Abstract
Brahma‐related gene 1 (BRG1) is one of two mutually exclusive ATPases that function as the catalytic subunit of human SWItch/Sucrose NonFermentable (SWI/SNF) chromatin remodeling enzymes. BRG1 has been identified as a tumor suppressor in some cancer types but has been shown to be expressed at elevated levels, relative to normal tissue, in other cancers. Using TCGA (The Cancer Genome Atlas) prostate cancer database, we determined that BRG1 mRNA and protein expression is elevated in prostate tumors relative to normal prostate tissue. Only 3 of 491 (0.6%) sequenced tumors showed amplification of the locus or mutation in the protein coding sequence, arguing against the idea that elevated expression due to amplification or expression of a mutant BRG1 protein is associated with prostate cancer. Kaplan‐Meier survival curves showed that BRG1 expression in prostate tumors inversely correlated with survival. However, BRG1 expression did not correlate with Gleason score/International Society of Urological Pathology (ISUP) Grade Group, indicating it is an independent predictor of tumor progression/patient outcome. To experimentally assess BRG1 as a possible therapeutic target, we treated prostate cancer cells with a biologic inhibitor called ADAADi (active DNA‐dependent ATPase A Domain inhibitor) that targets the activity of the SNF2 family of ATPases in biochemical assays but showed specificity for BRG1 in prior tissue culture experiments. The inhibitor decreased prostate cancer cell proliferation and induced apoptosis. When directly injected into xenografts established by injection of prostate cancer cells in mouse flanks, the inhibitor decreased tumor growth and increased survival. These results indicate the efficacy of pursuing BRG1 as both an indicator of patient outcome and as a therapeutic target.
Keywords: BRG1, chromatin remodeling, Gleason score, prostate cancer, SMARCA4, SWI/SNF
1. INTRODUCTION
Prostate cancer, the most common cancer in men, is the second leading cause of cancer death in men in the United States, and is the fifth leading cause of cancer death in men worldwide (http://globocan.iarc.fr/old/FactSheets/cancers/prostate-new.asp; https://www.cancer.org/cancer/prostate-cancer.html). Though treatable, definitive understanding of the molecular origins of prostate cancer is lacking, and there is considerable controversy about screening methods (Dong & Ji, 2017; Kinsella et al., 2018; Lee & Shen, 2015; Packer & Maitland, 2016; Tabayoyong & Abouassaly, 2015; Wang, Toivanen, Bergren, Chambon, & Shen, 2014). Continued efforts to identify molecular markers that distinguish between tumors that will remain latent, progress slowly, or progress aggressively are needed (Arriaga‐Canon et al., 2018; Filella, Fernandez‐Galan, Fernandez Bonifacio, & Foj, 2018).
There are two closely related, mutually exclusive ATPases that function as the catalytic subunits of the human/mammalian SWItch/Sucrose NonFermentable (SWI/SNF) chromatin remodeling enzymes, termed Brahma‐related gene 1 (BRG1) and Brahma (BRM; Imbalzano, Kwon, Green, & Kingston, 1994; Khavari, Peterson, Tamkun, Mendel, & Crabtree, 1993; Kwon, Imbalzano, Khavari, Kingston, & Green, 1994; Muchardt & Yaniv, 1993; W. Wang et al., 1996). These enzymes are widely utilized in the cell to regulate gene expression, replication, repair, recombination, and higher‐order genome organization. Not surprisingly, both enzymes have been implicated in diverse types of cancer (Hodges, Kirkland, & Crabtree, 2016; Savas & Skardasi, 2018; Wu et al., 2017). There is considerable evidence that both the BRG1 and the BRM enzymes are mutated or exhibit altered expression in many cancers, but to date, the evidence suggests that the consequences of these changes vary widely depending on the type of cancer.
Loss of BRG1 function has been shown in a number of cancers, most notably small cell carcinoma of the ovary, hypercalcemic type (Jelinic et al., 2014; Kupryjanczyk et al., 2013; Ramos et al., 2014; Witkowski et al., 2014), and non–small‐cell lung cancers (Fukuoka et al., 2004; Medina et al., 2004; Reisman, Sciarrotta, Wang, Funkhouser, & Weissman, 2003), and results in loss of a number of cell functions related to tumor suppression. BRM loss has been implicated in a number of tumor types, and the idea of targeting BRM in BRG1‐deficient cancers has received attention as a potential therapeutic strategy (Hohmann & Vakoc, 2014; Oike et al., 2013; Wilson et al., 2014). In contrast, there is emerging evidence that BRG1 is expressed at elevated levels in some tumors relative to normal tissue. This includes disparate tumor types such as breast cancer, melanoma, neuroblastoma, colorectal cancer, and prostate cancer (Bai et al., 2013; Do et al., 2016; Jubierre et al., 2016; Lin, Wong, Martinka, & Li, 2010; Saladi et al., 2010; Watanabe, Semba, & Yokozaki, 2011; Wu et al., 2015). Though increased expression of mutated proteins could be consistent with a tumor suppressive function, there is no evidence of prevalent BRG1 or BRM mutation in breast cancer or melanoma (Cancer Genome Atlas, 2012; Cancer Genome Atlas, 2015), where such analysis has been reported. Elevated BRG1 expression has been linked to numerous pathways converging on cell proliferation and survival, including sonic hedgehog (SHH) and WNT signaling, the PI3K/AKT pathway, and regulation of lipogenesis and ATP‐binding cassette (ABC) transporter induction (reviewed in [Wu et al., 2017]).
These results indicate that, if properly delivered, inhibitors of BRG1 function may represent a potential therapeutic approach to certain cancers. PFI‐3, a small molecule bromodomain inhibitor with structural specificity for three bromodomain containing subunits of the mammalian SWI/SNF enzymes (BRG1, BRM, and polybromo‐1, also called BAF180; Vangamudi et al., 2015) has been shown to modulate certain cell differentiation transitions, (Fedorov et al., 2015; Gerstenberger et al., 2016) but has no effect whatsoever on cancer cell proliferation (Vangamudi et al., 2015; Wu, Sharma et al., 2016). However, promising results for inhibition of cancer cell proliferation have resulted from studies of an as yet structurally undefined inhibitor of the SNF2 family of ATPases called ADAADi (active DNA‐dependent ATPase A Domain inhibitor). ADAADi is a chromatographically separable byproduct of the bacterial aminoglycoside‐phosphotransferase (APH) action upon aminoglycosides (Dutta et al., 2012; Muthuswami et al., 2000). ADAADi has largely been used as a biochemical probe to help define enzymatic activities of SNF2 ATPases in vitro (Dutta et al., 2012; Muthuswami et al., 2000; Sharma, Bansal, Haokip, Goel, & Muthuswami, 2015), and it therefore functions as an inhibitor of other adenosine triphosphate (ATP)‐dependent activities of these enzymes, such as chromatin remodeling (Muthuswami et al., 2000). In tissue culture, ADAADi inhibits cancer cell proliferation and survival (Dutta et al., 2012; Wu, Madany et al., 2016) and there appears to be some specificity of ADAADi for BRG1, as both ADAADi and shRNA‐mediated knockdown of BRG1 inhibited proliferation, but there was no additive effect of the inhibitor plus BRG1 knockdown (Wu, Madany et al., 2016). This result suggests that even if other related ATPases are contributing to cell proliferation and survival, their contributions are relatively minor. In addition to effects on cancer cell proliferation, ADAADi phenocopies BRG1 knockdown in demonstrating essential functional roles for BRG1 in cancer cell metabolism and in drug‐induced activation of ABC transporter proteins linked to chemoresistance (Wu, Madany et al., 2016; Wu, Sharma et al., 2016). A specific BRG1‐targeting molecule may therefore be of therapeutic value in the treatment of certain cancers.
In this report, we interrogated the The Cancer Genome Atlas (TCGA) Prostate Cancer database for links between human prostate tumors and BRG1 expression. We found that BRG1 expression, but not the expression of the related ATPase, BRM, is elevated in prostate cancer biopsies relative to normal prostate tissue. The mutation rate for BRG1 and BRM in prostate cancer is less than 1% for each protein. Stratifying tumor samples by expression level revealed an inverse relationship between BRG1 expression and patient outcome, indicating BRG1 is a prognostic indicator for prostate cancer. Interestingly, there was no correlation between BRG1 expression and the Gleason score of the tumor samples, indicating that BRG1 is a prognostic indicator that is independent of the microscopic features of the tumor. Challenging prostate cancer cells in tissue culture with ADAADi led to decreased proliferation and survival, with evidence of apoptosis among the dying cells. When prostate cancer cells were used to orthotopically seed tumors in mouse flanks, direct injection of ADAADi, but not other compounds, inhibited tumor growth and improved mouse survival.
2. MATERIALS AND METHODS
2.1. Analysis of TCGA prostate cancer patient data
Patient data was from TCGA prostate adenocarcinoma (PRAD) data set (Cancer Genome Atlas, 2015). Almost all prostate cancer is adenocarcinoma. The last update for the TCGA PRAD data is May 31, 2016 and includes data from 498 tumors and 52 normal. Various online programs were used to analyze this data. TCGA PRAD expression data was analyzed using Xena, functional genomics and analysis platform developed by the University of California at Santa Cruz (https://xena.ucsc.edu; Goldman et al., 2018). Correlations between BRG1 and BRM messenger RNA (mRNA) levels in patient tumors were plotted using GEPIA (Gene Expression Profiling Interactive Analysis), a web‐based tool for analysis of TCGA datasets (gepia.cancer‐pku.cn; Tang et al., 2017). GEPIA was also used to draw boxplots and to correlate disease‐free survival (also called relapse‐free survival) with gene expression. GEPIA uses the Log‐rank test, or the Mantel‐Cox test for survival hypothesis testing. The Cox proportional hazard ratio and the 95% confidence interval information are also calculated. BRG1 and BRM mutations in TCGA prostate tumors were analyzed by CBioPortal (http://www.cbioportal.org; Cerami et al., 2012; Gao et al., 2013). Correlations between Gleason Scores and gene expression were done using Betastasis (www.betastasis.com/prostate_cancer/tcga_prad_from_gdc). Conclusions about human subjects are derived from data in public databases where patient information is deidentified.
2.2. Synthesis and purification of ADAADi
ADAADi for cell culture studies was synthesized and purified as described (Dutta et al., 2012). ADAADi for animal studies was synthesized and purified as described (Muthuswami et al., 2000).
2.3. Cell lines
Human PRAD cell line PC3 was purchased from ATCC or from NCCS, Pune, India and maintained in Dulbecco’s modified Eagle’s Medium supplemented with 10% fetal bovine serum, 1% penicillin‐streptomycin Amphotericin cocktail at 37°C in the presence of 5% CO2.
2.4. Cell viability assay
5000 cells were seeded in each well of a 96‐well plate containing 200 µl media and incubated overnight. The media was replaced with media containing varying concentrations of ADAADi. The cells were incubated at 37°C for the indicated time point. The media in each well was then replaced with media containing 0.45 mg/ml 3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide (MTT) and incubated for 2 hr at 37°C in a CO2 incubator. The media containing MTT solution was discarded and the purple precipitate formed was dissolved in 100 µl isopropanol. The plate was incubated for 15 min at room temperature and the absorbance was measured at 570 nm.
2.5. Annexin V‐FITC apoptosis detection
PC3 cells were grown in 60‐mm dish to 60–70% confluency before treatment with sublethal concentration (5 μM) of ADAADi for 48 hr. The cells were stained with Annexin V and Propidium iodide using Annexin V‐FITC Apoptosis Detection Kit (Catalog # BMS500FI/100; eBiosciences/Thermo Fisher India, Mumbai, India). The samples were analyzed using BD FACS Calibur 4C flow cytometer (Becton Dickinson India, Gurgaon, India).
2.6. Mouse xenograft studies
All animal experiments were performed according to protocols approved by the Institutional Animal Care and Use Committee at the University of Virginia School of Medicine. CRL: CD‐1nu/nu Br mice were obtained from Charles River Animal Resources Facility. PC3 cells were grown in RPMI 1640 Media containing 10% serum. 200 μl of this cell culture was diluted 1:1 with Matrigel (Collaborative Biomedical Products, Bedford, MA) such that the total number of cells after dilution were ~2 × 106. The cells were then injected on the underside of the flank and the development of subcutaneous tumor size was monitored by measurement with Vernier calipers.
Control and ADAADi treatments were started when the tumor size reached 200 mm3. ADAADi was solubilized in phosphate‐buffered saline (PBS) and pH was adjusted to 7.2 using phosphoric acid and filter sterilized before injection. Fifty microlitre of the drug was administered by direct injection into the tumor. In two independent experiments, injections were repeated every other day for 2 weeks. Similarly, a third experiment utilized every other day protocol for 2 weeks and after a 1 week break from treatment, an additional set of injections was performed, again, every other day for 2 weeks. The mice were euthanized when the tumor size exceeded 1000 mm3 or when the weight of the mice decreased by more than 15% of their starting weight. The tumor size was calculated using the following formula: volume = (length × [width]2)/2.
3. RESULTS
3.1. BRG1 expression is elevated in prostate tumors relative to normal tissue
BRG1 and BRM, two closely related ATPases that are mutually exclusive catalytic subunits of mammalian SWI/SNF chromatin remodeling enzymes, have different functional roles in different types of cancer (Hodges et al., 2016; Savas & Skardasi, 2018; Wu et al., 2017). Prior reports about BRG1/BRM in prostate cancer have utilized immunohistochemistry (IHC) of patient tumor samples and determined that BRG1 protein levels were higher in tumor than in normal tissue. BRM protein levels were described as heterogeneous, with an average value suggesting BRM expression in tumors was lower than in normal tissue. cDNA microarray analysis was generally consistent with the evaluation of protein expression by IHC (Y. Li et al., 2006; Sun et al., 2007). We took a complementary approach to evaluate BRG1/BRM expression in prostate cancer by interrogating the TCGA database. Data from 45 patients with matched normal tissue and prostate tumors were evaluated. BRG1 expression, on average, was significantly elevated in adenomas relative to normal tissue, while average BRM mRNA expression was significantly decreased in the tumor relative to normal tissue (Table 1). As controls for our analyses, we evaluated prostate serum antigen (PSA; KLK3), which showed a significant elevation in expression in tumors, and SUN1, an inner nuclear envelope protein not previously linked to prostate cancer, which showed no significant difference (Table 1). Boxplots showing the ranges for BRG1 and BRM mRNA expression in all TCGA samples (n = 492 for tumors, n = 52 for normal) reinforced the conclusion that BRG1 mRNA expression is elevated in tumor samples compared with normal tissue while the converse is true for BRM mRNA (Figure 1a). The UCSC Xena tool was used to determine whether any correlation between BRG1 and BRM mRNA expression in prostate cancer exists; the results clearly show no correlation across the data set (Figure 1b). To evaluate protein expression, we queried published proteomic data for BRG1 from 28 prostate tumor and eight normal prostate tissue samples (Iglesias‐Gato et al., 2016). We were unable to identify similar proteomic data for BRM. The data show a statistically significant increase in BRG1 protein expression in the tumor relative to normal tissue (Table 2) that matches the magnitude of the increase in BRG1 mRNA determined from analyzing the TCGA data set (Table 1; Figure 1a).
Table 1.
mRNA expression in matched prostate tumor and normal tissue samples from TCGA
| Normalized | t Test | ||||||
|---|---|---|---|---|---|---|---|
| n | Mean | SD | Mean | SD | Two‐tailed, paired | ||
| BRG1 | Tumor | 45 | 385784 | 84255 | 1.501 | 0.328 | <2.537E−07 |
| Normal | 45 | 257021 | 7600 | 1.000 | 0.030 | ||
| BRM | Tumor | 45 | 488619 | 9436 | 0.896 | 0.017 | <0.029 |
| Normal | 45 | 545535 | 11773 | 1.000 | 0.022 | ||
| KLK3 (PSA) | Tumor | 45 | 89336621 | 46271108 | 2.370 | 1.227 | <0.002 |
| Normal | 45 | 37702588 | 3594923 | 1.000 | 0.095 | ||
| SUN1 | Tumor | 45 | 195982 | 17977 | 1.008 | 0.294 | 0.856 |
| Normal | 45 | 194474 | 3015 | 1.000 | 0.008 | ||
Note. BRG1: Brahma‐related gene 1; BRM: Brahma; mRNA: messenger RNA; KLK3: kallikrein related peptidase 3; PSA: prostate‐specific antigen; SD: standard deviation; TGCA: The Cancer Genome Atlas.
Figure 1.
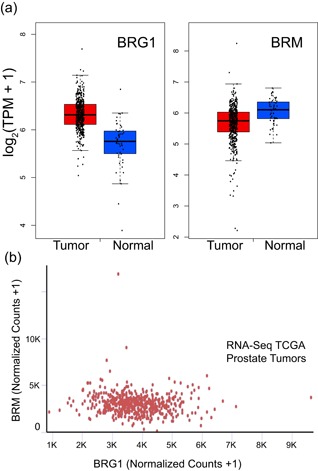
BRG1 mRNA expression in prostate cancer patient tissue is elevated compared with that in normal prostate tissue while BRM mRNA expression in prostate cancer patient tissue is reduced compared with that in normal prostate tissue. All data were extracted from the prostate adenocarcinoma TCGA data set. Plots were generated with GEPIA software (Tang et al., 2017). (a) Boxplots for BRG1 (left) and BRM (right) mRNA expression in prostate tumors (red) compared with normal prostate tissue (blue). Boxes enclose the middle two quartiles of mRNA expression with a centerline at the median. (b) BRG1 and BRM mRNA levels are not correlated in prostate tumors. BRG1: Brahma‐related gene 1; BRM: Brahma; GEPIA: Gene Expression Profiling Interactive Analysis; mRNA: messenger RNA; TCGA: The Cancer Genome Atlas; TPM: transcripts per kilobase million [Color figure can be viewed at wileyonlinelibrary.com]
Table 2.
BRG1 protein levels in prostate tumor and normal tissue samples extracted from (Iglesias‐Gato et al., 2016)
| Mean | t Test | ||||
|---|---|---|---|---|---|
| n | Mean | SD | Normalized | Two‐tailed, paired | |
| Normal | 8 | 0.716 | 0.180 | 1.000 | |
| Tumor | 28 | 1.054 | 0.583 | 1.473 | <0.013 |
Note. BRG1: Brahma‐related gene 1; SD: standard deviation.
We examined sequence data for the BRG1 and BRM coding sequences to determine whether any of the patient tumors were mutated for either or both ATPases. Only two of 491 patients had BRG1 mutations, one with a V698I and G775D double substitution, and one with an I1214L substitution (Figure 2a). In addition, one patient showed amplification of the BRG1 locus. No patients with deletions in the BRG1 coding sequence were identified. Three patients had BRM mutations, one had amplification of the locus, and 10 patients with homodeletions were identified (Figure 2b). One BRM mutation was an R1159Q substitution; another was a frameshift after amino acid 1253. Interestingly, the patient with the double substitution in BRG1 also had an R524M substitution in BRM. The significance of the mutation in both proteins is unknown. Regardless, the data indicate that a small percentage of prostate tumors contain mutations in either BRG1 or BRM. Thus the possibility that elevated expression of mutant BRG1 or BRM protein correlates with the prostate tumor phenotype is not supported.
Figure 2.
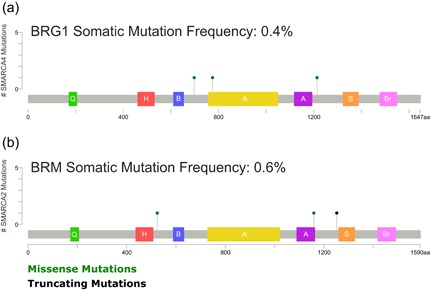
Schematic diagram of the location of BRG1 and BRM somatic mutations present in TCGA prostate tumor patients relative to known domains of BRG1 and BRM proteins. CBioPortal (Cerami et al., 2012; Gao et al., 2013) was used for data analysis. Domains indicated: Q (green) – QLQ domain, H (red) – HSA domain, B (blue) – BRK domain, A (yellow and purple) – bipartite SNF2 ATPase domain, S (orange) – SnAC domain, Br (pink) – bromodomain. BRG1: Brahma‐related gene 1; BRM: Brahma [Color figure can be viewed at wileyonlinelibrary.com]
3.2. BRG1 is a prognostic indicator for prostate cancer patient outcome
Despite the links between increased BRG1 expression and prostate tumors, there is presently no understanding of whether BRG1 expression correlates with patient outcome. We stratified patient BRG1 mRNA expression data from the TCGA database and compared patient survival among those in the highest quartile of BRG1 mRNA expression and those in the lowest quartile. Kaplan‐Meier plots demonstrate a significant difference in the two patient populations, with BRG1 expression inversely correlating with survival (Figure 3a). In contrast, a similar analysis of BRM expression in prostate cancer patients showed no correlation between BRM expression and patient outcome (Figure 3b). The results indicate that BRG1 is a prognostic indicator of prostate cancer patient outcome.
Figure 3.
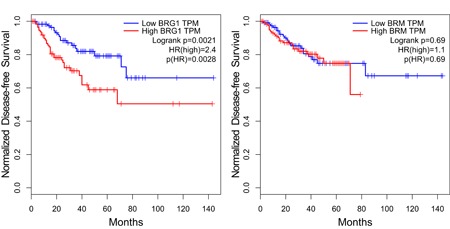
BRG1 mRNA levels (a) but not BRM mRNA levels (b) inversely correlate with prostate tumor patient survival. Kaplan‐Meier plots shown are based on an analysis of TCGA prostate patient data using GEPIA (Tang et al., 2017). The blue line labeled “Low” is the patients with the lowest quartile of mRNA levels; the red line labeled “High” is the patients with the highest quartile of mRNA levels. A Log‐rank test, the Mantel‐Cox test, was used to determine p values. HR, hazard ratio. BRM: Brahma; GEPIA: Gene Expression Profiling Interactive Analysis; mRNA: messenger RNA; TCGA: The Cancer Genome Atlas; TPM: transcripts per kilobase million [Color figure can be viewed at wileyonlinelibrary.com]
Traditionally, prostate tumors have been graded on the Gleason scale (Gleason, 1966; Gleason & Mellinger, 1974; Mellinger, Gleason, & Bailar, 1967). In this microscopic evaluation, the two most dominant patterns in the tumor biopsy are graded relative to normal prostate tissue, with the sum representing the Gleason score. Scores can range from 2 to 10, with 10 representing the least differentiated tumor cells that are most distinct from normal prostate tissue cells and that generally signify the worst prognosis. In practice, Gleason scores for prostate cancer patients typically range from 6 to 10. More recently, the International Society of Urological Pathology has further refined the diagnostic system (Epstein, Allsbrook, Amin, Egevad, & Committee, 2005). We plotted BRG1 and BRM mRNA expression as a function of Gleason score (Figure 4a,b) and show the data in a heat map format as well (Figure 4c). Average values are presented in Figure 4d. The data clearly indicate no correlation between BRG1 mRNA expression and Gleason score. Not surprisingly, there is also no correlation with BRM mRNA expression. Thus BRG1 mRNA expression is a prognostic indicator of prostate cancer patient outcome that is independent of Gleason score.
Figure 4.
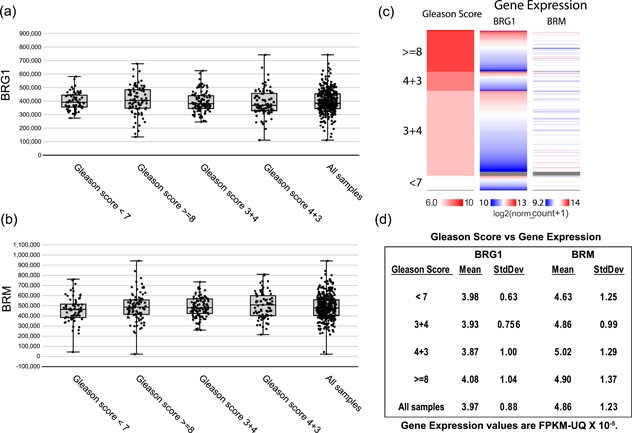
Neither BRG1 nor BRM mRNA levels correlate with prostate tumor Gleason score. (a) the ranges of BRG1 or (b) BRM mRNA levels from prostate tumor expression data in TCGA organized by the Gleason score for each tumor. The Project Betastasis website was used for data analysis and figure generation. FPKM, Fragments Per Kilobase of transcript per Million mapped reads, upper quartile normalized. (c) Heat map of BRG1 and BRM mRNA expression in prostate tumor samples with Gleason scores of <7, 7, or >7. (d) Mean BRG1 and BRM mRNA expression in prostate tumor samples with the indicated Gleason scores. BRG1: Brahma‐related gene 1; BRM: Brahma; mRNA: messenger RNA; SD: standard deviation [Color figure can be viewed at wileyonlinelibrary.com]
3.3. A BRG1 inhibitor diminishes prostate cancer cell survival in culture and in xenografts
The data suggest that targeting BRG1 may be of therapeutic benefit for prostate cancer. ADAADi is a biologic preparation isolated as a byproduct of the bacterial aminoglycoside‐3′‐phosphotransferase (APH (3′)‐III) enzyme reaction (Dutta et al., 2012; Muthuswami et al., 2000). It demonstrates preference for targeting BRG1 over BRM in cell culture experiments (Wu, Sharma et al., 2016) and phenocopies BRG1 knockdown in inhibiting lipid synthesis and in blocking drug‐induced activation of ABC transporter gene expression in breast cancer cells (Wu, Madany et al., 2016; Wu, Sharma et al., 2016). We therefore asked whether ADAADi might be used to inhibit prostate cancer cell proliferation and/or survival.
The PC3 prostate cancer cell line was derived from a patient’s metastatic PRAD and is capable of anchorage‐independent growth in culture and of generating tumors in athymic nude mice (Kaighn, Narayan, Ohnuki, Lechner, & Jones, 1979). Treatment of PC3 cells proliferating in culture with increasing concentrations of ADAADi gave a dose‐dependent decrease in cell viability (Figure 5a). Annexin V staining indicated that the observed cell death in the presence of a sublethal concentration of ADAADi was due to apoptosis (Figure 5b).
Figure 5.
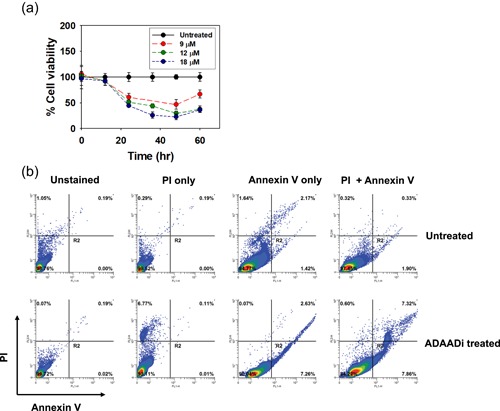
(a) ADAADi treatment inhibits PC3 cell proliferation. Cell viability was determined by MTT assay after 48 hr in the presence of the indicated concentration of inhibitor. (b) ADAADi treatment of PC3 cells increases the frequency of apoptosis. Cells were treated with 5 µM ADAADi for 48 hr and stained as indicated before FACS analysis. MMT: 3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide [Color figure can be viewed at wileyonlinelibrary.com]
Next, PC3 cells were injected into the flanks of athymic mice and subcutaneous tumor growth was monitored. ADAADi or PBS was injected directly into the subcutaneous tumor when the tumor reached a size of 200 mm3. In two independent experiments, every‐other‐day injections were executed for 2 weeks (Figure 6a,b and 6c,d). A third experiment used the every‐other‐day protocol for 2 weeks and, after a 1 week break from treatment, an additional set of injections were performed, again, every other day for 2 weeks (Figure 6e,f). The latter experiment also included a direct injection of the parent aminoglycoside (kanamycin) as a control in addition to PBS (Figure 6e). Tumor size was monitored during and after treatment. For clarity of presentation, tumor size data is presented for each of the three separate experiments that were performed (Figure 6a–f). Mice injected with PBS or kanamycin showed consistently increasing tumor size and were euthanized when the maximum allowable tumor burden was observed, in accordance with IACUC protocols (Figure 6a,c,e). ADAADi‐injected tumors showed a range of results. Some injected tumors expanded with similar kinetics to the control tumors (Figure 6b,d,f). Others showed delayed expansion but nevertheless expanded to the point where the animal was sacrificed (e.g., blue solid triangle in Experiment #2, green open and solid circles in Experiment #3). In contrast, some tumors failed to expand after ADAADi‐injection, and a subset of these tumors completely dissipated (e.g., red open and solid circles in Experiment #1). We conclude that ADAADi inhibited tumor growth in a subset of the treated animals. We also plotted survival data (Figure 7), which showed a clear survival advantage for the treated animals and highlighted the fact that approximately 30% of the treated animals survived for up to 7.5 months without evidence of tumor or any observable physical or behavioral anomaly, at which time these remaining animals were euthanized. Necropsy of these “survivors” revealed no tumors in other tissues, with the kidneys, lungs, lymph nodes proximal and distal to the injection site, or spleen specifically examined in each individual. The data indicate that direct injection of ADAADi into the xenografts resulted in tumor growth inhibition and a survival benefit, suggesting that this inhibitor has potential as a therapeutic agent.
Figure 6.
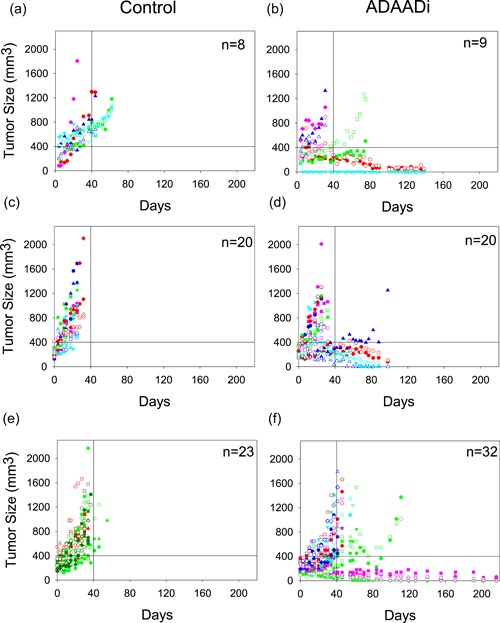
ADAADi injection into PC3‐induced mouse xenografts reduces, and in some cases, inhibits tumor growth. Three independent xenograft experiments were initiated by injection of PC3 cells. Tumor size was plotted as a function of time following injection of ADAADi or the control solution directly into the tumors. The number of xenografts (n) in each cohort is indicated. Data points for each individual mouse are indicated by specific shapes and colors. In Experiment 1 (a,b) and Experiment 2 (c,d), tumors were injected with PBS or ADAADi every other day for two weeks. In Experiment 3 (e,f), tumors were injected with ADAADi or with kanamycin instead of PBS as a control every other day for two weeks and after a one week break from treatment, an additional set of injections were performed every other day for two weeks. ADAADi: active DNA‐dependent ATPase A Domain inhibitor [Color figure can be viewed at wileyonlinelibrary.com]
Figure 7.
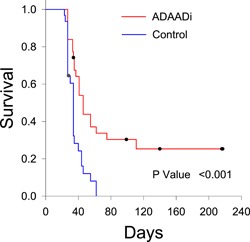
ADAADi injection into PC3‐induced xenografts extends survival. Kaplan‐Meier plots showing survival curves for all mice in the xenograft experiments presented in Figure 6. A Logrank test was used for statistical analysis. ADAADi: active DNA‐dependent ATPase A Domain inhibitor [Color figure can be viewed at wileyonlinelibrary.com]
4. DISCUSSION
The ATPases that are the catalytic subunits of human SWI/SNF enzymes have been linked to many types of cancer, but their functional contributions to oncogenesis are context dependent. Here we probed the TCGA database for expression and mutation frequency of BRG1 and BRM in prostate cancer. The analyses indicated elevated BRG1 mRNA and elevated BRG1 protein expression in prostate tumors relative to normal tissue. This is consistent with prior IHC studies of patient tumors (Y. Li et al., 2006). Although the TCGA data set did not contain information on BRM protein levels, a prior report revealed that BRM protein levels did not correlate with increased BRG1 protein in patient samples (Sun et al., 2007). A recent survey of multiple tumor types indicated that BRG1 expression was elevated in tumors whereas BRM was not (Guerrero‐Martinez & Reyes, 2018). Mechanistically, it appears that there is a direct correlation between BRG1 mRNA and protein expression, while posttranscriptional regulation likely plays a role in determining overall BRM protein expression.
Elevated expression of a regulatory protein can suggest tumor promoting activity but could instead indicate tumor suppressive activity if there is elevated expression of a mutant protein. To address this question, we examined the 491 available sequences in the TCGA prostate cancer database for mutations in BRG1 and BRM. Neither BRG1 nor BRM is frequently mutated in the patient tumors. This result is consistent with and extends a prior report finding no mutations in BRG1 coding sequences from 21 patient tumors (Valdman et al., 2003). Of the mutations identified in the TCGA database, a G775D alteration in BRG1 indicates a mutation in motif I of the conserved Snf2 ATPase domain (Flaus, Martin, Barton, & Owen‐Hughes, 2006). This residue is not conserved amongst the Snf2 family proteins. Nevertheless, we would predict that this mutation would render the protein inactive based on our prior structure‐function studies (Nongkhlaw, Gupta, Komath, & Muthuswami, 2012). The I1214L mutation identified in BRG1 is outside motif VI of the ATPase domain, but the residue is conserved in many closely related Snf2 family members, and analysis of the Sulfolobus solfataricus SWI2/SNF2 ATPase core complexed with DNA (Durr, Korner, Muller, Hickmann, & Hopfner, 2005) suggests that this mutation likely impairs ATPase activity. The BRM R1159Q mutation identified in one patient lies in motif VI and is conserved across the entire Snf2 ATPase family. Mutation of this residue in the SMARCAL1 ATPase resulted in the loss of ATPase activity (Bansal, Arya, Sethy, Rakesh, & Muthuswami, 2018). The other identified mutants are outside the conserved SNF2 ATPase domain, and the potential functional consequences are unknown.
Stratification of BRG1 expression amongst prostate tumor samples revealed that elevated BRG1 mRNA expression correlated with poor patient outcome. BRG1 expression is therefore a new marker for prostate cancer survival. This finding provides additional evidence that elevated BRG1 expression, not BRG1 mutation, is a marker for poor prognosis in an increasing number of cancer types. Prior work has demonstrated that high BRG1 mRNA expression inversely correlates with patient survival in breast cancer (Bai et al., 2013; Do et al., 2016; Wu, Sharma et al., 2016), colorectal cancer (S. Lin et al., 2016; Pyo, Son, Oh, & Kim, 2018), and neuroblastoma (Jubierre et al., 2016). In addition, BRG1 is required for various aspects of cancer cell survival, proliferation, and function in HeLa cells (Naidu, Love, Imbalzano, Grossman, & Androphy, 2009), leukemia cells (Buscarlet et al., 2014; J. Shi et al., 2013), breast cancer (Wu et al., 2015; Wu et al., 2016), hepatocarcinoma (Kaufmann et al., 2017), colorectal cancer (G. Wang et al., 2017), neuroblastoma (Jubierre et al., 2016), melanoma (Keenen, Qi, Saladi, Yeung, & de la Serna, 2010; H. Lin et al., 2010; Vachtenheim, Ondrusova, & Borovansky, 2010), and certain medulloblastoma tumors (Shi, Wang, Gu, Xuan, & Wu, 2016). Recent evidence indicates that the fusion between the TMPRSS2 gene and the ETS family transcription factor, ERG, that occurs in half of the prostate cancers, mediates its oncogenic effect at least in part by interacting with the human SWI/SNF enzymes and redirecting its chromatin interactions across the genome (Sandoval et al., 2018). Thus the chromatin remodeling enzyme and presumably its enzymatic function is a required component contributing to prostate oncogenesis. Collectively, the data contrast with the idea of BRG1 acting as a tumor suppressor in all cancer types, support the idea of context‐dependent function of SWI/SNF ATPases in cancer, and support the idea that BRG1 and/or BRM can, in some contexts, be drivers of oncogenesis (Wu et al., 2017).
We compared BRG1 expression to Gleason score, the commonly used staging system for prostate tumors (Mellinger et al., 1967), and found no correlation. The conclusion was based on analysis of 568 patient samples in the TCGA prostate cancer database. Our results indicate that BRG1 expression and Gleason score are therefore independent prognostic indicators of patient outcome. This point has been debated in the past, with one prior report finding a correlation (Sun et al., 2007) while another found no correlation (Y. Li et al., 2006). The smaller size of the respective sample pools in these studies (46 and 64 patient samples, respectively) may have contributed to the differing results.
In this report, we demonstrate that an inhibitor that shows specificity for BRG1 can be an effective tool in inhibiting prostate cancer cell survival in tissue culture and in xenografted prostate tumors. Prior studies have demonstrated that ADAADi is effective against numerous cancer cell types in culture (Dutta et al., 2012; Wu, Sharma et al., 2016), but here we show that it is effective in an animal tumor model. As this study was being completed, Ding et al. (2018) published a report identifying a synthetically lethal relationship in prostate cancer between BRG1 and the PTEN tumor suppressor (J. Li et al., 1997; Steck et al., 1997). They determined that phosphatase and tensin homolog (PTEN) loss sensitized the prostate cancer cells to BRG1 depletion in culture and in mouse tumor models with PTEN deficiency. Of particular note, PFI‐3, a bromodomain‐targeting drug specific for BRG1, BRM, and another SWI/SNF subunit called Polybromo (Gerstenberger et al., 2016) that has no effect on the proliferation of various cancer cell types (Vangamudi et al., 2015; Wu, Sharma et al., 2016), inhibited PTEN‐deficient prostate cancer cells in culture and compromised tumor growth in vivo when introduced by oral gavage to mice containing xenografts seeded by PTEN‐deficient prostate cancer cells. Our work and the work of Ding et al. (2018) therefore extend the proof‐of‐principle that targeting BRG1 can be an effective strategy for cancer treatment.
Specifically targeting the BRG1 enzyme as a therapeutic approach for cancer is an emerging idea. There are always drawbacks to targeting an essential protein or biological process, but we submit that differential effectiveness in cancerous versus normal cells will ultimately dictate the success of this strategy. Differential effectiveness is the reason that classical chemotherapy drugs that target cancers based on increased rate of cell division in tumor versus normal cells have been utilized for decades. So, despite BRG1 being ubiquitously expressed and functional in normal cells, there can be, and in fact are, cancer‐specific roles for BRG1, as shown by the enhanced requirement for BRG1 in PTEN‐deficient tumors (Ding et al., 2018). In addition, in triple negative breast cancer, BRG1 is specifically required for the upregulation of lipid and fatty acid synthesis enzymes that are required to produce elevated levels of these building blocks for rapid cell division; knockdown or inhibition of BRG1 reduced overall de novo lipid synthesis in the cancer cells but not in breast epithelial cells (Wu, Madany et al., 2016). Another example of a cancer‐specific role for BRG1 comes for other studies in triple negative breast cancer cells where it was shown that BRG1 mediates the induction of ABC transporters in response to treatment of cells with chemotherapeutic drugs. Knockdown or inhibition of BRG1 prevented induction of ABC transporters and resulted in increased intracellular retention of the drugs and increased chemosensitivity, raising the possibility that a BRG1 inhibitor could be an effective adjuvant treatment to classical chemotherapy drugs (Wu, Sharma et al., 2016). The data support the idea that inhibition or reduction of BRG1 could preferentially impair cancer cell growth and function relative to normal cells. Delivery, like with many inhibitory drugs, will be an issue that will require further development. However, it is apparent based on the work presented here and elsewhere that direct delivery of the inhibitor to the tumor can be effective in tumor reduction without impairing lifespan or apparent health of the treated individual. Presumably, this means that any effects of the treatment on the functions of BRG1 in normal cells exposed to the inhibitor were minimal or nonexistent.
Finally, this study will be greatly advanced once the inhibitory molecule(s) present in the ADAADi preparation are identified. Work in our labs continues to address this problem. In addition to better defining ADAADi, our work indicates that screens of chemical libraries should be performed to identify novel inhibitors of BRG1, which we would predict would have immediate preclinical relevance for future therapeutic approaches to cancers where BRG1 expression is elevated relative to normal tissue.
CONFLICTS OF INTEREST
The authors declare that there are no conflicts of interest.
AUTHOR CONTRIBUTIONS
Experiments were conceived by R. M., L. B., and J. W. H. and performed by R. M., L. B., R. R., and J. W. H. Bioinformatic analyses conceived by J. A. N. and A. N. I. and were performed by J. A. N. All authors analyzed and reviewed data and conclusions. A. N. I. wrote the manuscript with input from all other authors.
ACKNOWLEDGEMENTS
The authors thank Peter Pryciak for discussion and Hanna Witwicka for technical assistance and discussion. This study was partially supported by NIH grants (GM56244 to A. N. I. and EB014869 to J. A. N.). R. R. was supported by UGC non‐net fellowship. R. M. also acknowledges funding from DST‐PURSE (PAC‐JNU‐DST‐PURSE‐462 [Phase II]).
Muthuswami R, Bailey L, Rakesh R, Imbalzano AN, Nickerson JA, Hockensmith JW. BRG1 is a prognostic indicator and a potential therapeutic target for prostate cancer. J Cell Physiol. 2019;234:15194–15205. 10.1002/jcp.28161
Contributor Information
Anthony N. Imbalzano, Email: anthony.imbalzano@umassmed.edu.
Joel W. Hockensmith, Email: jwh6f@virginia.edu
References
REFERENCES
- Arriaga‐Canon, C. , De la rosa‐Velázquez, I. A. , González‐Barrios, R. , Montiel‐Manríquez, R. , Oliva‐Rico, D. , Jiménez‐Trejo, F. , … Herrera, L. A. (2018). The use of long noncoding RNAs as prognostic biomarkers and therapeutic targets in prostate cancer. Oncotarget, 9(29), 20872–20890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, J. , Mei, P. , Zhang, C. , Chen, F. , Li, C. , Pan, Z. , … Zheng, J. (2013). BRG1 is a prognostic marker and potential therapeutic target in human breast cancer. PLOS One, 8(3), e59772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal, R. , Arya, V. , Sethy, R. , Rakesh, R. , & Muthuswami, R. (2018). RecA‐like domain 2 of DNA‐dependent ATPase A domain, a SWI2/SNF2 protein, mediates conformational integrity and ATP hydrolysis. Bioscience Reports, 38(3), BSR20180568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscarlet, M. , Krasteva, V. , Ho, L. , Simon, C. , Hebert, J. , Wilhelm, B. , … Lessard, J. A. (2014). Essential role of BRG, the ATPase subunit of BAF chromatin remodeling complexes, in leukemia maintenance. Blood, 123(11), 1720–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network (2012). Comprehensive molecular portraits of human breast tumours. Nature, 490(7418), 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network (2015). Genomic classification of cutaneous melanoma. Cell, 161(7), 1681–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami, E. , Gao, J. , Dogrusoz, U. , Gross, B. E. , Sumer, S. O. , Aksoy, B. A. , … Schultz, N. (2012). The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discovery, 2(5), 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, Y. , Li, N. , Dong, B. , Guo, W. , Wei, H. , Chen, Q. , … Qin, J. (2018). Chromatin remodeling ATPase BRG1 and PTEN are synthetic lethal in prostate cancer. The Journal of Clinical Investigation. Advance online publication. 10.1172/JCI123557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do, S. I. , Yoon, G. , Kim, H. S. , Kim, K. , Lee, H. , Do, I. G. , … Sohn, J. H. (2016). Increased Brahma‐related gene 1 expression predicts distant metastasis and shorter survival in patients with invasive ductal carcinoma of the breast. Anticancer Research, 36(9), 4873–4882. [DOI] [PubMed] [Google Scholar]
- Dong, D. X. , & Ji, Z. G. (2017). Current progress and controversies in prostate cancer management. Chinese Medical Journal (Beijing), 130(24), 2991–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durr, H. , Körner, C. , Müller, M. , Hickmann, V. , & Hopfner, K. P. (2005). X‐ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell, 121(3), 363–373. [DOI] [PubMed] [Google Scholar]
- Dutta, P. , Tanti, G. K. , Sharma, S. , Goswami, S. K. , Komath, S. S. , Mayo, M. W. , … Muthuswami, R. (2012). Global epigenetic changes induced by SWI2/SNF2 inhibitors characterize neomycin‐resistant mammalian cells. PLOS One, 7(11), e49822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein, J. I. , Allsbrook, W. C., Jr. , Amin, M. B. , & Egevad, L. L. (2005). The 2005 International Society of Urological Pathology (ISUP) consensus conference on Gleason grading of prostatic carcinoma. American Journal of Surgical Pathology, 29(9), 1228–1242. [DOI] [PubMed] [Google Scholar]
- Fedorov, O. , Castex, J. , Tallant, C. , Owen, D. R. , Martin, S. , Aldeghi, M. , … Müller, S. (2015). Selective targeting of the BRG/PB1 bromodomains impairs embryonic and trophoblast stem cell maintenance. Science Advances, 1(10), e1500723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filella, X. , Fernández‐Galán, E. , Fernández Bonifacio, R. , & Foj, L. (2018). Emerging biomarkers in the diagnosis of prostate cancer. Pharmacogenomics and Personalized Medicine, 11, 83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaus, A. , Martin, D. M. , Barton, G. J. , & Owen‐Hughes, T. (2006). Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Research, 34(10), 2887–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuoka, J. , Fujii, T. , Shih, J. H. , Dracheva, T. , Meerzaman, D. , Player, A. , … Jen, J. (2004). Chromatin remodeling factors and BRM/BRG1 expression as prognostic indicators in non‐small cell lung cancer. Clinical Cancer Research, 10(13), 4314–4324. [DOI] [PubMed] [Google Scholar]
- Gao, J. , Aksoy, B. A. , Dogrusoz, U. , Dresdner, G. , Gross, B. , Sumer, S. O. , … Schultz, N. (2013). Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling, 6(269), pl1 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstenberger, B. S. , Trzupek, J. D. , Tallant, C. , Fedorov, O. , Filippakopoulos, P. , Brennan, P. E. , … Owen, D. R. (2016). Identification of a chemical probe for family VIII bromodomains through optimization of a fragment hit. Journal of Medicinal Chemistry, 59(10), 4800–4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason, D. F. (1966). Classification of prostatic carcinomas. Cancer Chemotherapy Reports, 50(3), 125–128. [PubMed] [Google Scholar]
- Gleason, D. F. , & Mellinger, G. T. (1974). Prediction of prognosis for prostatic adenocarcinoma by combined histological grading and clinical staging. Journal of Urology, 111(1), 58–64. [DOI] [PubMed] [Google Scholar]
- Goldman, M , Craft, B , Kamath, A , Brooks, A , Zhu, J , Haussler, D. 2018. The UCSC Xena Platform for cancer genomics data visualization and interpretation. bioRxiv 10.1101/326470 [DOI]
- Guerrero‐Martínez, J. A. , & Reyes, J. C. (2018). High expression of SMARCA4 or SMARCA2 is frequently associated with an opposite prognosis in cancer. Scientific Reports, 8(1), 2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges, C. , Kirkland, J. G. , & Crabtree, G. R. (2016). The many roles of BAF (mSWI/SNF) and PBAF complexes in cancer. Cold Spring Harbor Perspectives in Medicine, 6(8), a026930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann, A. F. , & Vakoc, C. R. (2014). A rationale to target the SWI/SNF complex for cancer therapy. Trends in Genetics, 30(8), 356–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias‐Gato, D. , Wikström, P. , Tyanova, S. , Lavallee, C. , Thysell, E. , Carlsson, J. , … Flores‐Morales, A. (2016). The proteome of primary prostate cancer. European Urology, 69(5), 942–952. [DOI] [PubMed] [Google Scholar]
- Imbalzano, A. N. , Kwon, H. , Green, M. R. , & Kingston, R. E. (1994). Facilitated binding of TATA‐binding protein to nucleosomal DNA. Nature, 370(6489), 481–485. [DOI] [PubMed] [Google Scholar]
- Jelinic, P. , Mueller, J. J. , Olvera, N. , Dao, F. , Scott, S. N. , Shah, R. , … Levine, D. A. (2014). Recurrent SMARCA4 mutations in small cell carcinoma of the ovary. Nature Genetics, 46(5), 424–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jubierre, L. , Soriano, A. , Planells‐Ferrer, L. , París‐Coderch, L. , Tenbaum, S. P. , Romero, O. A. , … Segura, M. F. (2016). BRG1/SMARCA4 is essential for neuroblastoma cell viability through modulation of cell death and survival pathways. Oncogene, 35(39), 5179–5190. [DOI] [PubMed] [Google Scholar]
- Kaighn, M. E. , Narayan, K. S. , Ohnuki, Y. , Lechner, J. F. , & Jones, L. W. (1979). Establishment and characterization of a human prostatic carcinoma cell line (PC‐3). Investigative Urology, 17(1), 16–23. [PubMed] [Google Scholar]
- Kaufmann, B. , Wang, B. , Zhong, S. , Laschinger, M. , Patil, P. , Lu, M. , … Hartmann, D. (2017). BRG1 promotes hepatocarcinogenesis by regulating proliferation and invasiveness. PLOS One, 12(7), e0180225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenen, B. , Qi, H. , Saladi, S. V. , Yeung, M. , & de la Serna, I. L. (2010). Heterogeneous SWI/SNF chromatin remodeling complexes promote expression of microphthalmia‐associated transcription factor target genes in melanoma. Oncogene, 29(1), 81–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khavari, P. A. , Peterson, C. L. , Tamkun, J. W. , Mendel, D. B. , & Crabtree, G. R. (1993). BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature, 366(6451), 170–174. [DOI] [PubMed] [Google Scholar]
- Kinsella, N. , Helleman, J. , Bruinsma, S. , Carlsson, S. , Cahill, D. , Brown, C. , & Van Hemelrijck, M. (2018). Active surveillance for prostate cancer: A systematic review of contemporary worldwide practices. Translational Andrology and Urology, 7(1), 83–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupryjańczyk, J. , Dansonka‐Mieszkowska, A. , Moes‐Sosnowska, J. , Plisiecka‐Hałasa, J. , Szafron, Ł. , Podgórska, A. , … Śpiewankiewicz, B. (2013). Ovarian small cell carcinoma of hypercalcemic type ‐ evidence of germline origin and SMARCA4 gene inactivation. a pilot study. Polish Journal of Pathology, 64(4), 238–246. [DOI] [PubMed] [Google Scholar]
- Kwon, H. , Imbalzano, A. N. , Khavari, P. A. , Kingston, R. E. , & Green, M. R. (1994). Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature, 370(6489), 477–481. [DOI] [PubMed] [Google Scholar]
- Lee, S. H. , & Shen, M. M. (2015). Cell types of origin for prostate cancer. Current Opinion in Cell Biology, 37, 35–41. [DOI] [PubMed] [Google Scholar]
- Li, J. , Yen, C. , Liaw, D. , Podsypanina, K. , Bose, S. , Wang, S. I. , … Parsons, R. (1997). PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science, 275(5308), 1943–1947. [DOI] [PubMed] [Google Scholar]
- Li, Y. , Shi, Q. L. , Jin, X. Z. , Meng, K. , Zhou, X. J. , & Sun, L. P. (2006). BRG1 expression in prostate carcinoma by application of tissue microarray. Zhonghua Nan ke Xue, 12(7), 629–632. [PubMed] [Google Scholar]
- Lin, H. , Wong, R. P. C. , Martinka, M. , & Li, G. (2010). BRG1 expression is increased in human cutaneous melanoma. The British Journal of Dermatology, 163(3), 502–510. [DOI] [PubMed] [Google Scholar]
- Lin, S. , Jiang, T. , Ye, L. , Han, Z. , Liu, Y. , Liu, C. , … Fan, J. (2016). The chromatin‐remodeling enzyme BRG1 promotes colon cancer progression via positive regulation of WNT3A. Oncotarget, 7(52), 86051–86063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina, P. P. , Carretero, J. , Fraga, M. F. , Esteller, M. , Sidransky, D. , & Sanchez‐Cespedes, M. (2004). Genetic and epigenetic screening for gene alterations of the chromatin‐remodeling factor, SMARCA4/BRG1, in lung tumors. Genes, Chromosomes and Cancer, 41(2), 170–177. [DOI] [PubMed] [Google Scholar]
- Mellinger, G. T. , Gleason, D. , & Bailar, J., 3rd (1967). The histology and prognosis of prostatic cancer. Journal of Urology, 97(2), 331–337. [DOI] [PubMed] [Google Scholar]
- Muchardt, C. , & Yaniv, M. (1993). A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. The EMBO Journal, 12(11), 4279–4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthuswami, R. , Mesner, L. D. , Wang, D. , Hill, D. A. , Imbalzano, A. N. , & Hockensmith, J. W. (2000). Phosphoaminoglycosides inhibit SWI2/SNF2 family DNA‐dependent molecular motor domains. Biochemistry, 39(15), 4358–4365. [DOI] [PubMed] [Google Scholar]
- Naidu, S. R. , Love, I. M. , Imbalzano, A. N. , Grossman, S. R. , & Androphy, E. J. (2009). The SWI/SNF chromatin remodeling subunit BRG1 is a critical regulator of p53 necessary for proliferation of malignant cells. Oncogene, 28(27), 2492–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nongkhlaw, M. , Gupta, M. , Komath, S. S. , & Muthuswami, R. (2012). Motifs Q and I are required for ATP hydrolysis but not for ATP binding in SWI2/SNF2 proteins. Biochemistry, 51(18), 3711–3722. [DOI] [PubMed] [Google Scholar]
- Oike, T. , Ogiwara, H. , Tominaga, Y. , Ito, K. , Ando, O. , Tsuta, K. , … Kohno, T. (2013). A synthetic lethality‐based strategy to treat cancers harboring a genetic deficiency in the chromatin remodeling factor BRG1. Cancer Research, 73(17), 5508–5518. [DOI] [PubMed] [Google Scholar]
- Packer, J. R. , & Maitland, N. J. (2016). The molecular and cellular origin of human prostate cancer. Biochimica et Biophysica Acta, 1863(6 Pt A), 1238–1260. [DOI] [PubMed] [Google Scholar]
- Pyo, J. S. , Son, B. K. , Oh, D. , & Kim, E. K. (2018). BRG1 is correlated with poor prognosis in colorectal cancer. Human Pathology, 73, 66–73. [DOI] [PubMed] [Google Scholar]
- Ramos, P. , Karnezis, A. N. , Craig, D. W. , Sekulic, A. , Russell, M. L. , Hendricks, W. P. D. , … Trent, J. M. (2014). Small cell carcinoma of the ovary, hypercalcemic type, displays frequent inactivating germline and somatic mutations in SMARCA4. Nature Genetics, 46(5), 427–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisman, D. N. , Sciarrotta, J. , Wang, W. , Funkhouser, W. K. , & Weissman, B. E. (2003). Loss of BRG1/BRM in human lung cancer cell lines and primary lung cancers: Correlation with poor prognosis. Cancer Research, 63(3), 560–566. [PubMed] [Google Scholar]
- Saladi, S. , Keenen, B. , Marathe, H. G. , Qi, H. , Chin, K. V. , & de la Serna, I. L. (2010). Modulation of extracellular matrix/adhesion molecule expression by BRG1 is associated with increased melanoma invasiveness. Molecular Cancer, 9, 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval, G. J. , Pulice, J. L. , Pakula, H. , Schenone, M. , Takeda, D. Y. , Pop, M. , … Kadoch, C. (2018). Binding of TMPRSS2‐ERG to BAF chromatin remodeling complexes mediates prostate oncogenesis. Molecular Cell, 71(4), 554–566.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savas, S. , & Skardasi, G. (2018). The SWI/SNF complex subunit genes: Their functions, variations, and links to risk and survival outcomes in human cancers. Critical Reviews in Oncology/Hematology, 123, 114–131. [DOI] [PubMed] [Google Scholar]
- Sharma, T. , Bansal, R. , Haokip, D. T. , Goel, I. , & Muthuswami, R. (2015). SMARCAL1 negatively regulates C‐Myc transcription by altering the conformation of the promoter region. Scientific Reports, 5, 17910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, J. , Whyte, W. A. , Zepeda‐Mendoza, C. J. , Milazzo, J. P. , Shen, C. , Roe, J. S. , … Vakoc, C. R. (2013). Role of SWI/SNF in acute leukemia maintenance and enhancer‐mediated Myc regulation. Genes and Development, 27(24), 2648–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, X. , Wang, Q. , Gu, J. , Xuan, Z. , & Wu, J. I. (2016). SMARCA4/Brg1 coordinates genetic and epigenetic networks underlying Shh‐type medulloblastoma development. Oncogene, 35(44), 5746–5758. [DOI] [PubMed] [Google Scholar]
- Steck, P. A. , Pershouse, M. A. , Jasser, S. A. , Yung, W. K. A. , Lin, H. , Ligon, A. H. , … Tavtigian, S. V. (1997). Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nature Genetics, 15(4), 356–362. [DOI] [PubMed] [Google Scholar]
- Sun, A. , Tawfik, O. , Gayed, B. , Thrasher, J. B. , Hoestje, S. , Li, C. , & Li, B. (2007). Aberrant expression of SWI/SNF catalytic subunits BRG1/BRM is associated with tumor development and increased invasiveness in prostate cancers. The Prostate, 67(2), 203–213. [DOI] [PubMed] [Google Scholar]
- Tabayoyong, W. , & Abouassaly, R. (2015). Prostate cancer screening and the associated controversy. Surgical Clinics of North America, 95(5), 1023–1039. [DOI] [PubMed] [Google Scholar]
- Tang, Z. , Li, C. , Kang, B. , Gao, G. , Li, C. , & Zhang, Z. (2017). GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Research, 45(W1), W98–W102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vachtenheim, J. , Ondrušová, L. , & Borovanský, J. (2010). SWI/SNF chromatin remodeling complex is critical for the expression of microphthalmia‐associated transcription factor in melanoma cells. Biochemical and Biophysical Research Communications, 392(3), 454–459. [DOI] [PubMed] [Google Scholar]
- Valdman, A. , Nordenskjöld, A. , Fang, X. , Naito, A. , Al‐Shukri, S. , Larsson, C. , … Li, C. (2003). Mutation analysis of the BRG1 gene in prostate cancer clinical samples. International Journal of Oncology, 22(5), 1003–1007. [PubMed] [Google Scholar]
- Vangamudi, B. , Paul, T. A. , Shah, P. K. , Kost‐Alimova, M. , Nottebaum, L. , Shi, X. , … Andersen, J. N. (2015). The SMARCA2/4 ATPase domain surpasses the bromodomain as a drug target in SWI/SNF‐mutant cancers: insights from cDNA rescue and PFI‐3 inhibitor studies. Cancer Research, 75(18), 3865–3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G. , Fu, Y. , Hu, F. , Lan, J. , Xu, F. , Yang, X. , & Hu, J. (2017). Loss of BRG1 induces CRC cell senescence by regulating p53/p21 pathway. Cell Death and Disease, 8(2), e2607 10.1038/cddis.2017.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W. , Côté, J. , Xue, Y. , Zhou, S. , Khavari, P. A. , Biggar, S. R. , … Crabtree, G. R. (1996). Purification and biochemical heterogeneity of the mammalian SWI‐SNF complex. The EMBO Journal, 15(19), 5370–5382. [PMC free article] [PubMed] [Google Scholar]
- Wang, Z. A. , Toivanen, R. , Bergren, S. K. , Chambon, P. , & Shen, M. M. (2014). Luminal cells are favored as the cell of origin for prostate cancer. Cell Reports, 8(5), 1339–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, T. , Semba, S. , & Yokozaki, H. (2011). Regulation of PTEN expression by the SWI/SNF chromatin‐remodelling protein BRG1 in human colorectal carcinoma cells. British Journal of Cancer, 104(1), 146–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, B. G. , Helming, K. C. , Wang, X. , Kim, Y. , Vazquez, F. , Jagani, Z. , … Roberts, C. W. M. (2014). Residual complexes containing SMARCA2 (BRM) underlie the oncogenic drive of SMARCA4 (BRG1) mutation. Molecular and Cellular Biology, 34(6), 1136–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkowski, L. , Carrot‐Zhang, J. , Albrecht, S. , Fahiminiya, S. , Hamel, N. , Tomiak, E. , … Foulkes, W. D. (2014). Germline and somatic SMARCA4 mutations characterize small cell carcinoma of the ovary, hypercalcemic type. Nature Genetics, 46(5), 438–443. [DOI] [PubMed] [Google Scholar]
- Wu, Q. , Lian, J. B. , Stein, J. L. , Stein, G. S. , Nickerson, J. A. , & Imbalzano, A. N. (2017). The BRG1 ATPase of human SWI/SNF chromatin remodeling enzymes as a driver of cancer. Epigenomics, 9(6), 919–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Q. , Madany, P. , Akech, J. , Dobson, J. R. , Douthwright, S. , Browne, G. , … Imbalzano, A. N. (2015). The SWI/SNF ATPases are required for triple negative breast cancer cell proliferation. Journal of Cellular Physiology, 230(11), 2683–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Q. , Madany, P. , Dobson, J. R. , Schnabl, J. M. , Sharma, S. , Smith, T. C. , … Nickerson, J. A. (2016). The BRG1 chromatin remodeling enzyme links cancer cell metabolism and proliferation. Oncotarget, 7(25), 38270–38281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Q. , Sharma, S. , Cui, H. , LeBlanc, S. E. , Zhang, H. , Muthuswami, R. , … mbalzano, A. N. (2016). Targeting the chromatin remodeling enzyme BRG1 increases the efficacy of chemotherapy drugs in breast cancer cells. Oncotarget, 7(19), 27158–27175. [DOI] [PMC free article] [PubMed] [Google Scholar]


