14 |
† | β = 183.25 (-36.39–402.88) p = 0.10 |
| ABCC2 | 2316.67 (560.03) n = 3 |
2304.67 (672.21) n = 30 |
2595.00 (683.64) n = 16 |
2238.33 (353.52) n = 6 |
β = 58.99 (-172.31–290.30) p = 0.61 |
| ABCG2 | – n = 0 |
2233.75 (517.19) n = 8 |
2416.25 (660.23) n = 48 |
† | β = -182.50 (-675.17–310.17) p = 0.46 |
| SLCO1B1 | 2407.06 (725.44) n = 17 |
2382.82 (610.46) n = 39 |
† | β = 24.24 (-352.55–401.03) p = 0.90 |
|
| SLCO1B3 | 2390.00 (699.39) n = 38 |
2432.86 (565.22) n = 14 |
2143.33 (230.29) n = 3 |
† | β = -44.27 (-346.45–257.91) p = 0.77 |
*Statisticaly significant.
†Denotes phenotype groups that did not exist for that gene (See Supplementary Table 2).
SD: Standard deviation.
Figure 2. . Paclitaxel Tc >0.05 stratified by genotype-predicted metabolic activity phenotype for CYP2C8 and SLCO1B1.
Paclitaxel Tc >0.05 stratified by metabolic activity phenotype (PM/IM and NM) for CYP2C8 (left) and SLCO1B1 (right). Lower activity CYP2C8 genetic phenotype (PM/IM) was associated with shorter Tc >0.05 and lower OATP1B1 (SLCO1B1) activity was associated with longer Tc >0.05 in the univariate analyses.
IM: Intermediate metabolizer; NM: Normal metabolizer; PM: Poor metabolizer.
Table 3. . Univariate association of clinical variables with paclitaxel Tc >0.05 and Cmax.
| Clinical variables | Associations with Tc >0.05 | Associations with Cmax | ||
|---|---|---|---|---|
| B coefficient (95% CI) | p-value | B coefficient (95% CI) | p-value | |
| Age (years) | 1.00 (0.99–1.00) | 0.90 | 15.15 (1.88–28.42) | 0.026 |
| BSA (m2) | 1.27 (0.96–1.68) | 0.10 | -262.77 (-1085.39–559.85) | 0.52 |
| Caucasian race (vs other) | 1.22 (0.97–1.53) | 0.09 | -387.50 (-1051.95–276.95) | 0.25 |
| Elevated alkaline phosphatase (vs normal) | 0.90 (0.76–1.06) | 0.22 | -83.33 (-577.99–411.32)) | 0.74 |
| Elevated ALT or AST (vs normal) | 1.09 (0.90–1.32) | 0.38 | 261.13 (-294.55–816.81) | 0.35 |
| Elevated albumin (vs normal) | 0.86 (0.55–1.35) | 0.52 | 356.18 (-948.61–1660.98) | 0.59 |
| Prior or concurrent chemotherapy (vs none) | 1.03 (0.82–1.30) | 0.79 | -503.46 (-1260.63–253.71) | 0.19 |
| Prior or concurrent HER2 therapy (vs none) | 1.02 (0.91–1.15) | 0.75 | 136.26 (-208.52–481.03) | 0.43 |
BSA: Body surface area; SD: Standard deviation.
In a post-hoc analysis to understand the contribution of individual genotypes, the association for low-activity CYP2C8 genetic phenotype was attributable to the CYP2C8*3 polymorphism (CYP2C8*1/*3 or CYP2C8*3/*3 mean = 8.92 h, CYP2C8*1/*1 mean = 11.03 h, p = 0.0006). Although the numbers for analysis was quite small, there was no association between CYP2C8*4 and Tc >0.05 (CYP2C8*1/*4 or CYP2C8*4/*4 mean = 10.25 h, *1/*1 mean = 11.03 h, p = 0.58).
In the secondary analysis of genes relevant to paclitaxel metabolism or transport, patients with lower low-activity OATP1B1 genetic phenotype had longer Tc >0.05 (PM/IM = 12.12 h, NM = 10.15 h, β = 1.17, 95% CI: 1.03–1.33, p = 0.014, Table 2 & Figure 2 right) and this association maintained significance after adjustment for relevant clinical covariates (β = 0.85, 95% CI: 0.75–0.96, p = 0.012). A model containing both CYP2C8 and SLCO1B1 maintained independent effects for SLCO1B1 (SLCO1B1: p = 0.01, CYP2C8: p = 0.09). In the hypothesis generating screen of genes with no known relevance to paclitaxel PK, patients with lower UGT2B17 activity had shorter Tc >0.05 (β = 1.13, 95% CI: 1.04–1.22, p = 0.003) and patients with lower CYP2C9 activity had shorter Tc >0.05 (β = 1.14, 95% CI: 1.03–1.26, p = 0.014, Supplementary Table 3). None of the other genes were associated with Tc >0.05.
Associations of clinical & genetic variables with Cmax
Increasing age was associated with higher Cmax (β = 15.15, 95% CI: 1.88–28.42, p = 0.026, Table 3). None of the genes from the candidate list or hypothesis generating screen, including CYP2C8, were associated with Cmax (Supplementary Table 3), although the association for patients with lower activity SLC15A2 genetic phenotype and higher Cmax approached significance (β = -237.36, 95% CI: -473.72 to -1.00, p = 0.054).
Discussion
Patients carrying putatively diminished-activity CYP2C8 variants, including CY2C8*3, have been reported to have slower paclitaxel clearance [9,10] and greater risk of paclitaxel-induced PN [10,17,18,23]. In this cohort we previously reported that patients with longer Tc >0.05 had greater risk of PN [5]. Based on these findings, we hypothesized that patients with low-activity CYP2C8 genetic phenotype would have longer Tc >0.05. Contrary to our primary hypothesis, low-activity CYP2C8 genetic phenotype was associated with significantly shorter Tc >0.05.
Two human PK studies have reported decreased paclitaxel clearance in patients carrying CYP2C8 *3 [9,10], one of which only detected the association in patients that were also ABCB1 2677G/T heterozygotes [10]. Other studies have found no association between CYP2C8 genotype or metabolizer phenotype and paclitaxel PK [24,25]. The two alleles classified as low-activity in our cohort were CYP2C8*3 (rs10509681 and rs11572080) and CY2C8*4 (rs1058930), with the majority of PM and IM patients carrying the CYP2C8*3 alleles. CYP2C8*3 genotype causes two non-synonymous changes (R139K and K399R) and has previously been characterized as diminished paclitaxel metabolic activity in multiple in vitro studies [8,11–14]. It is believed that the effect of CYP2C8*3 may be substrate dependent, as it is well established that CYP2C8*3 has higher metabolic activity toward rosiglitazone [19,20] and pioglitazone [26]. Contrary to our hypothesis that CYP2C8*3 confers decreased activity based on the in vitro and limited previous in vivo studies, our data suggest that CYP2C8*3 may in fact confer increased activity for paclitaxel metabolism, similar to other substrates. This further complicates interpretation of previous work from our group and others showing that CYP2C8*3 is associated with greater risk of paclitaxel-induced PN [10,17,18,23], however, these results have not been validated and may be false-positive findings.
In vitro CYP2C8*4 has been shown to confer variable levels of decreased paclitaxel metabolic activity [13,15,16]. One previous clinical PK study reported decreased paclitaxel clearance [9], whereas others have found no association, possibly due to the small number of CYP2C8*4 carriers [10,24,25]. Our study included only four carriers of CYP2C8*4, so the marginally shorter exposure was not significantly different from wild-type patients. Considering the CYP2C8*3 and CYP2C8*4 results together, it seems that the effect of CYP2C8 polymorphisms on human paclitaxel exposure is modest, at best.
In our analysis, patients with lower OATP1B1 activity, which is encoded by SLCO1B1, had longer Tc >0.05. Low-activity SLCO1B1 variants indirectly affect hepatic metabolism of various drugs by decreasing OATP1B1 expression at the cell membrane [27] and, consequently, diminishing hepatic uptake of drugs for metabolism [28–30]. Unlike our findings for paclitaxel, longer exposure in patients with low-activity OATP1B1 was as predicted. In vitro studies have suggested that SLCO1B1 likely has modest effects on paclitaxel transport [31,32], but SLCO1B1 has not been previously examined in humans for its impact on paclitaxel PK. In the hypothesis generating screen, lower CYP2C9 activity was associated with shorter Tc >0.05, but this association is likely due to linkage disequilibrium between the lower activity CYP2C9*2 and CYP2C8*3 alleles [33]. The association between lower UGT2B17 activity and shorter Tc >0.05 has not been previously observed, and is likely an artifact of the number of statistically uncorrected association tests run.
Our results indicate a modest effect of CYP2C8 and SLCO1B1 genotypes on paclitaxel PK in humans. In a prospective clinical trial in patients with non-small-cell lung cancer, randomization to utilizing Tc >0.05-guided paclitaxel dosing reduced PN without compromising efficacy [34]. Initial dose recommendations in the PK-guided arm were based on BSA, age and sex; incorporating pharmacogenetics into these personalized dosing algorithms could improve their precision and enable further optimization of therapeutic outcomes.
Age has been negatively correlated with clearance in patients receiving a 1-h paclitaxel infusion [35], but not Cmax. However, in this study we identified a relatively large effect on Cmax, which was unexpected since age related changes in paclitaxel clearance would not be expected to affect Cmax following the short 1-h infusions used in this cohort. Further research is needed to determine whether there is some other age-related change that explains the relatively large effect on Cmax detected in this study. The trend for Caucasian patients to have longer Tc >0.05 was likely due to the higher allele frequency of CYP2C8*3 in Caucasian populations compared with Asian and African–American populations, where it is typically rare or absent [36]. Consistent with the trend seen in this cohort, BSA has previously been positively correlated with paclitaxel elimination in a population-PK analysis [37].
The primary limitation of this analysis is its modest size, which may have resulted in reduced statistical power for many of these analyses of genetic and clinical associations. Due to sparse sampling, this analysis used Tc >0.05, a clinically relevant exposure parameter, rather than the more conventional PK parameters clearance or area under the curve (AUC) [9,10,24,25]. However, Tc >0.05, is predictive of PN [2,3]. Consequently, determining pharmacogenomic and clinical factors that impact Tc >0.05 has great potential to positively impact patient outcomes. Tc >0.05 was chosen as the primary end point due to the expectation that metabolism and active transport would have minimal effect on Cmax during a 1-h infusion, which is supported by the lack of any pharmacogenetic association in this analysis. Additionally, concomitant medications that induce or inhibit CYP2C8 or other enzymes or transporters could have impacted PK, but information on concomitant medication is not available for this cohort and could not be accounted for in this analysis.
A panel genotyping approach was selected to comprehensively investigate functionally consequential SNPs in genes relevant to drug metabolism and transport, and all genes with known or suspected functional consequence for paclitaxel metabolism or transport were included. However, enzyme activity was not measured directly, and our system for translating genotype to genetic phenotype is limited by the SNPs included on this panel and the current understanding of their functional consequence. Finally, the subjects included in this cohort were primarily Caucasian, which limits the diversity of polymorphisms and patient characteristics represented in the analysis.
Conclusion
In this secondary analysis of female patients with breast cancer receiving weekly 1-h paclitaxel infusions, patients with low-activity CYP2C8 genetic phenotype had shorter Tc >0.05 and patients with low-activity OATP1B1 genetic phenotype had longer Tc >0.05. Our findings suggest that CYP2C8*3 confers increased paclitaxel metabolism, which is contrary to prior studies but is consistent with its established functional impact on for other CYP2C8 substrates. If definitive associations between clinical and pharmacogenetic factors and paclitaxel PK are elucidated, they could potentially be incorporated into personalized dosing strategies to achieve optimal exposure levels that avoid toxicities and/or maximize treatment efficacy.
Future perspective
Clinical, genetic and PK information will be ideally integrated in the future to personalize paclitaxel dosing regimens to reduce the occurrence of dose-limiting toxicities while ensuring drug efficacy. To achieve that goal, discovery of genetic predictors of paclitaxel PK and toxicity are pivotal. Future research is needed to fully elucidate the impact of drug transporters and metabolizing enzymes on paclitaxel exposure. These efforts should be integrated with on-going efforts to determine predictors of toxicity to most accurately create a paclitaxel dosing model.
Summary points.
Based on prior findings, low CYP2C8 activity is expected to lead to longer systemic paclitaxel exposure or time above threshold, Tc >0.05.
A comprehensive CYP2C8 genotyping approach was taken including CYP2C8 *2, *3, *4, *5, *7 and *8.
Contrary to what was expected, patients with low-activity CYP2C8 genetic phenotype had shorter Tc >0.05.
CYP2C8*3 may confer increased activity to metabolize paclitaxel.
Patients with low-activity CYP2C9 genetic phenotype had shorter Tc >0.05 likely due to linkage disequilibrium between the low-activity CYP2C8*3 and CYP2C9*2 variants.
In a novel association, low-activity OATP1B1 genetic phenotype patients had longer Tc >0.05.
OATP1B1 and CYP2C8 pharmacogenomics may explain some variability in paclitaxel exposure.
Future studies that verify pharmacogenomic associations with paclitaxel exposure could lead to personalized dosing strategies to optimize efficacy and reduce toxicity.
Supplementary Material
Footnotes
Supplementary data
To view the supplementary data that accompany this paper please visit the journal website at: https://www.futuremedicine.com/doi/suppl/10.2217/pgs-2018-0162
Financial & competing interests disclosure
This work was supported by the National Center for Advancing Translational Sciences under award number KL2TR000434 and 2UL1TR000433 (DL Hertz) and the National Cancer Institutes of Health under Award Number P30CA046592 (KM Kidwell).
DL Hertz has an informal, unpaid collaborative relationship with Saladax Biomedical Inc., a company that offers CLIA-approved paclitaxel measurement. Saladax was not involved in the design, conduct, analysis, or sponsorship of this trial, and had no contribution to the writing of this manuscript. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The study was approved by the University of Michigan IRBMed and conducted in accordance with recognized ethical guidelines. All enrolled subjects signed written informed consent.
Author contributions
DL Hertz, ML Burness, JJ Griggs, C Van Poznak, AF Schott, DF Hayes, NL Henry contributed to the conception and design of the work. DL Hertz, K Vangipuram, ML Burness, JJ Griggs, C Van Poznak, AF Schott, DF Hayes and NL Henry contributed to the acquisition of the work. DL Hertz, KM Kidwell, AC Robinson, K Vangipuram, LA Marcath contributed to the analysis or interpretation of the data. All authors have contributed to the drafting of the work or revising it critically for content. All authors have reviewed and approve of this manuscript for submission. All authors agree to be accountable for all aspects of the work.
Disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. Additional support was provided by the Fashion Footwear Charitable Foundation of New York/QVC Presents Shoes on Sale™ (DF Hayes).
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Perez EA. Paclitaxel in breast cancer. Oncologist. 1998;3(6):373–389. [PubMed] [Google Scholar]
- 2.Mielke S, Sparreboom A, Steinberg SM, et al. Association of paclitaxel pharmacokinetics with the development of peripheral neuropathy in patients with advanced cancer. Clin. Cancer Res. 2005;11(13):4843–4850. doi: 10.1158/1078-0432.CCR-05-0298. [DOI] [PubMed] [Google Scholar]; •• Utilizes their previously collected clinical and paclitaxel pharmacokinetic data to show that patients that developed peripheral neuropathy had greater paclitaxel exposure, including the parameter time above threshold.
- 3.De Graan AJ, Elens L, Sprowl JA, et al. CYP3A4*22 genotype and systemic exposure affect paclitaxel-induced neurotoxicity. Clin. Cancer Res. 2013;19(12):3316–3324. doi: 10.1158/1078-0432.CCR-12-3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mielke S, Sparreboom A, Behringer D, Mross K. Paclitaxel pharmacokinetics and response to chemotherapy in patients with advanced cancer treated with a weekly regimen. Anticancer Res. 2005;25(6C):4423–4427. [PubMed] [Google Scholar]; • Determines that the parameter, time above threshold, was associated with paclitaxel treatment response.
- 5.Hertz DL, Kidwell KM, Vangipuram K, et al. Paclitaxel plasma concentration after the first infusion predicts treatment-limiting peripheral neuropathy. Clin. Cancer Res. 2018;24(15):3602–3610. doi: 10.1158/1078-0432.CCR-18-0656. [DOI] [PMC free article] [PubMed] [Google Scholar]; • In the cohort of patients included in the manuscript, increased paclitaxel exposure was predictive of peripheral neuropathy-induced treatment disruption.
- 6.Mross K, Hollander N, Hauns B, Schumacher M, Maier-Lenz H. The pharmacokinetics of a 1-h paclitaxel infusion. Cancer Chemother. Pharmacol. 2000;45(6):463–470. doi: 10.1007/s002800051020. [DOI] [PubMed] [Google Scholar]
- 7.Caudle KE, Dunnenberger HM, Freimuth RR, et al. Standardizing terms for clinical pharmacogenetic test results: consensus terms from the Clinical Pharmacogenetics Implementation Consortium (CPIC) Genet Med. 2017;19(2):215–223. doi: 10.1038/gim.2016.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai D, Zeldin DC, Blaisdell JA, et al. Polymorphisms in human CYP2C8 decrease metabolism of the anticancer drug paclitaxel and arachidonic acid. Pharmacogenetics. 2001;11(7):597–607. doi: 10.1097/00008571-200110000-00006. [DOI] [PubMed] [Google Scholar]; • Describes the alleles, CYP2C8*3 and CYP2C8*2, as decreased function for paclitaxel metabolism.
- 9.Bergmann TK, Brasch-Andersen C, Green H, et al. Impact of CYP2C8*3 on paclitaxel clearance: a population pharmacokinetic and pharmacogenomic study in 93 patients with ovarian cancer. Pharmacogenomics J. 2011;11(2):113–120. doi: 10.1038/tpj.2010.19. [DOI] [PubMed] [Google Scholar]; •• Conducts a prospective pharmacogenetic study on paclitaxel pharmacokinetics, which first described association between lower activity CYP2C8 and decreased paclitaxel clearance in a clinical study.
- 10.Green H, Soderkvist P, Rosenberg P, et al. Pharmacogenetic studies of paclitaxel in the treatment of ovarian cancer. Basic Clin. Pharmacol. Toxicol. 2009;104(2):130–137. doi: 10.1111/j.1742-7843.2008.00351.x. [DOI] [PubMed] [Google Scholar]; • Conducts a study similar but smaller to the prospective pharmacogenetic study paclitaxel pharmacokinetics published by Bergmann et al. CYP2C8*3 carriers had lower paclitaxel clearance when stratified for ABCB1 G2677T/A genotype.
- 11.Tsukada C, Saito T, Maekawa M, et al. Functional characterization of 12 allelic variants of CYP2C8 by assessment of paclitaxel 6alpha-hydroxylation and amodiaquine N-deethylation. Drug Metab. Pharmacokinet. 2015;30(5):366–373. doi: 10.1016/j.dmpk.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 12.Soyama A, Saito Y, Hanioka N, et al. Non-synonymous single nucleotide alterations found in the CYP2C8 gene result in reduced in vitro paclitaxel metabolism. Biol. Pharm. Bull. 2001;24(12):1427–1430. doi: 10.1248/bpb.24.1427. [DOI] [PubMed] [Google Scholar]
- 13.Yu L, Shi D, Ma L, Zhou Q, Zeng S. Influence of CYP2C8 polymorphisms on the hydroxylation metabolism of paclitaxel, repaglinide and ibuprofen enantiomers in vitro . Biopharm. Drug Dispos. 2013;34(5):278–287. doi: 10.1002/bdd.1842. [DOI] [PubMed] [Google Scholar]
- 14.Lee MY, Apellaniz-Ruiz M, Johansson I, et al. Role of cytochrome P450 2C8*3 (CYP2C8*3) in paclitaxel metabolism and paclitaxel-induced neurotoxicity. Pharmacogenomics. 2015;16(9):929–937. doi: 10.2217/pgs.15.46. [DOI] [PubMed] [Google Scholar]
- 15.Gao Y, Liu D, Wang H, Zhu J, Chen C. Functional characterization of five CYP2C8 variants and prediction of CYP2C8 genotype-dependent effects on in vitro and in vivo drug-drug interactions. Xenobiotica. 2010;40(7):467–475. doi: 10.3109/00498254.2010.487163. [DOI] [PubMed] [Google Scholar]
- 16.Bahadur N, Leathart JB, Mutch E, et al. CYP2C8 polymorphisms in Caucasians and their relationship with paclitaxel 6alpha-hydroxylase activity in human liver microsomes. Biochem. Pharmacol. 2002;64(11):1579–1589. doi: 10.1016/s0006-2952(02)01354-0. [DOI] [PubMed] [Google Scholar]
- 17.Leskela S, Jara C, Leandro-Garcia LJ, et al. Polymorphisms in cytochromes P450 2C8 and 3A5 are associated with paclitaxel neurotoxicity. Pharmacogenomics J. 2011;11(2):121–129. doi: 10.1038/tpj.2010.13. [DOI] [PubMed] [Google Scholar]
- 18.Hertz DL, Roy S, Motsinger-Reif AA, et al. CYP2C8*3 increases risk of neuropathy in breast cancer patients treated with paclitaxel. Ann. Oncol. 2013;24(6):1472–1478. doi: 10.1093/annonc/mdt018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aquilante CL, Bushman LR, Knutsen SD, Burt LE, Rome LC, Kosmiski LA. Influence of SLCO1B1 and CYP2C8 gene polymorphisms on rosiglitazone pharmacokinetics in healthy volunteers. Hum. Genomics. 2008;3(1):7–16. doi: 10.1186/1479-7364-3-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirchheiner J, Thomas S, Bauer S, et al. Pharmacokinetics and pharmacodynamics of rosiglitazone in relation to CYP2C8 genotype. Clin. Pharmacol. Ther. 2006;80(6):657–667. doi: 10.1016/j.clpt.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 21.Kraff S, Nieuweboer AJ, Mathijssen RH, et al. Pharmacokinetically based dosing of weekly paclitaxel to reduce drug-related neurotoxicity based on a single sample strategy. Cancer Chemother. Pharmacol. 2015;75(5):975–983. doi: 10.1007/s00280-015-2724-9. [DOI] [PubMed] [Google Scholar]
- 22.Marcath LA, Deal AM, Van Wieren E, et al. Comprehensive assessment of cytochromes P450 and transporter genetics with endoxifen concentration during tamoxifen treatment. Pharmacogenet. Genomics. 2017;27(11):402–409. doi: 10.1097/FPC.0000000000000311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hertz DL, Roy S, Jack J, et al. Genetic heterogeneity beyond CYP2C8*3 does not explain differential sensitivity to paclitaxel-induced neuropathy. Breast Cancer Res. Treat. 2014;145(1):245–254. doi: 10.1007/s10549-014-2910-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henningsson A, Marsh S, Loos WJ, et al. Association of CYP2C8, CYP3A4, CYP3A5, and ABCB1 polymorphisms with the pharmacokinetics of paclitaxel. Clin. Cancer Res. 2005;11(22):8097–8104. doi: 10.1158/1078-0432.CCR-05-1152. [DOI] [PubMed] [Google Scholar]
- 25.Marsh S, Soml o G, Li X, et al. Pharmacogenetic analysis of paclitaxel transport and metabolism genes in breast cancer. Pharmacogenomics J. 2007;7(5):362–365. doi: 10.1038/sj.tpj.6500434. [DOI] [PubMed] [Google Scholar]
- 26.Aquilante CL, Kosmiski LA, Bourne DW, et al. Impact of the CYP2C8 *3 polymorphism on the drug–drug interaction between gemfibrozil and pioglitazone. Br. J. Clin. Pharmacol. 2013;75(1):217–226. doi: 10.1111/j.1365-2125.2012.04343.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kameyama Y, Yamashita K, Kobayashi K, Hosokawa M, Chiba K. Functional characterization of SLCO1B1 (OATP-C) variants, SLCO1B1*5, SLCO1B1*15 and SLCO1B1*15+C1007G, by using transient expression systems of HeLa and HEK293 cells. Pharmacogenet. Genomics. 2005;15(7):513–522. doi: 10.1097/01.fpc.0000170913.73780.5f. [DOI] [PubMed] [Google Scholar]
- 28.Wilke RA, Ramsey LB, Johnson SG, et al. The clinical pharmacogenomics implementation consortium: CPIC guideline for SLCO1B1 and simvastatin-induced myopathy. Clin. Pharmacol. Ther. 2012;92(1):112–117. doi: 10.1038/clpt.2012.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramsey LB, Panetta JC, Smith C, et al. Genome-wide study of methotrexate clearance replicates SLCO1B1. Blood. 2013;121(6):898–904. doi: 10.1182/blood-2012-08-452839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chigutsa E, Visser ME, Swart EC, et al. The SLCO1B1 rs4149032 polymorphism is highly prevalent in South Africans and is associated with reduced rifampin concentrations: dosing implications. Antimicrob. Agents Chemother. 2011;55(9):4122–4127. doi: 10.1128/AAC.01833-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Durmus S, Van Hoppe S, Schinkel AH. The impact of organic anion-transporting polypeptides (OATPs) on disposition and toxicity of antitumor drugs: insights from knockout and humanized mice. Drug Resist. Updat. 2016;27:72–88. doi: 10.1016/j.drup.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 32.Nieuweboer AJ, Hu S, Gui C, et al. Influence of drug formulation on OATP1B-mediated transport of paclitaxel. Cancer Res. 2014;74(11):3137–3145. doi: 10.1158/0008-5472.CAN-13-3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yasar U, Lundgren S, Eliasson E, et al. Linkage between the CYP2C8 and CYP2C9 genetic polymorphisms. Biochem. Biophys. Res. Commun. 2002;299(1):25–28. doi: 10.1016/s0006-291x(02)02592-5. [DOI] [PubMed] [Google Scholar]
- 34.Joerger M, Von Pawel J, Kraff S, et al. Open-label, randomized study of individualized, pharmacokinetically (PK)-guided dosing of paclitaxel combined with carboplatin or cisplatin in patients with advanced non-small-cell lung cancer (NSCLC) Ann. Oncol. 2016;27(10):1895–1902. doi: 10.1093/annonc/mdw290. [DOI] [PubMed] [Google Scholar]
- 35.Smorenburg CH, Ten Tije AJ, Verweij J, et al. Altered clearance of unbound paclitaxel in elderly patients with metastatic breast cancer. Eur. J. Cancer. 2003;39(2):196–202. doi: 10.1016/s0959-8049(02)00611-1. [DOI] [PubMed] [Google Scholar]
- 36.Garcia-Martin E, Martinez C, Ladero JM, Agundez JA. Interethnic and intraethnic variability of CYP2C8 and CYP2C9 polymorphisms in healthy individuals. Mol. Diagn. Ther. 2006;10(1):29–40. doi: 10.1007/BF03256440. [DOI] [PubMed] [Google Scholar]
- 37.Joerger M, Huitema AD, Van Den Bongard DH, Schellens JH, Beijnen JH. Quantitative effect of gender, age, liver function, and body size on the population pharmacokinetics of paclitaxel in patients with solid tumors. Clin. Cancer Res. 2006;12(7 Pt 1):2150–2157. doi: 10.1158/1078-0432.CCR-05-2069. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.