Summary
Roots form highly complex systems varying in growth direction and branching pattern to forage for nutrients efficiently. Here mutations in the KAI2 (KARRIKIN INSENSITIVE) α/β‐fold hydrolase and the MAX2 (MORE AXILLARY GROWTH 2) F‐box leucine‐rich protein, which together perceive karrikins (smoke‐derived butenolides), caused alteration in root skewing in Arabidopsis thaliana. This phenotype was independent of endogenous strigolactones perception by the D14 α/β‐fold hydrolase and MAX2. Thus, KAI2/MAX2 effect on root growth may be through the perception of endogenous KAI2‐ligands (KLs), which have yet to be identified. Upon perception of a ligand, a KAI2/MAX2 complex is formed together with additional target proteins before ubiquitination and degradation through the 26S proteasome. Using a genetic approach, we show that SMAX1 (SUPPRESSOR OF MAX2‐1)/SMXL2 and SMXL6,7,8 (SUPPRESSOR OF MAX2‐1‐LIKE) are also likely degradation targets for the KAI2/MAX2 complex in the context of root skewing. In A. thaliana therefore, KAI2 and MAX2 act to limit root skewing, while kai2's gravitropic and mechano‐sensing responses remained largely unaffected. Many proteins are involved in root skewing, and we investigated the link between MAX2 and two members of the SKS/SKU family. Though KLs are yet to be identified in plants, our data support the hypothesis that they are present and can affect root skewing.
Keywords: karrikin, strigolactone, Arabidopsis thaliana, waving, root, skewing
Significance Statement
Arabidopsis thaliana mutants deficient in karrikins/KAI2‐ligand (KL) perception display an exaggerated root‐skewing phenotype, which is not readily explained by impairments in their mechano‐sensing and gravitropic responses. Strigolactones (SLs) belong to a recently characterised set of phytohormones. Their role in root skewing in Arabidopsis has been hypothesized, but no evidence supporting a role for SL in influencing root skewing was found here. Rather, results support a role for as‐yet unidentified endogenous KLs.
Introduction
Roots grow in complex patterns that are highly relevant to their adaptation to different soil conditions and yet very difficult to investigate in this complex medium. Arabidopsis thaliana roots grown vertically on solid medium produce specific surface‐dependent growth patterns described as skewing (deviation from vertical) and waving (Roy and Bassham, 2014). Established differences amongst Arabidopsis ecotypes suggest that these patterns may reflect an adaptive response relevant to natural soil conditions (Vaughn and Masson, 2011; Schultz et al., 2017).
Root skewing has been widely reported (Darwin and Darwin, 1880; Migliaccio and Piconese, 2001; Oliva and Dunand, 2007; Roy and Bassham, 2014; Shih et al., 2014), but the model describing its mechanism remains complex and incomplete. As Arabidopsis roots grow on the surface of solid agar, they follow the gravitropic vector (Figure 1a). Arabidopsis mutants impaired in the gravitropic response (Okada and Shimura, 1990) show a root‐skewing phenotype. On Earth, gravitropism is one component of the root‐skewing response, while under micro‐gravity as in the International Space Station directional light can also provide a vector directing growth (Paul et al., 2012; Roux, 2012). While gravitropism and negative phototropism can essentially be described in two dimensions, a third dimension must also be considered (z), which corresponds to the distance away from the growth surface (Figure 1a). This also allows for root movement or circumnutation along the z‐axis (Migliaccio and Piconese, 2001; Simmonds et al., 2005), and for this movement to be impaired when the roots touch the surface of the solid medium (Thompson and Holbrook, 2004). Arabidopsis mutants deficient in mechano‐sensing, such as feronia (Shih et al., 2014) or cml24 (Wang et al., 2011), show a root‐skewing phenotype, which supports a role for thigmotropism (change in growth direction in response to mechanical stimulation from surface contact) in root skewing. While root movement in the z‐dimension can affect root skewing, the root‐skewing response is measured as the deviation of root growth along the x‐axis that can only be seen when roots are grown on a surface. According to the current model, root skewing represents the integrated root tip response to gravity, negative phototropism, circumnutation and thigmotropism. Thus, the root growth patterns generated are dependent on the forces applied at the root tip and the characteristics of the mature roots, such as size and rigidity.
Figure 1.
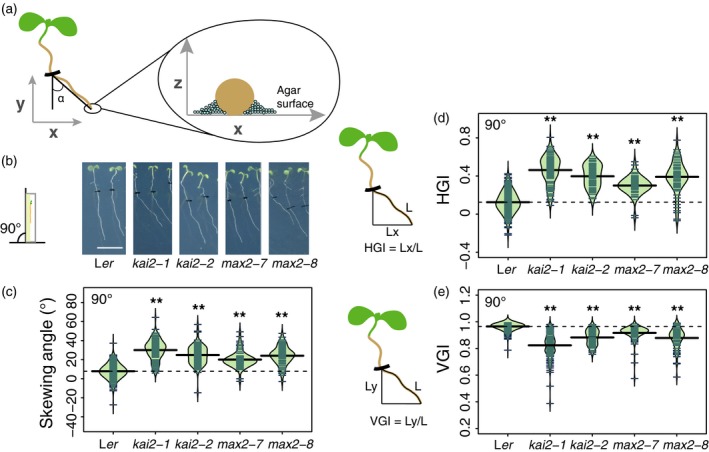
kai2 and max2 mutants display an exaggerated rightward root‐skewing phenotype.
(a) Arabidopsis roots grown on the surface of agar medium display a root‐skewing pattern that is the result of movement along the x‐, y‐ and z‐axes.
(b) Seedlings of kai2‐1, kai2‐2, max2‐7 and max2‐8 displayed an exaggerated rightward skew when grown at 90°. Scale bar: 1 cm.
(c) The root‐skewing angle (α) was measured as the deviation from the vertical for plants grown at a 90° angle.
(d) The increased root skewing can also be measured as an increase in horizontal growth index (HGI) or (e) a decrease in vertical growth index (VGI). Data for each genotype are displayed as a beanplot with the skewing angle of individual roots shown as dark green horizontal lines, while the mean is represented by a thick black horizontal line. The estimated density of the distribution is illustrated by the shaded colour. The dashed line corresponds to the mean for the wild‐type. Positive values are rightward skews. Significant differences compared with wild‐type (Tukey HSD) are shown: *P < 0.05, **P < 0.01. For each genotype, n > 65 in three separate experiments.
The role of plant hormones in root skewing and waving is poorly understood, but auxins (Okada and Shimura, 1990), ethylene (Buer et al., 2000, 2003), cytokinins (Kushwah et al., 2011) and brassinosteroids (Lanza et al., 2012) are implicated. Little is known of the role of a recently characterised set of phytohormones, strigolactones (SLs; Roy and Bassham, 2014), and related smoked‐derived butenolides, karrikins (KARs; Flematti et al., 2015), or the as‐yet unidentified endogenous ligands of the KAI2 (KARRIKIN INSENSITIVE) KAR receptor [KAI2‐ligand (KL); Sun et al., 2016]. Given the role of SLs in regulating root system architecture (Ruyter‐Spira et al., 2011; Mayzlish‐Gati et al., 2012; Rasmussen et al., 2012, 2013; Kapulnik and Koltai, 2014; Sun et al., 2014, 2015; Jiang et al., 2015; Matthys et al., 2016) and affecting auxin transport (Crawford et al., 2010; Shinohara et al., 2013), it would be interesting to test their role in root skewing.
Many elements of the SL perception pathway have been elucidated, and are either shared or related to components of the KAR/KL perception pathway. The current model suggests that SLs bind a related α/β‐fold hydrolase called D14 (Hamiaux et al., 2012; Chevalier et al., 2014; de Saint Germain et al., 2016; Yao et al., 2016), while KARs and KLs are perceived by binding the α/β‐fold hydrolase KAI2/D14‐like protein (Waters et al., 2012; Bythell‐Douglas et al., 2013; Sun et al., 2018). D14 can form a complex with MAX2 (MORE AXILLARY GROWTH2), a leucine‐rich repeat F‐box protein (Zhao et al., 2015; Yao et al., 2016), while physical interaction between KAI2 and MAX2 was demonstrated using yeast two‐hybrid (Toh et al., 2014). The KAR‐dependent degradation of KAI2 can also occur independently from MAX2, independently of ubiquitination or the activity of the 26S proteasome (Waters et al., 2015a). More recently, heat‐shock‐related proteins have been identified as degradation targets of MAX2 in rice (Jiang et al., 2013; Zhou et al., 2013) and Arabidopsis (SMXL, SUPPRESSOR OF MAX2‐1‐LIKE; Stanga et al., 2013; Soundappan et al., 2015; Moturu et al., 2018). Thus far, a dichotomy has been proposed with SMAX1 (SUPPRESSOR OF MAX2‐1) suppressing KAR‐related max2 phenotypes (e.g. germination and hypocotyl elongation), while other members of the SMXL family, namely SMXL6, SMXL7 and SMXL8, suppress SL‐related phenotypes [e.g. shoot branching and lateral root density (LRD); Waters et al., 2017]. Additional members SMXL3, 4 and 5 regulate phloem development in a SL‐ and KAR‐independent manner (Wallner et al., 2017). While some specificity of SL or KAR/KL signalling is established through the receptors, additional specificity is reinforced through the degradation targets. These have been described not merely as suppressors of signalling but also as growth regulators, the activities of which are modulated via SL or KAR/KL signalling (Jiang et al., 2013).
In this study, we asked whether SL and KAR/KL have a role in regulating root skewing. Using mutants impaired in proteins that are likely receptors for these compounds, we showed that while SL has no effect on root skewing, mutants deficient in KAR/KL perception, max2 and kai2, display an enhanced root‐skewing phenotype. We also investigated the mechanism by which KAI2 and MAX2 modulate root skewing.
Results
Mutation in kai2 and max2 increases root rightward skew
If KLs or KARs were involved in root skewing, then insensitive Arabidopsis mutants would display an aberrant root‐skewing phenotype. Vertically grown kai2‐1 and kai2‐2 mutants showed significantly increased rightward root skewing compared with the Ler wild‐type (α, root tip displacement, viewed from the back of the plate: Figure 1a–c; Tukey HSD, P < 0.01). The root‐skewing angle of kai2‐2 mutant in the Col‐0 background [kai2‐2 (6x Col‐0)] was also significantly higher than that of the wild‐type (Figure S1a; Tukey HSD, P < 0.01). Vertically grown max2‐7 and max2‐8 mutants showed a significant increase in rightward root skewing compared with their Ler parental wild‐type (Figure 1b,c; Tukey HSD, P < 0.01).
Horizontal growth index (HGI; ratio of root tip displacement along the x‐axis to root length; Grabov et al., 2004; Vaughn and Masson, 2011) was also significantly higher in kai2‐1, kai2‐2, max2‐7 and max2‐8 compared with wild‐type (Figure 1d; Tukey HSD, P < 0.01), supporting the skewing angle data and showing increased deviation from vertical by mutant roots. Similarly, the vertical growth index (VGI; ratio of root tip displacement along the y‐axis to root length; Grabov et al., 2004; Vaughn and Masson, 2011) was significantly smaller for kai2‐1, kai2‐2, max2‐7 and max2‐8 compared with wild‐type (Figure 1e; Tukey HSD, P < 0.01). In separate experiments, two complemented kai2‐2 lines (driven by the native promoter KAI2:KAI2 kai2‐2; Waters et al., 2015b) showed a significantly decreased root‐skewing angle compared with kai2‐2 (Figure S1b; Tukey HSD, P < 0.01). Overall these data suggest a role for both KAI2 and MAX2 in preventing exaggerated root skewing in Arabidopsis.
KAI2 and MAX2 affect root skewing and waving on a tilted surface
Positioning plates at a 45° angle from the vertical rather than vertically increases the root‐skewing angle. A significant increase in rightward root‐skewing angle was observed here for the Ler wild‐type grown at a 45° plate angle (Figure 2a,b; anova, F 1,510 = 134.9, P < 0.001), while kai2‐1, kai2‐2, max2‐7 and max2‐8 also showed a significantly increased rightward root‐skewing angle compared with Ler (Figure 2a,b; Tukey HSD, P < 0.01, except for max2‐7 where P < 0.05). The increase in mutant root skew relative to wild‐type was maintained at the 45° plate angle compared with growth at 90°, indicating that loss of KAI2 or MAX2 did not affect the mutant's ability to sense and respond to the tilt.
Figure 2.
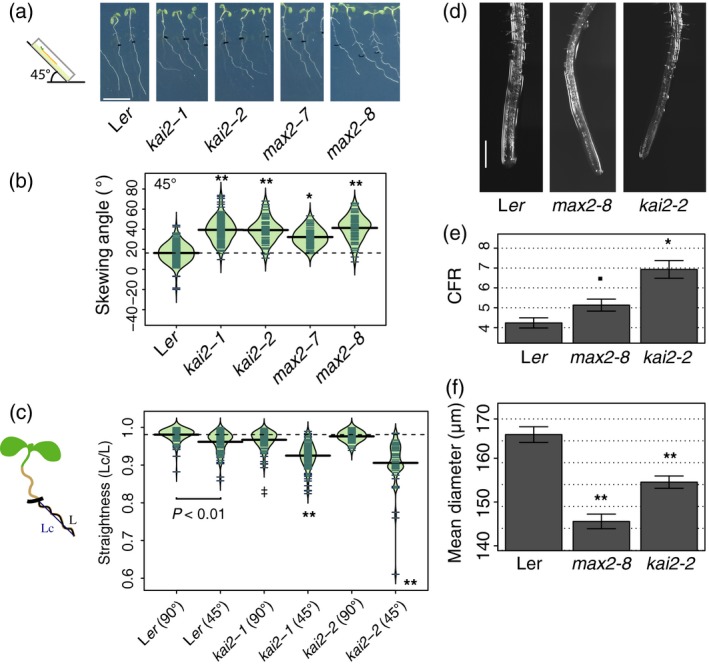
kai2 and max2 increased rightward root skewing and cell file rotation (CFR) when placed at 45°.
(a) Seedlings of kai2‐1, kai2‐2, max2‐7 and max2‐8 were grown vertically for 6 days, then placed at 45° for 3 days (indicated by a tick on the roots). Scale bar: 1 cm.
(b) The root‐skewing angle (α) was measured as the deviation from the vertical for plants grown at a 45° angle for 3 days.
(c) The straightness (measured as the ratio of the chord Lc to root length L; Grabov et al., 2004; Vaughn and Masson, 2011) of seedling roots from wild‐type, kai2‐1 and kai2‐2 decreased when plants were grown at 45° compared with 90° (shown in brackets after genotype). Data for each genotype are displayed as a beanplot with the straightness of individual roots shown as dark green horizontal lines, while the mean is represented by a thick black horizontal line. The estimated density of the distribution is illustrated by the shaded colour. The dashed line corresponds to the mean for the wild‐type. Significant differences compared with wild‐type grown at 45° (Tukey HSD) are shown as **P < 0.01, while comparisons with wild‐type grown at 90° are indicated by ‡ P < 0.05. For each genotype, n > 58 in three separate experiments.
(d) Both max2‐8 and kai2‐2 mutants show increased CFR, indicating that the root epidermal cells were twisting more compared with those of the wild‐type. CFR was measured as the number of epidermal cells that crossed a 1‐mm line 1.5–2 mm from the root tip. Plants were grown at 45°.
(e) Data shown as mean ± se, n = 28–42 plants obtained in four separate experiments, *P < 0.05, • P < 0.1. Scale bar: 500 μm.
(f) The root diameter of max2‐7 and kai2‐2 plants was lower than that of wild‐type. Data shown as mean ± SEM, n > 36 per genotype in a total of five experiments, **P < 0.01 (Tukey HSD).
Increased root skewing is often also accompanied by increased root waving (Roy and Bassham, 2014) – a decrease in root straightness calculated as the ratio of the cord over the root length (i.e. straight roots have a ratio of 1 and the lower the ratio the less straight/more wavy the root; Grabov et al., 2004; Vaughn and Masson, 2011). Growth on a tilted surface can also decrease straightness (Roy and Bassham, 2014). When grown at 45°, both kai2‐1 and kai2‐2 showed a decreased straightness (Figure 2c; Tukey HSD, P < 0.01). Ler was significantly less straight when grown at 45° compared with 90° (Tukey HSD, P < 0.01). When grown at a 90° plate angle, kai2‐1 (Tukey HSD, P < 0.05) but not kai2‐2 (Tukey HSD, n.s.) showed a significantly decreased straightness compared with wild‐type Ler (Figure 2d). These data show that KAI2 is involved in the negative control of both skewing and waving when plants are grown at an angle, but only skewing when grown vertically.
KAI2 and MAX2 affect epidermal cell file rotation and root diameter
Although mechanistic models for root skewing vary (Roy and Bassham, 2014), the rotation of epidermal cell files is considered to be an important feature (Sedbrook et al., 2002; Oliva and Dunand, 2007; Wang et al., 2011). Right‐handed cell file rotation (CFR) was increased in both kai2‐2 (mean ± se: 6.93 ± 0.44 cell mm−1; Tukey HSD, P < 0.01) and, to a lesser extent, max2‐8 (5.13 ± 0.30 cell mm−1; Tukey HSD, P = 0.08) compared with Ler wild‐type (4.24 ± 0.25 cell mm−1; Figure 2d,e).
Furthermore, the mean root diameter of the mutants was significantly narrower than that of wild‐type (Figure 2f; Ler: 166.43 ± 1.79 μm; kai2‐2: 155.57 ± 1.41 μm; max2‐8: 146.59 ± 1.67 μm; Tukey HSD, P < 0.001), suggesting that root radial expansion may be restricted.
KAI2 and MAX2 operate through the same genetic pathway
Genetic studies of the elongated hypocotyl phenotypes (Waters et al., 2012) have suggested that KAI2 and MAX2 are in the same signalling pathway, and physical interaction between these two proteins has been demonstrated in a yeast two‐hybrid assay (Toh et al., 2014). Here we use a similar genetic approach to show that the double‐mutant kai2‐2 max2‐8 has a significantly increased rightward root skew compared with wild‐type (Figure 3a,b; Tukey HSD, P < 0.01), which was not significantly different from that of kai2‐2 (Figure 3a,b; Tukey HSD, n.s.). That the skewing angle of the kai2‐2 max2‐8 double‐mutant was not greater than that of the kai2‐2 single‐mutant suggests that KAI2 and MAX2 operate in the same genetic pathway.
Figure 3.
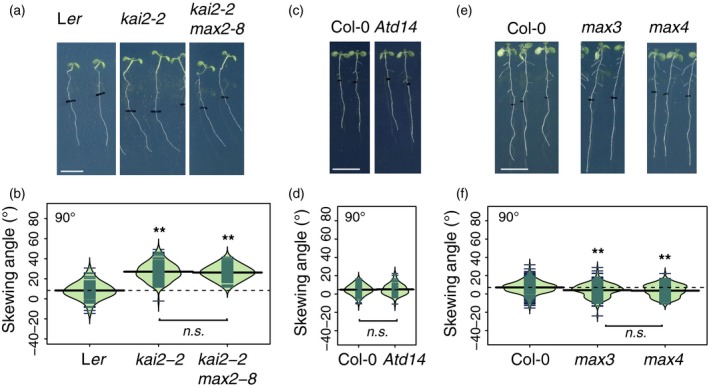
KAI2 and MAX2 regulate root skewing through the same genetic pathway, which does not involve D14.
(a, b) Seedlings for the double‐mutant kai2‐2 max2‐8 showed no further increase in root‐skewing angle compared with kai2‐2. Scale bar: 1 cm. Data for each genotype are displayed as a beanplot with the skewing angle of individual roots shown as dark green horizontal lines, while the mean is represented by a thick black horizontal line. The estimated density of the distribution is illustrated by the shaded colour. The dashed line corresponds to the mean for the wild‐type. **Indicates significant difference compared with wild‐type (Tukey HSD, P < 0.01). For each genotype, n > 66 in five separate experiments.
(c) Seedlings for the strigolactone (SL)‐insensitive mutant Atd14 showed no increased rightward root skewing, and the measured skewing angle was not significantly different from that of the wild‐type (d). For each genotype, n > 73 from three experiments.
(e, f) Seedlings for the SL synthesis mutants max3 and max4 show a slight decrease in rightward root skewing, n > 142 from three experiments.
Karrikin reduces root skewing, but is a poor analogue of KAI2‐ligand
The data demonstrate that in Arabidopsis an impairment in KAR/KL perception leads to greater rightward root skewing. This suggests that perhaps the abundance of KAR or KL in the roots may affect root skewing, and the hypothesis that an increased availability of KL or its analogue KAR2 might compensate for a lowered sensitivity of the system and decrease the rightward root skewing. In the absence of purified and identified KL compounds, the effect of KAR on root skewing was tested using the potent KAR2 (Nelson et al., 2009; Waters et al., 2015a). There was a significant effect of KAR2 in reducing rightward root skewing of Ler wild‐type plants with concentrations of 5 and 10 μm (Figure S2a; Tukey HSD, P < 0.01). However, a significant inhibitory effect on primary root elongation of Ler plants was evident at 10 μm KAR2 (Figure S2b; Tukey HSD, P < 0.01).
The presence of 2.5 and 5 μm KAR2 in the medium also significantly decreased the root‐skewing angle of kai2‐2 (Figure S2b; Tukey HSD, P < 0.01). The KAI2‐independent effect of KAR2 on root skewing may also be linked to reduced root elongation, as this was significantly lower in the presence of 5 μm KAR2 (Figure S2b; Tukey HSD, P < 0.01), but not at 2.5 μm (Figure S2b; Tukey HSD, n.s.). Similarly, the presence of 5 μm KAR2 in the medium significantly decreased the root‐skewing angle of max2‐8 (Figure S2e; Tukey HSD, P < 0.01) as well as primary root elongation (Figure S2f; Tukey HSD, P < 0.01). A negative effect of KAR1 on rightward root skewing (Figure S2g) could also be measured in kai2‐2 and max2‐8 plants, while Ler plants remained insensitive. KAR could significantly reduce rightward root skewing; however, this may be an unspecific effect as kai2 plants also responded and at lower KAR concentrations compared with Ler plants.
Strigolactones do not affect root skewing
Given the root‐skewing phenotype of max2 mutants, and the role of MAX2 in SL perception, we also investigated the role of SL in root skewing. To demonstrate a role for SL in regulating root skewing, we looked for evidence for a phenotype in SL perception (d14) and synthesis (max3, max4) mutants. Max3 and max4 mutants are impaired in carotenoid cleavage dioxygenase 7 and 8, respectively, key enzymes involved in SL synthesis (Sorefan et al., 2003; Booker et al., 2004; Schwartz et al., 2004). We also report here the effect of GR24, a widely used analogue of SL. Under our growth conditions, we have found no evidence supporting a role for SL in regulating root skewing. d14 mutants that are insensitive to SL but not KAR (Waters et al., 2012) showed no significant increase in root skewing compared with wild‐type (Figure 3c,d; anova, n.s.). In addition, both max3 and max4 show no significant increase in root‐skewing angle, rather they showed a small but significant reduction in root‐skewing angle compared with the wild‐type Col‐0 (Tukey HSD, P < 0.01, for both max3 and max4).
A racemic mix of GR24 (GR24rac) that was shown to regulate root growth (Kapulnik et al., 2011; Ruyter‐Spira et al., 2011; Rasmussen et al., 2012) and that can also be perceived by KAI2 (Scaffidi et al., 2014; Waters et al., 2015a) was tested at 1 and 5 μm, as greater concentrations tended to have a toxicity effect on root growth (Ruyter‐Spira et al., 2011). Treatment with GR24rac led to a small increase in rightward root skewing in Ler plants at 1 μm (Figure S3a; Tukey HSD, P < 0.05) but not at 5 μm GR24rac (Tukey HSD, n.s.), whereas only kai2‐2 and not kai2‐1 responded with a decrease in rightward root skewing at 5 μm GR24rac (Tukey HSD, P < 0.05). There was no significant effect of 1 or 5 μm GR24rac on the root skewing of Col‐0 plants (Figure S3b; anova, F 2,261 = 1.26, n.s.), whereas max2, but not d14 (anova, F 2,184 = 1.31, n.s.), showed a small but significant increase in root‐skewing angle under 1 μm GR24rac (Tukey HSD, P < 0.05) but not 5 μm GR24rac (Tukey HSD, n.s.).
GR24rac chemically complements SL‐deficient mutants, max3 and max4 (Ruyter‐Spira et al., 2011; Rasmussen et al., 2012). If SL could affect root skewing, max3 and max4 mutants (which showed a reduced root‐skewing phenotype compared with wild‐type; Figure 3e,f) should show a slight increase in rightward root skewing in the presence of GR24. However, both mutants showed a further decrease in the skewing angle, in the presence of 5 μm GR24 (Tukey HSD, P < 0.01).
Moreover, the root skewing of mutants deficient in DLK2 (D14‐LIKE 2) proteins (Waters et al., 2012) was not significantly different to wild‐type (Figure S1c; Tukey HSD, n.s.). As the DLK2 protein is related to both KAI2 and D14, overall these data demonstrate a specific role for KAI2 and MAX2 in modulating root skewing and thus implicate KL/KAR, and not SL, sensing through these proteins.
MAX2 effect on root skewing requires SMAX1, SMXL2 and SMXL6,7,8
The effect of MAX2 degradation targets, SMAX1 (SUPPRESSOR OF MAX2‐1) and SMXLs (SUPPRESSOR OF MAX2‐1‐LIKE, Stanga et al., 2013), on root skewing was examined, thus testing the hypothesis that the MAX2‐dependent regulation of protein abundance for members of the SMAX/SMXL family is relevant to the root‐skewing phenotype. The current mechanistic model for Arabidopsis is that SMAX1 and SMXL2 are important for the KL part of the signalling pathway, whereas SMXL6,7,8 are more relevant to the SL part of the pathway (Soundappan et al., 2015). Here we report that there was no significant difference between Col‐0 and max2 smax1‐2 (Tukey HSD, n.s.), Col‐0 and max2‐1 smxl2 (Tukey HSD, n.s.), Col‐0 and smax1‐2 smxl2 max2‐1 (Tukey HSD, n.s.) or Col‐0 and smxl6,7,8 max2‐1 (Tukey HSD, n.s.), thus it seems that the absence of SMAX1, SMXL2 or SMXL6,7,8 suppresses the max2 phenotype. Interestingly, both smax1‐2 and smxl6,7,8 mutants, but not smxl2 (Tukey HSD, n.s.), showed a significant decrease in root‐skewing phenotype compared with wild‐type (Figure 4a,b; Tukey HSD, P < 0.01), thus reinforcing the idea that the abundance of these proteins affects root skewing. Interestingly, while SMXL3, 4 and 5 are central regulators for phloem formation, and mutants deficient in at least two members of this subclade display a short and thin root phenotype, no skewing phenotype was reported (Wallner et al., 2017).
Figure 4.
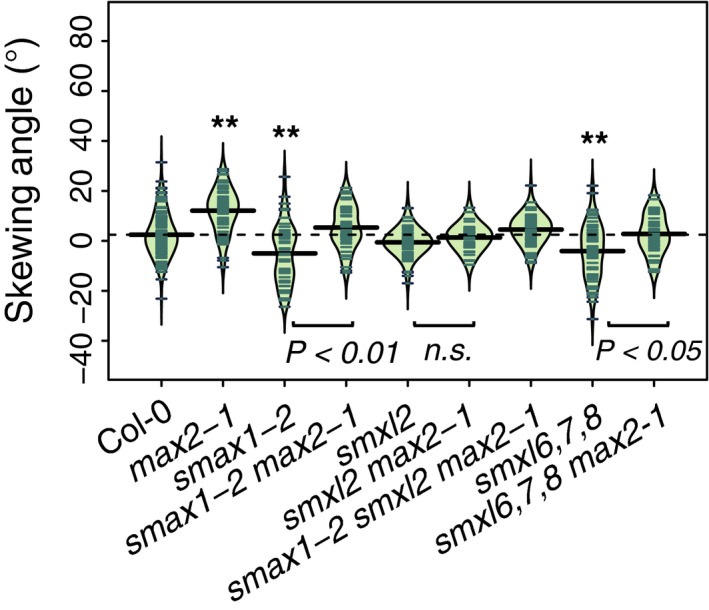
MAX2 effect on root skewing requires SMAX1, SMXL2 and SMXL6,7,8.
Root‐skewing phenotypes of Col‐0, max2‐1, smax2‐1, smax2‐1 max2‐1, smxl2, smxl2 max2‐1, smax1‐2 smxl2 max2‐1, smxl6,7,8 and smxl6,7,8 max2‐1 while grown at 90°. Data for each genotype are displayed as a beanplot with the skewing angle of individual roots shown as dark green horizontal lines, while the mean is represented by a thick black horizontal line. The estimated density of the distribution is illustrated by the shaded colour. The dashed line corresponds to the mean for the wild‐type. Significant differences compared with wild‐type (Tukey HSD) are shown: *P < 0.05 and **P < 0.01. For each genotype, n > 38 from three experiments.
kai2 and max2 can support a near‐normal mechano‐sensing response
The growth responses of the kai2 mutants on tilted plates suggested that the mutation does not affect the root tip's ability to sense the increased mechanical impedance afforded by the inclined growth medium. Rather, that the kai2 mutants have an exaggerated root skew when grown on a tilted surface suggests that downstream responses are impaired. To test for a role for KAI2 in mechano‐sensing responses, seedlings were subjected to mechanical stress prior to determination of root transcript levels of CML12 and CML24 (CALMODULIN‐LIKE PROTEIN; Figure 5a). These transcripts are known to increase upon mechanical stimulation (Braam and Davis, 1990). These tests also addressed max2 and d14 in the Col‐0 background (Figure 5b). Mechanical stimulation caused significant upregulation of CML12 and CML24 transcript in roots of all genotypes tested (anova, P < 0.01), but no mutants responded significantly differently to the wild‐type. Thus, the data suggest that root transcriptional mechano‐responsiveness is not drastically altered in either KL‐ or SL‐insensitive mutants.
Figure 5.
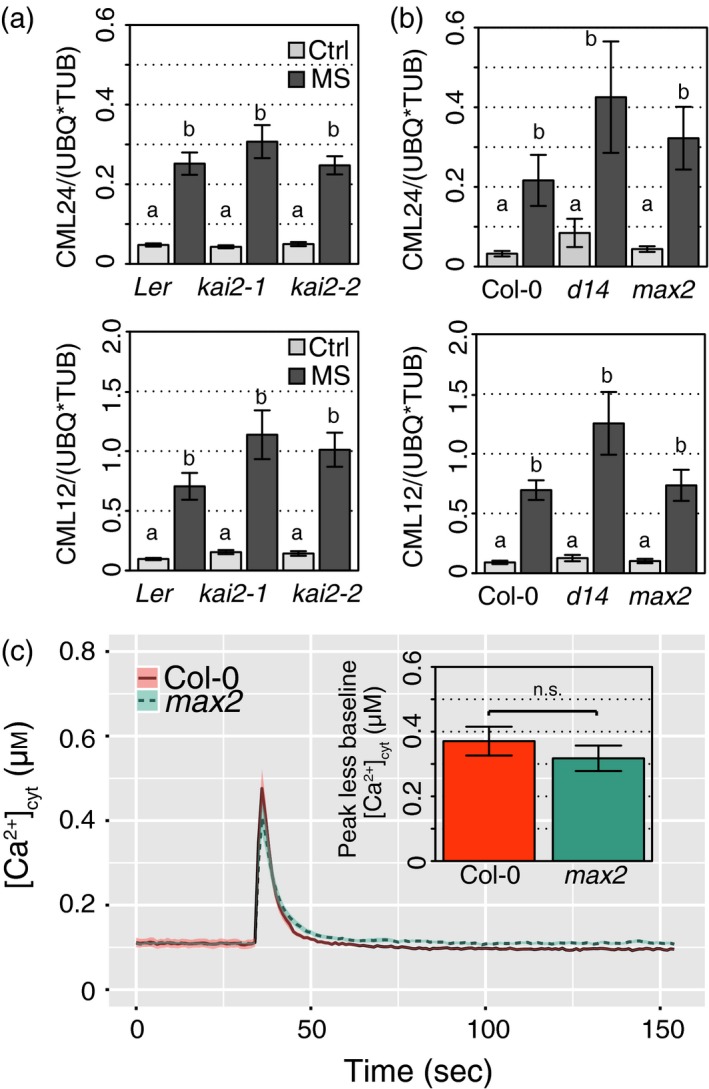
kai2, max2, d14 mutants support a near‐normal response to mechano‐stimulus.
Karrikin (KAR)‐ and strigolactone (SL)‐insensitive mutants showed a normal upregulation of touch response genes, in response to mechanical stimulation. Nine‐day‐old seedlings of wild‐type and mutants (a) kai2‐1 and kai2‐2, and (b) d14, max2‐1 were mechanically stimulated (MS) for 30 sec, then collected 30 min later for transcript analysis of touch‐sensitive genes CML12 and CML24, relative to housekeeping genes Tubulin 4 and Ubiquitin 10. The means of six–nine replicates from three independent experiments are shown, each replicate based on the RNA extracted from roots of 30–40 seedlings. Data are shown as mean ± se, letters indicate significant differences (Tukey HSD, P < 0.01).
(c) Mechano‐stimulated [Ca2+]cyt increase in max2 root tips. Individual excised root tips of Col‐0 and max2 expressing (apo)aequorin as a [Ca2+]cyt reporter were mechanically stimulated by addition of buffer at 35 sec. The mean ± SEM of 40–67 roots in five independent trials are shown. Inset: mean ± SEM maximal [Ca2+]cyt increment in response to stimulus (peak response minus baseline).
As a final test for alteration in mechano‐sensing and response, max2 (as the common lesion in KL‐ and SL‐pathways) was transformed to express (apo)aequorin as a reporter of cytosolic free Ca2+ ([Ca2+]cyt). [Ca2+]cyt increases transiently in response to mechano‐stimulation, acting as a second messenger (Knight et al., 1991; Shih et al., 2014). There was no significant difference between baseline level pre‐injection and post‐injection for Col‐0 (t‐test, n.s.) or max2 (t‐test, n.s.). There was no significant difference in the amplitude of the touch‐induced peak increase in [Ca2+]cyt between genotypes (Figure 5c; t‐test, n.s.). However, the total Ca2+ mobilised over the recording period (excluding the discharge) for max2 (33.99 ± 0.57 μm) was significantly higher than that for Col‐0 (29.91 ± 0.49 μm; t‐test, P < 0.01). This is in contrast to the feronia plasma membrane receptor‐like kinase mutant that fails to support normal touch‐induced [Ca2+]cyt elevation but, in common with max2 and kai2, has rightward‐skewing roots (Shih et al., 2014). Therefore, there is no clear link between calcium handling and root‐skewing phenotype of max2.
kai2 but not max2 has a slower early gravitropic response
Agravitropic mutants can also show an increased root skewing (Okada and Shimura, 1990). To investigate whether an aberrant gravitropic response of kai2‐2 plants contributed to their skewing phenotype, root tip orientation was monitored every 10 min after gravistimulation for 10 h. Both kai2‐2 and wild‐type responded significantly with a change in tip orientation over time (Figure 6; anova, F 1,4022 = 46.8, P < 0.01). Comparisons of the responses (normalised for elongation rate) using anova showed that there was a significant interaction between time and genotype (anova, F 1,4022 = 40.9, P < 0.01), indicating a difference in gravitropic response between genotypes. kai2‐2 has a dampened initial response as root tip angle started to decrease later than Ler. After 100 min, the angle of kai2‐2 was significantly higher than that of Ler (anova, F 1,64 = 4.4, P < 0.01), but at 600 min there was no significant difference (anova, F 1,64 = 0.24, n.s.). Overall, the difference in gravitropic response between kai2‐2 and Ler may be a small contributory factor to root skewing, but occurring only in the early stages of the response.
Figure 6.
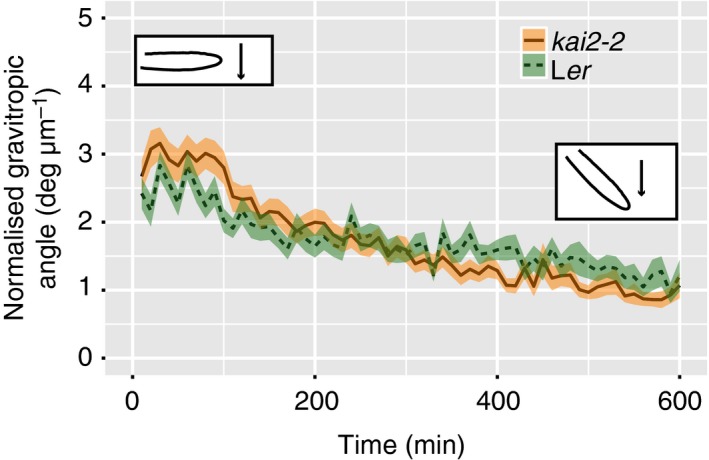
Gravitropic response of kai2 is slower than that of wild‐type.
The tip orientation of roots from wild‐type and kai2‐2 was recorded every 10 min and for 10 h after a change in gravitropic orientation. The change in tip orientation was normalised to the tip displacement to take into account differences in growth rate between genotypes. Data are shown as mean ± se, n = 16–22 plants obtained in five experiments.
The gravitropic response of SL synthesis and perception mutants was also monitored (Figure S4). All genotypes responded to the shift in gravitropic angle with a change in tip orientation over time (anova, F 1,2188 = 1343.1, P < 0.01). There was a significant difference amongst genotypes (anova, F 3,2188 = 30.1, P < 0.01), with SL‐insensitive and synthesis mutants showing earlier change in normalised gravitropic angle, compared with wild‐type. Although both kai2 and max2 were impaired in their initial gravitropic responses, their contrasting responses (delayed and earlier, respectively) are inconsistent with both skewing rightward.
sks3 suppresses max2 root skewing but not lateral root density phenotype
Similarly to the kai2 and max2 mutants, mutant plants deficient in the SKU5 protein that is linked to the plasma membrane by a glycosylphosphatidylinositol anchor also showed an increased rightward root‐skewing phenotype, increased CFR with no change in gravitropic response (Sedbrook et al., 2002). In our experiments, sku5 also displayed a rightward skew when grown vertically that was significantly greater than the wild‐type (Figure 7a,b; Tukey HSD, P < 0.05). We further investigated the phenotype of a related mutant, deficient in sks3 (sku5 similar 3), as well as multiple mutants deficient in members of the SKS/SKU family (Zhou, 2013). The sks3 mutant skewed to the left (Tukey HSD, P < 0.05). Interestingly, the root‐skewing phenotype of sks3 was maintained even in the absence of MAX2 (comparison sks3: sks3 max2‐1, Tukey HSD, n.s.), suggesting that sks3 completely suppresses the max2 phenotype. The skewing angle of sku5 max2 was not significantly higher than that of max2 (Tukey HSD, n.s.). sks3 and sku5 do not suppress the high LRD phenotype (Figure S5) or the decreased germination rate of max2 mutants (Figure S5). These data suggest that the abundance of SKS3 protein may itself affect root skewing, and support the previous report of a root‐skewing phenotype in sku5.
Figure 7.
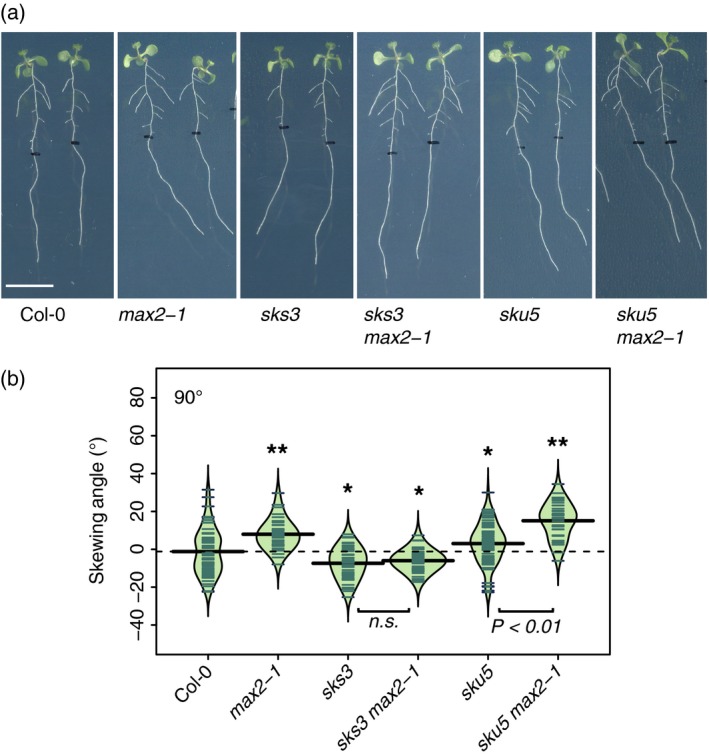
MAX2 regulation of root skewing involves SKS3.
(a) Seedlings of Col‐0, max2‐1, sks3, sks3/max2‐1, sku5, sku5/max2‐1 mutants grown at 90°. Scale bar: 1 cm.
(b) Data for each genotype are displayed as a beanplot with the skewing angle of individual roots shown as dark green horizontal lines, while the mean is represented by a thick black horizontal line. The estimated density of the distribution is illustrated by the shaded colour. The dashed line corresponds to the mean for the wild‐type. *Indicates a significant difference compared with wild‐type (Tukey HSD, P < 0.05). For each genotype, n > 34 in three separate experiments.
The cellular localisation of SKS3 (At5g48450) and SKU5 (At4g12420) was surveyed using available online high‐resolution root expression patterns (Brady et al., 2007). SKS3 is expressed in the cortex in the meristematic zone (Figure S6a), and at a lower level compared with SKU5 (Figure S6c,f). SKU5 mRNA could be detected in the endodermis in the meristematic and elongation zones, as well as the vasculature, particularly procambium of the meristematic zone (Figure S6d). KAI2 (At4g37470) transcripts occur in the vasculature (procambium) throughout the root, MAX2 (At2g42620) is also expressed in the vasculature but in the phloem companion cells and mostly in the elongation and maturation zones (Figure S7; Winter et al., 2007). Besides the overlap between SKU5 and KAI2 in the procambium in the meristematic zone, there is little overlap between KAI2, MAX2, SKU5 and SKS3 expression patterns.
Discussion
The characterisation of phenotypes for kai2 and max2 mutants that are unrelated to the presence of smoke, as well as the presence of these receptors in non‐fire following plants have led to the hypothesis that endogenous KLs are present in plants and act as phytohormones (Conn and Nelson, 2016). Evidence supporting their presence in the water‐soluble fraction of Arabidopsis shoot extract has now been reported (Sun et al., 2016). Here we report evidence demonstrating a role for KAI2 and MAX2 in preventing exaggerated root skewing in Arabidopsis, thus providing additional support for a hypothetical endogenous KL.
Phytohormones such as auxins, ethylene and cytokinins have been implicated in the regulation of root skewing, but thus far the role of SLs has remained unknown (Roy and Bassham, 2014). In Arabidopsis, SLs are primarily produced in the roots and transported in the xylem (Goldwasser et al., 2008; Kohlen et al., 2011), though some SL production could be shown in the shoots (for review, see Al‐Babili and Bouwmeester, 2015). They have been shown to regulate LRD, primary root elongation, root hair elongation and adventitious root growth (Koltai et al., 2009, 2010; Ruyter‐Spira et al., 2011; Rasmussen et al., 2012, 2013; Shinohara et al., 2013; Kapulnik and Koltai, 2014), but root skewing or waving phenotypes have not been reported. We investigated the root‐skewing phenotype of SL mutants, and found no evidence supporting a role for endogenous SLs in regulating root skewing. While max2 mutants displayed an increased root‐skewing phenotype, this is not the case for d14, and we can attribute the phenotype of max2 to its role in KAR/KL sensing. Mutants deficient in SL synthesis (max3, max4) show a reduced root‐skewing angle compared with wild‐type, but this is not chemically complemented by providing exogenous GR24rac. Thus, we propose that SL has no role in regulating root skewing in Arabidopsis, at least under our growth conditions.
Mutants have proved useful in identifying new components of the machinery regulating root skewing in Arabidopsis. Here, the increased root‐skewing phenotype of kai2 and max2 suggests that both KAI2 and MAX2 negatively regulate root skewing. Because these two proteins are involved in the perception of KAR/KL, this provides evidence supporting a role for KL in regulating root skewing, though it is possible that the effects of KAI2 and MAX2 may be ligand independent. These two proteins have been implicated in promoting germination, hypocotyl elongation, light response, reducing hyponasty, establishing arbuscular mycorrhizal fungal (AMF) symbiosis, affecting leaf morphology and drought resistance (Nelson et al., 2011; Sun and Ni, 2011; Waters et al., 2012; Stanga et al., 2013; Gutjahr et al., 2015; Li et al., 2017; Lee et al., 2018). Root‐related phenotypes reported for any mutant deficient in KAI2 include the absence of AMF symbiosis in rice (Gutjahr et al., 2015) and a lower large LRD in rice (Chiu et al., 2017). The AMF symbiosis seems to be impaired in the early stages of the interactions as no physical contact between plant and fungus or gene expression changes generally triggered by fungal signals could be found. Our data provide additional evidence of the functionality of KAI2 and MAX2 proteins in roots. In common with the symbiosis phenotype, it is currently difficult to resolve a phenotype that appears to relate to a response at the epidermal level (e.g. change in CFR in skewing) with the vascular localisation of max2 and kai2 expression, although the KAI2 and MAX2 protein localisation may differ from that of the mRNA. Interestingly, KAI2 light‐induced expression in the short term is hy5‐dependent (Waters and Smith, 2013), with the HY5 transcription factor binding to both a C/G‐box and a G‐box in the KAI2 promoter (Sun and Ni, 2011). The hy5 mutant was shown to have a root‐skewing phenotype (Oyama et al., 1997). Though the KAI2 and HY5 act largely independently to regulate hypocotyl length (Waters and Smith, 2013), it is unclear whether they might operate in the same pathway to regulate root skewing.
In the absence of endogenous purified KL compounds, KARs have been used as analogues to KLs. For phenotypes such as elongated hypocotyls or increased seed dormancy (Waters et al., 2012), KAR2 acts as a good synthetic analogue for KL (Conn and Nelson, 2016). However, this is not the case for root skewing. A high concentration of KAR2 or KAR1 was necessary to induce a root‐skewing phenotype, and this effect was KAI2 and MAX2‐independent. Interestingly, KAR2 also failed to induce significant changes in gene expression in rice roots (Gutjahr et al., 2015). Many SL compounds, which may be structurally related to KL, have been purified thus far (Bouwmeester et al., 2007), and perhaps there is also some structural diversity amongst KL compounds. Given that neither KAR nor GR24rac can affect the root‐skewing phenotype in a MAX2‐ and KAI2‐dependent way, we argue that they are poor KL analogues with regards to the regulation of root skewing, and that they have limited use in the study of KAI2/MAX2 regulation of this phenotype.
In common with the perception systems for phytohormones such as auxins, giberellins or jasmonate (Gray et al., 2001; Dill et al., 2004; Thines et al., 2007), the current model proposes that SL and KAR/KL are perceived by binding to an α/β hydrolase receptor (D14 or KAI2), which is then recruited by a SCFMAX2 complex that also includes proteins targeted for ubiquitination and degradation through the 26S proteasome. In Arabidopsis, degradation targets for the D14/MAX2 complex were first identified through a genetic screen for suppressors of the max2's low germination phenotype (Stanga et al., 2013). This study identified SMAX1 (Stanga et al., 2013), with additional members of the SMAX/SMXL family subsequently identified due to sequence similarity (Soundappan et al., 2015). Furthermore, the MAX2‐ and D14‐dependent degradation of SMXL7 upon treatment with GR24 demonstrated that this protein was effectively a target for degradation in Arabidopsis (Soundappan et al., 2015). In rice, D53 protein (orthologue to SMAX1) was identified as a suppressor of the SL pathway (Jiang et al., 2013; Zhou et al., 2013). Detailed analyses demonstrated the degradation of D53 in a GR24rac‐ as well as a D3‐ and D14‐dependent manner. Polyubiquination of D53 was also demonstrated upon treatment with GR24rac but not KAR1. Demonstrating the SMAX/SMXL degradation or interaction with MAX2 or D14 has relied a lot (but not exclusively) on the use of GR24rac as ligand. Interestingly, thus far the KAR/KL‐dependent degradation of members of the SMAX/SMXL family has not been shown yet. Perhaps it is that the KARs currently used are poor mimics of the endogenous ligand that could trigger the complex assembly, ubiquitination and degradation of the target proteins, and that the unavailability of such endogenous ligands is impeding these experiments. It is also possible that the KAR/KL signalling pathway operates following a different model.
The current model proposes a dichotomy between SMAX1‐KAI2‐KAR/KL‐, SMXL6,7,8‐ and D14‐SL‐signaling pathways (Soundappan et al., 2015), while SMXL3, 4 and 5 are regulated independently from KAR and SL (Wallner et al., 2017). However, our data do not support this idea, and support a role for MAX2 in regulating root skewing in a D14‐independent manner through SMXL6,7,8 as well as SMAX1/SMXL2. Thus, the dichotomy in terms of degradation targets may hold only for some phenotypes. Much may depend on the spatial localisation of proteins. SMAX1 is expressed in the root cap, while SMXL6, 7 and 8 are also present in the vasculature of mature roots (Soundappan et al., 2015). KAI2 expression could be found preferentially in the vasculature (Brady et al., 2007) potentially favouring interaction with SMXL6, 7 or 8.
It is unclear how changes in abundance of SMAX/SMXL proteins may affect root skewing, besides targeted changes in gene expression through their interaction with the TOPLESS/TOPLESS‐RELATED family of transcriptional co‐repressors (Jiang et al., 2013; Soundappan et al., 2015). Furthermore, it is likely that members of the SMAX/SMXL family are not the only proteins targeted for degradation through MAX2. Out of 117 proteins shown to be differentially abundant in max2 roots compared with wild‐type, the abundance of only nine of these was different between max2 and wild‐type in the presence of GR24rac (Walton et al., 2016). This suggests that MAX2 can lead to change in the abundance of many more proteins beyond SL signalling. Thus, we also considered additional proteins that may regulate the root‐skewing phenotype. Members of the SKS/SKU family, especially SKU5 and SKU6, have been shown to have a root‐skewing phenotype, though the sku6/spr1‐6 mutant also showed a twisted petiole phenotype that was not noted in kai2 or max2 mutants (Sedbrook et al., 2002; Sedbrook, 2004). Given the proposed role for the KAI2/MAX2 in targeting protein for degradation and given the phenotypes of multiple mutants (Figure 7), it is fair to hypothesise that SKS3 may be a target for degradation via the KAI2/MAX2 complex in the context of the root‐skewing phenotype. Though genetic data have been used in the past to demonstrate such links (Stanga et al., 2013; Soundappan et al., 2015), further data supporting a protein–protein interaction or evidence of degradation would be necessary. As discussed above, such experiments may be difficult to execute in the absence of an appropriate ligand. Interestingly, SKU5 could be detected and remain unchanged in the root proteome of max2 compared with Col‐0 plants (Walton et al., 2016). SKS3 protein could not be detected perhaps because it is expressed at much lower levels, compared with SKU5 (Figure S6). Thus far, germination, hypocotyl length and leaf shape have been standard phenotypes used to investigate the KAR/KL signalling pathway. We propose that the root‐skewing phenotype may be useful to identify new proteins targeted for degradation and gain new information on the signalling pathway for KAR/KL.
Here we demonstrated that functional KAI2 and MAX2 proteins could prevent an exaggerated root skewing. Given the known role for KAI2 and MAX2 in plants, it is currently difficult to provide a functional explanation for this phenotype. Furthermore, the mechanism by which KAI2 and MAX2 regulates root skewing remains elusive. We found no evidence supporting a role for KAI2 and MAX2 in regulating the root touch‐dependent upregulation of CML12 and CML24, suggesting that the mechano‐sensitive transcriptional response may be unaltered. The mechano‐stimulated [Ca2+]cyt response appears overall similar in max2 compared with wild‐type, and this does not correlate well with feronia's impaired mechano‐stimulated [Ca2+]cyt response but similar rightward skew (Shih et al., 2014). In addition, the kai2 and max2 mutants show opposite response to change in the gravitropic vector. Thus, an altered gravitropic response does not help explain the root‐skewing phenotype. The enhanced CFR and lower root diameter measured in both max2 and kai2 mutants may lead us to an explanation. Perhaps thinner roots encounter less friction with the underlying medium, leading to greater rotation and skewing. However, further work is needed to firmly establish the mechanism of root skewing and the role of KAI2 and MAX2.
The link established here between MAX2 and the SKU/SKS family suggested an interesting possibility that the max2 skewing phenotype is linked to cell wall modification or integrity. A case supporting a role for cell wall in affecting root skewing is already well established in the literature, with root‐skewing mutants showing clear cell wall phenotype (Nakashima et al., 2013; Lim et al., 2014; Roy and Bassham, 2014; Van der Does et al., 2017) as well as microarrays of roots showing different skewing angles (Vaughn and Masson, 2011; Schultz et al., 2017). For example, amongst the 11 highly probable skew gene candidates identified in Arabidopsis roots using microarrays, three were associated to the cell wall either because of their physical location (PAP24), or because of their role in cell wall integrity (DIN2) or formation (MIOX4; Schultz et al., 2017).
Several lines of evidence suggest that KL and KAR affect cell wall composition, though the sugar composition of max2 and kai2 mutants cell wall appears normal (J. Mortimer, personal communication). Amongst the 133 genes that are differentially regulated 24 h post‐imbibition with 1 μm KAR1, 11 relate to the cell wall, and genes belonging to the ‘plant‐cell type cell wall’ category of the GO cellular components were significantly enriched in the set of genes regulated by KAR1 (Nelson et al., 2010). Genes involved in cell wall organisation were also significantly enriched within a set of upregulated genes in the shoots of kai2‐2 compared with wild‐type (27 genes out of 680 significantly upregulated in kai2‐2; Li et al., 2017). Interestingly, two fatty acid reductase genes (At3g44560, At5g22500) that are involved in suberin biosynthesis were upregulated in both kai2 and max2 mutants compared with wild‐type (Li et al., 2017). In addition, metabolomic analyses showed reduced levels of phenylpropanoid contributing to lignin composition (including ρ‐coumaric acids and ferulic acids) in max2 roots compared with wild‐type roots under control conditions (Walton et al., 2016). These are also good indicators of lower levels of cutin monomer, which signals in the AMF‐root symbiosis (Wang et al. 2012). Furthermore, transmission electron microscopy images of kai2 mutants revealed a thinner cuticle, while wild‐type and over‐expressor lines showed a thicker cuticle (Li et al., 2017). Thus, an altered cell wall would fit with the impairment in the early events leading to the establishment of KAI2‐dependent AMF symbiosis in host species (Gutjahr et al., 2015), and could feasibly influence root skewing and waving.
Experimental Procedures
Plant material and growth conditions
Wild‐type Arabidopsis seeds Columbia‐0 (Col‐0) and Landsberg erecta (Ler) were the parental backgrounds for the mutants tested. Seeds for max2 (max2‐1; Stirnberg et al., 2002), max3 (max3‐9; Booker et al., 2004), max4 (max4‐1; Sorefan et al., 2003), Atd14 (Atd14‐1; Waters et al., 2012), as well as sku5 (salk_070056), sks3 (salk_0677925), sks3 max2‐1 and sku5 max2‐1 were provided by Prof. Dame Ottoline Leyser (SLCU). Seeds for max2‐7, max2‐8, kai2‐1, kai2‐2, dlk2‐1, dlk2‐2, dlk2‐3 and KAI2:KAI2 (kai2‐2) were a gift from Dr Mark Waters (University of Western Australia; Waters et al., 2012, 2015b). The Kai2‐2 allele was backcrossed six times to Col‐0 [kai2‐2 (6 × Col‐0)] and was a gift from Dr Mark Waters. Seeds were surface‐sterilised by treatment with 70% (v/v) ethanol, followed by a rinse with sterile distilled water, then incubation in 10% (v/v) sodium hypochlorite, 0.05% (v/v) Triton X‐100 for 5 min at 20°C with shaking (1250 rpm). After a further five washes with sterile distilled water, seeds were placed on the surface of 0.8% (w/v) agar (BD, UK) supplemented with ½ MS (Murashige and Skoog including vitamins, pH 5.6; Duchefa, The Netherlands). Arabidopsis seeds were stratified in the dark for 2 days at 4°C, before transfer to a growth cabinet under controlled conditions at 23°C, 16 h light: 8 h dark, and 80 μmol m−2 sec−1 irradiance. Growth plates were vertical unless stated otherwise.
Root‐skewing assay
After 9 days, images were taken by scanning plates from the back (i.e. roots were imaged through the agar) using a flat‐bed scanner (300 dpi), and root‐skewing angles were measured in ImageJ (Schneider et al., 2012) using the angle tool. NeuronJ (Meijering et al., 2004) was used to record the x‐ and y‐coordinates of the root tips and a marked section of the root. These coordinates were then used to calculate the HGI and VGI as previously described (Grabov et al., 2004; Vaughn and Masson, 2011). Waviness was measured as the ratio of the cord to the root length (Grabov et al., 2004; Vaughn and Masson, 2011).
GR24rac and karrikins
Plants were grown for 6 days on the surface of control medium [0.8% (w/v) agar supplemented with ½ MS, including vitamins, pH 5.6], then transferred to medium containing racemic GR24rac (LeadGen Labs, Orange, CT, USA), KAR2 or KAR1 (Toronto Research Chemicals, North York, ON, Canada), or only the carrier for the test compound as a control [sterile distilled water for KAR2 and KAR1, and 0.02% (v/v) acetone for GR24rac]. Plants were then grown for a further 3 days before scanning.
Cell file rotation and root diameter analysis
Images of the root tips from plants grown vertically for 6 days, then placed at a 45° angle from the vertical for a further 3 days, were taken using a Leica DFC365FX camera attached to a Leica M205FA stereo microscope (Leica Microsystems, Cambridge, UK) with a Planapo × 1.6 objective set to magnification of × 80.5. Images were stitched using the LAS X software platform (Leica Microsystems). Following Wang et al. (2011), CFR was defined as the number of epidermal cell files that crossed a 1‐mm‐long straight line drawn down the longitudinal axis of the root from 1.5 to 2.5 mm from the root apex. Using the same images as for CFR measurements, root diameter was measured approximately 2 mm from the root apex using ImageJ (Schneider et al., 2012), three measurements were done per individual root.
Mechanical stimulation assays for transcriptional response
Plants grown vertically on the surface of control plates for 9 days were transferred to a sterile buffer solution (0.1 mm KCl, 10 mm CaCl2 and 2 mm bis‐Tris propane, pH 5.8, adjusted with 0.5 m MES). A total of 30–40 seedlings per genotype were transferred into a Petri dish (3 cm in diameter), containing 3 ml of buffer solution, and left to acclimatize on the bench for 3 h with additional light (15W/865 Lumilux Daylight, maximum intensity: 86 μmol m−2 sec−1). Mechanical stimulation was applied by shaking vigorously for 30 sec, while control plants remained on the bench. Plants were then left untouched for a further 30 min after stimulation before being immersed in RNALater (Sigma Aldrich, Gillingham, UK) for sample collection as described previously. For both assays, RNA was extracted from roots using the RNeasy Plant Mini kit (Qiagen, Manchester, UK) per manufacturer's instructions, including an additional DNase digestion step. A LiCl precipitation step was used to purify and concentrate the RNA before downstream quantitative polymerase chain reaction (qPCR) analysis.
cDNA synthesis and transcript abundance measurement
Complementary DNA (cDNA) was synthesized from 500 ng RNA using the RT QuantiTect reverse transcription kit (Qiagen), following manufacturer's instructions, except that incubation time was lengthened for the gDNA Wipeout step (3 min at 42°C) and the cDNA synthesis (25 min at 42°C). cDNA was used as template in a quantitative real‐time PCR using the SYBR GREEN PCR kit (Qiagen) and the Rotor‐Gene 3000 thermocycler (Qiagen) to determine transcript abundance of the genes of interest Calmodulin‐like (CML) 12 and CML24. qPCR amplification cycle consisted of 5 min at 95°C followed by 40 cycles of 5 sec at 95°C and 10 sec at 60°C. Melting curves (ramping from 55°C to 95°C rising 1°C each step, with a 5‐sec delay between steps) were checked for unspecific amplification. qPCR traces were analysed using the R qpcR package (relevant parameters: data were normalised and the background subtracted; starting fit model: l4; efficiency estimation: cpD2; refmean: True; baseline subtraction using the average of the first five cycles; Ritz and Spiess, 2008; R package version 1.4‐0. 2015) to calculate Ct values. Efficiencies (all > 92%) were calculated using the calibration curve method. For each gene, the expression was calculated following the formula E = (eff−Ct). Expression of the genes of interest was normalised against two housekeeping genes Ubiquitin 10 (UBQ10) and Tubulin 4 (TUB4), as followed RGene of Interest = EGene of Interest/(sqrt(EUBQ10* ETUB4)). qPCR primers are listed in Table S1.
Measurements of cytosolic Ca2+ concentration ([Ca2+]cyt) in response to mechanical stimulation
Col‐0 and max2 [transformed using floral dip with Agrobacterium tumefaciens to express (apo)aequorin under a 35S promoter; Dodd et al., 2006] were used at T3 or T4 generation to determine cytosolic free Ca2+ concentration ([Ca2+]cyt). Equivalence of aequorin levels were determined by discharge assay of luminescence (> 4 million luminescence counts for both Col‐0 and max2). Plants were grown vertically on solid medium for 7–8 days as described above. Excised root tips (1 cm) were placed in the wells (one root per well) of a white 96‐well plate (Greiner Bio‐One, Stonehouse, UK) and incubated in 100 μl of bathing solution (10 μm coelentrazine; Lux Biotechnology, Edinburgh, UK 0.1 mm KCl, 10 mm CaCl2 and 2 mm bis‐Tris propane, pH 5.8 adjusted with 0.5 m MES) for 2 h in the dark, at room temperature. Luminescence was then recorded every second in a plate‐reading luminometer (FLUOstar Optima, BMG labtech, Ortenberg, Germany). After 35 sec, 100 μl of bathing solution (without coelentrazine) was injected into the well at 200 μl sec−1 to cause a mechanical stimulus to the root resulting in a sudden increase in luminescence (‘touch response’). The signal was monitored for a further 120 sec, when 100 μl of discharge solution [3 m CaCl2, in 30% (v/v) ethanol] was delivered to normalize the luminescence data and calculate [Ca2+]cyt (Laohavisit et al., 2012). The [Ca2+]cyt touch response of Col‐0 and max2 were then compared.
Root gravitropism assays
Arabidopsis plants were grown vertically for 14 days on the surface of control medium. On the day of the experiment, roots were positioned by aligning their root tips so that they could be imaged together. Plates were then placed vertically in the growth incubator but rotated through a 90° angle, thus inducing a 90° change in gravitropic orientation. Root tips were imaged using a Raspberry Pi camera module (http://www.raspberrypi.org/). Images were acquired every 10 min for 10 h. Image analysis was conducted using ARTT (Russino et al., 2013), which tracked the root tip growth and gave the tip orientation and displacement as output. Tip orientation was normalised to the displacement to take into account differences in growth rate.
Data representation and statistical analysis
Root‐skewing data were represented using beanplots constructed in the R environment (R Core Team, 2012) using the beanplot package (Kampstra, 2014), to show the variability in root‐skewing angle. Statistical analyses were also conducted in the R environment. Normal distribution of the data and equality of variance were verified using Shapiro and Levene tests (Lawstat package; Gastwirth et al., 2017), respectively. Significant differences amongst genotypes were verified using one‐way anova, followed by Tukey HSD. anovas were conducted on rank values as a non‐parametric method, when data did not uphold the assumptions of normality and homoscedasticity. All experiments were repeated at least three times.
Authors’ contribution
SMS and JMD planned and designed the research. SMS, YG, EM and FJ performed experiments and analysed data. SMS and JMD wrote the manuscript.
Conflict of interest
The authors declare no conflicts of interest.
Supporting information
Figure S1. Root‐skewing phenotype of kai2‐2 mutant in the Col‐0 background, complemented mutants and dlk2 mutants.
Figure S2. Effect of KAR on root skewing and primary root elongation in Ler, kai2 and max2.
Figure S3. Effect of GR24 on root skewing in kai2, max2 and d14.
Figure S4. Gravitropic response of max2, max3 and max4 is faster than wild‐type.
Figure S5. sks3 and sku5 do not suppress the high LRD in max2.
Figure S6. SKS3 and SKU5 transcript cellular localisation in the root.
Figure S7. MAX2 and KAI2 transcript cellular localisation in the root.
Table S1. Primer sequences used in qPCR analysis.
Acknowledgements
The authors thank Dr Mark Waters and Prof. Dame Ottoline Leyser for providing seeds and commenting on the manuscript. The authors also thank Prof. David Nelson for providing seeds, and Daniel Safka for support in setting up the raspberry Pi system. The authors are grateful to Dr Uta Paszkowski, Prof. Alex Webb, Dr Siobhan Braybrook, Prof. Sidney Shaw and Dr Jenny Mortimer for interesting discussions. This work was supported by the Broodbank Trust, the Newton Trust, the Gatsby Foundation, and the BBSRC Doctoral Training Programme (BB/J014540/1).
References
- Al‐Babili, S. and Bouwmeester, H.J. (2015) Strigolactones, a novel carotenoid‐derived plant hormone. Annu. Rev. Plant Biol. 66, 161–186. [DOI] [PubMed] [Google Scholar]
- Booker, J. , Auldridge, M. , Wills, S. , McCarty, D. , Klee, H. and Leyser, O. (2004) MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Curr. Biol. 14, 1232–1238. [DOI] [PubMed] [Google Scholar]
- Bouwmeester, H.J. , Roux, C. , Lopez‐Raez, J.A. and Bécard, G. (2007) Rhizosphere communication of plants, parasitic plants and AM fungi. Trends Plant Sci. 12, 224–230. [DOI] [PubMed] [Google Scholar]
- Braam, J. and Davis, R.W. (1990) Rain‐, wind‐, and touch‐induced expression of calmodulin and calmodulin‐related genes in Arabidopsis. Cell 60, 357–364. [DOI] [PubMed] [Google Scholar]
- Brady, S.M. , Orlando, D.A. , Lee, J.Y. , Wang, J.Y. , Koch, J. , Dinneny, J.R. , Mace, D. , Ohler, U. and Benfey, P.N. (2007) A high‐resolution root spatiotemporal map reveals dominant expression patterns. Science 318, 801–806. [DOI] [PubMed] [Google Scholar]
- Buer, C.S. , Masle, J. and Wasteneys, G.O. (2000) Growth conditions modulate root‐wave phenotypes in Arabidopsis. Plant Cell Physiol. 41, 1164–1170. [DOI] [PubMed] [Google Scholar]
- Buer, C.S. , Wasteneys, G.O. and Masle, J. (2003) Ethylene modulates root‐wave responses in Arabidopsis. Plant Physiol. 132, 1085–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bythell‐Douglas, R. , Waters, M.T. , Scaffidi, A. , Flematti, G.R. , Smith, S.M. and Bond, C.S. (2013) The structure of the karrikin‐insensitive protein (KAI2) in Arabidopsis thaliana . PLoS ONE 8, e54758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier, F. , Nieminen, K. , Sanchez‐Ferrero, J.C. , Rodriguez, M.L. , Chagoyen, M. , Hardtke, C.S. and Cubas, P. (2014) Strigolactone promotes degradation of DWARF14, an α/β hydrolase essential for strigolactone signaling in Arabidopsis. Plant Cell 26, 1134–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu, C.H. , Choi, J. and Paszkowski, U. (2017) Independent signalling cues underpin arbuscular mycorrhizal symbiosis and large lateral root induction in rice. New Phytol. 217, 552–557. [DOI] [PubMed] [Google Scholar]
- Conn, C.E. and Nelson, D.C. (2016) Evidence that KARRIKIN‐INSENSITIVE2 (KAI2) receptors may perceive an unknown signal that is not karrikin or strigolactone. Front. Plant Sci. 6, 1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford, S. , Shinohara, N. , Sieberer, T. , Williamson, L. , George, G. , Hepworth, J. , Muller, D. , Domagalska, M.A. and Leyser, O. (2010) Strigolactones enhance competition between shoot branches by dampening auxin transport. Development 137, 2905–2913. [DOI] [PubMed] [Google Scholar]
- Darwin, C. and Darwin, F. (1880) The Power of Movement in Plants. New York: D. Appleton and Company. [Google Scholar]
- Dill, A. , Thomas, S.G. , Hu, J. , Steber, C.M. and Sun, T.‐P. (2004) The Arabidopsis F‐box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin‐induced degradation. Plant Cell 16, 1392–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd, A.N. , Jakobsen, M.K. , Baker, A.J. , Telzerow, A. , Hou, S.‐W. , Laplaze, L. , Barrot, L. , Scott Poethig, R. , Haseloff, J. and Webb, A.A.R. (2006) Time of day modulates low‐temperature Ca2+ signals in Arabidopsis. Plant J. 48, 962–973. [DOI] [PubMed] [Google Scholar]
- Flematti, G.R. , Dixon, K.W. and Smith, S.M. (2015) What are karrikins and how were they ‘discovered’ by plants? BMC Biol. 13, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gastwirth, J.L. , Gel, Y.R. , Hui, W. , Lyubchich, V. and Miao, W. (2017) Tools for Biostatistics, Public Policy and Law. R package version, pp. 1–44.
- Goldwasser, Y. , Yoneyama, K. , Xie, X. and Yoneyama, K. (2008) Production of strigolactones by Arabidopsis thaliana responsible for Orobanche aegyptiaca seed germination. Plant Growth Regul. 55, 21–28. [Google Scholar]
- Grabov, A. , Ashley, M.K. , Rigas, S. , Hatzopoulos, P. , Dolan, L. and Vicente‐Agullo, F. (2004) Morphometric analysis of root shape. New Phytol. 165, 641–652. [DOI] [PubMed] [Google Scholar]
- Gray, W.M. , Kepinski, S. , Rouse, D. , Leyser, O. and Estelle, M. (2001) Auxin regulates SCFTIR1‐dependent degradation of AUX/IAA proteins. Nature 414, 271–276. [DOI] [PubMed] [Google Scholar]
- Gutjahr, C. , Gobbato, E. , Choi, J. et al (2015) Rice perception of symbiotic arbuscular mycorrhizal fungi requires the karrikin receptor complex. Science 350, 1516–1521. [DOI] [PubMed] [Google Scholar]
- Hamiaux, C. , Drummond, R.S.M. , Janssen, B.J. , Ledger, S.E. , Cooney, J.M. , Newcomb, R.D. and Snowden, K.C. (2012) DAD2 is an alpha/beta; hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr. Biol. 22, 2032–2036. [DOI] [PubMed] [Google Scholar]
- Jiang, L. , Liu, X. , Xiong, G. et al (2013) DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504, 401–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, L. , Matthys, C. , Marquez‐Garcia, B. , De Cuyper, C. , Smet, L. , De Keyser, A. , Boyer, F.‐D. , Beeckman, T. , Depuydt, S. and Goormachtig, S. (2015) Strigolactones spatially influence lateral root development through the cytokinin signaling network. J. Exp. Bot. 67, 379–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampstra, P. (2014) Beanplot: a Boxplot alternative for visual comparison of distributions. J. Stat. Softw. 28, 1–9. [Google Scholar]
- Kapulnik, Y. and Koltai, H. (2014) Strigolactone involvement in root development, response to abiotic stress, and interactions with the biotic soil environment. Plant Physiol. 166, 560–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapulnik, Y. , Delaux, P.‐M. , Resnick, N. et al (2011) Strigolactones affect lateral root formation and root‐hair elongation in Arabidopsis . Planta 233, 209–216. [DOI] [PubMed] [Google Scholar]
- Knight, M. , Campbell, A.K. , Smith, S.M. and Trewavas, A. (1991) Transgenic plant aequorin reports the effects of touch and cold‐shock and elicitors on cytoplasmic calcium. Nature 352, 524–526. [DOI] [PubMed] [Google Scholar]
- Kohlen, W. , Charnikhova, T. , Liu, Q. , Bours, R. , Domagalska, M.A. , Beguerie, S. , Verstappen, F. , Leyser, O. , Bouwmeester, H. and Ruyter‐Spira, C. (2011) Strigolactones are transported through the xylem and play a key role in shoot architectural response to phosphate deficiency in nonarbuscular mycorrhizal host Arabidopsis. Plant Physiol. 155, 974–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltai, H. , Dor, E. , Hershenhorn, J. et al (2009) Strigolactones’ effect on root growth and root‐hair elongation may be mediated by auxin‐efflux carriers. J. Plant Growth Regul. 29, 129–136. [Google Scholar]
- Koltai, H. , Lekkala, S.P. , Bhattacharya, C. et al (2010) A tomato strigolactone‐impaired mutant displays aberrant shoot morphology and plant interactions. J. Exp. Bot. 61, 1739–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushwah, S. , Jones, A.M. and Laxmi, A. (2011) Cytokinin interplay with ethylene, auxin, and glucose signaling controls arabidopsis seedling root directional growth. Plant Physiol. 156, 1851–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanza, M. , Garcia‐Ponce, B. , Castrillo, G. et al (2012) Role of actin cytoskeleton in brassinosteroid signaling and in its integration with the auxin response in plants. Dev. Cell 22, 1275–1285. [DOI] [PubMed] [Google Scholar]
- Laohavisit, A. , Shang, Z. , Rubio, L. et al (2012) Arabidopsis annexin1 mediates the radical‐activated plasma membrane Ca2+‐ and K+‐permeable conductance in root cells. Plant Cell 24, 1522–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, I. , Kim, K. , Lee, S. et al (2018) A missense allele of KARRIKIN‐INSENSITIVE2 impairs ligand‐binding and downstream signaling in Arabidopsis thaliana . J. Exp. Bot. 69, 3609–3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. , Nguyen, K.H. , Chu, H.D. et al (2017) The karrikin receptor KAI2 promotes drought resistance in Arabidopsis thaliana . PLoS Genet. 13, e1007076–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, M.H. , Wu, J. , Yao, J. et al (2014) Apyrase suppression raises extracellular ATP levels and induces gene expression and cell wall changes characteristic of stress responses. Plant Physiol. 164, 2054–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthys, C. , Walton, A. , Struk, S. , Stes, E. , Boyer, F.‐D. , Gevaert, K. and Goormachtig, S. (2016) The Whats, the Wheres and the Hows of strigolactone action in the roots. Planta 243, 1327–1337. [DOI] [PubMed] [Google Scholar]
- Mayzlish‐Gati, E. , De Cuyper, C. , Goormachtig, S. et al (2012) Strigolactones are involved in root response to low phosphate conditions in Arabidopsis. Plant Physiol. 160, 1329–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijering, E. , Jacobm, M. , Sarria, J.C. , Steiner, P. , Hirling, H. and Unser, M. (2004) Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry 58, 167–176. [DOI] [PubMed] [Google Scholar]
- Migliaccio, F. and Piconese, S. (2001) Spiralizations and tropisms in Arabidopsis roots. Trends Plant Sci. 6, 561–565. [DOI] [PubMed] [Google Scholar]
- Moturu, T.R. , Thula, S. , Singh, R.K. , Nodzyński, T. , Vařeková, R.S. , Friml, J. and Simon, S. (2018) Molecular evolution and diversification of the SMXL gene family. J. Exp. Bot. 69, 2367–2378. [DOI] [PubMed] [Google Scholar]
- Nakashima, J. , Liao, F. , Sparks, J.A. , Tang, Y. and Blancaflor, E.B. (2013) The actin cytoskeleton is a suppressor of the endogenous skewing behaviour of Arabidopsis primary roots in microgravity. Plant Biol. 16, 142–150. [DOI] [PubMed] [Google Scholar]
- Nelson, D.C. , Riseborough, J.A. , Flematti, G.R. , Stevens, J. , Ghisalberti, E.L. , Dixon, K.W. and Smith, S.M. (2009) Karrikins discovered in smoke trigger Arabidopsis seed germination by a mechanism requiring gibberellic acid synthesis and light. Plant Physiol. 149, 863–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, D.C. , Flematti, G.R. , Riseborough, J.A. , Ghisalberti, E.L. , Dixon, K.W. and Smith, S.M. (2010) Karrikins enhance light responses during germination and seedling development in Arabidopsis thaliana . Proc. Natl Acad. Sci. USA 107, 7095–7100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, D.C. , Scaffidi, A. , Dun, E.A. , Waters, M.T. , Flematti, G.R. , Dixon, K.W. , Beveridge, C.A. , Ghisalberti, E.L. and Smith, S.M. (2011) F‐box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA 108, 8897–8902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada, K. and Shimura, Y. (1990) Reversible root tip rotation in Arabidopsis seedlings induced by obstacle‐touching stimulus. Science 250, 274–276. [DOI] [PubMed] [Google Scholar]
- Oliva, M. and Dunand, C. (2007) Waving and skewing: how gravity and the surface of growth media affect root development in Arabidopsis . New Phytol. 176, 37–43. [DOI] [PubMed] [Google Scholar]
- Oyama, T. , Shimura, Y. and Okada, K. (1997) The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus‐induced development of root and hypocotyl. Genes Dev. 11, 2983–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul, A.‐L. , Amalfitano, C.E. and Ferl, R.J. (2012) Plant growth strategies are remodeled by spaceflight. BMC Plant Biol. 12, 1–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen, A. , Mason, M.G. , De Cuyper, C. et al (2012) Strigolactones suppress adventitious rooting in Arabidopsis and pea. Plant Physiol. 158, 1976–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen, A. , Depuydt, S. , Goormachtig, S. and Geelen, D. (2013) Strigolactones fine‐tune the root system. Planta 238, 615–626. [DOI] [PubMed] [Google Scholar]
- Ritz, C. and Spiess, A.N. (2008) qpcR: an R package for sigmoidal model selection in quantitative real‐time polymerase chain reaction analysis. Bioinformatics 24, 1549–1551. [DOI] [PubMed] [Google Scholar]
- Roux, S.J. (2012) Root waving and skewing – unexpectedly in micro‐g. BMC Plant Biol. 12, 1–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, R. and Bassham, D.C. (2014) Root growth movements: waving and skewing. Plant Sci. 221–222, 42–47. [DOI] [PubMed] [Google Scholar]
- Russino, A. , Ascrizzi, A. , Popova, L. , Tonazzini, A. , Mancuso, S. and Mazzolai, B. (2013) A novel tracking tool for the analysis of plant‐root tip movements. Bioinspir. Biomim. 8, 025004–025016. [DOI] [PubMed] [Google Scholar]
- Ruyter‐Spira, C. , Kohlen, W. , Charnikhova, T. et al (2011) Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: another belowground role for strigolactones? Plant Physiol. 155, 721–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Saint Germain, A. , Clavé, G. , Badet‐Denisot, M.‐A. et al (2016) An histidine covalent receptor and butenolide complex mediates strigolactone perception. Nat. Chem. Biol. 12, 787–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi, A. , Waters, M.T. , Sun, Y.K. , Skelton, B.W. , Dixon, K.W. , Ghisalberti, E.L. , Flematti, G.R. and Smith, S.M. (2014) Strigolactone hormones and their stereoisomers signal through two related receptor proteins to induce different physiological responses in Arabidopsis. Plant Physiol. 165, 1221–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, C.A. , Rasband, W.S. and Eliceiri, K.W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, E.R. , Zupanska, A.K. , Sng, N.J. , Paul, A.‐L. and Ferl, R.J. (2017) Skewing in Arabidopsis roots involves disparate environmental signaling pathways. BMC Plant Biol. 17, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, S.H. , Qin, X. and Loewen, M.C. (2004) The biochemical characterization of two carotenoid cleavage enzymes from Arabidopsis indicates that a carotenoid‐derived compound inhibits lateral branching. J. Biol. Chem. 279, 46 940–46 945. [DOI] [PubMed] [Google Scholar]
- Sedbrook, J.C. (2004) The Arabidopsis SKU6/SPIRAL1 gene encodes a plus end‐localized microtubule‐interacting protein involved in directional cell expansion. Plant Cell 16, 1506–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedbrook, J.C. , Carroll, K.L. , Hung, K.F. , Masson, P.H. and Somerville, C. (2002) The Arabidopsis SKU5 gene encodes an extracellular glycosyl phosphatidylinositol‐anchored glycoprotein involved in directional root growth. Plant Cell 14, 1635–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih, H.‐W. , Miller, N.D. , Dai, C. , Spalding, E.P. and Monshausen, G.B. (2014) The receptor‐like kinase FERONIA is required for mechanical signal transduction in Arabidopsis seedlings. Curr. Biol. 24, 1887–1892. [DOI] [PubMed] [Google Scholar]
- Shinohara, N. , Taylor, C. and Leyser, O. (2013) Strigolactone can promote or inhibit shoot branching by triggering rapid depletion of the auxin efflux protein PIN1 from the plasma membrane. PLoS Biol. 11, e1001474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds, C. , Söll, D. and Migliaccio, F. (2005) Circumnutation and gravitropism cause root waving in Arabidopsis thaliana . J. Exp. Bot. 46, 143–150. [Google Scholar]
- Sorefan, K. , Booker, J. , Haurogné, K. et al. (2003) MAX4 and RMS1 are orthologous dioxygenase‐like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev. 17, 1469‐1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soundappan, I. , Bennett, T. , Morffy, N. , Liang, Y. , Stanga, J.P. , Abbas, A. , Leyser, O. and Nelson, D.C. (2015) SMAX1‐like/D53 family members enable distinct MAX2‐dependent responses to strigolactones and karrikins in Arabidopsis . Plant Cell 27, 3143–3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanga, J.P. , Smith, S.M. , Briggs, W.R. and Nelson, D.C. (2013) SUPPRESSOR OF MORE AXILLARY GROWTH2 1 controls seed germination and seedling development in Arabidopsis. Plant Physiol. 163, 318–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnberg, P. , van de Sande, K. and Leyser, H.M.O. (2002) MAX1 and MAX2 control shoot lateral branching in Arabidopsis . Development 129, 1131–1141. [DOI] [PubMed] [Google Scholar]
- Sun, X.‐D. and Ni, M. (2011) HYPOSENSITIVE TO LIGHT, an alpha/beta fold protein, acts downstream of ELONGATED HYPOCOTYL 5 to regulate seedling de‐etiolation. Mol. Plant 4, 116–126. [DOI] [PubMed] [Google Scholar]
- Sun, H. , Tao, J. , Liu, S. , Huang, S. , Chen, S. , Xie, X. , Yoneyama, K. , Zhang, Y. and Xu, G. (2014) Strigolactones are involved in phosphate‐ and nitrate‐deficiency‐induced root development and auxin transport in rice. J. Exp. Bot. 65, 6735–6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, H. , Tao, J. , Gu, P. , Xu, G. and Zhang, Y. (2015) The role of strigolactones in root development. Plant Signal. Behav. 11, e1110662–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y.K. , Flematti, G.R. , Smith, S.M. and Waters, M.T. (2016) Reporter gene‐facilitated detection of compounds in Arabidopsis leaf extracts that activate the karrikin signaling pathway. Front. Plant Sci. 7, 164–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y.K. , Scaffidi, A. , Yao, J. , Melville, K. , Smith, S.M. , Flematti, G.R. and Waters, M.T. (2018) Divergent receptor proteins confer responses to different karrikins in two ephemeral weeds. BiorXiv https://doi.org/10/1101/376939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines, B. , Katsir, L. , Melotto, M. , Niu, Y. , Mandaokar, A. , Liu, G. , Nomura, K. , He, S.Y. , Howe, G.A. and Browse, J. (2007) JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 448, 661–665. [DOI] [PubMed] [Google Scholar]
- Thompson, M.V. and Holbrook, N.M. (2004) Root‐gel interactions and the root waving behavior of Arabidopsis. Plant Physiol. 135, 1822–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh, S. , Holbrook‐Smith, D. , Stokes, M.E. , Tsuchiya, Y. and McCourt, P. (2014) Detection of parasitic plant suicide germination compounds using a high‐throughput Arabidopsis HTL/KAI2 strigolactone perception system. Chem. Biol. 21, 988–998. [DOI] [PubMed] [Google Scholar]
- Van der Does, D. , Boutrot, F. , Engelsdorf, T. et al (2017) The Arabidopsis leucine‐rich repeat receptor kinase MIK2/LRR‐KISS connects cell wall integrity sensing, root growth and response to abiotic and biotic stresses. PLoS Genet. 13, e1006832–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn, L.M. and Masson, P.H. (2011) A QTL study for regions contributing to Arabidopsis thaliana root skewing on tilted surfaces. G3 1, 105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallner, E.‐S. , López‐Salmerón, V. , Belevich, I. et al (2017) Strigolactone‐ and karrikin‐independent SMXL proteins are central regulators of phloem formation. Curr. Biol. 27, 1241–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton, A. , Stes, E. , Goeminne, G. et al (2016) The response of the root proteome to the synthetic strigolactone GR24 in Arabidopsis . Mol. Cell Proteomics 15, 2744–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, E. , Schornack, S. , Marsh, J.F. , Gobbado, E. , Schwessinger, B. , Eastmond, P. , Schultze, M. , Kamoun, S. and Oldroyd, G.E.D. (2012) A common signaling process that promotes mycorrhizal and oomycete colonization of plants. Current Biol. 22, 2242–2246. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Wang, B. , Gilroy, S. , Wassim Chehab, E. and Braam, J. (2011) CML24 is involved in root mechanoresponses and cortical microtubule orientation in Arabidopsis . J. Plant Growth Regul. 30, 467–479. [Google Scholar]
- Waters, M.T. and Smith, S.M. (2013) KAI2‐ and MAX2‐mediated responses to karrikins and strigolactones are largely independent of HY5 in Arabidopsis seedlings. Mol. Plant 6, 63–75. [DOI] [PubMed] [Google Scholar]
- Waters, M.T. , Nelson, D.C. , Scaffidi, A. , Flematti, G.R. , Sun, Y.K. , Dixon, K.W. and Smith, S.M. (2012) Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 139, 1285–1295. [DOI] [PubMed] [Google Scholar]
- Waters, M.T. , Scaffidi, A. , Flematti, G. and Smith, S.M. (2015a) Substrate‐induced degradation of the α/β‐fold hydrolase KARRIKIN INSENSITIVE2 requires a functional catalytic triad but is independent of MAX2. Mol. Plant 8, 814–817. [DOI] [PubMed] [Google Scholar]
- Waters, M.T. , Scaffidi, A. , Moulin, S.L.Y. , Sun, Y.K. , Flematti, G.R. and Smith, S.M. (2015b) A Selaginella moellendorffii ortholog of KARRIKIN INSENSITIVE2 functions in Arabidopsis development but cannot mediate responses to karrikins or strigolactones. Plant Cell 27, 1925–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters, M.T. , Gutjahr, C. , Bennett, T. and Nelson, D.C. (2017) Strigolactone signaling and evolution. Annu. Rev. Plant Biol. 68, 291–322. [DOI] [PubMed] [Google Scholar]
- Winter, D. , Vinegar, B. , Nahal, B. , Ammar, R. , Wilson, G.V. and Provart, N.J. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large‐scale biological data sets. PLoS ONE 2(8), e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, R. , Ming, Z. , Yan, L. et al (2016) DWARF14 is a non‐canonical hormone receptor for strigolactone. Nature 536, 469–473. [DOI] [PubMed] [Google Scholar]
- Zhao, L.‐H. , Zhou, X.E. , Yi, W. et al (2015) Destabilization of strigolactone receptor DWARF14 by binding of ligand and E3‐ligase signaling effector DWARF3. Cell Res. 25, 1219–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, K. (2013) Functional characterisation of GPI‐anchored proteins in the SKU5/SKS gene family. Université Paris Sud‐ Paris XI, NNT:2013PA112096 tel‐01066888.
- Zhou, F. , Lin, Q. , Zhu, L. et al (2013) D14‐SCFD3‐dependent degradation of D53 regulates strigolactone signalling. Nature 504, 406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Root‐skewing phenotype of kai2‐2 mutant in the Col‐0 background, complemented mutants and dlk2 mutants.
Figure S2. Effect of KAR on root skewing and primary root elongation in Ler, kai2 and max2.
Figure S3. Effect of GR24 on root skewing in kai2, max2 and d14.
Figure S4. Gravitropic response of max2, max3 and max4 is faster than wild‐type.
Figure S5. sks3 and sku5 do not suppress the high LRD in max2.
Figure S6. SKS3 and SKU5 transcript cellular localisation in the root.
Figure S7. MAX2 and KAI2 transcript cellular localisation in the root.
Table S1. Primer sequences used in qPCR analysis.


