Abstract
Copy-number alterations Yielding Cancer Liabilities Owing to Partial losS (CYCLOPS) genes have been recently identified as the most enriched class of copy-number associated gene dependencies in human cancer. These genes are cell essential and render tumor cells highly sensitive to the expression of the remaining copy. Chromophobe renal cell carcinoma (chRCC) is characterized by frequent chromosomal deletions, but the relevance of CYCLOPS genes in this tumor subtype is unclear. We found 39 (31%) of 124 recently published candidate CYCLOPS genes (B. Paolella et al., eLife 2017;6:e23268) located on 7 autosomes that are frequently lost in chRCC. GISTIC and RNA-seq data obtained from the TCGA-KICH database showed that 62% of these CYCLOPS genes had significantly lower expression levels in samples with deletion of the respective gene. As copy number (CN) loss of the CYCLOPS gene SF3B1 (Splicing factor 3B subunit 1) has been recently reported in 71% chRCC, we explored the relevance of SF3B1 CN alteration and SF3B1 expression in a set of chRCC and additional oncocytic renal neoplasms. The frequency of SF3B1 CN loss (65%) was similar to that obtained from the TCGA-KICH database and correlated significantly with both lower SF3B1 mRNA (P < .05) and protein expression (P < .001). Other tumor subtypes with oncocytic cytoplasm had normal SF3B1 CN and displayed strong SF3B1 protein expression. These results suggest that CN loss of CYCLOPS genes is a characteristic feature in chRCC. Since many CYCLOPS genes code for components of proteasomes and transcriptional regulation, their alteration could make chRCC vulnerable to targeted drugs.
Introduction
In a recent study with 86 cancer cell lines, a class of genes was identified, which render cells with hemizygous loss highly dependent on the expression of the remaining copy [1]. As partial, but not complete inactivation was shown to be compatible with cancer cell survival, these genes were termed Copy number alterations Yielding Cancer Liabilities Owing to Partial losS (CYCLOPS) genes, which predominantly code for proteasome, spliceosome, and ribosome components [1], [2]
Chromophobe renal cell carcinoma (chRCC) is a distinct histological entity of RCC, which was first described by Thoenes et al. [3] and accounts for approximately 5–7% of RCC [4]. In contrast to other renal cancer subtypes, e.g. clear cell renal cell carcinoma (ccRCC) and papillary RCC (pRCC), chRCCs are characterized by frequent loss of chromosomes 1, 2, 6, 10, 13, 17, 21 and Y [4]. This leads to gene copy number alterations that affect tumor suppressor genes, such as PTEN, RB1 and p53, but also a high number of additional genes whose role is considered non-oncogenic. chRCC have a relatively indolent biologic behavior compared to ccRCC and pRCC [4], [5], [6]. However, several studies have demonstrated that some patients with chRCC die of metastatic disease and the survival of metastatic chRCC patients is comparable to metastatic ccRCC [5], [7]. It can be challenging to distinguish chRCC from other renal cell neoplasms with oncocytic cells, especially from renal oncocytoma and hybrid oncocytic/chromophobe tumors (HOCT) [4].
Given the high frequency of chromosomal losses observed in approximately 70% of chRCC [4], we hypothesized that CYCLOPS genes play a critical role in the pathogenesis of this tumor subtype. We, therefore, used The Cancer Genome Atlas (TCGA) and the data published by Paolella et al. [1] to determine copy number (CN) and mRNA expression levels of all CYCLOPS gene candidates being relevant for chRCC. As CN loss of the CYCLOPS gene SF3B1, which code Splicing Factor 3B subunit 1, was reported in 71% chRCC [1], we analyzed SF3B1 CN alterations, mRNA and protein expression levels in our own set of chRCC, hybrid oncocytic/chromphobe tumors (HOCT) and oncocytic papillary renal cell carcinoma (opRCC).
Materials and Methods
Data Sets and Databases
Corresponding clinical information of TCGA-KICH samples were obtained from TCGA Data Portal (https://portal.gdc.cancer.gov/). In the TCGA-KICH dataset, there are 66 primary chRCCs with copy number variation and RNA-seq data [8]. Digital whole slide images of TCGA cases were reviewed by using the Cancer Digital Slide Archive (http://cancer.digitalslidearchive.net/). The demographic and clinical characteristics for the selected 66 patients are summarized in Table 1.
Table 1.
Clinical data from two chRCC patient cohorts.
| Characteristics | Swiss | TCGA-KICH |
|---|---|---|
| Patient no. | 37 | 66 |
| Age range (median) | 18–86 (61) | 17–86 (50) |
| Female | 12 (32.4%) | 27 (40.9%) |
| Male | 25 (67.6%) | 39 (59.1%) |
| pT or T Stage* | ||
| 1 | 24 (64.9%) | 21 (31.8%) |
| 2 | 7 (18.9%) | 25 (37.9%) |
| 3 | 6 (16.2%) | 18 (27.3%) |
| 4 | 0 (0%) | 2 (3.0%) |
Swiss dataset: pT stage, TCGA-KICH: T stage.
A list of 124 CYCLOPS gene candidates identified by Paolella et al. [1] was used to identify those genes with frequent CN loss in chRCC. The manually curated Kyoto Encyclopedia of Genes and Genomes (KEGG) database was used via STRING in order to determine the enrichment of the genes of interest in KEGG pathway maps [9].
Publically available Level 3 TCGA data were downloaded from the FIREBROWSE database (http://firebrowse.org/) including the GISTIC2 (level 4) CN analysis data and NGS-based RNA-sequencing (RNA-seq) data as previously described [10]. The gene-level table consisting of discrete values indicating loss (<0) or no loss (≥0) for each CYCLOPS gene for each sample was obtained from the GISTIC2 data. This was used as a grouping on which the expression levels were compared correcting for multiple comparisons.
Swiss Tumor Samples
We selected 10 opRCCs, 37 chRCCs, 14 renal oncocytomas, and 6 HOCTs from the archive of the Department of Pathology and Molecular Pathology of the University Hospital Zurich, Switzerland. Tumors were staged according to the TNM staging system [11]. The demographic and clinicopathological characteristics for the 37 chRCC are summarized in Table 1. The study was approved by the Cantonal Ethics Committee of Zurich (KEK-ZH-Nr. 2013–0629; KEK-ZH-Nr. 2011–72/4). The retrospective use of normal and tumor tissues of RCC patients is in accordance with the Swiss Law (“Humanforschungsgesetz”), which, according to Article 34, allows the use of biomaterial and patient data for research purposes without informed consent under certain conditions that include the present cases. Law abidance of this study was reviewed and approved by the ethics commission of the Canton Zurich.
All tumors were reviewed by two pathologists (R.O. and H.M.) and histologically classified according to the World Health Organization guidelines [4]. ChRCCs were defined as tumors composed of large polygonal cells with clarified, so-called “pale cell” or eosinophilic cytoplasm with distinct cell border, perinuclear halo and irregular (raisinoid) nuclei. All chRCC were positive for CK7, succinate dehydrogenase subunit B (SDHB) and fumarate hydratase (FH) and negative for vimentin except for focal sarcomatoid area of one chRCC. Renal oncocytomas were defined as tumors composed of oncocytes (round nuclei with prominent nucleoli, eosinophilic granular cytoplasm) without raisinoid nuclear irregularity, and Cytokeratin 7 (CK7) negative or focal expression in central scar area. HOCTs were defined as tumors with overlapping histology between oncocytoma and chRCC. opRCCs were defined as papillary RCC with voluminous, finely granular, deeply eosinophilic cytoplasm and oncocytoma-like round to oval, regular, low grade nuclei. Their nuclei are single-layered and linearly aligned.
OncoScan® CN Assay
Tumor areas displaying >80% cancer cell portion were marked on the hematoxylin and eosin slides. DNA from formalin-fixed paraffin-embedded (FFPE) tumor tissue samples was obtained by punching 4 to 6 tissue cylinders (diameter 0.6 mm) from each sample. DNA extraction from FFPE tissue was done as described [12]. DNA was quantified by the fluorescence-based Qubit dsDNA HS Assay Kit. Genome-wide DNA copy-number alterations and allelic imbalances of 37 chRCC, 4 HOCTs and 8 opRCCs were analyzed by Affymetrix OncoScan® CNV FFPE Assay Kit (Affymetrix). The samples were processed by IMGM Laboratories GmbH (Martinsried, Germany) for CNV determination. The data were analyzed by the OncoScan Console (Affymetrix) and Nexus Express (Biodiscovery, Inc. CA, USA) softwares using Affymetrix TuScan algorithm. All array data were also manually reviewed for subtle alterations not automatically called by the software.
Immunohistochemistry (IHC)
FFPE sections (2 μm) were transferred to glass slides and treated using Ventana Benchmark XT and Bondmax (Leica Microsystems) automated systems. Antibodies and protocols are listed in Supplementary Table 1. Immunohistochemical evaluation was conducted by two pathologists (R.O. and H.M.) blinded to the clinical data. Immunostained tissue sections were scanned using the NanoZoomer Digital Slide Scanner (Hamamatsu Photonics K.K., Shizuoka, Japan). Non-neoplastic cells (e.g. proximal and distal renal tubular epithelial cells, inflammatory cells, fibroblasts, and endothelial cells) were used as internal positive control. SF3B1 expression was evaluated based on the percentage of positive cells and staining intensity using the Histoscore (H-score) as described previously [13]. The percentage of cells at different staining intensities was determined by visual assessment, thereafter the score was calculated using the formula 1 × (% of 1+ cells) + 2 × (% of 2+ cells) + 3 × (% of 3+ cells). The final score is on a continuous scale between 0 and 300. Samples were grouped in tumors with low (H-score ≤ 200) or high (H-score> 200) SF3B1 expression.
siRNA knockdown of SF3B1 in HEK293T cells was used for antibody validation. siRNAs were transfected into HEK293T cells using Lipofectamine-RNAiMAX reagent (Invitrogen) according to the manufacture's protocol. In brief, 250′000 HEK293T cells were seeded into a 6 well-plate. After 24 hours, reverse transient transfection was done using 100 nM siRNA Allstars control and siSF3B1 (SI04161766) (Qiagen) with 5ul Lipofectamine-RNAiMAX per well. Two days later the cells were harvested. Cell blocks were prepared as described previously [14] and subjected to immunohistochemistry.
Taqman Assay
RNA extraction from 19 FFPE chRCC was performed using Maxwell® 16 Tissue DNA Purification Kit (Promega). RNA quality was measured with RNA Qubit™ RNA HS Assay Kit (ThermoFisher). cDNA was prepared using Superscript IV Vilo Mastermix 50 Rxns (Thermo Fisher). The quantitative measurements were performed using the Taqman Fast Advanced master mix (Thermo Fisher) with 20 ng/μL ng of cDNA in each technical duplicate and the cycling parameters according to the protocol on a ViiA7 (Thermo Fisher). The thermal cycler profile was as follows: 20 seconds at 95 °C, 40 cycles of 1 second at 95 °C and 20 seconds at 60 °C. All reactions were performed in duplicates. Primer and probe set assay IDs for the TaqMan assays were Hs00961640_g1 for SF3B1 and Hs03929097 for GAPDH, (ThermoFisher). Normal tissue was used to normalize the quantitative analysis of all samples. The Ct value for each sample was calculated with the ΔΔCt-method, and the fold expression changes (tumor versus normal) were calculated using the 2−ΔΔCT method.
Statistical Analysis
All statistical analyses were done using R, 3.4.1 (R Foundation for Statistical Computing, Vienna, Austria) and the plugin EZR (Saitama Medical Center, Jichi Medical University, Saitama, Japan). The Welch two sample t-test was used when comparing the expression of CYLOPS genes between samples with loss and no loss. The P-values obtained where then corrected for multiple testing using the Benjamini & Hochberg method. The Welch's t-test, the Mann–Whitney U test and Pearson's correlation coefficient were used to assess associations between continuous and categorical variables. The paired t-test was used for paired normal and tumor data. P values <.05 were considered statistically significant.
Results
CYCLOPS Genes in Chromophobe Renal Cell Carcinoma
By focusing on genome-wide CN-associated gene dependencies (cell viability after gene suppression), Paolella et al. [1] identified 124 CYCLOPS genes, which fulfilled this criterion. Thirty-nine of the 124 CYCLOPS genes were located on the seven most frequently lost chromosomes in chRCC: 1, 2, 6, 10, 13, 17, and 21. These 39 genes were deemed to be the most relevant for this chRCC. The list of the genes shown in Supplementary Table 2 is a modified extract of the gene list shown in supplementary file 1B of Paolella et al. [1]. Protein functions and cellular pathways were from the National Center for Biotechnology Information (NCBI) database.
CYCLOPS Copy Number and Gene Expression
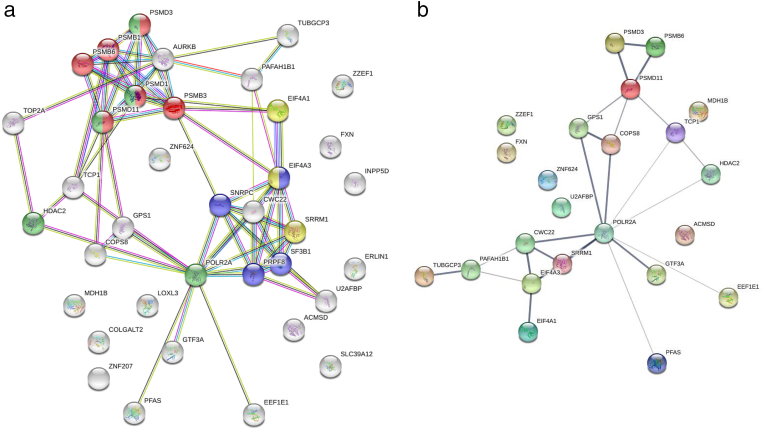
Two of the 39 CYCLOPS genes were read-through transcripts and removed from further analysis. The proteasome and spliceosome KEGG pathways were enriched when considering the remaining 37 genes (Figure 1A). Notably, the gene products of 19 (51%) genes are involved in RNA transcription/processing and proteasomes. Of the 37 CYCLOPS genes, 23 (62%) had significantly lower (adjusted P < .05) mRNA expression when comparing samples experiencing a single allele loss of the respective gene and those with no loss. The result of the CYCLOPS gene expression analysis is shown in Supplementary Table 3. When taking the 23 genes showing a significantly reduced expression, mRNA transport and surveillance pathways were enriched along with the proteasome pathway (Figure 1B).
Figure 1.
a) Protein–protein interaction of 37 CYCLOPS genes in chRCC constructed using KEGG enriched pathway and STRINGR databases. Red: Proteasome; blue: Spliceosome; green and yellow: Gene/RNA regulation and processing. b) Protein–protein interaction of 23 CYCLOPS genes lower expressed due to CN loss.
SF3B1 Copy Number and Expression Analysis in chRCC and Other Oncocytic Renal Neoplasms
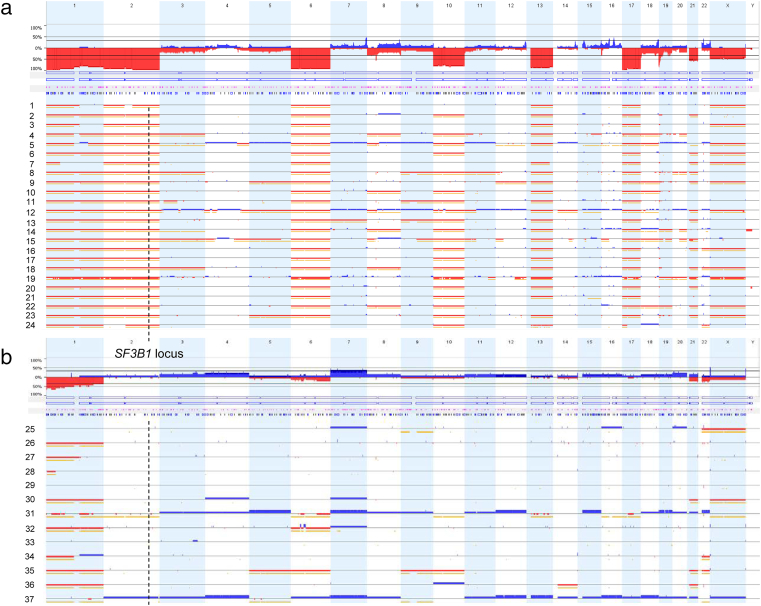
Although being frequently affected by CN loss [1], its influence on expression level of SF3B1 RNA and protein in chRCC have not been investigated thus far. For this purpose we first analyzed 37 chRCC using OncoScan FFPE assay. As expected, the majority of cases revealed loss of autosomes 1, 2, 6, 10, 13, 17, and 21 (Figure 2). Twenty-four of 37 (65%) tumors showed loss of SF3B1, which is located on chromosome 2. Neither 4 HOCT nor 8 oncocytic papillary RCC showed SF3B1 CN alteration (Supplementary Figure 1). The proportion of CN losses in the TCGA-KICH dataset was similar with 47 of 66 (71%) chRCC samples experiencing a SF3B1 CN loss.
Figure 2.
Chromosomal copy number (CN) alterations detected by OnsoScan analysis of 37 chRCCs. a) Tumors with CN loss and b) without CN loss. The location of SF3B1 on chromosome 2q33.1 is indicated by a dashed line. Blue: copy-number gain (Probe mean log2 threshold: 0.3); red: copy-number loss (Probe mean log2 threshold: −0.3); yellow signals: copy-neutral, loss-of-heterozygosity.
Association of CN Loss and Reduced SF3B1 mRNA Expression
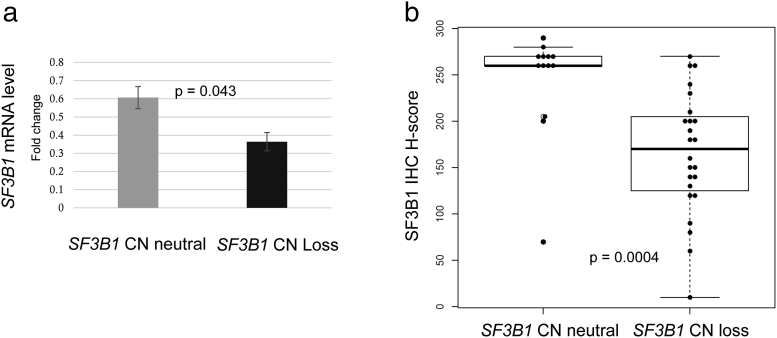
We evaluated SF3B1 mRNA expression in 19 matched normal kidney and cancer tissue samples from the Swiss cohort by Taqman assay. Nine chRCCs had SF3B1 CN loss and 10 chRCCs were without SF3B1 CN loss. The effect of CN on gene expression was analyzed by comparing the mean mRNA fold-changes in chRCC with and without SF3B1 CN loss. SF3B1 mRNA transcription level was significantly correlated with SF3B1 CN status (Figure 3A; Welch's t-test, P = .043). This data were comparable with those obtained from TCGA-KICH (see Supplementary Table 3). The mRNA levels in tumors with SF3B1 CN loss were 50% reduced.
Figure 3.
Association of SF3B1 mRNA and protein expression levels with SF3B1 copy number in Swiss chRCC cohort. a) Normalized mean mRNA expression of SF3B1 in tumors with SF3B1 CN neutral and SF3B1 CN loss (Welch's t-test, P = .043; bars indicate standard deviation). b). SF3B1 H-scores of chRCC without SF3B1 CN loss (mean 243.5). and with SF3B1 CN loss (mean165.4; Mann–Whitney U test; P = .0004).
Association of CN Loss and Reduced SF3B1 Protein Expression
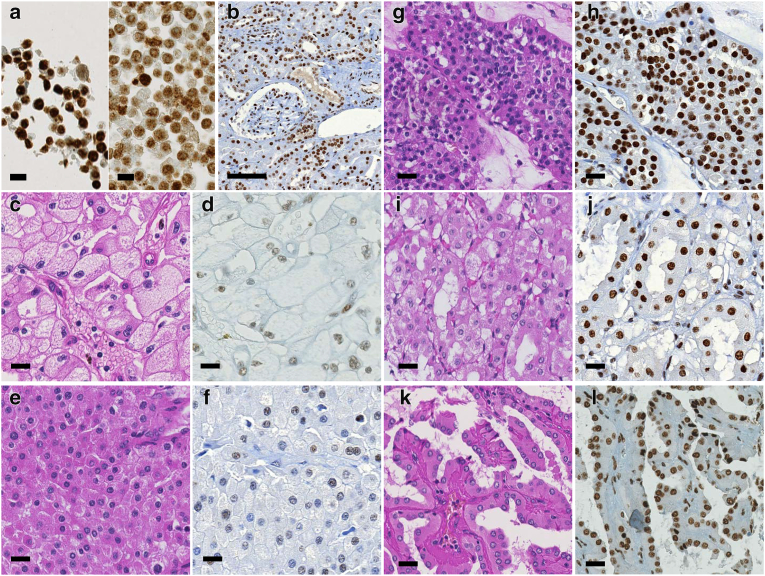
We evaluated SF3B1 protein expression by immunohistochemistry of 10 normal kidney tissue samples and in 37 Swiss chRCC samples for which we had available the SF3B1 CN status via OncoScan. To check whether this antibody was applicable for immunostaining, we performed siRNA-based knockdown of SF3B1 expression. Compared to control siRNA transfected HEK293 cells, which have two intact chromosomes 2 [15], the knockdown cells showed reduction of SF3B1 protein expression (Figure 4A).
Figure 4.
Immunohistochemistry of SF3B1. a) Left: strong expression of SF3B1 in HEK293 cells transfected with control siRNA (siControl); right: reduced expression in SF3B1-specific siRNA (siSF3B1); scale bar, 100 μm. b) Strong SF3B1 expression in proximal and distal renal tubules, glomeruli, smooth muscle cells of vascular wall and endothelial cells in non-neoplastic renal tissue. c-l) Hematoxylin/eosin (left) and immunohistochemically stained (right) tumor sections. c-d) chRCC with SF3B1 CN loss composed of many typical pale cells with distinct cell border and decreased expression of SF3B1 protein. e-f) chRCC with SF3B1 CN loss, purely composed of eosinophilic cells, and reduced SF3B1 expression. g-h) Strong SF3B1 expression in renal oncocytoma, i-j) in HOCT and k-l) in opRCC; scale bars, 20 μm.
In adjacent normal kidney tissue of 37 chRCC patients, SF3B1 was strongly positive in nuclei of all normal kidney cells including podocytes, mesangial cells, renal tubules, endothelial cells, interstitial fibroblasts and immune cells (Figure 4B). The SF3B1 protein level assessed by continuous H-score and binary evaluation using H-score with cutpoint 200 was significantly lower in the tumors with SF3B1 CN loss than in the remaining chRCC (Figure 3B; Mann–Whitney U test, P = .0004). Fifteen of 24 (63%) tumors with CN loss had reduced SF3B1 positivity (immunoreactivity of 1+; H-score ≤200) and 12 of 13 (92%) tumors with neutral CN had normal SF3B1 expression levels (immunoreactivity of 2+; H-score >200). No correlation was found between SF3B1 protein expression and pT stage. Examples of hematoxylin/eosin stained and immunostained chRCC are shown in Figure 4, C–F. All renal oncocytomas, HOCTs and opRCCs showed strong nuclear expression of SF3B1 (Figure 4, G–L).
Discussion
In this study, we demonstrate that copy number loss of CYCLOPS genes is a characteristic and unique feature of chRCC. We show that the 7 autosomes, which are lost in the majority of chRCC, harbor one-third of 124 CYCLOPS genes [1]. About half of these genes belong to pathways regulated by proteasomes and the transcription machinery. Gene expression analysis using TCGA-KICH data demonstrated that CN loss led to significantly lower transcription levels of more than 60% of the genes. This data suggest that in chRCC CN loss of CYCLOPS genes accompanied by a reduction of expression makes tumor cells even more vulnerable than CN loss alone.
CN loss of the CYCLOPS gene SF3B1 occurs most frequently in chRCC [1]. As the influence of CN loss to SF3B1 protein expression is yet unclear, we decided to analyze our own set of chRCC by immunohistochemistry. Our comprehensive CN study of chRCC demonstrated SF3B1 genomic loss in a large fraction of chRCC (65%), which is consistent with TCGA data demonstrating CN losses of SF3B1 in 71% chRCC [1], [8]. Reduced SF3B1 mRNA and protein expression was significantly associated with SF3B1 CN loss in chRCC. These findings indicate that SF3B1 CN loss causes reduced expression of SF3B1 in most chRCC. Potentially, additional mechanisms of SF3B1 downregulation exist, since we observed a few chRCC without SF3B1 CN loss but with reduced SF3B1 expression. As SF3B1 mutations do not exist in chRCC [1], [8], SF3B1 mRNA and protein expression may be influenced by post-transcriptional, translational and protein degradation regulation in these tumors [16].
SF3B1 is a core component of the U2 small nuclear ribonucleoprotein at the catalytic center of the spliceosome and contributes to intron removal by anchoring pre-mRNA onto the spliceosome [17]. Previous studies have indicated that mutations or aberrant splicing patterns in spliceosome components, including SF3B1, are associated with different cancer phenotypes [1], [17], [18], [19], [20]. Interestingly, in patients with myelodysplastic syndrome SF3B1 mutations lead to deregulated expression and splicing of several DNA repair and DNA damage response genes as well as of RNA-processing factors [21]. In regards to chRCC, the decrease of CN loss mediated SF3B1 expression may affect the splicing of transcripts involved in chromatin structure, DNA repair and DNA damage response, thereby possibly providing an explanation for the accumulation of elevated somatic mutation rate and mutation signature of DNA mismatch repair deficiency seen in this tumor type [8], [22].
It was recently shown that SF3B1 is a HIF1α target [23]. Mechanistic and functional linkages between HIF1α, SF3B1, and fructose metabolism by production of splice isoform ketohexokinase-C (KHK-C) were shown in cardiac hypertrophy. KHK-C is the central fructose-metabolizing enzyme and KHK-C expression through the HIF1α-SF3B1 axis promotes conversion of fructose carbon to lipids, suppresses mitochondrial oxidative phosphorylation and increases glycolysis [16]. In the TCGA-KICH dataset of chRCC we identified a highly significant positive correlation between HIF1α and SF3B1 mRNA expression (Supplementary Figure 3). It is therefore tempting to speculate that a SF3B1/HIF1α pathway exists in chRCC.
ChRCC, oncocytoma, and HOCT represent a spectrum of tumors with oncocytic cells. A rare group of oncocytic papillary tumors has also been described [24], [25], [26]. The biological behavior of these oncocytic neoplasms ranges from benign (oncocytoma), low malignant behavior (HOCT), to malignant (chRCC). The oncocytic phenotype is mainly due to the accumulation of mitochondria. The differential diagnosis between these oncocytic neoplasms is sometimes extremely difficult on the basis of morphology alone. Several studies reported that chromosome 2 loss is not present in oncocytoma [27], [28], [29], [30]. Our CN analysis showed no SF3B1 CN alteration in HOCT and opRCC. For this reason, we expected generally stronger SF3B1 protein expression in oncocytoma, HOCT and opRCC than in the majority of chRCC in which only one gene copy of SF3B1 exists. In our current study, strongly positive SF3B1 protein expression by IHC was observed in all renal oncocytomas, HOCTs and opRCCs. Therefore, SF3B1 expression may help in the differential diagnosis of oncocytic neoplasms.
As chromosome 2 loss hardly occur in clear cell and papillary RCC [31], [32], we used TCGA data to compare SF3B1 expression levels in these two tumor subtypes with those in chRCC. As expected, SF3B1 mRNA was significantly more abundant in clear cell and papillary RCC (Supplementary Table 4) suggesting that attenuated SF3B1 expression due to chromosome 2 loss in chRCC is unique for renal neoplasms.
In conclusion, we identified frequent CN loss combined with reduced expression of many CYCLOPS genes as characteristic feature for chRCC. Further studies taking advantage of the compromised integrity of spliceosomes and RNA processing may reveal novel strategies for the treatment of chRCC.
Acknowledgments
Acknowledgements
This manuscript is dedicated to the memory of Prof. W. Krek (Department of Biology, Institute of Molecular Health Sciences, ETH Zurich, Zurich, Switzerland), who died in 2018. He was a great scientist in the field of cell biology and he was involved in conception and design of this study. The authors thank Susanne Dettwiler and Fabiola Prutek for their outstanding technical assistance. This work was supported in part by Niigata Foundation for Promoting of Medicine (2015) and the Swiss National Science Foundation grant to H.M. (No. S-87701-03-01).
Disclosure/conflict of interest
The authors have no conflict of interest and nothing to disclose.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tranon.2019.05.005.
Contributor Information
Riuko Ohashi, Email: riuko@med.niigata-u.ac.jp.
Peter Schraml, Email: peter.schraml@usz.ch.
Aashil Batavia, Email: Aashil.batavia@usz.ch.
Silvia Angori, Email: silvia.angori@usz.ch.
Patrik Simmler, Email: patrik.simmler@biol.ethz.ch.
Niels Rupp, Email: niels.rupp@usz.ch.
Yoichi Ajioka, Email: ajioka@med.niigata-u.ac.jp.
Esther Oliva, Email: EOLIVA@mgh.harvard.edu.
Holger Moch, Email: holger.moch@usz.ch.
Appendix A. Supplementary data
Supplementary figures
Supplementary tables
References
- 1.Paolella BR, Gibson WJ, Urbanski LM, Alberta JA, Zack TI, Bandopadhayay P, Nichols CA, Agarwalla PK, Brown MS, Lamothe R. Copy-number and gene dependency analysis reveals partial copy loss of wild-type SF3B1 as a novel cancer vulnerability. elife. 2017;(6) doi: 10.7554/eLife.23268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nijhawan D, Zack TI, Ren Y, Strickland MR, Lamothe R, Schumacher SE, Tsherniak A, Besche HC, Rosenbluh J, Shehata S. Cancer vulnerabilities unveiled by genomic loss. Cell. 2012;150:842–854. doi: 10.1016/j.cell.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thoenes W, Storkel S, Rumpelt HJ. Human chromophobe cell renal carcinoma. Virchows Arch B Cell Pathol Incl Mol Pathol. 1985;48:207–217. doi: 10.1007/BF02890129. [DOI] [PubMed] [Google Scholar]
- 4.Moch H, Humphrey PA, Ulbright TM, Reuter VE. Lyon; France: 2016. WHO Classification of Tumours of the Urinary System and Male Genital Organs. International Agency for Research on Cancer. [Google Scholar]
- 5.Volpe A, Novara G, Antonelli A, Bertini R, Billia M, Carmignani G, Cunico SC, Longo N, Martignoni G, Minervini A. Chromophobe renal cell carcinoma (RCC): oncological outcomes and prognostic factors in a large multicentre series. BJU Int. 2012;110:76–83. doi: 10.1111/j.1464-410X.2011.10690.x. [DOI] [PubMed] [Google Scholar]
- 6.Cheville JC, Lohse CM, Zincke H, Weaver AL, Blute ML. Comparisons of outcome and prognostic features among histologic subtypes of renal cell carcinoma. Am J Surg Pathol. 2003;27:612–624. doi: 10.1097/00000478-200305000-00005. [DOI] [PubMed] [Google Scholar]
- 7.Casuscelli J, Weinhold N, Gundem G, Wang L, Zabor EC, Drill E, Wang PI, Nanjangud GJ, Redzematovic A, Nargund AM. Genomic landscape and evolution of metastatic chromophobe renal cell carcinoma. JCI Insight. 2017;2 doi: 10.1172/jci.insight.92688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis CF, Ricketts CJ, Wang M, Yang L, Cherniack AD, Shen H, Buhay C, Kang H, Kim SC, Fahey CC. The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell. 2014;26:319–330. doi: 10.1016/j.ccr.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:447–452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun M, Tong P, Kong W, Dong B, Huang Y, Park IY, Zhou L, Liu XD, Ding Z, Zhang X. HNF1B Loss Exacerbates the Development of Chromophobe Renal Cell Carcinomas. Cancer Res. 2017;77:5313–5326. doi: 10.1158/0008-5472.CAN-17-0986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bertero L, Massa F, Metovic J, Zanetti R, Castellano I, Ricardi U, Papotti M, Cassoni P. Eighth Edition of the UICC classification of malignant tumours: an overview of the changes in the pathological TNM classification criteria-What has changed and why? Virchows Arch. 2018;472:519–531. doi: 10.1007/s00428-017-2276-y. [DOI] [PubMed] [Google Scholar]
- 12.Deml KF, Schildhaus HU, Comperat E, von Teichman A, Storz M, Schraml P, Bonventre JV, Fend F, Fleige B, Nerlich A. Clear cell papillary renal cell carcinoma and renal angiomyoadenomatous tumor: two variants of a morphologic, immunohistochemical, and genetic distinct entity of renal cell carcinoma. Am J Surg Pathol. 2015;39:889–901. doi: 10.1097/PAS.0000000000000456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCarty KS, Jr., Szabo E, Flowers JL, Cox EB, Leight GS, Miller L, Konrath J, Soper JT, Budwit DA, Creasman WT. Use of a monoclonal anti-estrogen receptor antibody in the immunohistochemical evaluation of human tumors. Cancer Res. 1986;46:4244s–4248s. [PubMed] [Google Scholar]
- 14.Struckmann K, Mertz K, Steu S, Storz M, Staller P, Krek W, Schraml P, Moch H. pVHL co-ordinately regulates CXCR4/CXCL12 and MMP2/MMP9 expression in human clear-cell renal cell carcinoma. J Pathol. 2008;214:464–471. doi: 10.1002/path.2310. [DOI] [PubMed] [Google Scholar]
- 15.Stepanenko A, Andreieva S, Korets K, Mykytenko D, Huleyuk N, Vassetzky Y, Kavsan V. Step-wise and punctuated genome evolution drive phenotype changes of tumor cells. Mutat Res. 2015;771:56–69. doi: 10.1016/j.mrfmmm.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 16.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonnal S, Vigevani L, Valcarcel J. The spliceosome as a target of novel antitumour drugs. Nat Rev Drug Discov. 2012;11:847–859. doi: 10.1038/nrd3823. [DOI] [PubMed] [Google Scholar]
- 18.Webb TR, Joyner AS, Potter PM. The development and application of small molecule modulators of SF3b as therapeutic agents for cancer. Drug Discov Today. 2013;18:43–49. doi: 10.1016/j.drudis.2012.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dvinge H, Kim E, Abdel-Wahab O, Bradley RK. RNA splicing factors as oncoproteins and tumour suppressors. Nat Rev Cancer. 2016;16:413–430. doi: 10.1038/nrc.2016.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harbour JW, Roberson ED, Anbunathan H, Onken MD, Worley LA, Bowcock AM. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat Genet. 2013;45:133–135. doi: 10.1038/ng.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dolatshad H, Pellagatti A, Fernandez-Mercado M, Yip BH, Malcovati L, Attwood M, Przychodzen B, Sahgal N, Kanapin AA, Lockstone H. Disruption of SF3B1 results in deregulated expression and splicing of key genes and pathways in myelodysplastic syndrome hematopoietic stem and progenitor cells. Leukemia. 2015;29:1092–1103. doi: 10.1038/leu.2014.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mirtschink P, Krishnan J, Grimm F, Sarre A, Horl M, Kayikci M, Fankhauser N, Christinat Y, Cortijo C, Feehan O. HIF-driven SF3B1 induces KHK-C to enforce fructolysis and heart disease. Nature. 2015;522:444–449. doi: 10.1038/nature14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hes O, Brunelli M, Michal M, Cossu Rocca P, Hora M, Chilosi M, Mina M, Boudova L, Menestrina F, Martignoni G. Oncocytic papillary renal cell carcinoma: a clinicopathologic, immunohistochemical, ultrastructural, and interphase cytogenetic study of 12 cases. Ann Diagn Pathol. 2006;10:133–139. doi: 10.1016/j.anndiagpath.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 25.Masuzawa N, Kishimoto M, Nishimura A, Shichiri Y, Yanagisawa A. Oncocytic renal cell carcinoma having papillotubular growth: rare morphological variant of papillary renal cell carcinoma. Pathol Int. 2008;58:300–305. doi: 10.1111/j.1440-1827.2008.02227.x. [DOI] [PubMed] [Google Scholar]
- 26.Xia QY, Rao Q, Shen Q, Shi SS, Li L, Liu B, Zhang J, Wang YF, Shi QL, Wang JD. Oncocytic papillary renal cell carcinoma: a clinicopathological study emphasizing distinct morphology, extended immunohistochemical profile and cytogenetic features. Int J Clin Exp Pathol. 2013;6:1392–1399. [PMC free article] [PubMed] [Google Scholar]
- 27.Yap NY, Rajandram R, Ng KL, Pailoor J, Fadzli A, Gobe GC. Genetic and Chromosomal Aberrations and Their Clinical Significance in Renal Neoplasms. Biomed Res Int. 2015;2015(476508) doi: 10.1155/2015/476508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brunelli M, Eble JN, Zhang S, Martignoni G, Delahunt B, Cheng L. Eosinophilic and classic chromophobe renal cell carcinomas have similar frequent losses of multiple chromosomes from among chromosomes 1, 2, 6, 10, and 17, and this pattern of genetic abnormality is not present in renal oncocytoma Modern pathology : an official journal of the United States and Canadian Academy of Pathology. Inc. 2005;18:161–169. doi: 10.1038/modpathol.3800286. [DOI] [PubMed] [Google Scholar]
- 29.Yusenko MV, Kuiper RP, Boethe T, Ljungberg B, van Kessel AG, Kovacs G. High-resolution DNA copy number and gene expression analyses distinguish chromophobe renal cell carcinomas and renal oncocytomas. BMC Cancer. 2009;9(152) doi: 10.1186/1471-2407-9-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan MH, Wong CF, Tan HL, Yang XJ, Ditlev J, Matsuda D, Khoo SK, Sugimura J, Fujioka T, Furge KA. Genomic expression and single-nucleotide polymorphism profiling discriminates chromophobe renal cell carcinoma and oncocytoma. BMC Cancer. 2010;10(196) doi: 10.1186/1471-2407-10-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang F, Richter J, Schraml P, Bubendorf L, Gasser T, Sauter G, Mihatsch MJ, Moch H. Chromosomal imbalances in papillary renal cell carcinoma: genetic differences between histological subtypes. Am J Pathol. 1998;153:1467–1473. doi: 10.1016/S0002-9440(10)65734-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moch H, Presti JC, Sauter G, Buchholz N, Jordan P, Mihatsch MJ, Waldman FM. Genetic aberrations detected by comparative genomic hybridization are associated with clinical outcome in renal cell carcinoma. Cancer Res. 1996;56:27–30. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures
Supplementary tables