Key Points
Question
What is the prevalence of carriers of intermediate and pathological ranges of polyglutamine disease–associated alleles among the general population?
Findings
In a cross-sectional study that included 14 196 participants from 5 European population-based cohort studies without an established polyglutamine disease diagnosis from 5 large European population–based cohorts, 10.7% of the participants had a CAG repeat number within the intermediate range of at least 1 polyglutamine disease–associated gene, and up to 1.3% of the participants had a CAG repeat number within the disease-causing range.
Meaning
These results indicate a considerably higher prevalence of carriers of intermediate and pathological ranges of polyglutamine disease–associated alleles among the general population than previously estimated.
This cross-sectional study of 5 European cohort studies determines the prevalence of carriers of intermediate and pathological polyglutamine disease–associated alleles among the general population.
Abstract
Importance
Nine hereditary neurodegenerative diseases are known as polyglutamine diseases, including Huntington disease, 6 spinocerebellar ataxias (SCAs) (SCA1, SCA2, SCA3, SCA6, SCA7, and SCA17), dentatorubral-pallidoluysion atrophy, and spinal bulbar muscular atrophy.
Objective
To determine the prevalence of carriers of intermediate and pathological polyglutamine disease–associated alleles among the general population.
Design, Setting, and Participants
This observational cross-sectional study included data from 5 large European population–based cohorts that were compiled between 1997 and 2012, and the analyses were conducted in 2018. In total, 16 547 DNA samples were obtained from participants of the 5 cohorts. Individuals with a lifetime diagnosis of major depression were excluded (n = 2351). In the remaining 14 196 participants without an established polyglutamine disease diagnosis, the CAG repeat size in both alleles of all 9 polyglutamine disease–associated genes (PDAGs) (ie, ATXN1, ATXN2, ATXN3, CACNA1A, ATXN7, TBP, HTT, ATN1, and AR) was determined.
Exposure
The number of CAG repeats in the alleles of the 9 PDAGs.
Main Outcomes and Measures
The number of individuals with alleles within the intermediate or pathological range per PDAG, as well as differences in sex, age, and body mass index between individuals carrying alleles within the normal or intermediate range and individuals carrying alleles within the pathological range of PDAGs.
Results
In the 14 196 analyzed participants (age range, 18-99 years; 56.3% female), 10.7% had a CAG repeat number within the intermediate range of at least 1 PDAG. Moreover, up to 1.3% of the participants had a CAG repeat number within the disease-causing range, predominantly in the lower pathological range associated with elderly onset. No differences in sex, age, or body mass index were found between individuals with CAG repeat numbers within the pathological range and individuals with CAG repeat numbers within the normal or intermediate range.
Conclusions and Relevance
These results indicate a high prevalence of individuals carrying intermediate and pathological ranges of polyglutamine disease–associated alleles among the general population. Therefore, a substantially larger proportion of individuals than previously estimated may be at risk of developing a polyglutamine disease later in life or bearing children with a de novo mutation.
Introduction
Nine hereditary neurodegenerative diseases are known as polyglutamine diseases, including Huntington disease (HD), 6 spinocerebellar ataxias (SCAs) (SCA1, SCA2, SCA3, SCA6, SCA7, and SCA17), dentatorubral-pallidoluysion atrophy, and spinal bulbar muscular atrophy. All are caused by a CAG triplet repeat expansion in the protein coding regions of different genes.1,2 The number of CAG repeats that result in the development of a polyglutamine disease differs per disease and its associated gene (ie, ATXN1, ATXN2, ATXN3, CACNA1A, ATXN7, TBP, HTT, ATN1, and AR) (Table 1).3,4,5,6,7,8,9,10,11 In addition to a specific pathological range, most polyglutamine disease–associated genes (PDAGs) also have an intermediate range, defined as the range in CAG repeat numbers just below the pathological range for which expansion into the fully pathological range has been observed on intergenerational transmission.3,4,5,6,7,8,9,10,11 Polyglutamine diseases are rare. Huntington disease is the most prevalent, affecting 10.3 to 13.7 individuals per 100 000 among the Western population.12 However, these prevalence estimates are based on cases that undergo genetic testing, either due to the presence of characteristic neurological symptoms or the existence of a family member with a polyglutamine disease. Therefore, by genotyping a large number of individuals from different population-based studies, we aimed to obtain a better estimate of the true prevalence of carriers of intermediate and pathological PDAG alleles among the general population.
Table 1. Polyglutamine Disease–Associated Gene (PDAG) Characteristics.
| Disease | Age at Onset, Mean (Range), y | PDAG | OMIM Accession No. | Locus | Protein | Repeat | Normal | Intermediate | Pathological [Mean] |
|---|---|---|---|---|---|---|---|---|---|
| SCA1 | 38 (11-75) | ATXN1 | 601556 | 6p23 | Ataxin 1 | (CAG)n(CAT)n(CAG)na | 6-35 (6-44)b | 36-38 | 39-91 (45-91b) [47] |
| SCA2 | 36 (2-71) | ATXN2 | 601517 | 12q24 | Ataxin 2 | [(CAG)n(CAA)n(CAG)n]nc | 14-31d | 32d | 33-500 [39]d |
| SCA3 | 40 (10-78) | ATXN3 | 607047 | 14q24-q31 | Ataxin 3 | (CAG)2CAA AAG CAG CAA(CAG)n | 11-44 | 45-59 | 60-87 [68] |
| SCA6 | 53 (24-77) | CACNA1A | 601011 | 19p13 | CACNA1A | (CAG)n | 4-18 | 19 | 20-33 [23] |
| SCA7 | 32 (0-93) | ATXN7 | 607640 | 3p21-p12 | Ataxin 7 | (CAG)n | 4-27 | 28-33 | 34-460 [50] |
| SCA17 | 35 (3-75) | TBP | 600075 | 6q27 | TBP | [(CAG)n(CAA)n(CAG)n] | 25-40d | NA | 41-66 [48]d |
| HD | 40 (4-70) | HTT | 143100 | 4p16.3 | Huntingtin | (CAG)n | 6-26 | 27-35 | 36-121 [42] |
| DRPLA | 32 (0-72) | ATN1 | 607462 | 12p13 | Atrophin 1 | (CAG)n | 3-38 | 39-47 | 48-93 [65] |
| SBMA | 45 (22-79) | AR | 313700 | Xq11-q12 | Androgen receptor | (CAG)n | 6-34 | NA | 36-73 [47] |
Abbreviations: CAA, cytosine-adenine-adenine; CACNA1A, calcium channel, voltage-dependent P/Q type, α1A subunit; CAG, cytosine-adenine-guanine; CAT, cytosine-adenine-thymine; DRPLA, dentatorubral-pallidoluysion atrophy; HD, Huntington disease; NA, not applicable; SBMA, spinal bulbar muscular atrophy; SCA, spinocerebellar ataxia; TBP, thymine-adenine-thymine-adenine (TATA) box-binding protein.
Could be interrupted by 1 to 4 CAT trinucleotide repeats.
Range if CAT trinucleotide repeat interruptions are present.
Could be interrupted by 1 to 4 CAA trinucleotide repeats.
Includes potential CAA trinucleotide repeat interruptions.
Methods
This observational cross-sectional study included data from 5 large European population–based cohorts that were compiled between 1997 and 2012, and the analyses were conducted in 2018. In total, 16 547 DNA samples were obtained from participants of the 5 cohorts. In these samples, we determined the CAG repeat number in the 9 PDAGs. The 5 cohorts formed a uniform European population and included the following large and well-characterized cohorts: the Netherlands Study of Depression and Anxiety (NESDA), the Netherlands Study of Depression in Older Persons (NESDO), the Netherlands Epidemiology of Obesity (NEO), the Prospective Study of Pravastatin in the Elderly at Risk (PROSPER), and the Leiden 85-plus Study. Together, these studies comprise a total of 14 196 individuals from the Netherlands, Scotland, and Ireland (after excluding 2351 patients with a lifetime diagnosis of major depression). None of these participants were suspected of being at risk of developing a polyglutamine disease. However, inclusion criteria per cohort varied between being 60 years or older (NESDO), being 18 to 65 years old (NESDA), being 85 years or older (Leiden 85-plus Study), having a high body mass index (BMI) (NEO), and being at risk of a cardiovascular disorder (PROSPER). Detailed descriptions of the study protocols have been published previously,13,14,15,16,17 and Table 2 summarizes the cohort characteristics. The institutional ethics review boards of all involved centers approved the 5 studies, and all participants of the NESDA, NESDO, NEO, and PROSPER gave written informed consent. Participants of the Leiden 85-plus Study gave verbal informed consent.
Table 2. Cohort Characteristics.
| Cohort | NESDA | NESDO | NEO | PROSPER | Leiden 85-plus Study | Total |
|---|---|---|---|---|---|---|
| No. (%) | 1008 (7.1) | 132 (0.9) | 6671 (47.0) | 5786 (40.8) | 599 (4.2) | 14 196 (100) |
| Sex, No. (%) | ||||||
| Male | 375 (37.2) | 51 (38.6) | 3156 (47.3) | 2798 (48.4) | 202 (33.7) | 6582 (46.4) |
| Female | 633 (62.8) | 81 (61.4) | 3515 (52.7) | 2988 (51.6) | 397 (66.3) | 7614 (53.6) |
| Age, mean (range), y | 41.5 (18-65) | 70.1 (60-93) | 55.8 (44-66) | 75.3 (69-83) | 85.0 | 64.1 (18-93) |
| BMI, mean (range) | 25.2 (15.8-47.3) | 27.0 (17.6-40.5) | 30.1 (17.2-61.2) | 26.8 (15.2-50.1) | 27.2 (14.6-43.1) | 28.3 (14.6-61.2) |
| Country, No. (%) | ||||||
| the Netherlands | 1008 (100) | 132 (100) | 6671 (100) | 1096 (18.9) | 599 (100) | 9506 (67.0) |
| Scotland | 0 | 0 | 0 | 2517 (43.5) | 0 | 2517 (17.7) |
| Ireland | 0 | 0 | 0 | 2173 (37.6) | 0 | 2173 (15.3) |
Abbreviations: BMI, body mass index (calculated as weight in kilograms divided by height in meters squared); NEO, the Netherlands Epidemiology of Obesity; NESDA, the Netherlands Study of Depression and Anxiety; NESDO, the Netherlands Study of Depression in Older Persons; PROSPER, Prospective Study of Pravastatin in the Elderly at Risk.
We applied similar genotyping as reported previously to determine the CAG repeat length in the 9 PDAGs for each individual.18 In brief, a polymerase chain reaction (PCR) was performed in a TProfessional thermocycler (Biometra; Westburg) with labeled primers flanking the CAG stretch of the PDAGs (Biolegio) (eTable 1 in the Supplement). Each PCR included a negative control without genomic DNA and a reference sample of CEPH 1347-02 genomic DNA. The PCR products were run on an automatic DNA sequencer (ABI 3730; Applied Biosystems) and analyzed using a software program (version 2.4.0; GeneMarker). For every analysis, we included 3 controls with known CAG repeat lengths for each PDAG to assure every run was performed reliably. All assessments were performed with cases and controls randomized on plates and masking with respect to disease status information.
To gain more insight into the individuals with CAG repeat numbers in the pathological ranges of the different PDAGs, we compared the mean age and BMI and the distributions of participants over cohort, sex, and country between the individuals with CAG repeat numbers within the pathological range and those with CAG repeat numbers within the normal or intermediate range for each PDAG. For the analyses of the continuous variables (ie, age and BMI), we used an independent-samples t test; for the categorical variables (ie, cohort, sex, and country), we used the χ2 test. We applied a false discovery rate correction19 to account for multiple testing assuming 9 independent tests with a 2-sided Q set at 0.05. All analyses were performed using statistical software (STATA/SE, version 14.2; StataCorp LLC).
Results
The age of all 14 196 included participants ranged between 18 and 99 years, and 56.3% were female. In total, we were able to genotype between 13 035 and 13 709 individuals for each PDAG. The missing samples were due to too little available DNA material and were absent completely at random. The mean, median, and range of the CAG repeat numbers per PDAG were similar over the cohorts (eTable 2 in the Supplement). In total, 1520 individuals had a CAG repeat number within the intermediate range in at least 1 PDAG, while 190 individuals had a CAG repeat number within the fully pathological range (Table 3, Figure, and eFigure in the Supplement). Therefore, the prevalence of individuals carrying intermediate and pathological alleles among this large sample from the general population can be estimated at around 10.7% and 1.3%, respectively. After correction for multiple testing, no differences in age, BMI, or the distribution of individuals over the cohorts, sexes, or countries were found between those with CAG repeat numbers within the pathological range and those with CAG repeat numbers within the normal or intermediate range for any of the PDAGs (eTable 3 in the Supplement).
Table 3. Number of Individuals Within the Intermediate and Pathological Ranges per Polyglutamine Disease–Associated Gene (PDAG).
| PDAG | Allele | Total No. | Range | No. | ||
|---|---|---|---|---|---|---|
| Normal | Intermediate | Pathologicala | ||||
| ATXN1 | Short | 13 668 | 17-36 | 13 664 | 4 | 0 |
| Long | 13 668 | 22-44 | 13 023 | 636b | 9 (39-44)b | |
| ATXN2 | Short | 13 536 | 11-30 | 13 536 | 0 | 0 |
| Long | 13 536 | 17-36 | 13 518 | 10 | 8 (33, 36) | |
| ATXN3 | Short | 13 545 | 13-35 | 13 545 | 0 | 0 |
| Long | 13 544 | 13-62 | 13 539 | 3 | 2 (61, 62) | |
| CACNA1A | Short | 13 615 | 4-14 | 13 615 | 0 | 0 |
| Long | 13 614 | 4-22 | 13 610 | 3 | 1 (22) | |
| ATXN7 | Short | 13 035 | 5-16 | 13 035 | 0 | 0 |
| Long | 13 035 | 7-30 | 13 033 | 2 | 0 | |
| TBP | Short | 13 585 | 23-40 | 13 585 | NA | 0 |
| Long | 13 585 | 30-47 | 13 442 | NA | 143 (41-45, 47) | |
| HTT | Short | 13 670 | 6-31 | 13 661 | 9 | 0 |
| Long | 13 670 | 10-40 | 12 785 | 866 | 19 (36-40) | |
| ATN1 | Short | 13 709 | 3-22 | 13 709 | 0 | 0 |
| Long | 13 709 | 8-28 | 13 709 | 0 | 0 | |
| AR | Allele male | 6236 | 7-36 | 6233 | NA | 1 (36) |
| Short female | 7214 | 7-30 | 7214 | NA | 0 | |
| Long female | 7214 | 15-39 | 7206 | NA | 7 (36-39) | |
Abbreviations: CAG, cytosine-adenine-guanine; CAT, cytosine-adenine-thymine; NA, not applicable.
Numbers in parentheses refer to the actual CAG repeat sizes.
Number of individuals assuming no CAT interruptions were present within the CAG repeat sequence.
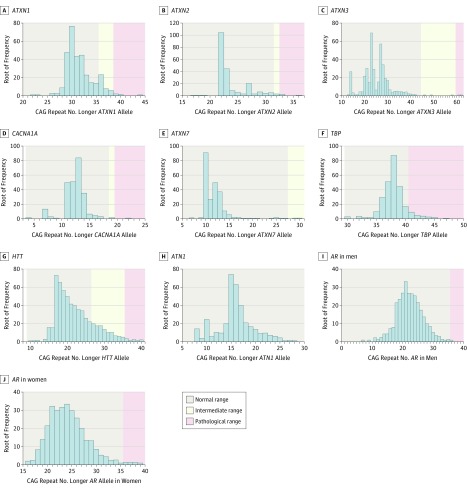
Figure. Frequency Distributions of the CAG Repeat Numbers in Polyglutamine Disease–Associated Genes.
The figures represent rootograms (ie, a frequency graph in which the vertical axis has been scaled by its square root to allow better depiction of wide frequency variations) for the longer allele of each polyglutamine disease–associated gene. A, A total of 636 individuals had at least 1 allele with a CAG repeat number within the intermediate range of ATXN1, and 9 individuals had CAG repeat numbers within the pathological range. B, For ATXN2, a total of 10 individuals had CAG repeat numbers within the intermediate range, and 8 had repeat numbers within the pathological range. C, Just 3 individuals had CAG repeat numbers within the intermediate range of ATXN3, and 2 individuals had CAG repeat numbers within the pathological range. D, Three individuals and 1 individual had CAG repeat numbers within the intermediate and pathological ranges of CACNA1A, respectively. E, No individuals had CAG repeat numbers within the pathological range of ATXN7, but 2 participants had CAG repeat numbers within its intermediate range. F, TBP has no known intermediate CAG repeat range, but 143 individuals had CAG repeat numbers within the pathological range of TBP. One of these individuals also had a pathological CAG repeat in ATXN2. G, As many as 866 individuals had at least 1 allele with a CAG repeat number within the intermediate range of HTT, and 19 individuals had pathological HTT CAG repeat numbers. H, All individuals had normal-range CAG repeat numbers in ATN1. I and J, Because AR is linked to the X chromosome, men and women are displayed separately, and AR has no known intermediate CAG repeat range. One man and 7 women had CAG repeat numbers within the pathological range.
Discussion
To our knowledge, this is the largest group of individuals from the general population, without a known polyglutamine disease diagnosis, in which CAG repeat sequences in all 9 PDAGs have been determined. In a uniform European population of approximately 14 000 individuals, we found a substantial number of people with CAG repeat numbers falling within the pathological range of at least 1 of the PDAGs. These results greatly exceeded the expected numbers based on the existing data on the prevalence of polyglutamine diseases (Table 4).3,4,5,6,7,8,9,10,11 Although not all individuals in our analyses were randomly selected, participation in the cohorts was not contingent on genotype or likely polyglutamine disease phenotype. Therefore, our estimates can be taken to reflect the prevalence of these genotypes among the general population. Furthermore, we found that a considerably high proportion of individuals had CAG repeat numbers within the intermediate ranges of PDAGs, indicating that a substantial number of people are at risk of bearing children with a polyglutamine disease due to a de novo mutation resulting from increased instability of elongated trinucleotide repeat sequences during gametogenesis.20 In addition, previous research has shown that even intermediate or large normal-range repeats in HTT, ATXN7, and TBP may increase the risk of cognitive, psychiatric, and motor abnormalities later in life.21,22,23,24 One must note that this study was performed in uniform cohorts of participants from northern Europe. Therefore, the prevalence estimates found herein cannot readily be assumed for individuals of a different descent, such as East Asian, African, or Portuguese. Therefore, future studies should aim to investigate more diverse cohorts.
Table 4. Known Prevalence of Polyglutamine Diseases Compared With the Prevalence Among Carriers of CAG Repeat Lengths Within the Disease-Causing Range.
The individuals with CAG repeat numbers within the pathological ranges of the PDAGs had CAG repeat numbers within the low or borderline pathological range. Because the age at onset in all polyglutamine diseases has an inverse association with the CAG repeat number in the mutated allele, we would expect most of these individuals to develop polyglutamine disease–associated symptoms at an old age. Unfortunately, we had no long-term follow-up data on our participants and thus were unable to assess whether any of the carriers of intermediate or pathological ranges of alleles would have developed disease symptoms later in life. In addition, several authors argue that CAG repeat ranges considered pathological also contain a range associated with reduced penetrance of the respective polyglutamine disease.3,7,8,10 In this study, all individuals with CAG repeat numbers within the pathological ranges of TBP and AR could be considered to belong to this range of reduced penetrance, defined as 41 to 48 repeats for TBP and 36 to 37 repeats for AR. Most individuals with CAG repeat numbers within the disease-associated range of TBP had 41 to 44 repeats (n = 137) for which the penetrance is estimated at 50%, leading to a new estimate of about 552 per 100 000 individuals being at risk of developing SCA17 during their lifetime.7 For HTT, 16 of the 19 participants with CAG repeat numbers within the pathological range also belonged to the reduced penetrance range (36-39 repeats). Consistent with this finding, a previous HD study25 revealed that 18 of 7315 individuals had CAG repeat numbers in the pathological range of HTT, with most of them belonging to the reduced penetrance range. The estimated penetrance rates of HTT alleles with 36 to 39 CAG repeats at age 65 years is 6% for 36 repeats, 10% for 37 repeats, 19% for 38 repeats, and 35% for 39 repeats.26 By age 65 years, 58% of individuals with 40 CAG repeats are estimated to have developed HD.26 The 3 individuals herein with 40 CAG repeats in HTT were aged 62, 63, and 64 years, respectively, at the time of assessment and could thus still develop HD at a later age. Furthermore, the penetrance of SCA1 is largely dependent on the presence of cytosine-adenine-thymine (CAT) interruptions within the CAG repeat sequence. The number of individuals considered to have CAG repeat numbers within the intermediate and pathological ranges of ATXN1 was based on ranges assuming no CAT trinucleotide interruptions were present within the CAG repeat sequence. However, our genotyping method did not allow us to determine the presence of such interruptions. The frequency of these interruptions in disease-associated alleles is about 11%.27 Therefore, the individuals with pathological CAG repeat numbers in ATXN1 could be asymptomatic due to CAT interruptions. To more accurately determine which of the individuals with CAG repeat numbers within the disease-associated ranges are actually at risk of developing a polyglutamine disease, we believe that the disease-associated alleles need to be explored in more detail. Specifically, future experiments should assess whether repeat interruptions, which could markedly influence disease penetrance, are more abundant in disease-associated alleles from the general population compared with those from clinical series.
In SCA1 and SCA2, the risk of alleles within the intermediate range expanding into the disease-associated range also depends on the presence of interruptions. For ATXN1, between 36 and 38 CAG repeats without CAT interruptions are considered at risk of elongation.3 Likewise, the stability of intermediate alleles in SCA2 is dependent on cytosine-adenine-adenine (CAA) interruptions. The actual frequency of meiotic expansions of intermediate alleles into the disease-associated range has been most intensively investigated in HD. A study by Semaka et al28 showed that 7.8% of the intermediate HTT alleles expanded into the pathological range. As the CAG repeat number increased, the frequency of these expansions also increased, with 1.6% expanding at 28 repeats and 21.0% expanding at 35 repeats. Most intermediate alleles expanded with 1 to 3 CAG repeats, and an expansion into the disease-associated range was not observed until 30 repeats.28 Little is known about the frequency of such de novo mutations for the other polyglutamine diseases. However, a close association exists between the prevalence of dominantly inherited SCAs, dentatorubral-pallidoluysion atrophy, and HD and the frequencies of intermediate alleles, supporting the notion that intermediate alleles contribute to the de novo production of disease-associated alleles. The distribution of CAG repeat numbers over ATXN1, ATXN2, ATXN3, CACNA1A, HTT, and ATN1 reported in these previous studies29,30 are similar to the distributions we found.
To determine the CAG repeat sizes in the PDAG, we applied targeted PCR amplification, followed by fragment analysis. This method is considered the criterion standard for determining numbers of short tandem repeats and is currently the primary methodology applied for genetic diagnosis of polyglutamine diseases in the clinic. However, this approach requires that target sites are predefined, which does not facilitate the pursuit of variations in short tandem repeats throughout the entire genome. Tang et al31 presented whole-genome sequencing along with the software package TREDPARSE that allows estimation of the length of short tandem repeats in many loci on the basis of one run. Using this method, they identified 138 at-risk individuals in a total of 12 632 genomes for 15 disease loci, including SCA1 (n = 26), SCA2 (n = 4), SCA6 (n = 2), SCA17 (n = 52), HD (n = 5), and spinal bulbar muscular atrophy (n = 1). The frequencies of these disease-associated alleles differ somewhat from our reported frequencies. However, apart from the difference in genotyping technique, their sample also consisted of a diverse population, including not only European but also African, South Asian, East Asian, and Native American individuals, making direct comparison with our cohort unreliable. Although novel techniques reported by Tang et al31 are promising for genome-wide estimation of repeat sequences at many loci, their validation remains crucial for clinical implementation.
Limitations
Limitations of this research include the fact that our study was performed in uniform cohorts of participants from northern Europe, meaning that the prevalence estimates found in this study cannot be assumed for individuals of a different descent. Furthermore, we did not possess long-term follow-up data on our participants; thus, we could not assess whether the carriers of intermediate or pathological ranges of alleles would have developed disease symptoms later in life. In addition, our genotyping method did not allow us to determine the presence of trinucleotide interruptions. Such interruptions could influence disease penetrance and perhaps provide an explanation for the fact that the individuals in our cohorts with expanded CAG repeat sequences did not carry a polyglutamine disease diagnosis. Therefore, we believe that the presence of such interruptions within the disease-associated alleles should be further explored.
Conclusions
A late age at onset, a reduced penetrance, or the presence of interruptions could all explain the asymptomatic status of our carriers of intermediate and pathological polyglutamine disease–associated alleles at the time of assessment. Nonetheless, the number of individuals with CAG repeat numbers within the intermediate and pathological ranges of the PDAGs in our population remains striking. Our findings suggest that a larger proportion of the population may be at risk of developing a polyglutamine disease than previously estimated. Conversely, a large part of the population might also possess characteristics that prevent expression of the polyglutamine disease phenotype. Further investigation of such characteristics could lead to new insights into the treatment and prevention of the polyglutamine diseases, as well as the behavior of CAG repeats within the human population in general.
eTable 1. Polyglutamine disease-associated genes and primers.
eTable 2. Polyglutamine disease-associated genes summary over cohorts.
eTable 3. Differences in characteristics between individuals with CAG repeat numbers within the pathological range and individuals with CAG repeat numbers within the normal range.
eFigure. The distribution of the CAG repeat numbers in the polyglutamine disease associated genes (PDAGs) per allele and cohort.
References
- 1.Orr HT. Polyglutamine neurodegeneration: expanded glutamines enhance native functions. Curr Opin Genet Dev. 2012;22(3):650-656. doi: 10.1016/j.gde.2012.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Durr A. Autosomal dominant cerebellar ataxias: polyglutamine expansions and beyond. Lancet Neurol. 2010;9(9):885-894. doi: 10.1016/S1474-4422(10)70183-6 [DOI] [PubMed] [Google Scholar]
- 3.Opal P, Ashizawa T. Spinocerebellar ataxia type 1 In: Adam MP, Ardinger HH, Pagon RA, et al. , eds. GeneReviews. Seattle: University of Washington; 1993. [PubMed] [Google Scholar]
- 4.Pulst SM. Spinocerebellar ataxia type 2 In: Adam MP, Ardinger HH, Pagon RA, et al. , eds. GeneReviews. Seattle: University of Washington; 1998. [Google Scholar]
- 5.Gomez CM. Spinocerebellar ataxia type 6 In: Adam MP, Ardinger HH, Pagon RA, et al. , eds. GeneReviews. Seattle: University of Washington; 1993. [PubMed] [Google Scholar]
- 6.Garden G. Spinocerebellar ataxia type 7 In: Adam MP, Ardinger HH, Pagon RA, et al. , eds. GeneReviews. Seattle: University of Washington; 1993. [Google Scholar]
- 7.Toyoshima Y, Onodera O, Yamada M, Tsuji S, Takahashi H. Spinocerebellar ataxia type 17 In: Adam MP, Ardinger HH, Pagon RA, et al. , eds. GeneReviews. Seattle: University of Washington; 1993. [PubMed] [Google Scholar]
- 8.Caron NS, Wright GEB, Hayden MR. Huntington disease In: Adam MP, Ardinger HH, Pagon RA, et al. , eds. GeneReviews. Seattle: University of Washington; 1998. [Google Scholar]
- 9.Veneziano L, Frontali M. DRPLA In: Adam MP, Ardinger HH, Pagon RA, et al. , eds. GeneReviews. Seattle: University of Washington; 1993. [Google Scholar]
- 10.La Spada A. Spinal and bulbar muscular atrophy In: Adam MP, Ardinger HH, Pagon RA, et al. , eds. GeneReviews. Seattle: University of Washington; 1993. [PubMed] [Google Scholar]
- 11.Paulson H. Spinocerebellar ataxia type 3 In: Adam MP, Ardinger HH, Pagon RA, et al. , eds. GeneReviews. Seattle: University of Washington; 1998. [Google Scholar]
- 12.Bates GP, Dorsey R, Gusella JF, et al. Huntington disease. Nat Rev Dis Primers. 2015;1:15005. doi: 10.1038/nrdp.2015.5 [DOI] [PubMed] [Google Scholar]
- 13.Penninx BW, Beekman AT, Smit JH, et al. ; NESDA Research Consortium . The Netherlands Study of Depression and Anxiety (NESDA): rationale, objectives and methods. Int J Methods Psychiatr Res. 2008;17(3):121-140. doi: 10.1002/mpr.256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Comijs HC, van Marwijk HW, van der Mast RC, et al. The Netherlands Study of Depression in Older Persons (NESDO): a prospective cohort study. BMC Res Notes. 2011;4:524. doi: 10.1186/1756-0500-4-524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Mutsert R, den Heijer M, Rabelink TJ, et al. The Netherlands Epidemiology of Obesity (NEO) study: study design and data collection. Eur J Epidemiol. 2013;28(6):513-523. doi: 10.1007/s10654-013-9801-3 [DOI] [PubMed] [Google Scholar]
- 16.Shepherd J, Blauw GJ, Murphy MB, et al. ; PROSPER Study Group. Prospective Study of Pravastatin in the Elderly at Risk . Pravastatin in elderly individuals at risk of vascular disease (PROSPER): a randomised controlled trial. Lancet. 2002;360(9346):1623-1630. doi: 10.1016/S0140-6736(02)11600-X [DOI] [PubMed] [Google Scholar]
- 17.der Wiel AB, van Exel E, de Craen AJ, et al. A high response is not essential to prevent selection bias: results from the Leiden 85-plus Study. J Clin Epidemiol. 2002;55(11):1119-1125. doi: 10.1016/S0895-4356(02)00505-X [DOI] [PubMed] [Google Scholar]
- 18.Gardiner SL, de Mutsert R, Trompet S, et al. Repeat length variations in polyglutamine disease–associated genes affect body mass index [published online August 17, 2018]. Int J Obes. doi: 10.1038/s41366-018-0161-7 [DOI] [PubMed] [Google Scholar]
- 19.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57(1):289-300. https://www.jstor.org/stable/2346101. Accessed February 18, 2019. [Google Scholar]
- 20.Wheeler VC, Persichetti F, McNeil SM, et al. ; US-Venezuela Collaborative Research Group . Factors associated with HD CAG repeat instability in Huntington disease. J Med Genet. 2007;44(11):695-701. doi: 10.1136/jmg.2007.050930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cubo E, Ramos-Arroyo MA, Martinez-Horta S, Martínez-Descalls A, Calvo S, Gil-Polo C; European HD Network . Clinical manifestations of intermediate allele carriers in Huntington disease. Neurology. 2016;87(6):571-578. [DOI] [PubMed] [Google Scholar]
- 22.Killoran A, Biglan KM, Jankovic J, et al. Characterization of the Huntington intermediate CAG repeat expansion phenotype in PHAROS. Neurology. 2013;80(22):2022-2027. doi: 10.1212/WNL.0b013e318294b304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gardiner SL, van Belzen MJ, Boogaard MW, et al. Large normal-range TBP and ATXN7 CAG repeat lengths are associated with increased lifetime risk of depression. Transl Psychiatry. 2017;7(6):e1143. doi: 10.1038/tp.2017.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gardiner SL, van Belzen MJ, Boogaard MW, et al. Huntingtin gene repeat size variations affect risk of lifetime depression. Transl Psychiatry. 2017;7(12):1277. doi: 10.1038/s41398-017-0042-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kay C, Collins JA, Miedzybrodzka Z, et al. Huntington disease reduced penetrance alleles occur at high frequency in the general population. Neurology. 2016;87(3):282-288. doi: 10.1212/WNL.0000000000002858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langbehn DR, Brinkman RR, Falush D, Paulsen JS, Hayden MR; International Huntington’s Disease Collaborative Group . A new model for prediction of the age of onset and penetrance for Huntington’s disease based on CAG length. Clin Genet. 2004;65(4):267-277. doi: 10.1111/j.1399-0004.2004.00241.x [DOI] [PubMed] [Google Scholar]
- 27.Menon RP, Nethisinghe S, Faggiano S, et al. The role of interruptions in polyQ in the pathology of SCA1. PLoS Genet. 2013;9(7):e1003648. doi: 10.1371/journal.pgen.1003648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Semaka A, Kay C, Doty C, et al. CAG size–specific risk estimates for intermediate allele repeat instability in Huntington disease. J Med Genet. 2013;50(10):696-703. doi: 10.1136/jmedgenet-2013-101796 [DOI] [PubMed] [Google Scholar]
- 29.Takano H, Cancel G, Ikeuchi T, et al. Close associations between prevalences of dominantly inherited spinocerebellar ataxias with CAG-repeat expansions and frequencies of large normal CAG alleles in Japanese and Caucasian populations. Am J Hum Genet. 1998;63(4):1060-1066. doi: 10.1086/302067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kay C, Collins JA, Wright GEB, et al. The molecular epidemiology of Huntington disease is related to intermediate allele frequency and haplotype in the general population. Am J Med Genet B Neuropsychiatr Genet. 2018;177(3):346-357. doi: 10.1002/ajmg.b.32618 [DOI] [PubMed] [Google Scholar]
- 31.Tang H, Kirkness EF, Lippert C, et al. Profiling of short-tandem-repeat disease alleles in 12,632 human whole genomes. Am J Hum Genet. 2017;101(5):700-715. doi: 10.1016/j.ajhg.2017.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eTable 1. Polyglutamine disease-associated genes and primers.
eTable 2. Polyglutamine disease-associated genes summary over cohorts.
eTable 3. Differences in characteristics between individuals with CAG repeat numbers within the pathological range and individuals with CAG repeat numbers within the normal range.
eFigure. The distribution of the CAG repeat numbers in the polyglutamine disease associated genes (PDAGs) per allele and cohort.