Abstract
Fusarium head blight (FHB), caused principally by the species belonging to the Fusarium graminearum species complex (FGSC), is an important disease in wheat, barley, and other small grain crops worldwide. Grain infected with species in the FGSC may be contaminated with trichothecene mycotoxins such as deoxynivalenol (DON) and nivalenol (NIV). In this study, we characterized the phylogenetic relationships, chemotype diversity, phenotypic characters, and aggressiveness of 150 strains in FGSC collected from eight different countries. Phylogenetic analysis based on portions of translation elongation factor 1-α (EF-1α) gene from 150 strains revealed six species in the FGSC, F. graminearum s.s, F. asiaticum, F. meridionale, F. cortaderiae, F. boothii, and F. austroamericanum. In this collection, 50% of the strains were 15-acetyldeoxynivalenol (15-ADON), 35% were 3-acetyldeoxynivalenol (3-ADON) and 15% were NIV. Evaluation of strains on moderately resistant (MR) wheat cultivar Carberry indicated that there is no significant difference among the species for FHB disease severity (DS), fusarium damaged kernel percentage (FDK%) and DON production. However, significant differences were observed among the chemotypes. Results showed significantly higher FHB DS, FDK%, DON production, growth rates, and macroconidia production for the 3-ADON strains than the 15-ADON and NIV strains. In addition, significant differences for FHB response variables were observed among the strains from different countries. Our results demonstrate that type and amount of trichothecene produced by a strain play a key role in determining the level of aggressiveness of that particular strain, regardless of the species.
Keywords: Fusarium Head Blight, mycotoxins, 3-ADON, 15-ADON, NIV, FGSC, trichothecene, deoxynivalenol, nivalenol, phylogenetics, wheat, F. graminearum s.s, F. asiaticum, F. meridionale, F. cortaderiae, F. boothii and F. austroamericanum, EF-1α gene
1. Introduction
FHB is economically one of the most important diseases of wheat throughout the world. Epidemics of FHB over the past 15 years in North America have had a devastating economic impact on agriculture [1,2]. Many FHB outbreaks have been reported in Canada, Asia, Europe, Australia, and South America suggesting that it is a major threat to world grain production [3,4]. One of the major concerns attributed with FHB is the contamination of grains with trichothecene mycotoxins and other estrogenic compounds. The F. graminearum species are capable of producing various B-trichothecenes, particularly, DON, its acetylated derivatives, 3-ADON and 15-ADON and NIV and its acetylated derivative, 4-acetyl nivalenol (4-ANIV) [5]. Based on the trichothecene profile, F. graminearum strains can be categorized into three main chemical groups or chemotypes namely, 3-ADON, 15-ADON, and NIV. A 3-ADON chemotype produces DON and 3-ADON, while a 15-ADON chemotype produces DON and 15-ADON and a NIV chemotype produces NIV and 4-ANIV [6,7,8]. In North America, DON is the primary mycotoxin in Fusarium infected grain, while in parts of Asia and Europe both DON and NIV are the common contaminants of grain [9]. These mycotoxins pose a significant threat to human and animal health [10]. Trichothecenes are also acutely phytotoxic and act as virulence factors on sensitive host plants [5].
Fusarium head blight is caused by several Fusarium spp. such as F. graminearum, F. sporotrichioides, F. culmorum, F. cerealis, F. avenaceum, F. equiseti, and F. poae [2]. Among these species of Fusarium, members of the FGSC are still considered to be the major etiological agents of FHB worldwide [9,11]. Other Fusarium species play a minor role in FHB development. Species of Fusarium have traditionally been classified based on the morphological characteristics such as the shape and size of the macroconidia, presence/absence of chlamydospores, and presence/absence, shape, and supporting structures of the macroconidia and microconidia [12].
Until the year 2000, members of the FGSC were considered a single cosmopolitan species as morphological approaches to species identification failed to accurately delimit species for this group. However, with the advances in DNA sequencing technology, sequence similarity at one or more diagnostic loci has become an important tool in determining the species limits with Fusarium spp. [12]. The sequences most widely used to identify species of Fusarium are portions of DNA sequences encoding EF-1α, β-tubulin, internally transcribed spacer (ITS) regions in the ribosomal DNA repeat region (ITS1 and ITS2), histone H3, and trichothecene biosynthesis genes, especially TRI101 [8,11]. O’Donnell et al. [11] first identified seven phylogenetic species within the FGSC, using genealogical concordance phylogenetic species recognition (GCPSR). Later, using a high-throughput multilocus genotyping (MLGT) assay of portions of 13 housekeeping genes, coupled with GCPSR, another nine phylogenetically distinct, cryptic species were identified within the FGSC [13,14,15,16]. The species designation F. graminearum was therefore referred to as sensu stricto (s. s.) and retained for the species most commonly associated with FHB worldwide.
So far, 16 monophyletic species have been identified within the FGSC and these include: F. austroamericanum (lineage 1), F. meridionale (lineage 2), F. boothii (lineage 3) F. mesoamericanum (lineage 4), F. acacia-mearnsii (lineage 5), F. asiaticum, F. graminearum sensu stricto (lineage 7), F. cortaderiae (lineage 8), and eight additional monotypic lineages, including: F. brasilicum, F. vorosii, F. gerlachii, F. aethiopicum, F. ussurianum, F. nepalense, F. louisianense, and U.S. Gulf Coast population of F. graminearum [8,13,14,15,16,17]. Among the members of the FGSC, F. graminearum s.s has been found worldwide, while the distribution of other species was found to be restricted to certain geographic areas.
The EF-1α gene that encodes an essential part of the protein translation machinery has been extensively used to differentiate Fusarium spp. It has been shown that EF-1α provides much better resolution of relationships among and within lineages than other loci such as β-tubulin, calmodulin and ITS region [8,18]. Additionally, the absence of non-orthologous copies of EF-1α in the genus makes this locus a better candidate to resolve phylogenetic relationships among species. EF-1α gene has been used to investigate the species limits within the F. graminearum clade.
Recent studies have shown the emergence of new species within the FGSC and rapid population shifts that are characterized by changes in chemotype frequency [15,19]. The displacement of native 15-ADON chemotypes by a population with 3-ADON chemotypes in North America and emergence of new NIV-type and NX-2 populations in the USA have shown the importance of continued assessment of population shifts in FGSC [15,19,20,21]. Pathogenic and toxigenic differences between F. graminearum populations in combination with environmental factors are hypothesized to be the major reason for rapid shifts in pathogen populations. In recent years, many studies have been performed to examine species and trichothecene chemotypes diversity among FGSC strains [3,4,22,23]. Most of these studies were restricted either to a country or a particular region in a country. Very few studies have been undertaken to examine and compare the genetic diversity and pathogenicity among strains of FGSC associated with FHB from different countries [2,24,25]. Additionally, the findings from genetic diversity studies have been useful in making decisions in breeding for FHB resistant wheat cultivars. It has been reported that aggressiveness of F. graminearum strains collected from different regions within a country and even within populations from individual fields are highly variable, and therefore, FHB resistant wheat cultivars that are resistant in one region may not give consistent results in other regions [24,26]. The performance of a resistant cultivar mainly relies on the pathogen profile, environmental conditions and the interaction between these two variables in a specific location. Variation in pathogenicity and aggressiveness in pathogen populations can lead to host resistance being overcome [27,28]. Therefore, a better understanding of the pathogen profile, chemotype diversity and aggressiveness is important to align plant protection to the existing and potentially changing pathogen population, further; this may lead to improved strategies for disease management [27].
The members of the FGSC are highly variable in a number of morphological traits, including the size and shape of their conidia, growth rate on standard media, pathogenicity on wheat cultivars and the type of mycotoxins produced. Acquiring knowledge of the trichothecene mycotoxins produced by predominant FGSC populations in a cereal production region and further analysing their role in the pathogenesis are important to understand the factors affecting plant-pathogen interactions. Additionally, this will offer new approaches to breeding for FHB resistance. Therefore, the objectives of this study were to: (1) Determine the trichothecene chemotypes of F. graminearum strains based on TRI3 and TRI12 gene-specific polymerase chain reaction (PCR) assays; (2) evaluate the molecular phylogenetic relationships of FGSC strains collected from different countries based on the EF-1α gene; (3) determine the phenotypic characteristics of the strains in the FGSC; and (4) determine the aggressiveness of strains from different species in the FGSC.
2. Results
2.1. Identification of Fusarium Strains to Species Level
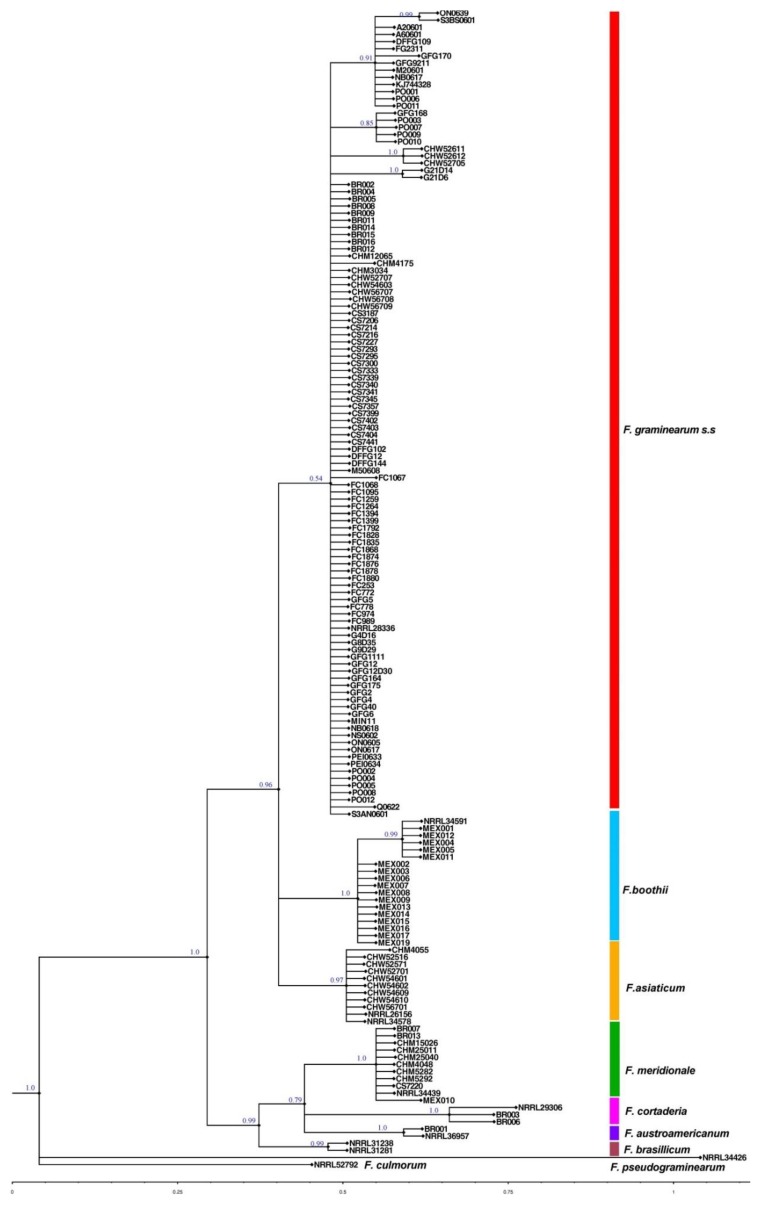
Genetic polymorphisms in the DNA sequence of the EF-1α have been used to assist in the identification of 150 strains to species level. In the EF-1α analysis, 111 strains formed a monophyletic clade with reference strain F. graminearum s.s NRRL28336 confirming the strains identity as F. graminearum s.s (Figure 1). This clade includes strains from Australia, Poland, UK, Germany, Brazil, China and Canada. Another 17 strains from Mexico formed a monophyletic clade including the F. boothii reference strain NRRL34591. Within this clade, five strains from Mexico formed an intra-clade structure with F. boothii reference strain NRRL34591. Nine strains from China formed a monophyletic clade with F. asiaticum reference strains NRRL 26156 and NRRL 34578. Ten strains from the collection formed a monophyletic clade with F. meridionale reference strain NRRL 34439. This clade consists of strains originating from Australia, Brazil, China and Mexico. Two strains from Brazil formed a monophyletic clade with F. cortaderiae reference strain NRRL 29306. One strain from Brazil also formed a monophyletic clade with F. austroamericarnum reference strain NRRL 36957.
Figure 1.
Inferred topology from the Bayesian analysis of EF-1α. Numbers at the nodes represent posterior probabilities.
2.2. Trichothecene Chemotypes
The PCR assays based on TRI3 and TRI12 genes amplified products of 840, 610 and 243 bp corresponding with NIV, 15-ADON and 3-ADON chemotypes, respectively. Among the 111 F. graminearum s.s strains, 50% of the strains were of 15-ADON chemotype, 35% were 3-ADON producers and 15% were NIV producers (Supplementary Table S1). All NIV producing strains belonging to F. graminearum s.s were from Germany. Among the nine Chinese F. asiaticum strains, three strains were determined to be 3-ADON, three were NIV and another three were 15-ADON chemotypes. Among the 18 strains from Mexico, 17 strains were identified as F. boothii and all were 15-ADON producers. One strain was identified as F. meridionale and it was a NIV producer. All the F. meridionale strains in the collection (from Brazil, China and Australia) were identified as NIV producers. Also, the two strains of F. cortaderiae (from Brazil) were identified as an NIV chemotype. One strain from Brazil, which identified as F. austroamericanum, was determined to be a NIV chemotype.
2.3. Aggressiveness of Strains on Spring Wheat
The aggressiveness of 133 strains listed in Supplementary Table S1, except for strains obtained from Mexico, was evaluated on the MR spring wheat cultivar Carberry. Analysis of variance on FHB DS indicated that there were no significant differences among species. However, there were significant differences among the chemotypes. In addition, the two-way interaction between species and chemotypes was not significant (Table 1). F. graminearum s.s and F. asiaticum 3-ADON strains showed a comparatively higher DS than other species and chemotypes (Table 2). When we analysed the DS data according to the country of origin of strains, and the trichothecene chemotypes, DS was significantly different among the strains originating from different countries. The interaction country*chemotype was also significant. The 3-ADON chemotypes from Germany, UK, Poland, and China showed higher DS followed by Canadian 3-ADON strains (Table 2). We observed a similar pattern for FDK%, in which FDK% was not significantly different among the species, however, different among the chemotypes. The species*chemotype interaction was not significant. However, F. graminearum 3-ADON and F. asiaticum 3-ADON strains showed a higher FDK% compared to other species and chemotypes. In contrast to FHB DS, no significant difference was observed among the countries for FDK%. The two-way interaction, country*chemotype was also not significantly different. However, 3-ADON chemotypes from UK, Germany, China, Canada, and Poland showed higher FDK% than other chemotypes.
Table 1.
Analysis of variance (ANOVA) table for fusarium head blight disease severity percentage (DS%), fusarium damaged kernel percentage (FDK%), deoxynivalenol (DON) or nivalenol (NIV) content in parts per million, radial growth in centimeters and macroconidia production in Fusarium graminearum species complex strains collected from different countries.
| Trait | Source | DF | MS | F Value | Pr >F |
|---|---|---|---|---|---|
| DS% | Species | 4 | 108.82 | 0.89 | 0.4695 |
| Country | 6 | 3358.81 | 27.47 | <0.0001 | |
| Chemotype | 2 | 12,181.00 | 99.61 | <0.0001 | |
| Species*Chemotype | 8 | 327.51 | 2.69 | 0.0687 | |
| Residual | 636 | 122.28 | |||
| FDK% | Species | 4 | 11.90 | 1.18 | 0.3253 |
| Country | 6 | 9.52 | 0.94 | 0.4694 | |
| Chemotype | 2 | 2481.38 | 245.05 | <0.0001 | |
| Species*Chemotype | 8 | 3.19 | 0.32 | 0.7303 | |
| Residual | 116 | 10.13 | |||
| DON/NIV (ppm) | Species | 4 | 37.08 | 0.34 | 0.8509 |
| Country | 6 | 28.80 | 4.6 | 0.0003 | |
| Chemotype | 2 | 395.18 | 63.19 | <0.0001 | |
| Species*Chemotype | 8 | 7.01 | 1.12 | 0.3292 | |
| Residual | 116 | 6.25 | |||
| Radial growth (cm) | Species | 4 | 4.00 | 6.35 | <0.0001 |
| Country | 6 | 5.70 | 9.04 | <0.0001 | |
| Chemotype | 2 | 98.63 | 156.36 | <0.0001 | |
| Species*Chemotype | 8 | 0.36 | 0.57 | 0.5670 | |
| Residual | 376 | 0.63 | |||
| Macroconidia | Species | 4 | 294.52 | 7.94 | <0.0001 |
| Country | 2 | 270.51 | 7.29 | <0.0001 | |
| Chemotype | 6 | 8772.54 | 236.57 | <0.0001 | |
| Species*Chemotype | 8 | 393.36 | 10.61 | 0.0077 | |
| Residual | 376 | 37.08 |
DF—Degree of Freedom; MS—Mean Square; Pr—Probability.
Table 2.
Country of origin of strains, trichothecene chemotypes, mean values for fusarium head blight disease severity (DS%), fusarium damaged kernel percentage (FDK%), deoxynivalenol (DON) or nivalenol (NIV) content in parts per million, radial growth in centimeters, and macroconidia production of strains used in the study.
| Country of Origin of Strains | Chemotype | DS% | FDK% | DON/NIV Content (ppm) | Radial Growth (cm) | Macroconidia (×104/mL) |
|---|---|---|---|---|---|---|
| Germany | 3-ADON | 53.2 a | 38.0 a | 17.3 a | 8.5 a | 69.1 ab |
| UK | 3-ADON | 48.1 ab | 35.7 a | 10.3 b | 8.1 ab | 66.2 b |
| Poland | 3-ADON | 46.9 abc | 35.6 a | 11.0 b | 7.2 c | 67.7 ab |
| China | 3-ADON | 44.8 abc | 36.0 a | 12.3 b | 6.8 c | 64.2 b |
| Canada | 3-ADON | 42.1 bcd | 35.5 a | 12.6 b | 7.6 bc | 70.9 a |
| China | 15-ADON | 39.1 cde | 14.1 b | 4.9 cd | 4.6 fg | 38.4 def |
| Germany | 15-ADON | 35.8 de | 13.2 b | 5.7 c | 4.8 efg | 43.0 cd |
| Canada | 15-ADON | 32.7 e | 15.7 b | 4.6 cd | 5.4 de | 43.6 cd |
| UK | 15-ADON | 23.5 fg | 13.3 b | 5.2 cd | 5.0 def | 45.0 c |
| Brazil | 15-ADON | 23.3 f | 16.1 b | 4.2 cd | 5.6 d | 38.4 def |
| Poland | 15-ADON | 18.8 fgh | 13.8 b | 4.0 cd | 5.7 d | 33.6 efg |
| Australia | 15-ADON | 16.6 gh | 13.6 b | 4.3 cd | 4.5 fg | 44.0 c |
| Brazil | NIV | 15.8 fgh | 5.8 c | 0.4 d | 4.1 gh | 32.8 fg |
| Germany | NIV | 14.3 fgh | 4.2 c | 0.3 d | 4.0 gh | 26.2 g |
| China | NIV | 13.0 h | 5.6 c | 1.1 d | 3.4 h | 40.0 cde |
| Australia | NIV | 9.8 fgh | 7.0 bc | 0.3 d | 2.8 h | 26.6 fg |
DS was recorded from wheat cultivar, Carberry and averaged from five replications, FDK and DON/NIV content were measured from a pooled sample obtained from five replications, radial growth and macroconidia production were obtained from three replications. Means with the same letter in the column for each variable are not significantly different.
2.4. Deoxynivalenol/Nivalenol Content in Infected Grains
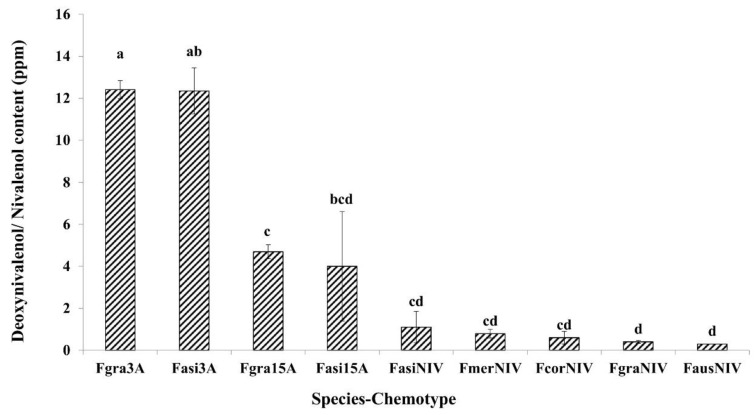
The ANOVA for total DON/NIV content in infected grains showed no significant difference among species, however, there were significant differences among the chemotypes (Table 1). The interaction species*chemotype was also not significant. However, F. gramineraum 3-ADON and F. asiaticum 3-ADON strains produced higher amounts of DON in infected grains compared to other strains in the collection (Figure 2). In terms of DON/NIV content, significant differences were detected among countries and the country*chemotype interaction. The 3-ADON strains from Germany produced higher amounts of DON, followed by the 3-ADON strains from Canada, China, Poland, and UK. NIV producing strains showed the lowest toxin contamination in infected grains (Table 2).
Figure 2.
Comparison of mean deoxynivalenol/nivalenol production by chemotype of different species in the Fusarium graminearum species complex on moderately resistant wheat cultivar Carberry. Means with the same letter for deoxynivalenol/nivalenol content are not significantly different. (Fgra3A—F. graminearum 3-ADON chemotype; Fasi3A—F. asiaticum 3-ADON chemotype; Fgra15A—F. graminearum 15-ADON chemotype; Fasi15A—F. asiaticum 15-ADON chemotype; FasiNIV—F. asiaticum NIV chemotype; FmerNIV—F. meridionale NIV chemotype; FcorNIV—F. cortaderiae NIV chemotype; FgraNIV—F. graminearum NIV chemotype; and FausNIV—F. austroamericanum NIV chemotype). Bars on the top of the each column represent the standard errors.
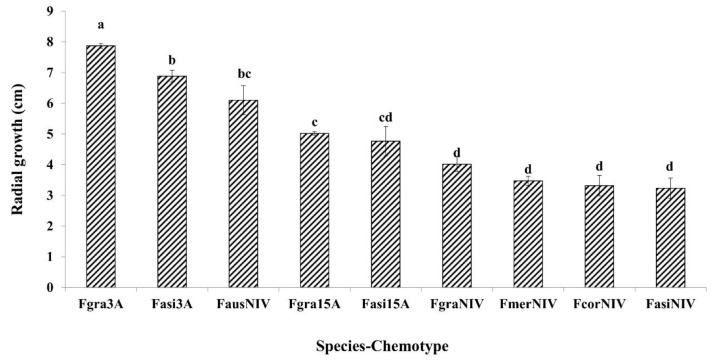
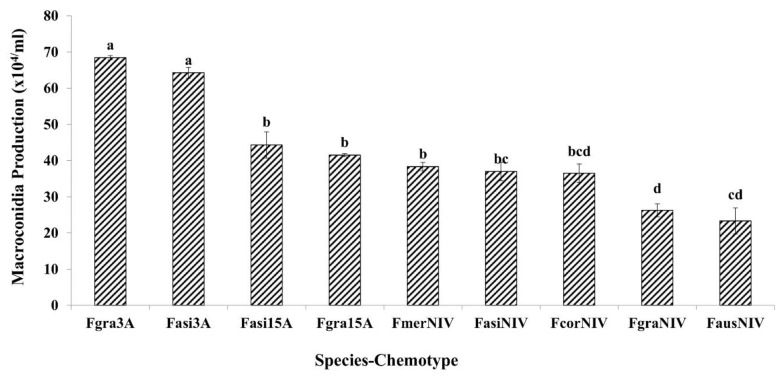
In the present study, significant differences were observed among the species and chemotypes in the FGSC for growth rate (Table 1). The two-way interaction species*chemotype was not significant. F. graminearum s.s 3-ADON, F. asiaticum 3-ADON and F. graminearum 15-ADON showed significantly higher growth rates than other species (Figure 3). Additionally, significant differences in growth rates were observed among the strains originated from different countries and the country*chemotype interaction was statistically significant (Table 2). The 3-ADON producing strains from Germany, UK, Canada, Poland, and China showed higher growth rates than the other chemotypes from the same countries. Similar results were observed for macroconidia production, except the two-way interaction species*chemotype was significantly different. F. graminearum s.s 3-ADON strains showed the highest macroconidia production on SNA media under in vitro conditions followed by F. asiaticum 3-ADON and F. graminearum 15-ADON strains (Figure 4). Canadian 3-ADON strains produced higher amounts of macroconidia under in vitro conditions followed by 3-ADON strains from Germany, Poland, UK and China (Table 2).
Figure 3.
Comparison of mean radial growth by chemotype of different species in the Fusarium graminearum species complex on potato dextrose agar (PDA) media, seven days post-inoculation. Means with the same letter for mean radial growth are not significantly different. (Fgra3A—F. graminearum 3-ADON chemotype; Fasi3A—F. asiaticum 3-ADON chemotype; Fasi15A—F. asiaticum 15-ADON chemotype; Fgra15A—F. graminearum 15-ADON chemotype; FausNIV—F. austroamericanum NIV chemotype; FgraNIV—F. graminearum NIV chemotype; FmerNIV—F. meridionale NIV chemotype; FcorNIV—F. cortaderiae NIV chemotype; and FasiNIV—F. asiaticum NIV chemotype). Bars on the top of each column represent the standard errors.
Figure 4.
Comparison of mean macroconidia production by chemotype of different species in the Fusarium graminearum species complex on synthetic nutrient deficient agar (SNA) media, seven days post-inoculation. Means with the same letter for macroconidia production are not significantly different. (Fgra3A—F. graminearum 3-ADON chemotype; Fasi3A—F. asiaticum 3-ADON chemotype; Fasi15A—F. asiaticum 15-ADON chemotype; Fgra15A—F. graminearum 15-ADON chemotype; FmerNIV—F. meridionale NIV chemotype; FasiNIV—F. asiaticum NIV chemotype; and FcorNIV—F. cortaderiae NIV chemotype, FgraNIV—F. graminearum NIV chemotype; FausNIV—F. austroamericanum NIV chemotype). Bars on the top of each column represent the standard errors.
2.5. Correlation of FHB Disease Variables
To study the association between FHB disease variables, data on 132 strains were included in the correlation analysis. A significant positive correlation was observed between the FHB disease variables (Table 3). A highly significant positive correlation existed between FDK and DON/NIV content (R = 0.84, P < 0.0001). A significant positive correlation was also identified between DS and FDK (R = 0.72, P < 0.0001). Among the examined FHB disease variables, the lowest correlation was observed between the DS and macroconidia production (R = 0.64, P < 0.0001).
Table 3.
Pearson correlation coefficients of fusarium head blight response variables measured under controlled environment conditions and in the laboratory.
| FHB DS% | FDK % 1 | Radial Growth | Macroconidia Production | DON/NIV Content 2 | |
|---|---|---|---|---|---|
| FHB DS% | 1.000 | 0.72 * <0.0001 |
0.67 * <0.0001 |
0.64 * <0.0001 |
0.66 * <0.0001 |
| % FDK | 1.000 | 0.84 * <0.0001 |
0.90 * <0.0001 |
0.84 * <0.0001 |
|
| Radial growth | 1.000 | 0.79 * <0.0001 |
0.75 * <0.0001 |
||
| Macroconidia production | 1.000 | 0.80 * <0.0001 |
|||
| DON/NIV content | 1.000 |
1 Fusarium Damaged Kernel Percentage, 2 Deoxynivalenol or Nivalenol Content * Means correlation coefficient is significant at p < 0.0001.
3. Discussion
In this study we conducted phylogenetic, in vitro and greenhouse experiments to compare the strains representing prevailing populations of the FGSC collected from different countries in relation to their trichothecene chemotype, phenotypic traits, and aggressiveness parameters. This type of comparative assessments for different species of the FGSC has not been exhaustively assessed since the subdivision of species in the FGSC has been proposed by O’Donnell et al. [11]. To our knowledge this is the first study that used Fusarium strains from multiple countries to analyze the phylogenetic relationships, chemotype diversity, and aggressiveness. Therefore, this study helps to understand the population subdivision in FGSC based on differences in geographic location, chemotype diversity, and aggressiveness.
Based on the EF-1α gene sequences six species in the FGSC could be identified in our Fusarium strain collection. All the strains examined from Canada, UK, Germany and Poland, most of the strains from Australia and Brazil and a few strains from China belong to the F. graminearum s.s clade. This finding agrees with other reports that show the wide geographical distribution of F. graminearum s.s among the other FHB species [2,9,17]. The strains from China belong to three different species, F. asiaticum, F. graminearum s.s, and F. meridionale. In Southern China, F. asiaticum is (approximately 97%) responsible for the major component of the FHB complex on wheat grown. However, in North and Northeast China, F. graminearum s.s (approximately 76%) was the most predominant species [29,30]. This uneven distribution of Fusarium species in the FGSC is believed to depend on the different crop rotation practices and temperatures in those regions [30,31]. Zhang et al. [32] reported the occurrence of F. meridionale strains on maize in China for the first time. All the examined strains from Mexico belong to the F. boothii group except for one strain which belongs to F. meridionale. The presence of F. boothii in Mexico has been previously reported by Malihipour et al. [33]. Similarly, all strains from Australia belong to the F. graminearum s.s except for one strain that was identified as F. meridionale (strain CS7220, isolated from wheat). Therefore, the discovery of NIV producing F. meridionale from Australia represents a potential risk for the Australian wheat industry.
Strains from Brazil consist of F. graminearum s.s, F. cortaderiae, F. austroamericanum and F. meridionale. Del Ponte et al. [4] reported the co-occurrence of different species in the F. graminearum species complex in Brazil. They identified five species within FGSC in a multiyear survey of >200 wheat fields in Paraná (PR) and Rio Grande do Sul (RS) states. These five species include F. gramineraum s.s, F. meridionale, F. asiaticum, F. cortderiae, and F. austroamericanum.
All the strains in this collection from European countries such as Germany, UK and Poland were identified as F. graminearum s.s. For example, in the Netherlands and Denmark, F. culmorum used to be the major causative agent of FHB before the year 2000; however, there was a population shift from F. culmorum to F. graminearum in Europe [34,35]. The dominance of F. graminearum s.s in European countries has also been reported by Jennings et al. [36] and Talas et al. [37]. Among the different species in FGSC, all strains of F. boothii were determined to be 15-ADON producers. The PCR assays and chemical analysis done by Malihipour et al. [33] and Sampietro et al. [38] also reported that F. boothii strains are capable of producing DON and 15-ADON. All strains of F. meridionale, F. cortaderiae, and F. austroamericanum were determined to be NIV producers. The trichothecene chemotypes detected in our collection are consistent with other reports of F. meridionale, F. cortaderiae, F. austroamericanum, and F. boothii from other parts of the world including Europe, China, Brazil, and Mexico [18,22,39].
Although many reports have been published on aggressiveness of Fusarium species and F. graminearum strains collected from a specific field or country, only a few studies have analyzed and compared the aggressiveness in species within the FGSC [25,40]. In our study we have evaluated and compared the aggressiveness of strains representing five phylogenetic species of the FGSC collected from different geographical regions. We used the spring wheat cultivar Carberry which expresses moderate resistance to FHB and has marker alleles associated with Fhb1 [41]. To date, most selections in FHB resistant wheat breeding programs worldwide are concentrated on the Fhb1 [42]. Fhb1 is a major FHB resistance gene that is essential to provide Type II resistance in wheat cultivars against F. graminearum [43]. Therefore, use of a cultivar having Fhb1 is important to evaluate the stability of resistance under the influence of different species and chemotypes of F. graminearum. In our study, in terms of F. graminearum species origin, no significant differences were observed for FHB DS, FDK% and DON/NIV content. However, significant differences were observed for radial growth and macroconidia production. Goswami & Kistler [2] also reported a large variation among different strains in terms of aggressiveness and trichothecene production. This variation appeared to be strain-specific rather than species-specific characteristics. In terms of chemotype of strains, significant differences were observed for all analysed FHB response variables. When we analyzed the data based on the country of origin of strains, significant differences were observed for all FHB response variables except for FDK%. Malihipour et al. [33] also compared the aggressiveness of F. graminearum strains from Canada, Iran, and Mexico and found significant differences in aggressiveness of the strains from different geographical regions. In the current study, country*chemotype interaction was also significantly different for FHB DS, DON/NIV content, radial growth and macroconidia production. However, country*chemotype interaction was not significantly different for FDK%. The 3-ADON producing strains from Europe (Germany, UK, and Poland) showed highest FHB DS in cultivar Carberry compared to other strains. When the total DON content in infected grains was considered, F. graminearum s.s 3-ADON showed the highest DON accumulation followed by F. asiaticum 3-ADON, F. graminearum s.s 15-ADON and F. asiaticum 15-ADON producing strains. Additionally, the 3-ADON producing strains from Germany showed the highest DON content followed by Canadian, Chinese and Poland strains. Apart from FHB DS and DON production, 3-ADON strains showed higher levels of FDK%, radial growth and macroconidia production compared to the 15-ADON and NIV strains. In our study, both F. graminearum s.s and F. asiaticum species showed higher aggressiveness than the other species of the FGSC. Other species in the FGSC such as F. meridionale, F. austroamericanum, and F. cortaderiae showed lower aggressiveness towards the MR wheat cultivar, Carberry. One of the reasons may be the production of less phytotoxic NIV by these species.
A study done by Eudes et al. [44] compared the phytotoxicity of eight different trichothecenes and reported that NIV is less phytotoxic than DON. Taken together, the results obtained from this study showed that, 3-ADON producing strains had the highest aggressiveness followed by 15-ADON and NIV producing strains, respectively. Similar results have been reported by Puri & Zhong [45], Malihipour et al. [33], Spolti et al. [46], and Zhang et al. [30] in which they also explained the higher aggressiveness of 3-ADON producing strains over 15-ADON and NIV strains. The lowest aggressiveness of NIV producing strains have also been reported in other studies [2,47]. The higher aggressiveness of 3-ADON producing strains in terms of DON production, mycelial growth, and macroconidia production may provide a fitness advantage over other chemotypes, suggesting that selection was driving the rapid spread of 3-ADON over 15-ADON and NIV chemotypes. Therefore, the higher aggressiveness and possibility of production of higher DON by 3-ADON strains in wheat cultivars introgressed with Fhb1, is a concern due to the rapid increase in 3-ADON producing strains in Canada and North America [21,48,49].
A significant correlation was observed between the amount of DON/NIV produced by each strain and its level of aggressiveness on wheat in terms of DS, FDK%, growth, and macroconidia production. Therefore, this study suggests that the type and amount of trichothecenes produced by a strain functions as a major determinant of aggressiveness on wheat. Additionally, our results are in agreement with previous reports showing that trichothecenes have a major role in determining the aggressiveness of the pathogen [37,50]. In terms of species origin of strains, we did not find a clear association between the species of a strain and its aggressiveness. However, the aggressiveness of strains may be partially based on species and population-specific features [51]. Therefore, systematic testing of many more strains representing different species, populations, and toxin types within the FGSC is required to understand this relationship in more detail.
In this study, we characterized the phylogenetic relationships, chemotype patterns, and aggressiveness of Fusarium strains in the FGSC collected from different regions, and evaluated the association between their phylogenetic and/or chemotype patterns with aggressiveness. The existence of high variability in Fusarium strains may explain the reasons for different reactions of wheat genotypes in different locations. Therefore, the results of the current study also suggest that screening for FHB resistance may require the use of highly aggressive strains or a mixture of strains representative of the FGSC diversity in order to develop durable FHB resistant wheat cultivars. Gaining knowledge of the phylogenetic relationships and trichothecene chemotype diversity in FGSC populations from major cereal producing regions/countries are also important to assess the sustainability of currently used resistant cultivars and fungicides. Further, it helps to establish quarantine regulations to limit pathogen spread among cereal growing regions.
4. Materials and Methods
4.1. Fusarium Strain Collection
One hundred and fifty Fusarium strains obtained from eight countries were included in this study (Supplementary Table S1). Strains were collected to represent different species in the FGSC from different regions in the world. All strains are stored in the culture collection maintained at Department of Plant Science, University of Manitoba, Canada.
4.2. DNA Extraction
All strains were grown on potato dextrose agar (PDA) (Difco Laboratories, ON, Canada) plates at 24–25 °C, under fluorescent light for 7 days and genomic DNA was extracted from the freeze-dried aerial mycelium using a modified cetyltrimethyl ammonium bromide (CTAB) based protocol described by Lodhiet al. [52]. The lyophilized mycelium (~200 mg) was ground in 600 µL of TES buffer (100 mM Tris, 10 mM EDTA, 2% SDS). Then 140 µL of 5M NaCl and 10% CTAB were added and vortexed. The mixture was incubated at 65 °C for 20 min. Protein was removed by adding 600 µL of a mixture of phenol:chloroform:isoamylalchohol (25:24:1). The mixture was centrifuged at 10,000 rpm for 15 min. This step was repeated twice. DNA was precipitated with 80 µL of 5M NaCl and 1000 µL of 95% ethanol. The mixture was centrifuged at 13,000 rpm for 5 min. The DNA pellet was washed using 200 µL of 80% ethanol. After the pellet dried, it was dissolved in 100 µL of autoclaved distilled water. DNA was treated with RNase (0.75% vol/vol). DNA was quantified using the NanoDrop3300 (Thermo Fisher Scientific Inc., Waltham, MA, USA). DNA was diluted using sterilized distilled water for a final concentration of 50 ng/μL and stored at −20 °C until further used.
4.3. PCR Assay and DNA Sequencing
Sequence analysis was performed for PCR-amplified fragments of EF-1α gene that has been used previously to discriminate between species in the FGSC. The primers used to amplify gene fragments are listed in Supplementary Table S2. The PCR reactions were performed in a 25 μL volume containing 20 ng of template DNA, 2.0 mM MgCl2, 50 mM KCl, 10 mM Tris HCl (pH 8.0), 0.2 mM each dNTP (Invitrogen Life Technologies, Carlsbad, CA, USA), 0.4 μM each primer, and 0.75 units of Taq DNA polymerase (Invitrogen Life Technologies, CA, USA). The PCR amplification protocol consisted of an initial denaturation at 94 °C for 2 min, followed by 30 cycles of 30 s at 94 °C, 60 s at annealing temperature, 1 min at 68 °C, and a final extension of 68 °C for 10 min. The annealing temperatures for the PCR amplification was 52 °C. For sequence analysis, PCR amplified DNA fragments were purified with Exosap-IT (Affymetrix Inc., Santa Clara, CA, USA) PCR product clean up kit according to the manufacturer’s instructions. Sequencing reactions were prepared using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA). Finally, the sequencing products were analyzed on an Applied Biosystems 3730xl DNA Analyzer (Applied Biosystems, CA, USA) at the University of Kentucky Advanced Genetic Technologies Center.
4.4. Molecular Phylogenetic Analysis
EF-1α gene sequences were assembled, trimmed and edited using GENEIOUS v. 5.4.5 [53]. Sequences were aligned manually using BIOEDIT v. 7.1.3 sequence alignment editor [54]. The final data set had an aligned length of 679 bp for EF-1α gene. Genetic distances and a test for base composition heterogeneity were performed using PAUP* v. 4.0 b10 [55]. A chi-square test of homogeneity of base frequencies across taxa was used to estimate the frequency distribution of observed number of substitutional changes per character. Base composition of aligned sequences was calculated in MEGA 5.1 beta version [56]. The Kimura 2-parameter plus Gamma (K80+G) and symmetrical (SYM) models were determined as the best fitting models of nucleotide substitution using the hierarchical likelihood ratio tests (hLRT) for EF-1α gene [57]. The model selection test was carried out using MODELTEST v. 3.7 [58]. Bayesian phylogenetic inference was performed using MrBayes v. 3.1.2 [59] with two independent runs with four chains each with default priors and run for 107generations. Chains were sampled every 1000th generation. Convergence of all parameters and correct mixing of chains were confirmed by examining the likelihood of plots for each run and when the average standard deviation of split frequencies was less than 0.02 [60]. Trees were summarized from the two independent searches using majority rule consensus after discarding 25% of the samples for burn-in. Calculation of the consensus tree and of the posterior probabilities of clades were done based upon the trees sampled after the burn-in, and trees were compiled and drawn using FIGTREE v. 1.3.1 [61]. F. pseudograminearum strain NRRL 34426 and F. culmorum strain NRRL 52792 were used as out-groups for EF-1α gene phylogenetic tree reconstruction. Additionally, sequences from Agricultural Research Services ARS (NRRL) culture collection reference strains were downloaded from GenBank and incorporated into each alignment.
4.5. PCR Assays to Determine Trichothecene Chemotypes
All primers used for the PCR-based identification of mycotoxin biosynthetic genes, along with references are presented in Supplementary Table S2. Chemotype identification of each strain was performed using multiplex PCR primers 3CON, 3NA, 3D3A, 3D15A described by Ward et al. [20]. The multiplex PCR primers amplified an 840 bp fragment from NIV producing strains, a 610 bp fragment from 15-ADON producers and a 243 bp fragment from 3-ADON producers, respectively. The PCR reactions were performed in 15 μL volumes containing 20 ng of template DNA, 2.0 mM MgCl2, 50 mM KCl, 10 mM Tris HCl (pH 8.0), 0.2 mM each dNTP (Invitrogen Life Technologies, Carlsbad, CA, USA), 0.4 μM each primer, and 0.75 units of Taq DNA polymerase (Invitrogen Life Technologies, CA, USA). The PCR cycling conditions for multiplex PCR consisted of an initial denaturation at 94 °C for 4 min, followed by 35 cycles of 1 min at 94 °C, 40 s at 52 °C, 40 s at 72 °C, and a final extension of 72 °C for 6 min. The PCR amplicons were separated on a 2% agarose gel stained with RedSafe nucleic acid staining solution (iNtRON Biotechnology Inc., FroggaBio, Toronto, ON, Canada).
4.6. Radial Growth and Macroconidia Production
The radial growth rate of each strain was evaluated in vitro on PDA plates in three replications. A small agar plug (5 mm in diameter) of each strain was placed in the center of a 9 cm PDA plate. The cultures were grown at 24 °C with a 24 h photoperiod. The average growth was measured every 48 h for six days from three replicates in two perpendicular directions. Average colony diameter was used to calculate the growth rate of the strain.
The macroconidia production was assessed based on the colonies growing on synthetic nutrient agar (SNA) media during seven days at room temperature and a 24 h photoperiod. After seven days of growth, conidia were harvested by adding 1 mL of sterilized distilled water and scraping the surface of SNA media with a sterilized scalpel. Number of conidia was determined using a haemocytometer and the conidial concentration was expressed in conidia/mL.
4.7. Aggressiveness Experiments and Mycotoxin Analysis
One hundred and thirty three strains listed in Supplementary Table S1 were individually inoculated on FHB MRwheat cultivar Carberry (except for the strains obtained from Mexico). Wheat plants were grown in plastic pots containing Sunshine Mix (Sun Gro Horticulture Ltd., Agawam, MA, USA). The pots were arranged in a completely randomized design with five replicates (one plant per pot). Inoculations were done using the dual floret point inoculation method. For each wheat plant, 4–5 spikes were inoculated once individual spikes were close to 50% anthesis. The point of inoculation on each spike was determined by calculating the total spikelets per spike and multiplied by two-thirds. Each spike was inoculated by injecting 10 µl of a macroconidial suspension (5 × 104 spores/mL) between the lemma and palea of a floret. To facilitate infection, high humidity (>80%) was maintained around the inoculated spikes by placing a glassine bag over the spike. The bag was removed 48 h post-inoculation. FHB DSwas rated 14 days post-inoculation using a pictorial scale reported by Stack and McMullen [62].
After maturity, inoculated spikes were hand harvested and threshed using a belt thresher. A 10 g random seed sample was selected from pooled replicates and FDK% was counted as the number of FDKs in the total number of seeds. A FDK was considered to be any seed that was shrivelled, had any mycelial growth, or a chalky white or pink discoloration.
DON and NIV analysis was carried out using the same sample used for FDK determination. The samples were ground to a powder of similar consistency using a coffee grinder (Cuisinart model no: DCG20BKNC) for 5–8 min. DON was extracted using 50 mL of deionized water, and then quantified using Veratox DON 5/5 kit (product no: 8331) supplied by Neogen (Lansing, MI, USA) with a DON quantification limit (LOQ) of 0.1 ppm. The amount of NIV (LOQ-0.06 ppm) in infected grains was analyzed by GC-MS according to the protocol described by Amarasinghe et al. [49].
4.8. Statistical Analysis
Analysis of variance (ANOVA) for FHB DS, FDK%, DON/NIV content, radial growth, and macroconidia production was performed using ‘PROC MIXED’ procedure of the SAS software (SAS version 9.3, SAS Institute Inc., Cary, NC, USA). Because this data set has an unequal distribution of sample sizes, it was not possible to analyze the data set including all effects and their interactions. Therefore, to determine the effect of species and chemotype on FHB disease variables, data were analyzed using the following model statement; species country chemotype rep species*chemotype. A second data analysis was performed to determine the effect of country of origin and chemotype on FHB disease variables. The model statement for this analysis consisted of species country chemotype rep country*chemotype. The species, country, chemotype and their interactions (species*chemotype and country*chemotype) were considered as fixed effects and replicated as a random effect. The correlation between FHB response variables were analyzed using the SAS PROC CORR (SAS version 9.3, SAS Institute Inc., Cary, NC, USA) procedure.
Acknowledgments
Authors would like to acknowledge all the researchers who provided strains for this study and James Tucker for critically reviewing the manuscript before submission.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6651/11/5/263/s1. Table S1: Strain code, species, geographic origin, host and trichothecene chemotype of Fusarium graminearum species complex strains used in the study; Table S2: List of primers used to identify the species and chemotypes in the Fusarium graminearum species complex strains.
Author Contributions
C.A. and W.G.D.F. conceptualized the hypothesis and work; C.A. conducted experiments and wrote the manuscript. B.S.; provided guidance in phylogenetic analysis and sequence analysis and revised the manuscript. W.G.D.F.; received the funding, supervised the project and revised the manuscript.
Funding
This research was funded by Growing Forward 2-National Wheat Improvement funding program (WGRF), The CAP Program (2018 to 2023) by WGRF, the Manitoba Growing Forward 2 funding program and the Natural Science and Engineering Research Council of Canada Discovery (NSERC-Discovery) funding to W.G.D.F.
Conflicts of Interest
The authors declare no conflict of interest.
Key Contribution
This study compared the aggressiveness of species within the F. graminearum species complex. No significant differences were observed for DS, FDK%, and DON/NIV content. However, significant differences were observed based on the type of trichothecenes produced by the strains. Strains producing 3-ADON showed higher levels of DS, FDK%, and DON content compared to 15-ADON and NIV producing strains.
References
- 1.McMullen M., Jones R., Gallenberg D., America S. Scab of Wheat and Barley: A Re-emerging Disease of Devastating Impact. Plant Dis. 1997;81 doi: 10.1094/PDIS.1997.81.12.1340. [DOI] [PubMed] [Google Scholar]
- 2.McMullen M., Bergstrom G., De Wolf E., Dill-Macky R., Hershman D., Shaner G., Van Sanford D. A unified effort to fight an enemy of wheat and barley: Fusarium head blight. Plant Dis. 2012;96:1712–1728. doi: 10.1094/PDIS-03-12-0291-FE. [DOI] [PubMed] [Google Scholar]
- 3.Obanor F., Neate S., Simpfendorfer S., Sabburg R., Wilson P., Chakraborty S. Fusarium graminearum and Fusarium pseudograminearum caused the 2010 head blight epidemics in Australia. Plant Pathol. 2013;62:79–91. doi: 10.1111/j.1365-3059.2012.02615.x. [DOI] [Google Scholar]
- 4.Del Ponte E., Spolti P., Ward T., Gomes L.B., Nicolli C.P., Kuhnem P.R., Silva C.N., Tessmann D.J. Regional and field-specific factors affect the composition of Fusarium head blight pathogens in subtropical no-till wheat agroecosystem of Brazil. Phytopathology. 2015;105:246–254. doi: 10.1094/PHYTO-04-14-0102-R. [DOI] [PubMed] [Google Scholar]
- 5.Foroud N.A., Eudes F. Trichothecenes in cereal grains. Int. J. Mol. Sci. 2009;10:147–173. doi: 10.3390/ijms10010147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller J.D., Greenhalgh R., Wang Y., Lu M., Mycologia S., Apr N.M. Trichothecene Chemotypes of Three Fusarium Species. Mycologia. 1991;83:121–130. doi: 10.1080/00275514.1991.12025988. [DOI] [Google Scholar]
- 7.Ward T.J., Bielawski J.P., Kistler H.C., Sullivan E., O’Donnell K. Ancestral polymorphism and adaptive evolution in the trichothecene mycotoxin gene cluster of phytopathogenic Fusarium. Proc. Natl. Acad. Sci. USA. 2002;99:9278–9283. doi: 10.1073/pnas.142307199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Donnell K., Ward T.J., Geiser D.M., Kistler H.C., Aoki T. Genealogical concordance between the mating type locus and seven other nuclear genes supports formal recognition of nine phylogenetically distinct species within the Fusarium graminearum clade. Fungal Genet. Biol. 2004;41:600–623. doi: 10.1016/j.fgb.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 9.Van der Lee T., Zhang H., van Diepeningen A., Waalwijk C. Biogeography of Fusarium graminearum species complex and chemotypes: A review. Food Addit. Contam. Part A. 2015;32:1–8. doi: 10.1080/19440049.2014.984244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ichinoe M., Kurata H., Sugiura Y., Ueno Y. Chemotaxonomy of Gibberella zeae with special reference to production of trichothecenes and zearalenone. Appl. Environ. Microbiol. 1983;46:1346–1369. doi: 10.1128/aem.46.6.1364-1369.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Donnell K., Kistler H.C., Tacke B.K., Casper H.H. Gene genealogies reveal global phylogeographic structure and reproductive isolation among lineages of Fusarium graminearum, the fungus causing wheat scab. Proc. Natl. Acad. Sci. USA. 2000;97:7905–7910. doi: 10.1073/pnas.130193297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leslie J.F., Summerell B.A. The Fusarium Laboratory Manual. Wiley-Blackwell; Ames, IA, USA: 2006. pp. 81–96. [Google Scholar]
- 13.Starkey D.E., Ward T.J., Aoki T., Gale L.R., Kistler H.C., Geiser D.M., Suga H., Toth B., Varga J., O’Donnell K. Global molecular surveillance reveals novel Fusarium head blight species and trichothecene toxin diversity. Fungal Genet. Biol. 2007;44:1191–1204. doi: 10.1016/j.fgb.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Yli-Mattila T., Gagkaeva T., Ward T.J., Aoki T., Kistler H.C., O’Donnell K. A novel Asian clade within the Fusarium graminearum species complex includes a newly discovered cereal head blight pathogen from the Russian Far East. Mycologia. 2009;101:841–852. doi: 10.3852/08-217. [DOI] [PubMed] [Google Scholar]
- 15.Gale L.R., Harrison S.A., Ward T.J., O’Donnell K., Milus E.A., Gale S.W., Kistler H.C. Nivalenol-type populations of Fusarium graminearum and F. asiaticum are prevalent on wheat in southern Louisiana. Phytopathology. 2011;101:124–134. doi: 10.1094/PHYTO-03-10-0067. [DOI] [PubMed] [Google Scholar]
- 16.Sarver B.A.J., Ward T.J., Gale L.R., Broz K., Kistler C.H., Aoki T., Nicholson P., Carter J., O’Donnell K. Novel Fusarium head blight pathogens from Nepal and Louisiana revealed by multilocus genealogical concordance. Fungal Genet. Biol. 2011;48:1096–1107. doi: 10.1016/j.fgb.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 17.Wang J.H., Ndoye M., Zhang J.B., Li H.P., Liao Y.C. Population Structure and Genetic Diversity of the Fusarium graminearum Species Complex. Toxins. 2011;3:1020–1037. doi: 10.3390/toxins3081020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desjardins A.E., Proctor R.H. Genetic diversity and trichothecene chemotypes of the Fusarium graminearum clade isolated from maize in Nepal and identification of a putative new lineage. Fungal Biol. 2011;115:38–48. doi: 10.1016/j.funbio.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 19.Varga E., Wiesenberger G., Hametner C., Ward T.J., Dong Y., Schöfbeck D., McCormick S., Broz K., Stückler R., Schuhmacher R., et al. New tricks of an old enemy: Isolates of Fusarium graminearum produce a type A trichothecene mycotoxin. Environ. Microbiol. 2015;17:2588–2600. doi: 10.1111/1462-2920.12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ward T.J., Clear R.M., Rooney A.P., O’Donnell K., Gaba D., Patrick S., Starkey D.E., Gilbert J., Geiser D.M., Nowicki T.W. An adaptive evolutionary shift in Fusarium head blight pathogen populations is driving the rapid spread of more toxigenic Fusarium graminearum in North America. Fungal Genet. Biol. 2008;45:473–484. doi: 10.1016/j.fgb.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 21.Liang J.M., Xayamongkhon H., Broz K., Dong Y., McCormick S.P., Abramova S., Ward T.J., Ma Z.H., Kistler H.C. Temporal dynamics and population genetic structure of Fusarium graminearum in the upper Midwestern United States. Fungal Genet. Biol. 2014;73:83–92. doi: 10.1016/j.fgb.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 22.Boutigny A.L., Ward T.J., Ballois N., Iancu G., Ioos R. Diversity of the Fusarium graminearum species complex on French cereals. Eur. J. Plant Pathol. 2014;138:133–148. doi: 10.1007/s10658-013-0312-6. [DOI] [Google Scholar]
- 23.Qiu J., Shi J. Genetic relationships, Carbendazim sensitivity and mycotoxin production of the Fusarium graminearum populations from maize, wheat and rice in eastern china. Toxins. 2014;6:2291–2309. doi: 10.3390/toxins6082291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miedaner T., Reinbrecht C. Trichothecene content of rye and wheat genotypes inoculated with a deoxynivalenol- and a nivalenol-producing isolate of Fusarium culmorum. J. Phytopathol. 2001;149:245–251. doi: 10.1046/j.1439-0434.2001.00608.x. [DOI] [Google Scholar]
- 25.Tóth B., Mesterházy Á., Horváth Z., Bartók T., Varga M., Varga J. Genetic variability of central European isolates of the Fusarium graminearum species complex. Eur. J. Plant Pathol. 2005;113:35–45. doi: 10.1007/s10658-005-0296-y. [DOI] [Google Scholar]
- 26.Miedaner T., Schilling A.G. Genetic variation of aggressiveness in individual field populations of Fusarium graminearum and Fusarium culmorum tested on young plants of winter rye. Eur. J. Plant Pathol. 1996;102:823–830. doi: 10.1007/BF01877051. [DOI] [Google Scholar]
- 27.Akinsanmi O.A., Backhouse D., Simpfendorfer S., Chakraborty S. Genetic diversity of Australian Fusarium graminearum and F. pseudograminearum. Plant Pathol. 2006;55:494–504. doi: 10.1111/j.1365-3059.2006.01398.x. [DOI] [Google Scholar]
- 28.Miedaner T., Cumagun C.J.R., Chakraborty S. Population genetics of three important head blight pathogens Fusarium graminearum, F. pseudograminearum and F. culmorum. J. Phytopathol. 2008;156:129–139. doi: 10.1111/j.1439-0434.2007.01394.x. [DOI] [Google Scholar]
- 29.Qu B., Li H.P., Zhang J.B., Huang T., Carter J., Liao Y.C., Nicolson P. Comparison of genetic diversity and pathogenicity of fusarium head blight pathogens from China and Europe by SSCP and seedling assays on wheat. Plant Pathol. 2008;57:642–651. doi: 10.1111/j.1365-3059.2008.01824.x. [DOI] [Google Scholar]
- 30.Zhang H., van der Lee T., Waalwijk C., Chen W., Xu J., Xu J., Zhang Y., Feng J. Population analysis of the Fusarium graminearum species complex from wheat in China show a shift to more aggressive isolates. PLoS ONE. 2012;7:e31722. doi: 10.1371/journal.pone.0031722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J.B., Li H.P., Dang F.J., Qu B., Xu Y.B., Zhao C.S., Liao C.A. Determination of the trichothecene mycotoxin chemotypes and associated geographical distribution and phylogenetic species of the Fusarium graminearum clade from China. Mycol. Res. 2007;111:967–997. doi: 10.1016/j.mycres.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 32.Zhang H., Luo W., Pan Y., Xu J., Xu S.J., Chen W.Q., Feng J. First report of Fusarium Maize Ear Rot caused by Fusarium meridionale in China. Plant Dis. 2014;98:1156. doi: 10.1094/PDIS-01-14-0108-PDN. [DOI] [PubMed] [Google Scholar]
- 33.Malihipour A., Gilbert J., Canada A., Rt M.B. Molecular Phylogenetic Analysis, Trichothecene Chemotype Patterns, and Variation in Aggressiveness of Fusarium Isolates Causing Head Blight in Wheat. Plant Dis. 2012;96:1016–1025. doi: 10.1094/PDIS-10-11-0866-RE. [DOI] [PubMed] [Google Scholar]
- 34.Nielsen L.K., Jensen J.D., Nielsen G.C., Jensen J.E., Spliid N.H., Thomsen I.K., Justesen A.F., Collinge D.B., Jørgensen L.N. Fusarium head blight of cereals in Denmark: Species complex and related mycotoxins. Phytopathology. 2011;101:960–969. doi: 10.1094/PHYTO-07-10-0188. [DOI] [PubMed] [Google Scholar]
- 35.Waalwijk C., Kastelein P., de Vries I., Kerényi Z., van Der Lee T., Hesselink T., Kohl J., Kema G. Major changes in Fusarium spp. in wheat in the Netherlands. Eur. J. Plant Pathol. 2003;109:743–754. doi: 10.1023/A:1026086510156. [DOI] [Google Scholar]
- 36.Jennings P., Coates M.E., Walsh K., Turner J.A., Nicholson P. Determination of deoxynivalenol- and nivalenol-producing chemotypes of Fusarium graminearum isolated from wheat crops in England and Wales. Plant Pathol. 2004;53:643–652. doi: 10.1111/j.0032-0862.2004.01061.x. [DOI] [Google Scholar]
- 37.Talas F., Kalih R., Miedaner T. Isolates for Aggressiveness and Deoxynivalenol Production in Wheat Head Blight. Phytopathology. 2012;102:128–134. doi: 10.1094/PHYTO-06-11-0162. [DOI] [PubMed] [Google Scholar]
- 38.Sampietro D.A., Aristimuño M.E., Jimenez C.M., Vattuone M.A., Catalán C.A. Trichothecene genotypes and chemotypes in Fusarium graminearum complex strains isolated from maize fields of northwest Argentina. Int. J. Food Microbiol. 2012;153:229–233. doi: 10.1016/j.ijfoodmicro.2011.10.029. [DOI] [PubMed] [Google Scholar]
- 39.Scoz L.B., Astolfi P., Reartes D.S., Schmale D.G., Moraes M.G., Del Ponte E.M. Trichothecene mycotoxin genotypes of Fusarium graminearum sensu stricto and Fusarium meridionale in wheat from southern Brazil. Plant Pathol. 2009;58:344–351. doi: 10.1111/j.1365-3059.2008.01949.x. [DOI] [Google Scholar]
- 40.Alvarez C.L., Somma S., Moretti A., Fernández Pinto V. Aggressiveness of Fusarium graminearum sensu stricto isolates in wheat kernels in Argentina. J. Phytopathol. 2010;158:173–181. doi: 10.1111/j.1439-0434.2009.01596.x. [DOI] [Google Scholar]
- 41.DePauw R.M., Singh A., Singh A.K., Knox R.E., Singh D., Bhavani S., Fetch T. The presence of SrCad and Sr2 influences reaction to stripe rust and Fusarium head blight; Proceedings of the BGRI Technical Workshop; St. Paul, MN, USA. 13–16 June 2011. [Google Scholar]
- 42.Rawat N., Pumphery M.O., Liu S., Zhang X., Tiwari V.K., Ando K., Trick H.N., Bockus W.W., Akhunov E., Anderson J.A., et al. Wheat Fhb1 encodes a chimeric lectin with agglutinin domains and a pore-forming toxin-like domain conferring resistance to Fusarium head blight. Nat. Genet. 2016;48:1576–1580. doi: 10.1038/ng.3706. [DOI] [PubMed] [Google Scholar]
- 43.Cuthbert P.A., Somers D.J., Thomas J., Cloutier S., Brule-babel A. Fine mapping Fhb1, a major gene controlling fusarium head blight resistance in bread wheat (Triticum aestivum L.) Theor. Appl. Genet. 2006;112:1465–1472. doi: 10.1007/s00122-006-0249-7. [DOI] [PubMed] [Google Scholar]
- 44.Eudes F., Comeau A., Rioux S., Collin J. Phytotoxicity of eight mycotoxins associated with the fusariosis of wheat spikelets. Can. J. Plant Pathol. 2000;22:286–292. doi: 10.1080/07060660009500477. [DOI] [Google Scholar]
- 45.Puri K.D., Zhong S. The 3ADON population of Fusarium graminearum found in North Dakota is more aggressive and produces a higher level of DON than the prevalent 15ADON population in spring wheat. Phytopathology. 2010;100:1007–1014. doi: 10.1094/PHYTO-12-09-0332. [DOI] [PubMed] [Google Scholar]
- 46.Spolti P., Barros N.C., Gomes L.B., dos Santos J., Del Ponte E.M. Phenotypic and pathogenic traits of two species of the Fusarium graminearum complex possessing either 15-ADON or NIV genotype. Eur. J. Plant Pathol. 2012;133:621–629. [Google Scholar]
- 47.Cumagun C.J.R., Bowden R.L., Jurgenson J.E., Leslie J.F., Miedaner T. Genetic mapping of pathogenicity and aggressiveness of Gibberella zeae Fusarium graminearum toward wheat. Phytopathology. 2004;94:520–526. doi: 10.1094/PHYTO.2004.94.5.520. [DOI] [PubMed] [Google Scholar]
- 48.Guo X.W., Fernando W.G.D., Seow-Brock H.Y. Population Structure, Chemotype Diversity, and Potential Chemotype Shifting of Fusarium graminearum in Wheat Fields of Manitoba. Plant Dis. 2008;92:756–762. doi: 10.1094/PDIS-92-5-0756. [DOI] [PubMed] [Google Scholar]
- 49.Amarasinghe C.C., Tittlemier S.A., Fernando W.G.D. Nivalenol-producing Fusarium cerealis associated with fusarium head blight in winter wheat in Manitoba, Canada. Plant Pathol. 2015;64:988–995. doi: 10.1111/ppa.12329. [DOI] [Google Scholar]
- 50.McCromick S. The role of DON in pathogenicity. In: Leonard K.J., Bushnell W.R., editors. Fusarium Head Blight of Wheat and Barley. The American Phytopathological Society; St. Paul, MN, USA: 2003. pp. 35–43. [Google Scholar]
- 51.Umpiérrez-failache M., Garmendia G., Pereyra S., Rodríguez-haralambides A., Ward T.J., Vero S. Regional differences in species composition and toxigenic potential among Fusarium head blight isolates from Uruguay indicate a risk of nivalenol contamination in new wheat production areas. Int. J. Food Microbiol. 2013;166:135–140. doi: 10.1016/j.ijfoodmicro.2013.06.029. [DOI] [PubMed] [Google Scholar]
- 52.Lodhi M.A., Ye G.-N., Weeden N.F., Reish B.I. A simple and Efficient Method for DNA Extraction from Grapevine Cultivars and Vitis Species. Plant Mol. Biol. Rep. 1994;12:6–13. doi: 10.1007/BF02668658. [DOI] [Google Scholar]
- 53.Drummond A.J., Ashton B., Buxton S., Cheung M., Cooper A., Duran C., Field M., Heled J., Kearse M., Markowitz S., et al. GENEIOUS v. 5.4.5. [(accessed on 12 February 2015)];2011 Available online: http://www.geneious.com.
- 54.Hall T.A. BIOEDIT: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 55.Swofford D.L. PAUP: Phylogenetic Analysis Using Parsimony (and Other Methods) Sinauer Associates; Sunderland, MA, USA: 2000. Version 4.0. [Google Scholar]
- 56.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA 5: Molecular genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tamura K., Nei M. Estimation of the number of base nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 58.Posada D., Crandall K.A. MODELTEST: Testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 59.Huelsenbeck J.P. Bayesian Phylogenetic Model Selection Using Reversible Jump Markov chain Monte Carlo. Mol. Biol. Evol. 2004;21:1123–1133. doi: 10.1093/molbev/msh123. [DOI] [PubMed] [Google Scholar]
- 60.Huelsenbeck J.P., Larget B., Miller R.E., Ronquist F. Potential applications and pitfalls of Bayesian inference of phylogeny. Syst. Biol. 2002;51:673–688. doi: 10.1080/10635150290102366. [DOI] [PubMed] [Google Scholar]
- 61.Rambaut A. FigTree v1.4.2. Molecular Evolution, Phylogenetics and Epidemiology. University of Edinburgh, Institute of Evolutionary Biology; Edinburgh, Scotland: 2014. [(accessed on 5 March 2015)]. [WWW document] Available online: http://tree.bio.ed.ac.uk/software/figtree/ [Google Scholar]
- 62.Stack R., McMullen M. A visual scale to estimate severity of Fusarium head blight of wheat. [(accessed on 7 September 2015)];1995 Available online: http://www.ag.ndsu.edu/pubs/plantsci/smgrains/pp1095.pdf.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.