Abstract
It is a matter of current debate whether the bone marrow is a hub for circulating memory T lymphocytes and/or the home of resident memory T lymphocytes. Here we demonstrate for CD69+ murine CD8+, and CD69+ murine and human CD4+ memory T lymphocytes of the bone marrow, making up between 30 and 60% of bone marrow memory T lymphocytes, that they express the gene expression signature of tissue‐resident memory T lymphocytes. This suggests that a substantial proportion of bone marrow memory T lymphocytes are resident. It adds to previous evidence that bone marrow memory T cells are resting in terms of mobility and proliferation, and maintain exclusive long‐term memory to distinct, systemic antigens.
Keywords: Bone marrow, CD69, Memory T cells, Spleen, Tissue‐resident signature genes
It is a matter of current debate, whether the bone marrow is a hub for circulating memory T lymphocytes, and/or the home of tissue‐resident memory T (TRM) lymphocytes. While several groups could not find evidence for tissue‐residency of bone marrow memory T cells 1, we have previously demonstrated the exclusive maintenance of T‐cell memory for distinct (systemic) antigens in the human and murine bone marrow, arguing that at least memory T cells maintaining those specificities were bone marrow‐resident 2, 3, 4. We have also shown that bone marrow memory T cells are resting in terms of proliferation 2, 3, 5 and are maintained independent of the circulation, i.e. their numbers are unaffected by treatment with FTY720 6, a drug that blocks egress of lymphocytes from secondary lymphoid organs by antagonizing of S1PR1. Moreover, in mice and humans approximately 30% to 60% of memory T cells of the bone marrow express the residency marker CD69, and for human bone marrow memory CD4+ T lymphocytes and murine bone marrow memory CD4+ and CD8+ T lymphocytes, we have shown that these cells do not express S1pr1 5, 7, reported to enable lymphocyte egress into the blood.
Here, we show that CD69 expressing murine memory CD4+ and CD8+ T cells, and human memory CD4+ T cells of the bone marrow also express a set of genes reported to be signature genes of TRM from other tissues 8, 9. We isolated human memory CD4+ T cells and murine CD8+ memory T cells from bone marrow in steady‐state situations and murine CD69+ and CD69− memory CD4 T cells from spleen and bone marrow 60 days after a secondary immunization with a defined antigen (LCMV GP61–80) 7. The isolation of the murine CD4+ memory T cells was carried out according to published guidelines 10 and is documented in Supporting Information Fig. 1. The isolation of murine memory CD8 and human memory CD4 has been documented earlier 3, 5. Transcriptomes of murine CD8+ and human CD4+ memory T cells were analyzed by Affymetrix microarrays, while transcriptomes of murine CD4 memory T cells were analyzed by next generation RNA‐Seq. Both CD4 (Fig. 1A) and CD8 (Fig. 1B) murine CD69+, but not CD69− memory T cells of the bone marrow, show the “universal” transcriptional signature of TRM cells reported by Mackay and colleagues for CD8 TRM cells of different tissues (not bone marrow) 9. It should be noted that the Affymetrix microarrays lacked probes for the genes Zfp683 (Hobit), Sidt1, and A430078G23Rik (Fig. 1B). Interestingly, the murine TRM signature genes were also expressed by the CD69+ CD4 memory T cells from the spleen (Fig. 1A). Human CD69+ but not CD69− bone marrow memory CD4+ T cells show the TRM signature previously reported for human CD4+ and CD8+ TRM by Kumar and colleagues 8 (Fig. 1C). These differences in relative gene expression are highly significant (p < 0.0001 to p < 0.0072), as shown by gene set enrichment analysis (Supporting Information Fig. 2). We validated the expression of several TRM signature genes by immunofluorescence and quantitative PCR. We had previously shown by qPCR that CD69+ memory T lymphocytes of murine bone marrow do not express S1pr1 5, 7, and show here that they also do not express Klf2 (Supporting Information Fig. 3A). In addition, human and murine CD69+ memory CD4+ T cells express CXCR6, but little CD62L (Supporting Information Fig. 3B and C). We also compared the expression of the human homologues from the murine TRM signature genes by human bone marrow memory CD4+ T cells by principal‐component analysis. The predominant principal component 1 (PC1, 63%)) separates CD69−CD4+ cells of peripheral blood, when activated, from both, CD69−CD4+ cells of blood and bone marrow and CD69+CD4+ resting cells from bone marrow. PC2 (14%) then separates bone marrow CD69+CD4+ cells from CD69−CD4+ cells from both blood and bone marrow (Fig. 1D). Taken together, murine and human CD69+ memory T cells of the bone marrow express the TRM signature genes reported for TRM of other tissues 8, 9, suggesting that memory T cells are indeed resident in the bone marrow. Whether or not the CD69− memory T cells of the bone marrow are also resident remains to be shown, e.g. by comparing their repertoire to that of circulating memory T cells from blood. The difference in gene expression between CD69+ and CD69− memory T cells probably also explains why Baliu‐Pique and colleagues were not able to detect any clear TRM signatures in total memory T cells isolated from goat bone marrow 1. Similarly, a lack of discrimination between resting memory T cells and their (proliferating) precursors may have impacted on their lifetime analysis by deuterium labeling. An alternative explanation would be that indeed the maintenance of T‐cell memory is differently organized in goats versus mice and humans.
Figure 1.
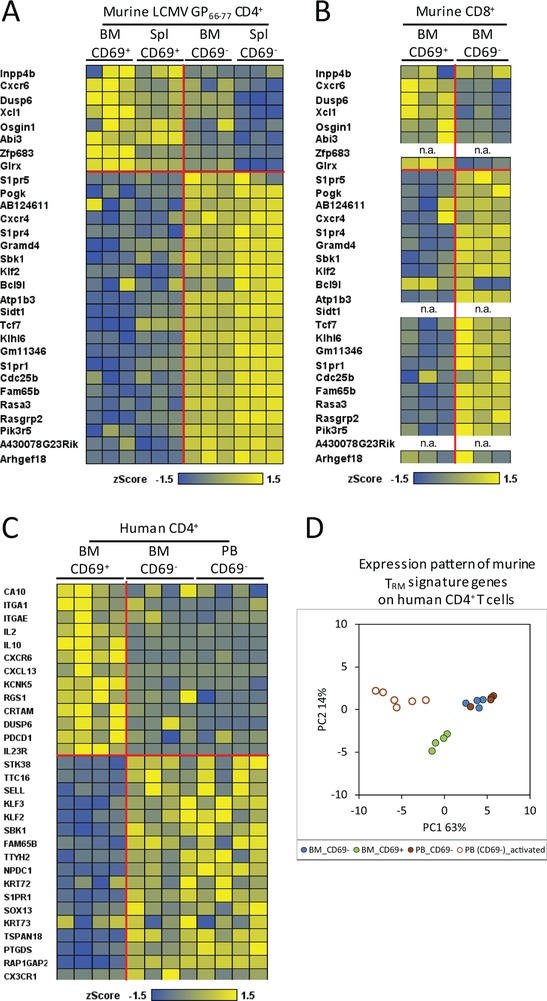
CD69 expression of bone marrow and spleen memory T cells defines a transcriptionally distinct population as reported earlier for TRM cells of other tissues. (A and B) Heatmaps showing reported murine TRM signature genes of other tissues for CD8 T cells 9, on bone marrow and spleen LCMV‐specific memory CD4+ T cells (A) and steady‐state bone marrow memory CD8+ T cells (B) isolated from C57BL/6 mice (Supporting Information Fig. 1; 5, 7). n.a., (probes) not present on the arrays, and thus gene expression not analyzed. (C) Heatmap of reported human TRM signature genes for both CD4+ and CD8+ T cells of other tissues 8 on human bone marrow and paired peripheral blood (>98% being CD69−) memory CD4+ T cells according to CD69 expression 3. (D) Principal‐component analysis (PCA) of described murine TRM signature genes 9 on the cells analyzed in C in comparison with recently activated blood effector/memory CD4+ T cells. In A–C, the horizontal and vertical red lines separate up‐ or downregulated genes as described for murine and human TRM signature genes of other tissues 8, 9. (A) Data shown are from individual mice with a total of three mice from one experiment; (B) data shown are pooled from three independent experiments with cells pooled from 8 to 10 mice per experiment; C/D, data shown were taken from our previously published data GSE50677 3.
Author contributions
Conceptualization, A.R. and J.D.; Methodology, F.S., M.A.M., Ö.S.A., and J.D.; Investigation, F.S., Ö.S.A., A.R., W.J.D., C.C., G.A.H. and M.F.M.; Formal Analysis, P.D.; Writing – Review & Editing, J.D. and A.R.; Visualization, P.D., F.S., W.J.D and J.D; Funding Acquisition, H.D.C., A.R. and J.D.
Conflict of interest
The authors declare no commercial or financial conflict of interest.
Data deposition
The data about resting murine memory CD4+ and CD8+ T lymphocytes discussed in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE124796).
Supporting information
Figure S1. Gating strategy for the isolation of LCMV‐specific memory CD4+ T cells, expressing or not CD69, from murine bone marrow (A) and spleen (B). Eight‐weekold C57BL/6 mice were twice immunized with LCMV GP61‐80, rested for 60 d, and LCMV.GP66‐77 tetramer‐specific CD69+ and CD69‐ memory CD4+ T cells were isolated by MACS enrichment of CD4+ cells and then by FACS. Data shown are representative of three different mice from one experiment.
Figure S2. Gene set enrichment analysis (GSEA) comparing gene sets of murine bone marrow/spleen CD69+ versus CD69‐ memory CD4+ T cells (A) and murine bone marrow CD69+ versus CD69‐ memory CD8+ T cells (B) using the published murine TRM signature genes (Mackay et al., 2016), and gene sets of human bone marrow CD69+ versus bone marrow/blood CD69‐ memory CD4+ T cells using published human TRM signature genes (Kumar et al., 2017). In each plot, the x axis shows the genes ranked with absolute value of log fold change between CD69+ versus CD69‐ cells, and the y axis shows the running enrichment score (ES), comparing the respective TRM genes with indicated p values. Up: up‐regulated TRM signature genes. Down: down‐regulated TRM signature genes.
Figure S3. Validation of differential expression of the transcription factor Klf2 and the surface proteins by memory CD4+ T cells of bone marrow, spleen, and/or blood, expressing CD69 or not. A, Klf2 relative expression by LCMV‐specific CD69+ and CD69‐ memory CD4+ T cells of the bone marrow. Data shown are one experiment from individual mice with n = 3. B, CXCR6 surface protein expression by LCMVspecific CD69+ bone marrow and spleen and CD69‐ bone marrow memory CD4+ T cells. Geometric mean of fluorescent intensity (gMFI) of each analyzed cell subset is indicated. Data shown are representative of three independent experiments. C, CXCR6 and CD62L surface protein expression by human memory CD4+ and CD8+ T cells, expressing or not CD69, form paired bone marrow and peripheral blood samples. Data shown are one representative of three independent experiments.
Acknowledgments
This work was supported by the European Research Council (ERC) Advanced Grant 268987 (to A.R.), by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)–‐Priority program 1468 IMMUNOBONE (to H.D.C and A.R.) and Project number 389687267 (to J.D and A.R.), and by the Leibniz Science Campus Chronic Inflammation (www.chronische‐entzuendung.org). F.S. was supported by Osteoimmune, a FP7 Marie Curie Initial Training Network (FP7‐PEOPLE‐2011‐ITN‐289150). A.R. was in part supported by International Max Planck Research School for Infectious Diseases and Immunology Generation 2011. W.J.D. was supported by the China Scholarship Council (CSC). C.C. was supported by the Leibniz Graduate School for Rheumatology (LGRh). G.A.H. and M.F.M. were supported by the state of Berlin and the “European Regional Development Fund” (ERDF 2014–2020, EFRE 1.8/11, Deutsches Rheuma‐Forschungszentrum).
References
- 1. Baliu‐Pique, M. et al., Front. Immunol. 2018. 9: 2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tokoyoda, K. et al., Immunity 2009. 30: 721–730. [DOI] [PubMed] [Google Scholar]
- 3. Okhrimenko, A. et al., Proc. Natl. Acad. Sci. USA 2014. 111: 9229–9234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chang, H. D. et al., Immunol. Rev. 2018. 283: 86–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sercan Alp, Ö. et al., Eur. J. Immunol. 2015. 45: 975–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Siracusa, F. et al., Eur. J. Immunol. 2017. 47: 1900–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Siracusa, F. et al., Proc. Natl. Acad. Sci. USA 2018. 115: 1334–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kumar, B. V. et al., Cell Rep. 2017. 20: 2921–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mackay, L. K. et al., Science 2016. 352: 459–463. [DOI] [PubMed] [Google Scholar]
- 10. Cossarizza, A. et al., Eur. J. Immunol. 2017. 47: 1584–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Gating strategy for the isolation of LCMV‐specific memory CD4+ T cells, expressing or not CD69, from murine bone marrow (A) and spleen (B). Eight‐weekold C57BL/6 mice were twice immunized with LCMV GP61‐80, rested for 60 d, and LCMV.GP66‐77 tetramer‐specific CD69+ and CD69‐ memory CD4+ T cells were isolated by MACS enrichment of CD4+ cells and then by FACS. Data shown are representative of three different mice from one experiment.
Figure S2. Gene set enrichment analysis (GSEA) comparing gene sets of murine bone marrow/spleen CD69+ versus CD69‐ memory CD4+ T cells (A) and murine bone marrow CD69+ versus CD69‐ memory CD8+ T cells (B) using the published murine TRM signature genes (Mackay et al., 2016), and gene sets of human bone marrow CD69+ versus bone marrow/blood CD69‐ memory CD4+ T cells using published human TRM signature genes (Kumar et al., 2017). In each plot, the x axis shows the genes ranked with absolute value of log fold change between CD69+ versus CD69‐ cells, and the y axis shows the running enrichment score (ES), comparing the respective TRM genes with indicated p values. Up: up‐regulated TRM signature genes. Down: down‐regulated TRM signature genes.
Figure S3. Validation of differential expression of the transcription factor Klf2 and the surface proteins by memory CD4+ T cells of bone marrow, spleen, and/or blood, expressing CD69 or not. A, Klf2 relative expression by LCMV‐specific CD69+ and CD69‐ memory CD4+ T cells of the bone marrow. Data shown are one experiment from individual mice with n = 3. B, CXCR6 surface protein expression by LCMVspecific CD69+ bone marrow and spleen and CD69‐ bone marrow memory CD4+ T cells. Geometric mean of fluorescent intensity (gMFI) of each analyzed cell subset is indicated. Data shown are representative of three independent experiments. C, CXCR6 and CD62L surface protein expression by human memory CD4+ and CD8+ T cells, expressing or not CD69, form paired bone marrow and peripheral blood samples. Data shown are one representative of three independent experiments.


