Abstract
Clinical symptom response to antipsychotic medications is highly variable. Genome-wide association studies (GWAS) provide a ‘hypothesis-free’ method of interrogating the genome for biomarkers of antipsychotic response. We performed a systematic review of GWAS findings for antipsychotic efficacy or effectiveness. 14 studies met our inclusion criteria, ten of which examined antipsychotic response using quantitative rating scales to measure symptom improvement. 15 genome-wide significant loci were identified, seven of which were replicated in other antipsychotic GWAS publications: CNTNAP5, GRID2, GRM7, 8q24 (KCNK9), PCDH7, SLC1A1 and TNIK. Notably, four replicated loci are involved in glutamatergic pathways. Additional validation and evaluation of the biological significance of these markers is warranted. These markers should also be evaluated for clinical utility, especially in the context of other validated pharmacogenomic variants (e.g., CYP450 genes). These findings may generate new avenues for development of novel antipsychotic treatments.
Keywords: : antipsychotic, genome-wide association study, GWAS, pharmacogenomics, psychopharmacology, schizophrenia, systematic review, treatment response
The role of antipsychotic medications in mental health treatment continues to grow. Between 1997 and 2007, the number of antipsychotic prescriptions rose 86% from 17.4 to 32.4 million in the USA [1]. This increase is partly related to a rising trend of antipsychotic use for conditions other than psychotic disorders, including indications as adjunctive therapy for major depressive disorder and off-label usage for a myriad of other psychiatric conditions. In the treatment of psychosis, response rates range from 47% for individuals who have received prior treatment to 66% for antipsychotic-naive individuals [2], while 1-year discontinuation rates may be as high as 74% due to poor tolerability or lack of efficacy [3]. Side effects are common and may include sedation, weight gain, movement disorder, hormone dysregulation and many others [4]. There is significant variability in patients’ response and tolerability to antipsychotics [5] that often makes it difficult for clinicians to prospectively assess risk–benefit tradeoffs for individual patients.
Biological predictors of antipsychotic response, regardless of condition, remain elusive. Pharmacogenetic studies may be useful in examining genetic contributions to variance in drug treatment outcomes. CYP450 variants may alter drug metabolism and impact serum concentrations, dosing strategies and dose-related outcomes of several antipsychotic medications [5]. Ultrarapid metabolizers of antipsychotic medications may have subtherapeutic serum levels of a medication at a given dose, leading to reduced efficacy. Poor metabolism of antipsychotic medications may increase exposure 30–60% [6]. This may indirectly impact drug efficacy as greater than expected serum levels may increase the risk for dose-related side effects, ostensibly resulting in lower adherence to the medication. Pharmacodynamic candidate gene studies have identified several promising associations, although none of these have been conclusively shown to reliably predict antipsychotic response [5].
Genome-wide association studies (GWAS) provide an unbiased or ‘hypothesis-free’ method of interrogating the genome for novel markers of antipsychotic treatment outcomes beyond known drug metabolism and pharmacodynamic genes. Though not without their limitations [7], GWAS are nevertheless a powerful hypothesis generating tool to gain insights into additional molecular mechanisms or pathways implicated in drug action. In this article, we review GWAS findings for antipsychotic efficacy or effectiveness and discuss future directions for the field.
Methods
Comprehensive searches of the electronic PubMed database and Google Scholar were performed (2009–July 2018) using the following search string: (‘GWAS’or ‘Genome wide association’) and (‘antipsychotic’ OR ‘dopamine antagonist’), utilizing an English language filter.
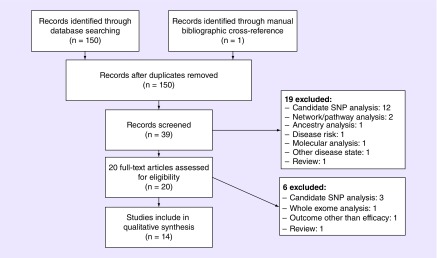
Results were manually screened. Studies that were included were those that evaluated antipsychotic efficacy or effectiveness in humans with schizophrenia or psychosis, excluding studies that evaluated antipsychotic tolerability or use in other disease states (e.g., bipolar disorder, major depressive disorder, etc.). All studies that performed genome-wide genotyping and association analyses were included. Studies described as candidate gene analyses and whole-exome sequencing studies were excluded. Original studies were identified (Figure 1) and manual bibliographic cross-reference was also performed. This approach yielded one additional full-text article that was accessed, but did not meet criteria for the review. Studies that utilized standardized quantitative measures of clinical response (e.g., Positive and Negative Symptom Scale [PANSS]) are reviewed in-depth; studies that used other measures of clinical response (i.e., clinically defined treatment resistance, neurocognitive measures, optimal antipsychotic dosage) are briefly discussed. If not specified in the statistical analysis methodology, genome-wide significance was assumed to be p < 5 × 10-8 [8].
Figure 1. . Flow chart depicting the selection process of studies included in the review.
Results
Fourteen studies met the inclusion criteria with a total sample size of 7366. Studies used a variety of antipsychotics including aripiprazole (n = 859), clozapine (n = 174), haloperidol n = (288), iloperidone (n = 210), lurasidone (n = 433), olanzapine (n = 1057), perphenazine (n = 312), quetiapine (n = 546), paliperidone (n = 1390), risperidone (n = 1223), ziprasidone (n = 498) and mixed antipsychotics (n = 418). Duration of observation ranged from 2 weeks to 18 months.
Of the 14 studies, ten (total n = 7108) used quantitative measures of clinical response (Table 1), while four used alternative measures of response (e.g., neuropsychological performance or clinically defined treatment resistance). The ten quantitative studies are reviewed in detail below, followed by a brief review of the four additional studies. Table 2 lists the top SNPs and genes that were identified in each of the reviewed studies.
Table 1. . Genome-wide association studies of antipsychotic response.
| Study | Year | Sample size | Source of study sample | Array platform | Ancestry | Medications (n) | Duration of illness | Period of observation | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| Lavedan et al. | (2009) | 210 | Phase III drug trials | Affymetrix 500k | Mixed | Iloperidone | Not specified | 28 days | [9] |
| Ikeda et al. | (2010) | 108 | JPN_1, JPN_2, UK GWAS | Illumina Sentrix BeadChip | Japanese | Risperidone | First episode | 8 weeks | [10] |
| McClay et al. (CATIE) | (2011) | 738 | CATIE (PANSS) | Affymetrix 500k + custom 164k | Mixed | Olanzapine (173), perphenazine (119), quetiapine (160), risperidone (159), ziprasidone (90) | >1 year of schizophrenia diagnosis | 18 months | [11] |
| McClay et al. (CATIE) | (2011) | 738 | CATIE (cognition) | Affymetrix 500k + custom 164k | Mixed | Olanzapine (173), perphenazine (119), quetiapine (160), risperidone (159), ziprasidone (90) | >1 year of schizophrenia diagnosis | 18 months | [22] |
| Clark et al. (CATIE) | (2013) | 738 | CATIE (CGI-S and PGI) | Affymetrix 500k + custom 164k | Mixed | Olanzapine (173), perphenazine (119), quetiapine (160), risperidone (159), ziprasidone (90) | >1 year of schizophrenia diagnosis | 18 months | [12] |
| Drago et al. | (2014) | Discovery: 96; replication: 169 | Independent sample with replication in CATIE severely ill subset | Discovery: Illumina Human Hap 300 and Illumina Human 610-Quad, replication: Affymetrix 500k and custom 164k | Discovery: White, replication: mixed | Discovery: haloperidol; replication: mixed | Not specified | 1 month | [13] |
| Li et al. | (2014) | 174 | Previously collected prospective clinical trial | Illumina 610K Quad BeadChip® | Caucasian | Clozapine | Not specified | n/a (retrospective) | [23] |
| Stevenson et al. | (2016) | 86 | Independent sample | Affymetrix Genome-wide Human SNP array 6.0 | Mixed | Primarily risperidone | Yes | 6 weeks | [14] |
| Sacchetti et al. | (2016) | 86 | GEAR study | Affymetrix Human Mapping GeneChip 6.0 | Italian | Risperidone | Not specified | 2 weeks | [15] |
| Koga et al. | (2016) | 79 | Independent sample | Illumina HumanOmni2.5-8 BeadChip Kit | European | Mixed | >3 years of schizophrenia diagnosis | n/a (retrospective) | [25] |
| Koga et al. | (2017) | 84 | Independent sample | Illumina HumanOmni2.5-8 BeadChip Kit | European | Mixed | >3 years of schizophrenia diagnosis | n/a (retrospective) | [24] |
| Li et al. | (2017) | 1390 | Phase III drug trials | Human1M-Duov3 or PsychArray | European | Paliperidone | >1 year of schizophrenia diagnosis | 6–13 weeks | [16] |
| Li et al. | (2018) | 435 | Lurasidone Phase III trials | Illumina Omni5Exome-4v1 BeadChip | Mixed | Lurasidone | Not specified | 6 weeks | [18] |
| Yu et al. | (2018) | Discovery: 2413, replication: 1379 | Chinese Antipsychotics Pharmacogenomics Consortium | Illumina Human Omni ZhongHua-8 BeadChips | Han Chinese | Discovery: aripiprazole (397), haloperidol (192), olanzapine (425), perphenazine (193), quetiapine (386), risperidone (412), ziprasidone (408) Replication: aripiprazole (462), olanzapine (459), risperidone (458) |
Both first episode and relapsed patients | 6 weeks | [20] |
CATIE: Clinical antipsychotic trials of intervention effectiveness; PANSS: Positive and Negative Symptom Scale; CGI-S: Clinical Global Impression - Severity; PGI: Patient Global Improvement; GEAR: Genes and Early Antipsychotic Response; GWAS: Genome-wide association studies
Table 2. . Top ten associations by study with p < 5 × 10-5.
| Study | Year | Dosing: fixed or flexible? | Efficacy measure | Medication | Primary findings† | Ref. | ||
|---|---|---|---|---|---|---|---|---|
| Gene | Polymorphism | p (or q)-value | ||||||
| Lavedan et al. | (2009) | Fixed | >20% improvement in PANSS total score | Iloperidone | NUDT9P1 | rs4528226 | 7.8 × 10-7 | [9] |
| Iloperidone | TNR | rs875326 | 2.1 × 10-5 | |||||
| Ikeda et al. | (2010) | Flexible | PANSS total score | Risperidone | ATP2B2 | rs2289273 | 1.60 × 10-5 | [10] |
| McClay et al. | (2011) | Flexible | PANSS positive, negative and excitement scores | Ziprasidone | Intergenic | rs17390445 | 9.8 × 10-8 | [11] |
| Ziprasidone | Intergenic | rs888219 | 2.3 × 10-7 | |||||
| Risperidone | ANKS1B | rs7968606 | 3.2 × 10-7 | |||||
| Olanzapine | CNTNAP5 | rs17727261 | 5.4 × 10-7 | |||||
| Risperidone | Intergenic | rs11722719 | 5.4 × 10-7 | |||||
| Olanzapine | Intergenic | rs10888501 | 1.4 × 10-6 | |||||
| Olanzapine | Intergenic | rs1040994 | 1.8 × 10-6 | |||||
| Olanzapine | Intergenic | rs10484256 | 2.7 × 10-6 | |||||
| Olanzapine | Intergenic | rs7635839 | 3.0 × 10-6 | |||||
| Risperidone | Intergenic | rs12526186 | 3.1 × 10-6 | |||||
| Clark et al. | (2013) | Flexible | CGI-S improvement | Olanzapine | Intergenic | rs8050896 | 3.9 × 10-8 | [12] |
| Risperidone | ATP1A2 | rs6688363 | 1.6 × 10-7 | |||||
| Risperidone | Intergenic | rs7395555 | 2.0 × 10-7 | |||||
| Risperidone | TNFRSF11A | rs2980976 | 3.0 × 10-7 | |||||
| Risperidone | PPA2 | rs2636697 | 3.7 × 10-7 | |||||
| PGI improvement | Olanzapine | PDE4D | rs17382202 | 4.2 × 10-8 | ||||
| Quetiapine | SPOPL | rs10170310 | 1.0 × 10-7 | |||||
| Quetiapine | PDE4D | rs17742120 | 1.6 × 10-7 | |||||
| Quetiapine | PDE4D | rs2164660 | 1.9 × 10-7 | |||||
| Risperidone | TJP1 | rs711355 | 2.3 × 10-7 | |||||
| Drago et al. | (2014) | Flexible | PANSS total score improvement | Haloperidol | Intergenic | rs7912580 | 1.4 × 10-6 | [13] |
| EIF2AK4 | rs2412459 | 8.9 × 10-6 | ||||||
| Stevenson et al. | (2016) | Flexible | BPRS total score | Primarily risperidone | GRID2 | rs9307122 | 1.1 × 10-8 | [14] |
| GRID2 | rs1875705 | 1.1 × 10-8 | ||||||
| APTX | rs3824457 | 1.3 × 10-6 | ||||||
| Intergenic | rs1992525 | 1.4 × 10-6 | ||||||
| GRID2 | rs994011 | 1.6 × 10-6 | ||||||
| MDGA1 | rs804855 | 1.8 × 10-6 | ||||||
| SMU1 | rs2274766 | 2.5 × 10-6 | ||||||
| LOC727677 (CASC8) | rs687279 | 2.9 × 10-6 | ||||||
| Intergenic | rs2288687 | 4.8 × 10-6 | ||||||
| NPY | rs16100 | 5.2 × 10-6 | ||||||
| Sacchetti et al. | (2016) | Flexible | PANSS positive score | Risperidone | GRM7 | rs2133450 | 4.3 × 10-8 | [15] |
| Li et al. | (2017) | Flexible | CGI-S, PANSS total score | Paliperidone | ADCK1 | rs56240334 | 8.0 × 10-8 | [16] |
| Intergenic | rs12915820 | 1.2 × 10-7 | ||||||
| Yu et al. | (2018) | Flexible | PANSS total score | Aripiprazole, olanzapine, quetiapine, risperidone, ziprasidone, haloperidol, perphenazine | MEGF10 | rs72790443 | 1.4 × 10-9 | [20] |
| SLC1A1 | rs1471786 | 2.3 × 10-9 | ||||||
| PCDH7 | rs9291547 | 3.2 × 10-9 | ||||||
| CNTNAP5 | rs12711680 | 2.1 × 10-8 | ||||||
| TNIK | rs6444970 | 4.9 × 10-8 | ||||||
| PCDH7 | rs77685948 | 6.7 × 10-8 | ||||||
| SPATA6L | rs301440 | 2.0 × 10-7 | ||||||
| MEGF10 | rs150192045 | 1.7 × 10-7 | ||||||
| CNTNAP5 | rs4848939 | 3.8 × 10-7 | ||||||
| NBEA | rs9544382 | 9.4 × 10-6 | ||||||
| Li et al. | (2018) | Fixed | PANSS total score | Lurasidone | KCNK9 | rs4736253 | 4.8 × 10-8 | [18] |
| DYNC2H1 | rs10895475 | 2.4 × 10-7 | ||||||
| CTNNA2 | rs10180106 | 4.9 × 10-7 | ||||||
| STXBP5L | rs511841 | 2.6 × 10-7 | ||||||
†Genome-wide significant findings (as defined by the source publications) are bolded.
BPRS: Brief Psychiatric Rating Scale; CGI-S: Clinical Global Impression - Severity; PANSS: Positive and negative symptom scale; PGI: Patient global improvement.
Lavedan et al. (2009): iloperidone response
Lavedan et al. [9] performed a GWAS in patients with schizophrenia from a Phase III clinical trial of iloperidone. The study enrolled 407 patients total (randomized to iloperidone 12-mg twice daily, ziprasidone 80-mg twice daily or placebo) and followed them for 28 days. The GWAS analysis was restricted to the 210 individuals who received iloperidone. Two separate analytical methods were used: a two-phase approach and a single-phase approach. In the two-phase approach, the study sample was randomly assigned to either a discovery set (n = 106) or a confirmatory set. In the discovery set, the analysis was restricted to test genotypic differences between ‘extreme responders’ (i.e., patients in the <30% percentile [n = 30] and >70% percentile [n = 32]). The findings from this analysis were then examined for replication in the full confirmatory set (n = 104). In a separate single-phase approach, the entire sample was analyzed together. The change in PANSS total score at 28 days from baseline was used as the primary outcome measure in both analyses.
The primary finding was a genome-wide significant association between iloperidone response in the two-phase analysis and rs4528226, located near the NUDT9P1 pseudogene, approximately 200kb away from the HTR7 gene. No findings exceeded GWAS significance using the single-phase approach. The authors also generated the top 100 SNPs identified by each method. Five top SNPs were found on both lists, all of which resided in regions with prior evidence for involvement in psychosis risk or neurotransmitter signaling (Table 2).
Ikeda et al. (2010): risperidone response
Ikeda et al. [10] performed a GWAS in participants enrolled in an open-label prospective trial of risperidone in 108 antipsychotic-naive, first episode patients with schizophrenia. Subjects were prescribed 0.5–4 mg of risperidone per day (mean 2.5 mg), and the dose was titrated to response over 8 weeks. Percent change in total PANSS score was used as the primary end point. The investigators then cross-referenced top genes from the GWAS (i.e., genes with at least one SNP p < 5 × 10-4) with gene expression data from mice who were exposed to risperidone compared with an untreated control group. Seven genes were identified that contained a top SNP in humans and also showed altered expression in risperidone-exposed mice.
No genome-wide significant SNPs were identified. The top hit was rs2289273, a synonymous variant in ATP2B2.
McClay et al. (2011): CATIE trial: analysis of PANSS scores
McClay et al. [11] performed a GWAS in 738 patients with schizophrenia who were enrolled in Phase I of the Clinical Antipsychotic Trials of Intervention Effectiveness (CATIE). The design of this trial has been described elsewhere [2]. Briefly, individuals with schizophrenia were enrolled from 57 sites and randomized to olanzapine (7.5–30 mg/day), perphenazine (8–32 mg/day), quetiapine (200–800 mg/day), risperidone (1.5–6 mg/day) or ziprasidone (40–160 mg/day) and followed for up to 18 months. In this analysis, change in Positive, Negative, Disorganization, Excitement and Emotional Distress subscales from the PANSS, as well as change in total PANSS score, were used as end points for the five medications used in Phase I (olanzapine, quetiapine, risperidone, ziprasidone and perphenazine). Polymorphisms were declared genome-wide significant if they were within the false discovery rate (FDR) threshold q < 0.10.
According to the FDR threshold for this study, one genome-wide SNP was identified. The rs17390445 SNP was associated with differential improvement in positive symptoms with ziprasidone, despite being over 1 Mb from the nearest gene. One of the genes in this general region of chromosome 4p15.1 includes the PCDH7 gene.
Clark et al. (2013): CATIE trial: analysis of global illness severity measures
Clark et al. [12] also performed a GWAS in 738 patients with schizophrenia from Phase I of the CATIE trial. Unlike the McClay et al. analysis that used the clinician administered PANSS score as the primary end point, this study used the Clinical Global Impression – Severity (CGI-S) and Patient Global Improvement (PGI) scales to assess global illness severity as rated by clinicians and patients, respectively. Analyses were stratified by antipsychotic drug. Polymorphisms were declared genome-wide significant if they were within the FDR threshold q < 0.10. According to this criterion, 13 genome-wide significant SNPs were identified.
Analysis of PGI-rated effectiveness in the quetiapine subgroup identified three SNPs (all in high linkage disequilibrium [LD]) in an intron of PDE4D: rs17382202, rs17742120 and rs2164660. Another trio of intronic SNPs in high LD were identified in TJP1 among risperidone-treated patients: rs711355, rs785423 and rs813676. Finally, an association was observed in olanzapine users between PGI outcome and rs10170310 in the SPOPL gene.
Analysis of CGI-S in the risperidone subgroup identified two intronic SNPs in high LD within the PPA2 gene, rs2636697 and rs2636719. Other top SNPs for risperidone included two intergenic SNPs: rs2980976 and rs8050896. CGI-S-measured response to olanzapine was associated with the intergenic rs6688363 SNP, while response to ziprasidone was associated with rs7395555, an intergenic SNP.
The authors attempted to validate findings by identifying significant SNPs that overlapped across the two scales. Four statistically significant SNPs were identified: rs8050986 and rs785423 with risperidone, and rs6688363 and rs10170310 with olanzapine. Clark et al. also replicated one marker that was associated with the PANSS negative subscale in the McClay CATIE analysis: rs17727261. The lack of considerable overlap between these three different measures may be, as the authors noted, due to subjective differences in conceptualization of illness between clinician and patient, a documented challenge in psychiatric research. It may also indicate false-positive associations.
Drago et al. (2014): haloperidol response
Drago et al. [13] performed a GWAS in 96 acutely psychotic individuals treated with the first-generation antipsychotic haloperidol (dosing not described). A replication sample of 169 participants was culled from the CATIE trial, specifically selecting those with a baseline PANSS score >75 in order to match the initial sample (mean baseline PANSS = 104.78). In both samples, mean percent decrease in PANSS over 1 month was used as the primary end point.
No SNP achieved genome-wide significance. Two SNPs (rs7912580 and rs2412459) were nominally associated with response in both samples; however, the direction of association was opposite in the two samples. The rs7912580 SNP, located in an intergenic region near ARID5B and RTKN2, was associated with better response to haloperidol in the initial sample, but was associated with worsened response to the mix of second-generation antipsychotics and the first-generation antipsychotic perphenazine that were used in the CATIE trial. Similarly, the rs2412459 SNP, located in an intron of EIF2AK4, was associated with improved response in the initial sample, and was associated with worsened response in the CATIE sample. Given that the first sample was treated with haloperidol and the second sample primarily second-generation antipsychotics, these SNPs may be biomarkers for differential response to antipsychotic classes. Additional evaluation would be necessary to support this hypothesis. However, the opposite findings in the two datasets may also indicate that these SNPs represent spurious associations.
Stevenson et al. (2016): first episode psychosis
Stevenson et al. [14] performed a GWAS in 86 individuals with first episode psychosis (primarily schizophrenia diagnoses). Participants were either antipsychotic-naive or had a limited history of antipsychotic exposure (i.e., <18 weeks lifetime exposure to antipsychotics) and were treated primarily with risperidone monotherapy (mean chlorpromazine equivalents = 249.2 ± 172.9 standard deviation [SD]) for 6 weeks. Change in the Brief Psychiatric Rating Scale score was used as the primary end point.
Genome-wide analysis identified two SNPs in the GRID2 gene (rs9307122 and rs1875705) that were associated with reduced response according to the Brief Psychiatric Rating Scale change score. Both were in complete LD with one another. Interestingly, separate genomic imaging data showed that both SNPs were significantly associated with GRID2 expression in the hippocampus.
Sacchetti et al. (2016): risperidone response
Sacchetti et al. [15] examined 86 European participants in a pharmacogenomic study of early response to risperidone. Participants had a diagnosis of schizophrenia or schizoaffective disorder and were evaluated for risperidone response (3–13 mg/day) using change in PANSS scores after 2 weeks. One SNP, rs2133450, located in an intron of the GRM7 gene, reached genome-wide significance for association with PANSS positive symptom improvement. C homozygotes of this SNP showed less improvement in positive symptoms, a finding that was validated in a secondary analysis of the risperidone arm of the aforementioned CATIE schizophrenia trial.
Li et al. (2017): paliperidone response
Li et al. [16] performed a GWAS in 1390 European individuals from 12 clinical studies of the second-generation antipsychotic paliperidone. All participants had a DSM-IV diagnosis of schizophrenia and were experiencing an acute psychotic episode. Participants were divided into three cohorts based on the formulation of paliperidone used in the trials: paliperidone extended release (ER) oral (3–15 mg/day, n = 4896) and paliperidone palmitate long-acting injection (25–150 mg, n = 576 and 350). A meta-analysis was performed across these three cohorts. Primary outcomes analyzed were CGI-S and PANSS total, positive and negative change scores from baseline after 6–13 weeks of treatment.
Two models were used: a general linear model and a mixed linear model. The top SNP, rs56240334, is located in an intron of ADCK1 and was significantly associated with CGI-S, total PANSS, PANSS positive and negative subscales using the mixed linear model. The authors also attempted to validate a number of candidate SNPs in GRM7, GRID2, HTR2A and DRD2 from previous risperidone trials, including markers that were previously identified to be genome-wide significant. All failed to replicate except rs6314 in HTR2A, which was nominally significant.
Li et al. (2018a & b): lurasidone response
Li et al. performed a GWAS in three Phase III trials of lurasidone (40–160 mg/day). The authors published results based on a combined analysis of the first two trials in 2018 [17], but did not include the third trial due to differences in baseline patient characteristics and medication doses. Later in 2018, the authors published a meta-analysis of all three studies [18]. Both analyses are discussed together.
The combined analysis included 302 (171 of European and 131 of predominantly African ancestry) acutely psychotic individuals who received 40, 80 or 120 mg of lurasidone. The meta-analysis added 133 (99 European ancestry, 34 African ancestry) lurasidone-treated patients. In both analyses, the primary outcome was the change in total PANSS score from baseline after 6 weeks of treatment. In the combined analysis, top hits (defined as p < 10-5) in either group included SNPs in the following seven genes: PTCH1, NGL1 (aka LRRC4C), RBFOX1, c18orf64, NTRK3, CAMTA1 and ZNF438. Top hits (defined as p < 10-4) for both groups combined included SNPs in three genes: PTPRD, MAGI1 and KCNK9. The combined analysis also validated several previously reported polymorphisms. Two top SNPs from the McClay et al. CATIE analysis, rs17390445 and rs11722719, near PCDH7, were nominally associated with change in PANSS Positive Subscale score in the lurasidone study sample.
The meta-analysis identified one genome-wide significant SNP in participants of European Ancestry, rs4736253 in KCNK9 gene. This polymorphism is in partial LD [19] (r2 = 0.498, D' = 0.795) with the rs7017126 SNP in KCNK9 that had been previously identified in the combined analysis, and demonstrated an association with KCNK9 expression in the temporal cortex in previous imaging genetics studies that were mined by the investigators. Other top SNPs in Europeans included rs10895475 near DYNC2H1 and rs101895475 in the CTNNA2 gene. The rs101895475 SNP is significantly associated with the expression of CTNNA2 in Europeans. In participants of African ancestry, rs13394481 was the top SNP in the region of CTNNA2. This SNP was also correlated with lower expression of CTNNA2 mRNA as identified by expression quantitative trait loci analyses (Braincloud) also performed by the authors. Thus, lower expression of CTNNA2 appeared to be associated with reduced response to lurasidone. The overall top SNP in participants of African ancestry was rs511841, near the 5′-UTR of STXBP5L. However, none of the identified SNPs in the STXBP5L gene were associated with differential expression in either European or African ancestry individuals.
Yu et al. (2018): large analysis of multiple drugs
Yu et al. [20] performed a GWAS in Han Chinese individuals with schizophrenia. The study enrolled 2413 patients in a discovery cohort and 1379 patients in a validation cohort. Participants were randomized (1:1:1:1:1:1) to olanzapine (mean 18.7 mg/day), risperidone (mean 4.8 mg/day), quetiapine (mean 646.8 mg/day), aripiprazole (mean 23.7 mg/day), ziprasidone (mean 118.2 mg/day), haloperidol (mean 13.8 mg/day) or perphenazine (mean 26.5 mg/day). The primary outcome was percent change in PANSS score from baseline after 6 weeks of treatment.
Three genome-wide significant SNPs were identified in the discovery cohort and subsequently confirmed in the validation cohort: rs72790443 located in an intron of MEGF10; rs1471786 located in an intron of SLC1A1 and rs9291547 located in an intron of PCDH7. When the samples were combined, two additional SNPs achieved genome-wide significance: rs12711680 located in an intron of CNTNAP5 and rs6444970, located in an intron of TNIK gene. Several medication-specific SNPs also achieved genome-wide significance in the combined sample. The rs2239063 SNP, located in an intron of the CACNA1C gene was associated with response to olanzapine. Notably, CACNA1C contains one of the most widely replicated markers of neuropsychiatric dysfunction (rs1006737) [21]. Risperidone treatment response was associated with the intronic rs16921385 SNP of SLC1A1. The rs17022006 SNP, located in an intron of CNTN4, was associated with aripiprazole response.
This study also validated many previously identified polymorphisms and/or genes. These included polymorphisms near PCDH7 previously identified in the McClay et al. analysis of the CATIE trial and the Li et al. study. Additionally, an SNP in CNTNAP5 (p = rs17727261) was previously identified by McClay et al. to be associated with PANSS negative symptom improvement with risperidone; it is in linkage equilibrium with the aforementioned rs12711680 (r2 = 0.0093, D' = 0.8865). The study also validated previously reported findings in DRD2, HTR2A, GRM7, CYP2D6, RELN, NRG1 and NRXN1, all of which were nominally associated with response.
Other studies
Other measures of response that have been assessed with a GWAS methodology include cognitive performance [22], clinically defined treatment resistance [23,24] and antipsychotic dosing [25]. None of these studies identified any SNPs exceeding the established p < 5 × 10-8 threshold of significance. However, some biologically plausible associations emerged that were consistent with analyses of symptom response. Most notably, top SNPs associated with working memory performance in olanzapine-treated subjects in the CATIE study were observed in DRD2.
Discussion
To date, there have been ten GWAS of antipsychotic efficacy or effectiveness for the treatment of schizophrenia and related disorders that used quantitative measures of symptomatology, and an additional four studies that used alternative measures that may also provide insight into other aspects of treatment outcomes. In total, these studies included 7108 participants and examined a number of first- and second-generation antipsychotics. In these studies, 20 variants, representing 15 genes/genetic loci, exceeded statistical significance thresholds for associations with response. These included variants in or near the ATP1A2, CNTNAP5, GRID2, GRM7, KCNK9, MEGF10, NUDT9P1, PCDH7, PDE4D, PPA2, SLC1A1, SPOPL, TJP1, TNFRSF11A and TNIK genes. Of these, several loci were replicated in multiple independent datasets (Table 3). These loci and their potential relevance to antipsychotic response are reviewed in further detail below.
Table 3. . Replicated genome-wide significant loci.
| Gene | Location | Gene function/significance | SNP | p-value | Study | Ref. |
|---|---|---|---|---|---|---|
| CNTNAP5 | 2q14.3 | • Protein is highly expressed in myelinated neurons • Involved in generation of action potentials • Implicated in GWAS of bipolar disorder |
rs12711680 | 2.12 × 10-8 | Yu et al. | [26] |
| rs4848939 | 3.79 × 10-7 | Yu et al. | [26] | |||
| rs17727261 | 5.4 × 10-7 | McClay et al. | [11] | |||
| GRM7 | 3p26.1 | • Encodes a metabotropic glutamate receptor • Plays a critical role in glutamate regulation • Implicated in schizophrenia and bipolar disorder |
rs2133450 | 4.33 × 10-8 | Sacchetti et al., CATIE trial‡ | [17] |
| rs2069062 | < 0.04 | Stevenson et al. | [16] | |||
| rs1532544 | < 0.04 | Stevenson et al. | [16] | |||
| rs1156328 | 4.50 × 10-5 | Yu et al. | [26] | |||
| TNIK | 3q26.2 | • Encodes a kinase that also has scaffolding properties • Modulates concentrations of GRIA1 at the cellular surface • Interacts with DISC1, a known schizophrenia risk marker |
rs6444970 | 4.85 × 10-8 | Yu et al. | [26] |
| rs2088885 | 0.035 | Xu et al. | [36] | |||
| PCDH7 | 4p15.1 | • Encodes a membrane-bound protein involved in cell adhesion • Modulates signal transduction of the dopamine D1 receptor |
rs9291547 | 3.24 × 10-9 | Yu et al. | [26] |
| rs77685948 | 6.69 × 10-8 | Yu et al. | [26] | |||
| rs17390445 | 9.8 × 10-8 | McClay et al. | [11] | |||
| rs17390445 | 0.02 | Li et al. | [20] | |||
| rs11722719 | 5.4 × 10-7 | McClay et al. | [11] | |||
| rs11722719 | 0.0015 | Li et al. | [20] | |||
| GRID2 | 4q22.2 | • Encodes an ionotropic glutamate receptor • Mediates excitatory synaptic transmission • Implicated in risk for OCD and psychosis |
rs9307122 | 1.10 × 10-8 | Stevenson et al. | [16] |
| rs1875705 | 1.10 × 10-8 | Stevenson et al. | [16] | |||
| rs76800659† | 5.10 × 10-4 | Yu et al. | [26] | |||
| rs3775003 | 2.20 × 10-4 | Ikeda et al. | [9] | |||
| KCNK9 | 8q24.3 | • Encodes a leak potassium channel • Plays a critical role in setting resting membrane potential and repolarizing neurons • 8q24 region has been heavily implicated in neuropsychiatric disorders |
rs4736253 | 4.78 × 10-8 | Li et al. | [20,21] |
| rs2288687 | 4.79 × 10-6 | Stevenson et al. | [16] | |||
| rs1992525 | 1.41 × 10-6 | Stevenson et al. | [16] | |||
| rs6981424 | 1.04 × 10-5 | Stevenson et al. | [16] | |||
| rs687279 | 2.89 × 10-6 | Stevenson et al. | [16] | |||
| SLC1A1 | 9p24.2 | • Encodes the glutamate reuptake transporter • Implicated in risk for OCD, antipsychotic-induced OCD and schizophrenia • Expression is decreased following chronic antipsychotic treatment in animals |
rs1471786 | 2.33 × 10-9 | Yu et al. | [26] |
| rs301440§ | 2.03 × 10-7 | Yu et al. | [26] | |||
| rs10974671 | 0.0035 | Stevenson et al. | [16] | |||
Replication defined as exceeding GWAS significance in one study and achieving at least nominal significance in a second.
†In perfect LD with rs1875705.
‡Sacchetti et al. validated this finding in a secondary analysis of the CATIE dataset.
§In the nearby SPATA6L gene.
CATIE: Clinical antipsychotic trials of intervention effectiveness; GWAS: Genome-wide association studies; LD: Linkage disequilibrium; OCD: Obsessive–compulsive disorder.
CNTNAP5
Variants in the CNTNAP5 gene were associated with antipsychotic response in three studies using three different measures of symptom improvement [11,12,20]. CNTNAP5, encoding Caspr 5 protein, is located on the long arm of chromosome 2. Very little is known about the specific function of Caspr 5, although the Caspr family of proteins are highly expressed in myelinated neurons, and other family members are involved in neuron formation, myelin stability and neuronal signaling [26]. Variants in CNTNAP5 have been identified in prior GWAS of bipolar disorder [27,28] and a microdeletion in CNTNAP5 was identified in a family study of autism spectrum disorder [29]. Links to disease risk and neuron development from prior studies are consistent with hypotheses that certain developmental abnormalities related to disease risk may also influence responsiveness to treatment. Additional studies are needed to elucidate the pharmacological relevance of these relationships.
GRM7
Variants in GRM7 were associated with antipsychotic response in three studies, primarily those involving risperidone or paliperidone treatment [14,15,26], and one SNP, rs2133450, was further validated in a secondary analysis of the CATIE dataset [17]. However, this SNP failed to replicate in the post hoc analysis by Li et al. in their paliperidone-treated population [18]. GRM7 encodes the mGluR7 receptor, a G-protein-coupled receptor that plays a critical role in glutamate regulation, preventing excitotoxicity presynaptically by inhibiting adenylate cyclase and postsynaptically by decreasing NMDA receptor activity [30]. GRM7 is located at chromosome 3p26.1, a region previously identified as being associated with schizophrenia [31–33] and bipolar disorder [34]. It is clear that GRM7 plays a role in the etiology of neuropsychiatric disorders and antipsychotic treatment response, and that this may involve mechanisms related to glutamate signaling in the brain. This may indicate that pharmacological manipulation of mGluR7 or other glutamate receptors may provide further insights into the development of new treatments. Additionally, whether GRM7 variants are useful as biomarkers for response to existing therapies warrants further assessment.
TNIK
SNPs in the TNIK gene were associated with antipsychotic response in one GWAS [26]. Variants in this gene were previously associated with clozapine response in a large candidate gene study of 995 Han Chinese patients with schizophrenia [35]. TNIK, located at chromosome 3q26.2, may modulate concentrations of GRIA1 at the cellular surface [36]. It also interacts with DISC1 (a known schizophrenia risk marker) to regulate synapse function [37], which itself has an impact on glutamate synaptic spine formation [38] and glutamate metabolism [39].
PCDH7
SNPs in PCDH7 were associated with antipsychotic response in three studies comprised of a number of different second-generation antipsychotics [11,17,26]. The PCDH7 gene is located on the short arm of chromosome 4. Protocadherin inhibits PP1α [40], a key component of the signal transduction pathway used by the dopamine D1 receptor [41]. Overexpression of protocadherin 7 inhibits neuronal survival [42]. As corroborating data to support findings from their GWAS analyses, Li et al. reanalyzed two independent gene expression datasets and found decreased expression levels of PCDH7 in postmortem tissue of the dorsolateral prefrontal cortex in patients with schizophrenia [17]. More data are needed to evaluate the genetic architecture of PCDH7 and the surrounding region, identify causative variants and elucidate their biological significance for antipsychotic response.
GRID2
Four SNPs in or near GRID2 were highly associated with response to risperidone, two intronic variants exceeding GWAS significance in the study by Stevenson et al. [14], a different intronic GRID2 SNP in the secondary analysis of subjects receiving risperidone in the study by Yu et al. [26], and finally a SNP approximately 2 Mb downstream of GRID2 was associated in the study by Ikeda et al. [10]. The three SNPs identified by the Stevenson and Yu groups are in nearly perfect LD [19]. However, the GRID2 SNPs identified by Stevenson et al. failed to replicate in the post hoc analysis by Li et al. in their paliperidone-treated population [18]. GRID2, located at chromosome 4q22.1, encodes the GluD2 receptor, which mediates excitatory synaptic transmission [43]. Dysfunction in GluD2 is associated with a host of psychiatric phenotypes including obsessive–compulsive disorder [44], schizophrenia endophenotypes [45] and psychosis secondary to Huntington's disease [46]. Stevenson et al. noted that the rs1875705 SNP was also associated with lower hippocampal expression in a publicly searchable brain expression quantitative trait loci database [47]. The GRID2 associations with risperidone in these three studies are compelling. However, an SNP in partial LD [19] with both SNPs identified by Stevenson et al. was not associated with haloperidol efficacy or in a candidate gene analysis of 101 acutely ill patients with schizophrenia [48].
SLC1A1
Three separate SNPs in SLC1A1 or the nearby SPATA6L gene exceeding or approaching genome-wide significance were identified in the studies by Yu et al. [26] and Stevenson et al. [14]. The two SNPs identified by Yu et al. appear to be in modest LD [22] and may thus represent a link to a region relevant to response. SLC1A1, located at chromosome 9p24.2, is one of a family of amino acid transporters with a high affinity for glutamate. It is expressed in neurons throughout the brain, primarily at postsynaptic terminals, where it is involved in glutamate reuptake [49]. It is considered one of the top candidate genes for obsessive compulsive disorder [50], and antipsychotic-induced obsessive compulsive disorder [51–55]. SLC1A1 has also been implicated in schizophrenia risk [56–62], and decreased expression following chronic antipsychotic treatment has been observed in animal models [63]. The findings by Yu et al. require validation, but provide additional evidence for the role of glutamate system genes in schizophrenia etiology and treatment response.
8q24 region (KCNK9)
While there are compelling genes discovered in GWAS of antipsychotic response, another broader pattern of association was observed across variants identified in genes harbored within the chromosome 8q24 region. The primary gene in this region is KCNK9, with SNPs associated with lurasidone response in the studies by Li et al. [17,18]. The KCNK9 gene is located at chromosome 8q24.3 and is a member of the two pore-domain potassium channel subfamily – a group of widely expressed potassium channels that play a critical role in setting the resting membrane potential and repolarizing neurons [64]. Variants in KCNK9 are associated with Birk–Barel syndrome, a paternally imprinted genetic disorder characterized by facial dysmorphism, intellectual disability, hyperactivity and hypotonia [65]. Approximately 10 Mb upstream, Stevenson et al. [14] identified several intergenic SNPs that approached genome-wide significance, plus two more located in the nearby CASC8 gene. Notably, another potassium channel, KCNQ3, is located approximately 3 Mb downstream from the SNPs identified by Stevenson et al. Variants in KCNQ3 have been associated with bipolar disorder [66,67] and this is a site of action for several anticonvulsants, including gabapentin [68]. In fact, the 8q24 region as a whole has been the subject of considerable interest for psychiatric genetic research. GWAS and linkage analyses have associated this region with bipolar disorder [69–76] and psychosis [77–79]. As a result, it is unclear if the KCNK9 finding suggests a causative role for KCNK9 in antipsychotic response, linkage with a causative gene elsewhere in the region or part of a phenotypic or mechanistic interaction. Nevertheless, this finding confirms an intriguing role for this region in the etiology of neuropsychiatric disease and psychopharmacologic response.
Challenges in GWAS of antipsychotic response
Across ten studies, 15 genome-wide significant genes/genomic regions were identified. Of these, seven were replicated in at least one study. However, only three specific SNPs (rs1875705 in GRID2 and rs17390445 and rs11722719 near PCDH7) were replicated. While the replication of loci enables further research regarding the etiology of psychotic disorders and may identify new drug-able genes, the lack of extensive replication of specific SNPs limits the development of pharmacogenomic tests that may assist providers in treatment selection. Elucidation of the causative SNPs of the identified genomic loci without replicated SNPs via fine mapping or other techniques is warranted.
Heterogeneity across studies was high and presents challenges reconciling differential findings, and lack of replication of some associations. For example, there was heterogeneity in participants' prior duration of illness, exposure to previous treatments and how treatment outcomes were measured. Two studies exclusively enrolled first-episode patients [10,14], three studies exclusively enrolled patients with a longer standing diagnosis of schizophrenia [11,12,16] and the remainder either enrolled a mixture or did not specify. With respect to medication efficacy or effectiveness, some studies defined treatment outcomes using clinician-rated assessments, while others used patient-rated surveys and others used measures such as treatment resistance. Heterogeneity was also seen in dosing protocols. Three out of the ten studies used fixed dosing, while the other seven allowed flexible dosing. As it relates to genetic variation related to drug exposure, flexibly dosed protocols are less likely to identify associations with drug metabolism genes or other genes related to dose sensitivity. Consistent with this, no drug metabolism genes were identified in the top hits in the studies reviewed here. We did not observe robust patterns indicating differential likelihood of detecting GWAS significant SNPs in studies utilizing different dosing strategies.
Studies reviewed herein used a wide variety of both typical and atypical antipsychotics. While some studies only enrolled patients taking one particular medication, others randomized patients to one of several medications. These latter studies often evaluated treatment response using all patients, performing subanalyses within each medication group. While grouping all antipsychotic medication arms together can increase sample size and may be helpful to identify biomarkers of general antipsychotic responsiveness, it is less helpful for identifying markers that can aid in the pharmacotherapy decision-making process. Conversely, biomarkers for individual drugs may represent the most clinically useful markers, but are likely the most difficult to identify due to limitations in sample size.
Opportunities in GWAS of antipsychotic response
While the dopamine dysfunction hypothesis of schizophrenia remains consistent with the common pharmacodynamic characteristic of all currently approved antipsychotic drugs, evidence has also pointed to a role for dysfunction in the glutamate system, the primary excitatory neurotransmitter in the CNS [80]. This is further supported by the identification of genome-wide significant variants in four glutamatergic genes: GRID2, GRM7, SLC1A1 and TNIK in relation to treatment response. Promising targets such as these require additional research to validate their association with antipsychotic response. While not the specific focus of this review, secondary pathway analyses performed as part of some of these studies further support the hypothesis that polymorphisms in glutamate system genes account for variability in antipsychotic response. Drug development efforts targeting the glutamate system have failed in Phase II or III studies [81]. However, these studies have targeted other receptors or transporters. Genetic associations identified in discovery GWAS reinforce the importance of continuing to examine targets within the glutamate system for further drug development. Though not strictly necessary for clinical implementation, research that helps to elucidate the biological significance of systems or pathways further our understanding of the etiology of psychotic illness and determinants of treatment response.
It is important to re-emphasize the need for clinical validation of reported findings. The GWAS done to date have largely not replicated candidate gene studies that have been conducted over the course of a number of years. Variants in many pharmacodynamic genes such as ANKK1/DRD2, CACNA1C and HTR2A are included in clinically available pharmacogenetic test panels [5], yet none of these variants appeared among the top hits in any of the identified GWAS. Moreover, the association status of these previously identified variants was rarely reported in any of the reviewed papers, which complicates the evaluation of the validity of these markers. We suggest that future studies explicitly report on the association status of these markers until their relevance can be conclusively determined.
Although significant strides have been made in recent years, enrollment numbers in antipsychotic GWAS remain low. Four out of ten studies using quantitative trait rating scale measures had sample sizes of less than 200. The most recent study published, which included nearly 4000 participants, identified disproportionately more genome-wide significant findings than all of the other studies combined. Similar large-scale individual efforts and other efforts using combined datasets are encouraged.
Conclusion
We reviewed GWAS studies that used quantitative measures of antipsychotic response. Although these studies identified a total of 20 genome-wide significant SNPs, only two (rs11722719 near PCDH7 and rs1875705 in GRID2), replicated across independent datasets. While perhaps the most parsimonious conclusion that can be drawn from these findings is largely a failure in replication, there is some room for optimism. Despite significant heterogeneity in study design and medications, seven genes/genomic regions showed independent replication, indicating regions of high interest that require further study. The findings also continue to support a role for glutamate system genes in treatment response to drugs with primary pharmacodynamic effects on D2 receptors. This reinforces the importance of glutamate neurotransmission in the pathophysiology of schizophrenia, and further highlights pathways beyond dopamine for investigating novel molecules to treat psychosis.
Future perspective in GWAS of antipsychotic response
Evolutions in our understanding of the genomic underpinnings of antipsychotic response will be driven by two primary factors: sample size and technological advancement. With greater sample sizes, we expect the field to move beyond individual SNP associations to polygenic algorithms that more accurately describe the biological reality of antipsychotic response. Machine learning and artificial intelligence are likely to be powerful tools to aid our understanding of interactions within a genomic network – a key limitation of the genome-wide association approach. Polygenic algorithms are also likely to include other forms of genetic variation (e.g., copy number variations, epigenetics) that may be informative. In this regard, we anticipate a move beyond single SNP predictors to a holistic view of genomic contributors and environmental exposures that may be used to improve the precision with which we can match medications to patients. We also expect that these efforts will help to better describe the etiology of psychotic disorders and the mechanisms by which our present medications help to alleviate symptoms of these conditions (albeit imperfectly). Advancements in our understanding of the molecular subtypes of cancer have enabled the development of highly effective therapies that target those molecular mechanisms. In the same way, we hope that a better elucidate the molecular mechanisms of psychotic disorders will drive further innovation in antipsychotic drug development, leading to more effective treatment for our patients.
Executive summary.
Genome-wide association studies of antipsychotic response
Ten genome-wide association studies (GWAS) of antipsychotic efficacy or effectiveness were identified, including a total of 7108 participants (7366 including participants from qualitative studies).
Replicated findings
Seven replicated genes/genomic regions were identified (CNTNAP5, GRID2, GRM7, 8q24 [KCNK9], PCDH7, SLC1A1 and TNIK), although replication of individual SNPs was rare.
Genes involved in glutamatergic pathways were overrepresented among replicated findings, comprising four of the seven replicated loci.
Challenges in GWAS of antipsychotic response
Heterogeneity across studies was high, with considerable differences in duration of illness among participants, medication selection, dosing schedule, period of observation and outcome measures.
Many studies had small-sample sizes. Larger studies were better powered to identify genome-wide significant markers.
Opportunities in GWAS of antipsychotic response
Replicated loci affirm the role of glutamate in schizophrenia pharmacotherapy and support continued efforts to develop novel therapeutic approaches targeting this system.
A number of genomic regions were replicated, but few specific polymorphisms were identified that were consistently associated with antipsychotic efficacy or effectiveness. Additional biological work is necessary to examine the functional relevance of specific gene variants.
Clinical validation and assessment of biological function of replicated SNPs is also required, and these markers should also be assessed for clinical utility in prospective trials alongside other validated markers (e.g., CYP450 variation).
Future perspective in GWAS of antipsychotic response
Evolutions in our understanding of the genomic underpinnings of antipsychotic response will be driven by two primary factors: increasing sample sizes in studies and technological advancement (e.g., machine learning, enhanced genomic-sequencing techniques).
Footnotes
Financial & competing interests disclosure
This work was in part supported by NIH grant MH083888 to JR Bishop. JR Bishop has no other financial conflicts to disclose. In the past 12 months, JD Allen has consulted for 23andMe, Clarigent Health, Prescient Medicine and Translational Software. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Author's contributions
JD Allen performed the systematic review, synthesis of results and was the primary author of the manuscript. JR Bishop conceptualized the project, provided overall direction, assisted with the interpretation of results and writing of the manuscript.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Stagnitti MN. Agency for Healthcare Research and Quality. Rockville, MD, USA: 2010. Trends in antipsychotics purchases and expenses for the U.S. civilian noninstitutionalized population, 1997 and 2007; pp. 1–6. Statistical Brief #275. Stagnitti, M. N. Stat. Br. #275. January 2010. [Google Scholar]
- 2.Haddad PM, Correll CU. The acute efficacy of antipsychotics in schizophrenia: a review of recent meta-analyses. Ther. Adv. Psychopharmacol. 2018;8(11):303–318. doi: 10.1177/2045125318781475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lieberman JA, Stroup TS, McEvoy JP, et al. Effectiveness of antipsychotic drugs in patients with chronic schizophrenia. N. Engl. J. Med. 2005;353(12):1209–1223. doi: 10.1056/NEJMoa051688. [DOI] [PubMed] [Google Scholar]
- 4.Correll CU, Rubio JM, Kane JM. What is the risk-benefit ratio of long-term antipsychotic treatment in people with schizophrenia? World Psychiatry. 2018;17(2):149–160. doi: 10.1002/wps.20516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eum S, Lee AM, Bishop JR. Pharmacogenetic tests for antipsychotic medications: clinical implications and considerations. Dialogues Clin. Neurosci. 2016;18(3):323–337. doi: 10.31887/DCNS.2016.18.3/jbishop. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Review of pharmacogenomic markers commonly found on commercial testing platforms.
- 6.Ravyn D, Ravyn V, Lowney R, Nasrallah HA. CYP450 pharmacogenetic treatment strategies for antipsychotics: a review of the evidence. Schizophr. Res. 2013;149(1–3):1–14. doi: 10.1016/j.schres.2013.06.035. [DOI] [PubMed] [Google Scholar]
- 7.Motsinger-Reif AA, Jorgenson E, Relling M V, et al. Genome-wide association studies in pharmacogenomics: successes and lessons. Pharmacogenet. Genomics. 2013;23(8):383–394. doi: 10.1097/FPC.0b013e32833d7b45. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Excellent review and discussion of the role of genome-wide association studies (GWAS) in pharmacogenomics.
- 8.Panagiotou OA, Ioannidis JP, Genome-Wide Significance Project What should the genome-wide significance threshold be? Empirical replication of borderline genetic associations. Int. J. Epidemiol. 2012;41(1):273–286. doi: 10.1093/ije/dyr178. [DOI] [PubMed] [Google Scholar]
- 9.Lavedan C, Licamele L, Volpi S, et al. Association of the NPAS3 gene and five other loci with response to the antipsychotic iloperidone identified in a whole genome association study. Mol. Psychiatry. 2009;14(8):804–819. doi: 10.1038/mp.2008.56. [DOI] [PubMed] [Google Scholar]; • First GWAS of antipsychotic response.
- 10.Ikeda M, Tomita Y, Mouri A, et al. Identification of novel candidate genes for treatment response to risperidone and susceptibility for schizophrenia: integrated analysis among pharmacogenomics, mouse expression, and genetic case–control association approaches. Biol. Psychiatry. 2010;67(3):263–269. doi: 10.1016/j.biopsych.2009.08.030. [DOI] [PubMed] [Google Scholar]
- 11.McClay JL, Adkins DE, Aberg K, et al. Genome-wide pharmacogenomic analysis of response to treatment with antipsychotics. Mol. Psychiatry. 2011;16(1):76–85. doi: 10.1038/mp.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]; • First GWAS analysis using the Clinical Antipsychotic Trials of Intervention Effectiveness dataset.
- 12.Clark SL, Souza RP, Adkins DE, et al. Genome-wide association study of patient-rated and clinician-rated global impression of severity during antipsychotic treatment. Pharmacogenet. Genomics. 2013;23(2):69–77. doi: 10.1097/FPC.0b013e32835ca260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drago A, Giegling I, Schäfer M, et al. Genome-wide association study supports the role of the immunological system and of the neurodevelopmental processes in response to haloperidol treatment. Pharmacogenet. Genomics. 2014;24(6):314–319. doi: 10.1097/FPC.0000000000000052. [DOI] [PubMed] [Google Scholar]
- 14.Stevenson JM, Reilly JL, Harris MSH, et al. Antipsychotic pharmacogenomics in first episode psychosis: a role for glutamate genes. Transl. Psychiatry. 2016;6(2):e739–e748. doi: 10.1038/tp.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sacchetti E, Magri C, Minelli A, et al. The GRM7 gene, early response to risperidone, and schizophrenia: a genome-wide association study and a confirmatory pharmacogenetic analysis. Pharmacogenomics J. 2017;17(2):146–154. doi: 10.1038/tpj.2015.90. [DOI] [PubMed] [Google Scholar]
- 16.Li Q, Wineinger NE, Fu DJ, et al. Genome-wide association study of paliperidone efficacy. Pharmacogenet. Genomics. 2017;27(1):7–18. doi: 10.1097/FPC.0000000000000250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J, Yoshikawa A, Brennan MD, Ramsey TL, Meltzer HY. Genetic predictors of antipsychotic response to lurasidone identified in a genome wide association study and by schizophrenia risk genes. Schizophr. Res. 2018;192:194–204. doi: 10.1016/j.schres.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 18.Li J, Loebel A, Meltzer HY. Identifying the genetic risk factors for treatment response to lurasidone by genome-wide association study: a meta-analysis of samples from three independent clinical trials. Schizophr. Res. 2018;199:203–213. doi: 10.1016/j.schres.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 19.Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31(21):3555–3557. doi: 10.1093/bioinformatics/btv402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu H, Yan H, Wang L, et al. Five novel loci associated with antipsychotic treatment response in patients with schizophrenia: a genome-wide association study. Lancet Psychiatry. 2018;5(4):327–338. doi: 10.1016/S2215-0366(18)30049-X. [DOI] [PubMed] [Google Scholar]; •• Largest antipsychotic GWAS analysis performed to date.
- 21.Nie F, Wang X, Zhao P, et al. Genetic analysis of SNPs in CACNA1C and ANK3 gene with schizophrenia: a comprehensive meta-analysis. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2015;168(8):637–648. doi: 10.1002/ajmg.b.32348. [DOI] [PubMed] [Google Scholar]
- 22.McClay JL, Adkins DE, Berg K, et al. Genome-wide pharmacogenomic study of neurocognition as an indicator of antipsychotic treatment response in schizophrenia. Neuropsychopharmacology. 2011;36(3):616–626. doi: 10.1038/npp.2010.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J, Meltzer HY. A genetic locus in 7p12.2 associated with treatment resistant schizophrenia. Schizophr. Res. 2014;159(2–3):333–339. doi: 10.1016/j.schres.2014.08.018. [DOI] [PubMed] [Google Scholar]
- 24.Koga A, Bani-Fatemi A, Hettige N, et al. GWAS analysis of treatment resistant schizophrenia: interaction effect of childhood trauma. Pharmacogenomics. 2017;18(7):663–671. doi: 10.2217/pgs-2016-0137. [DOI] [PubMed] [Google Scholar]
- 25.Koga AT, Strauss J, Zai C, Remington G, De Luca V. Genome-wide association analysis to predict optimal antipsychotic dosage in schizophrenia: a pilot study. J. Neural Transm. 2016;123(3):329–338. doi: 10.1007/s00702-015-1472-7. [DOI] [PubMed] [Google Scholar]
- 26.Zou Y, Zhang WF, Liu HY, et al. Structure and function of the contactin-associated protein family in myelinated axons and their relationship with nerve diseases. Neural Regen. Res. 2017;12(9):1551–1558. doi: 10.4103/1673-5374.215268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Djurovic S, Gustafsson O, Mattingsdal M, et al. A genome-wide association study of bipolar disorder in Norwegian individuals, followed by replication in Icelandic sample. J. Affect. Disord. 2010;126(1–2):312–316. doi: 10.1016/j.jad.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 28.Xu W, Cohen-Woods S, Chen Q, et al. Genome-wide association study of bipolar disorder in Canadian and UK populations corroborates disease loci including SYNE1 and CSMD1 . BMC Med. Genet. 2014;15(1):2. doi: 10.1186/1471-2350-15-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pagnamenta AT, Bacchelli E, De Jonge M V, et al. Characterization of a family with rare deletions in CNTNAP5 and DOCK4 suggests novel risk loci for autism and dyslexia. Biol. Psychiatry. 2010;68(4):320–328. doi: 10.1016/j.biopsych.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gu Z, Liu W, Wei J, Yan Z. Regulation of N-methyl-D-aspartic acid (NMDA) receptors by metabotropic glutamate receptor 7. J. Biol. Chem. 2012;287(13):10265–10275. doi: 10.1074/jbc.M111.325175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pulver AE, Lasseter VK, Kasch L, et al. Schizophrenia: a genome scan targets chromosomes 3p and 8p as potential sites of susceptibility genes. Am. J. Med. Genet. 1995;60(3):252–260. doi: 10.1002/ajmg.1320600316. [DOI] [PubMed] [Google Scholar]
- 32.Pulver AE, Mulle J, Nestadt G, et al. Genetic heterogeneity in schizophrenia: stratification of genome scan data using co-segregating related phenotypes. Mol. Psychiatry. 2000;5(6):650–653. doi: 10.1038/sj.mp.4000814. [DOI] [PubMed] [Google Scholar]
- 33.Irmansyah, Schwab SG, Heriani, et al. Genome-wide scan in 124 Indonesian sib-pair families with schizophrenia reveals genome-wide significant linkage to a locus on chromosome 3p26–21. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2008;147B(7):1245–1252. doi: 10.1002/ajmg.b.30763. [DOI] [PubMed] [Google Scholar]
- 34.Kerner B, Lambert CG, Muthén BO. Genome-wide association study in bipolar patients stratified by co-morbidity. PLoS ONE. 2011;6(12):e28477. doi: 10.1371/journal.pone.0028477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu Q, Wu X, Li M, et al. Association studies of genomic variants with treatment response to risperidone, clozapine, quetiapine and chlorpromazine in the Chinese Han population. Pharmacogenomics J. 2016;16(4):357–365. doi: 10.1038/tpj.2015.61. [DOI] [PubMed] [Google Scholar]
- 36.MacLaren EJ, Charlesworth P, Coba MP, Grant SGN. Knockdown of mental disorder susceptibility genes disrupts neuronal network physiology in vitro . Mol. Cell. Neurosci. 2011;47(2):93–99. doi: 10.1016/j.mcn.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Q, Charych EI, Pulito VL, et al. The psychiatric disease risk factors DISC1 and TNIK interact to regulate synapse composition and function. Mol. Psychiatry. 2011;16(10):1006–1023. doi: 10.1038/mp.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hayashi-Takagi A, Takaki M, Graziane N, et al. Disrupted-in-schizophrenia 1 (DISC1) regulates spines of the glutamate synapse via Rac1. Nat. Neurosci. 2010;13(3):327–332. doi: 10.1038/nn.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abazyan S, Yang EJ, Abazyan B, et al. Mutant disrupted-in-schizophrenia 1 in astrocytes: focus on glutamate metabolism. J. Neurosci. Res. 2014;92(12):1659–1668. doi: 10.1002/jnr.23459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshida K, Watanabe M, Kato H, Dutta A, Sugano S. BH-protocadherin-c, a member of the cadherin superfamily, interacts with protein phosphatase 1 alpha through its intracellular domain. FEBS Lett. 1999;460(1):93–98. doi: 10.1016/s0014-5793(99)01309-5. [DOI] [PubMed] [Google Scholar]
- 41.Muly EC, Greengard P, Goldman-Rakic PS. Distribution of protein phosphatases-1 alpha and -1 gamma 1 and the D(1) dopamine receptor in primate prefrontal cortex: evidence for discrete populations of spines. J. Comp. Neurol. 2001;440(3):261–270. doi: 10.1002/cne.1384. [DOI] [PubMed] [Google Scholar]
- 42.Xiao H, Sun Z, Wan J, Hou S, Xiong Y. Overexpression of protocadherin 7 inhibits neuronal survival by downregulating BIRC5 in vitro. Exp. Cell Res. 2018;366(1):71–80. doi: 10.1016/j.yexcr.2018.03.016. [DOI] [PubMed] [Google Scholar]
- 43.Yuan H, Low C, Moody OA, Jenkins A, Traynelis SF. Ionotropic GABA and glutamate receptor mutations and human neurologic diseases. Mol. Pharmacol. 2015;88:203–217. doi: 10.1124/mol.115.097998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.International Obsessive Compulsive Disorder Foundation Genetics Collaborative (IOCDF-GC); OCD Collaborative Genetics Association Studies (OCGAS) Revealing the complex genetic architecture of obsessive–compulsive disorder using meta-analysis. Mol. Psychiatry. 2018;23(5):1181–1188. doi: 10.1038/mp.2017.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Greenwood TA, Lazzeroni LC, Calkins ME, et al. Genetic assessment of additional endophenotypes from the Consortium on the Genetics of Schizophrenia Family study. Schizophr. Res. 2016;170(1):30–40. doi: 10.1016/j.schres.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsuang DW, Greenwood TA, Jayadev S, Davis M, Shutes-David A, Bird TD. A genetic study of psychosis in Huntington's disease: evidence for the involvement of glutamate signaling pathways. J. Huntingtons Dis. 2018;7(1):51–59. doi: 10.3233/JHD-170277. [DOI] [PubMed] [Google Scholar]
- 47.Ramasamy A, Trabzuni D, Guelfi S, et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat. Neurosci. 2014;17(10):1418–1428. doi: 10.1038/nn.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Giegling I, Drago A, Dolžan V, et al. Glutamatergic gene variants impact the clinical profile of efficacy and side effects of haloperidol. Pharmacogenet. Genomics. 2011;21(4):206–216. doi: 10.1097/FPC.0b013e32833efb18. [DOI] [PubMed] [Google Scholar]
- 49.Bjørn-Yoshimoto WE, Underhill SM. The importance of the excitatory amino acid transporter 3 (EAAT3) Neurochem. Int. 2016;98(0):4–18. doi: 10.1016/j.neuint.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rajendram R, Kronenberg S, Burton CL, Arnold PD. Glutamate genetics in obsessive–compulsive disorder: a review. J. Can. Acad. Child Adolesc. Psychiatry. 2017;26(3):205–213. [PMC free article] [PubMed] [Google Scholar]
- 51.Kwon JS, Joo YH, Nam HJ, et al. Association of the glutamate transporter gene SLC1A1 with atypical antipsychotics-induced obsessive–compulsive symptoms. Arch. Gen. Psychiatry. 2009;66(11):1233–1241. doi: 10.1001/archgenpsychiatry.2009.155. [DOI] [PubMed] [Google Scholar]
- 52.Ryu S, Oh S, Cho E-Y, et al. Interaction between genetic variants of DLGAP3 and SLC1A1 affecting the risk of atypical antipsychotics-induced obsessive–compulsive symptoms. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2011;156B(8):949–959. doi: 10.1002/ajmg.b.31242. [DOI] [PubMed] [Google Scholar]
- 53.Schirmbeck F, Nieratschker V, Frank J, et al. Polymorphisms in the glutamate transporter gene SLC1A1 and obsessive–compulsive symptoms induced by second-generation antipsychotic agents. Psychiatr. Genet. 2012;22(5):245–252. doi: 10.1097/YPG.0b013e328353fbee. [DOI] [PubMed] [Google Scholar]
- 54.Cai J, Zhang W, Yi Z, et al. Influence of polymorphisms in genes SLC1A1, GRIN2B, and GRIK2 on clozapine-induced obsessive–compulsive symptoms. Psychopharmacology (Berl). 2013;230(1):49–55. doi: 10.1007/s00213-013-3137-2. [DOI] [PubMed] [Google Scholar]
- 55.Schirmbeck F, Zink M. Comorbid obsessive–compulsive symptoms in schizophrenia: contributions of pharmacological and genetic factors. Front. Pharmacol. 2013;4:99. doi: 10.3389/fphar.2013.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nudmamud-Thanoi S, Piyabhan P, Harte MK, Cahir M, Reynolds GP. Deficits of neuronal glutamatergic markers in the caudate nucleus in schizophrenia. J. Neural Transm. Suppl. 2007;(72):281–285. doi: 10.1007/978-3-211-73574-9_34. [DOI] [PubMed] [Google Scholar]
- 57.Horiuchi Y, Iida S, Koga M, et al. Association of SNPs linked to increased expression of SLC1A1 with schizophrenia. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2012;159B(1):30–37. doi: 10.1002/ajmg.b.31249. [DOI] [PubMed] [Google Scholar]
- 58.Afshari P, Myles-Worsley M, Cohen OS, et al. Characterization of a novel mutation in SLC1A1 associated with schizophrenia. Mol. Neuropsychiatry. 2015;1(3):125–144. doi: 10.1159/000433599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Myles-Worsley M, Tiobech J, Browning SR, et al. Deletion at the SLC1A1 glutamate transporter gene co-segregates with schizophrenia and bipolar schizoaffective disorder in a 5-generation family. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2013;162B(2):87–95. doi: 10.1002/ajmg.b.32125. [DOI] [PubMed] [Google Scholar]
- 60.Rees E, Walters JTR, Chambert KD, et al. CNV analysis in a large schizophrenia sample implicates deletions at 16p12.1 and SLC1A1 and duplications at 1p36.33 and CGNL1 . Hum. Mol. Genet. 2014;23(6):1669–1676. doi: 10.1093/hmg/ddt540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sundararajan T, Manzardo AM, Butler MG. Functional analysis of schizophrenia genes using GeneAnalytics program and integrated databases. Gene. 2018;641:25–34. doi: 10.1016/j.gene.2017.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Demily C, Hubert L, Franck N, Poisson A, Munnich A, Besmond C. Somatic mosaicism for SLC1A1 mutation supports threshold effect and familial aggregation in schizophrenia spectrum disorders. Schizophr. Res. 2017 doi: 10.1016/j.schres.2017.11.028. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 63.Zink M, Englisch S, Schmitt A. Antipsychotic treatment modulates glutamate transport and NMDA receptor expression. Eur. Arch. Psychiatry Clin. Neurosci. 2014;264(1):67–82. doi: 10.1007/s00406-014-0534-4. [DOI] [PubMed] [Google Scholar]
- 64.Imbrici P, Camerino DC, Tricarico D. Major channels involved in neuropsychiatric disorders and therapeutic perspectives. Front. Genet. 2013;4(MAY):1–19. doi: 10.3389/fgene.2013.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barel O, Shalev SA, Ofir R, et al. Maternally inherited Birk Barel Mental Retardation Dysmorphism Syndrome caused by a mutation in the genomically imprinted potassium channel KCNK9. Am. J. Hum. Genet. 2008;83(2):193–199. doi: 10.1016/j.ajhg.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kaminsky Z, Jones I, Verma R, et al. DNA methylation and expression of KCNQ3 in bipolar disorder. Bipolar Disord. 2015;17(2):150–159. doi: 10.1111/bdi.12230. [DOI] [PubMed] [Google Scholar]
- 67.Zhang P, Xiang N, Chen Y, et al. Family-based association analysis to finemap bipolar linkage peak on chromosome 8q24 using 2,500 genotyped SNPs and 15,000 imputed SNPs. Bipolar Disord. 2010;12(8):786–792. doi: 10.1111/j.1399-5618.2010.00883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Manville RW, Abbott G. Gabapentin is a potent activator of KCNQ3 and KCNQ5 potassium channels. Mol. Pharmacol. 2018;94(4):1155–1163. doi: 10.1124/mol.118.112953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cichon S. A genome screen for genes predisposing to bipolar affective disorder detects a new susceptibility locus on 8q. Hum. Mol. Genet. 2001;10(25):2933–2944. doi: 10.1093/hmg/10.25.2933. [DOI] [PubMed] [Google Scholar]
- 70.McInnis MG, Lan TH, Willour VL, et al. Genome-wide scan of bipolar disorder in 65 pedigrees: supportive evidence for linkage at 8q24, 18q22, 4q32, 2p12, and 13q12. Mol. Psychiatry. 2003;8(3):288–298. doi: 10.1038/sj.mp.4001277. [DOI] [PubMed] [Google Scholar]
- 71.Avramopoulos D, Willour VL, Zandi PP, et al. Linkage of bipolar affective disorder on chromosome 8q24: follow-up and parametric analysis. Mol. Psychiatry. 2004;9(2):191–196. doi: 10.1038/sj.mp.4001388. [DOI] [PubMed] [Google Scholar]
- 72.Zandi PP, Avramopoulos D, Willour VL, et al. SNP fine mapping of chromosome 8q24 in bipolar disorder. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2007;144(5):625–630. doi: 10.1002/ajmg.b.30486. [DOI] [PubMed] [Google Scholar]
- 73.Gonzalez S, Camarillo C, Rodriguez M, et al. A genome-wide linkage scan of bipolar disorder in Latino families identifies susceptibility loci at 8q24 and 14q32. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2014;165(6):479–491. doi: 10.1002/ajmg.b.32251. [DOI] [PubMed] [Google Scholar]
- 74.Del Zompo M, Severino G, Ardau R, et al. Genome-scan for bipolar disorder with sib-pair families in the Sardinian population: a new susceptibility locus on chromosome 1p22-p21? Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2010;153(6):1200–1208. doi: 10.1002/ajmg.b.31092. [DOI] [PubMed] [Google Scholar]
- 75.Zandi PP, Zöllner S, Avramopoulos D, et al. Family-based SNP association study on 8q24 in bipolar disorder. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2008;147B(5):612–618. doi: 10.1002/ajmg.b.30651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Greenwood TA, Badner JA, Byerley W, et al. Heritability and linkage analysis of personality in bipolar disorder. J. Affect. Disord. 2013;151(2):748–755. doi: 10.1016/j.jad.2013.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kebir O, Chaumette B, Krebs MO. Epigenetic variability in conversion to psychosis: novel findings from an innovative longitudinal methylomic analysis. Transl. Psychiatry. 2018;8(1) doi: 10.1038/s41398-018-0138-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jones I, Hamshere M, Nangle JM, et al. Bipolar affective puerperal psychosis: genome-wide significant evidence for linkage to chromosome 16. Am. J. Psychiatry. 2007;164(7):1099–1104. doi: 10.1176/ajp.2007.164.7.1099. [DOI] [PubMed] [Google Scholar]
- 79.Jones I, Craddock N. Searching for the puerperal trigger: molecular genetic studies of bipolar affective puerperal psychosis. Psychopharmacol. Bull. 2007;40(2):115–128. [PubMed] [Google Scholar]
- 80.Howes O, McCutcheon R, Stone J. Glutamate and dopamine in schizophrenia: an update for the 21st century. J. Psychopharmacol. 2015;29(2):97–115. doi: 10.1177/0269881114563634. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Excellent review of glutamate and dopamine research in schizophrenia.
- 81.Girgis R, Zoghbi A, Javitt D, et al. The past and future of novel, non-dopamine-2 receptor therapeutics for schizophrenia: a critical and comprehensive review. J. Psychiatr. Res. 2019;108:57–83. doi: 10.1016/j.jpsychires.2018.07.006. [DOI] [PubMed] [Google Scholar]