Abstract
Early detection of metastatic colorectal cancer, at initial diagnosis or during routine surveillance, can improve survival outcomes. Current routine investigations, including CEA and CT, have limited sensitivity and specificity. Recent studies of colorectal cancer cohorts under post surgery surveillance indicate circulating tumor DNA (ctDNA) evidence of recurrence can occur many months before clinical detection. Another possible role for ctDNA is in the further assessment of indeterminate findings on standard CEA or CT investigations. To further explore this potential, we undertook a prospective study. Further investigation, including FDG-PET imaging, was at clinician discretion, blinded to ctDNA analysis. Forty-nine patients were enrolled. Analysed here are the 45 patients with an evaluable blood sample of whom 6 had an isolated elevated CEA, 30 had indeterminate CT findings, and 9 had both. FDG-PET scans were performed in 30 patients. Fourteen of 45 patients (31%) had detectable ctDNA. At completion of the planned 2 year follow-up, recurrence has occurred in 21 (47%) patients. Detectable ctDNA at study entry was associated with inferior relapse free survival (HR 4.85, p<0.0001). Where FDG-PET scan was normal/equivocal (n = 15, 50%) 1 of 1 with detectable ctDNA versus 3 of 14 with undetectable ctDNA ultimately had recurrence confirmed. In summary, for colorectal cancer patients with indeterminate findings on routine investigations, ctDNA detection increases the probability that the findings indicate metastatic disease, including in a non-predefined subset that also underwent FDG-PET imaging. Further studies of the value of ctDNA analysis during patient surveillance are warranted.
Keywords: ctDNA, colorectal cancer, recurrence detection, indeterminate findings
Introduction
Colorectal cancer is the third most common malignancy worldwide and the incidence is expected to continue to rise.1 Approximately 80% of patients will initially present with no clear evidence of metastatic disease.2 For these patients, standard tests at initial diagnosis and during routine surveillance will include computed tomography of the chest, abdomen and pelvis (CT CAP) and carcinoembryonic antigen (CEA) testing.3 These tests, whilst universally accepted as part of routine care, have their limitations. CEA has limited sensitivity and specificity and CT imaging is associated with radiation exposure and has a significant rate of false positivity.4
Where there is suspicion of metastatic disease due to an elevated CEA and/or abnormal CT imaging, further investigations can include 18Fluorodeoxyglucose positron emission tomography (FDG-PET) scans. These, however, can also be associated with false positive and negative findings.5 For example, colorectal cancers are not infrequently mucinous tumors where the associated rate of false negative FDG-PET is up to 41%.6 Scan findings are not infrequently equivocal, which can lead to further anxiety and investigation. This can include findings incidental to the colorectal cancer history. Additionally, in certain scenarios, such as the not uncommon finding of non-specific small lung nodules on CT imaging, FDG-PET imaging is clearly unhelpful.
Due to the limitations of available tests during follow-up of early stage colorectal cancer, a significant proportion of patients will have non-specific findings at diagnosis or during surveillance that ultimately do not represent recurrent disease. These will result in additional and/or repeated investigations and also create significant patient anxiety. In many instances it may not be possible to undertake a definitive investigation, resulting in patients having to wait months to undergo repeat investigation before they can be cleared. Again, small lung nodules are a particular challenge as colorectal cancer lung metastases can behave in a very indolent fashion.
The potential value of circulating tumor deoxyribonucleic acid (ctDNA) as a cancer biomarker has been demonstrated in multiple studies and scenarios. For patients with metastatic disease, ctDNA is detectable in around 90% of patients and has potential utility as a liquid biopsy and serial sampling can define early responses to chemotherapy.7, 8 ctDNA also has potential as a marker of minimal residual disease, with detection following curative intent surgery for colon or rectal cancer associated with a very high risk of later recurrence.9, 10 In a study where serial ctDNA analysis was performed during follow-up of a cohort of stage II colon cancer patients, ctDNA was detectable a median of 167 days prior to confirmed radiologic recurrence and overall was elevated in 85% of patients with recurrent disease. In contrast, CEA was only elevated in 41% of patients who recurred. Given the demonstrated sensitivity and specificity of ctDNA in the minimal residual disease and surveillance settings, we further evaluated the value of ctDNA in patients with non-specific findings on routine investigation, at colorectal cancer diagnosis or during surveillance.
Methods
Study Design and Participants
This prospective multi-centre study recruited patients with colorectal cancer managed at 3 Australian hospitals (Eastern Health, Melbourne Health and Western Health). Key eligibility criteria included: a histological diagnosis of colorectal cancer with indeterminate findings raising suspicion of metastatic disease or local recurrence on routine investigations performed within 8 weeks of enrolment. Indeterminate findings were defined as non-specific findings on CT scan or an elevated CEA (defined as CEA >5 μg/L). Patients needed to be suitable for further investigation/follow-up and have a representative tumour sample available for molecular testing, Patients with a previous malignancy within the last three years were excluded.
Clinicians documented their index of suspicion (high/intermediate/low) for recurrent/metastatic disease at study enrolment and were blinded to ctDNA results. All patients had undergone resection of the primary tumor prior to the blood draw for ctDNA analysis.
A single sample of up to 60mL of venous blood was collected within 10 weeks of the initial abnormal standard investigation. Blood was drawn into EDTA tubes, 2mL of whole blood transferred into cryovials and the rest processed and centrifuged twice at 1200g and 1800g within 3 hours into plasma. Samples were stored at −70°C. All plasma and tumor samples were sent for analysis at the Ludwig Center at Johns Hopkins.
Disease status was recorded at a minimum of 3 monthly intervals and patients underwent further investigations as per routine standard of care, including FDG-PET imaging and/or repeat CT or CEA investigation at clinician defined intervals. All patients were followed up until metastatic disease or local recurrence was confirmed, or up to 2 years from enrolment.
This study was approved by the Melbourne Health Human Research Ethics Committee (HREC/11/MH/320) and all participants provided written informed consent.
Procedures
Formalin-fixed paraffin-embedded tumor tissue was analysed for somatic mutations in 15 genes recurrently mutated in colorectal cancer, as previously described.9 Tumor sections were macro-dissected under a dissecting microscope to ensure a neoplastic cellularity of >30%. DNA was purified with a Qiagen FFPE Kit (Qiagen cat #56494). Primers were designed and sequencing results analyzed as previously described.7, 11
For each patient, one mutation identified in the tumor tissue was assessed in the plasma for the presence of ctDNA. When more than one somatic mutation was identified in the tumor tissue, the mutation with the highest mutant allele fraction (MAF) relative to the MAF in normal control DNA was selected for ctDNA analysis for that patient. Ten ml of plasma was purified from each patient using the QIAamp Circulating Nucleic Acid kit (Qiagen cat# 55114). To distinguish ctDNA in the plasma samples from artifactual variants arising during sequencing and sample preparation steps, we used Safe-SeqS, an error-reduction technology for detection of low frequency mutations.11 As per the standard SafeSeqS assay, plasma DNA was aliquoted into 24 wells of a 96-well plate, so that an average of 0.5 to 3ng DNA was contained in each well. The DNA from each well was then amplified (15 cycles) using primers containing unique identifier sequences (UIDs), which consisted of fourteen random bases with an equal probability of A, C, T, and G, to allow for the distinction of each template molecule. The amplified reactions were purified with AMPure XP beads (Beckman Coulter) and eluted in 250 μL of Buffer EB (Qiagen). One percent (2.5 μL) of purified PCR product was then amplified in a second round of PCR with universal primers. The PCR products were purified with AMPure and sequenced on an Illumina MiSeq instrument.
High quality sequence reads were selected based on quality scores, which were generated by the Illumina sequencing instrument to indicate the probability that an error was made in base calling. The template-specific portion of the reads was matched to reference sequences. Reads from a common template molecule were then grouped based on the UIDs that were incorporated as molecular barcodes. Artifactual mutations introduced during the sample preparation or sequencing steps were reduced by requiring a mutation to be present in >90% of reads in a UID family in order for that UID to be scored as a “supermutant”. Wells with fewer than 200 UIDs as a result of poor amplification were excluded. DNA from the peripheral blood lymphocytes of healthy individuals was used as a control in each experiment to identify potential false positive mutations.
ctDNA was classified as detectable (ctDNA-positive) or undetectable (ctDNA-negative) based on a permutation test that compared the mutation frequency in the sample of interest with the mutation frequencies in controls. First, the mutant allele fraction (MAF), defined as the ratio between the number of supermutants and the number of UIDs for the mutation of interest, was calculated for each well with >200 UIDs. The difference in the distributions of the MAFs between the sample of interest and the controls was then statistically evaluated via the permutation test, using the permTS function of the R package perm (R software version 3.2.3). The one-sided test was used to avoid attributing significance to a ctDNA-negative sample that has fewer supermutants than the associated control. A 0.1 p-value was then chosen as the threshold to classify a sample of interest as ctDNA-positive (p<0.1) or ctDNA-negative. Given the lack of a gold standard, a specificity of at least 0.90 was considered desirable, corresponding to a p-value equal to 0.1.
Statistical Analysis
Relapse-free survival (RFS), was measured from date of enrolment to documented first recurrence or death as a result of colorectal cancer, and was censored at last follow-up or non-colorectal cancer-related death. Survival analyses were estimated using the Kaplan-Meier method using GraphPad Prism version 6.07 (GraphPad Software Inc., California, USA), where P values <0.05 were considered significant.
Results
Between Jan 2014 and Dec 2015, 49 patients were enrolled. Of these, 45 (92%) were included in the analysis, with four patients excluded due to inadequate blood samples (n=2), or no plasma sample being available (n=2). Two patients (4%) had no mutation found in their tumor, but were included in the analysis. Baseline characteristics and outcomes are shown in Table 1. Median age was 67 years and 71% of patients were male. Seven (16%) were newly diagnosed and 38 were under surveillance after previous curative intent colorectal cancer surgery.
Table 1:
Baseline characteristics and outcomes
| % | |||
|---|---|---|---|
| Age | Median (range) | 67 years (48–85) | |
| >65 | 24 | 53 | |
| <65 | 21 | 47 | |
| Sex | M | 32 | 71 |
| F | 13 | 29 | |
| New diagnosis Surveillance | 7 | 16 | |
| 38 | 84 | ||
| Primary location | Right | 15 | 33 |
| Left | 13 | 29 | |
| Rectum | 17 | 38 | |
| Stage | 0 | 2 | 4 |
| 1 | 4 | 9 | |
| 2 | 18 | 40 | |
| 3 | 20 | 44 | |
| 4 | 1 | 2 | |
| CEA | High | 15 | 33 |
| Normal | 30 | 67 | |
| CT CAP | Likely metastases* | 3 | 7 |
| Abnormal | 41 | 91 | |
| Normal | 1 | 2 | |
| Clinical suspicion | High | 13 | 29 |
| Intermediate | 22 | 49 | |
| Low | 10 | 22 | |
| FDG-PET | Total | 30 | 67 |
| Likely metastases | 15 | ||
| Equivocal | 9 | ||
| Normal | 6 | ||
| ctDNA | Positive | 14 | 31 |
| Negative | 29 | 64 | |
| No mutation | 2 | 4 | |
| Metastases confirmed | 21 | 47 | |
| Metastatectomy | Total | 13 | 29 |
| Lung | 10 | ||
| Liver | 2 | ||
| Other | 1 (Brain) | ||
CT performed subsequent to enrollment
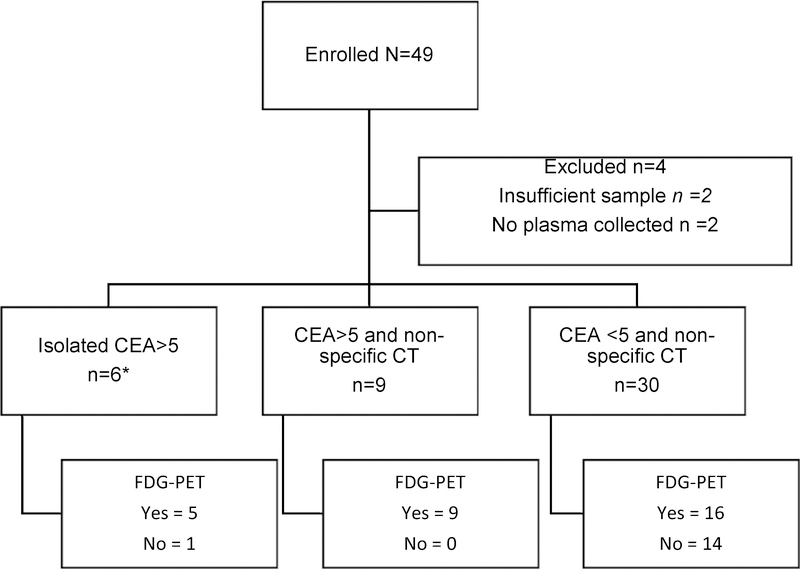
A consort diagram (Figure 1) illustrates patient features at diagnosis leading to enrolment and subsequent radiological investigations. At the time of enrolment, 6 patients had an isolated elevation of CEA (>5.0 μg/L), 9 had an elevated CEA and non-specific CT scan findings, and 30 had only non-specific CT scan findings. The median CEA level was 3 μg/L (range <0.5 – 36 μg/L).
Figure 1:
Patient enrolment and subsequent imaging
*1 patient had CT reported as normal shortly before the elevated CEA was detected, with the treating clinician electing not to repeat the CT. Remaining 5 patients had CT performed after enrolment.
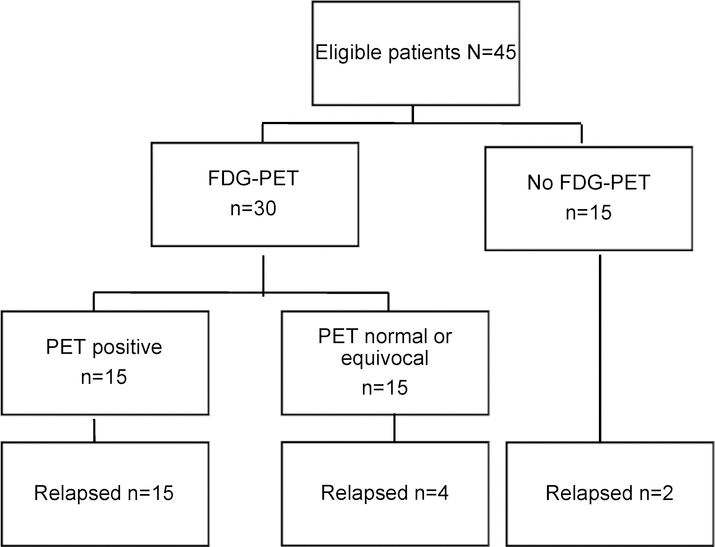
Five of the six patients with an isolated elevated CEA were enrolled and then underwent a CT scan, with findings considered by the treating clinician as diagnostic of recurrence (n=3) or non-specific (n=2). The remaining patient had had a CT scan just prior to enrolment that was reported as normal. Thirty of 45 patients (67%) underwent further investigation with FDG-PET scan at clinician discretion. Fifteen (50%) FDG-PET scans were deemed positive for malignancy; the remaining 15 were reported as normal (n=6) or equivocal (n=9). The outcomes for patients according to FDG-PET results are depicted in Figure 2.
Figure 2:
Patient outcomes according to FDG-PET result
Based on the available historical data (including stage of primary tumor, use of adjuvant therapy and time since initial diagnosis) and any biochemical and radiological results at the time of enrolment, at study entry the treating clinician assessed the likelihood that findings represented recurrent disease as: high in 13 (29%) patients, intermediate in 22 (49%) patients and low in 10 (22%) patients.
With all patients having either experienced an event or having completed the per protocol 2 years of follow-up, 21 of 45 (47%) have had clear progression of disease. Median time to confirmed relapse was 90 days (range 13–614 days). Thirteen (62%) of 21 patients ultimately underwent resection of their metastatic disease.
According to clinicians’ index of suspicion, metastases were confirmed in: 12/13 (92%) of high; 6/22 (27%) of intermediate; and 3/10 (30%) of low suspicion patients. The median time to recurrence was 88 days (range 13–172) in the high suspicion and 174 days (range 36–614 days) in the low-intermediate suspicion groups.
Table 2 shows the relationship between clinician index of suspicion, CEA, ctDNA result and relapse. ctDNA analysis was positive in 14 of 45 patients (31%). Twelve of these patients have relapsed, comprising 57% of the 21 patients with confirmed relapse. Two patients with positive ctDNA had not yet recurred at the completion of the 2 year follow-up period. Of the 15 patients with a high CEA at enrolment, 10 (67%) have relapsed including all 7 patients with positive ctDNA. Of the 30 patients with an initially normal CEA, 11 (37%) have relapsed. ctDNA was positive in 7 patients with an initially normal CEA, five of these patients have clearly developed metastatic disease.
Table 2:
Relationship between clinician index of suspicion, CEA ctDNA result and relapse
| Number N=45 | ctDNA positive (%) | ctDNA negative (%) | Number relapsed (%) | |
|---|---|---|---|---|
| Index of suspicion | ||||
| High | 13 | 9 (69) | 4 (31) | 12 (92) |
| Intermediate* | 22 | 2 (9) | 18 (82) | 6 (27) |
| Low | 10 | 3 (30) | 7 (70) | 3 (30) |
| CEA | ||||
| High | 15 | 7 (47) | 8 (53) | 10 (67) |
| Normal* | 30 | 7 (23) | 21 (70) | 11 (37) |
2 patients had no detectable mutation
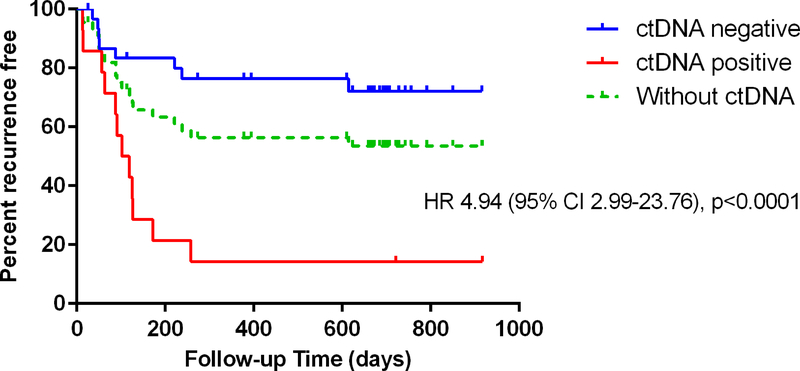
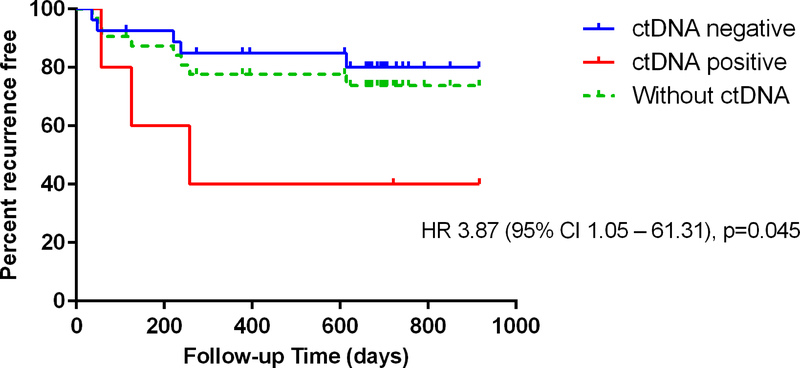
A positive ctDNA was associated with significantly inferior relapse free survival amongst all patients analysed (HR 4.94, p<0.0001) and amongst those with low to intermediate suspicion of recurrence (HR 3.87, p=0.045). Figure 3a and 3b.
Figure 3:
Relapse free survival in a) all analysed patients (n=45) and stratified by ctDNA result. Hazard ratio compares ctDNA positive and negative patients b) patients with intermediate or low clinical suspicion of recurrence (n=22). All patients and stratified by ctDNA result. Hazard ratio compares ctDNA positive and negative patients.
The relationship between FDG-PET findings, CEA and ctDNA results are shown in Table 3 for the 30 patients who underwent FDG-PET imaging. Of the 14 patients with an elevated CEA, 9 had positive FDG-PET findings, all of whom were later confirmed to have colorectal cancer metastases. Seven (50%) of the 14 CEA-elevated patients who had FDG-PET were ctDNA positive, and all have relapsed. Conversely, all 5 patients with an elevated CEA, but normal or equivocal FDG-PET findings were negative for ctDNA and remain in complete remission.
Table 3:
Outcomes of patients who had FDG-PET scans
| Number n=30 | ctDNA positive (%) | ctDNA negative (%) | Number relapsed (%) | |
|---|---|---|---|---|
| PET positive & CEA High | 9 | 7 (78) | 2 (22) | 9 (100) |
| PET N or equivocal & CEA High | 5 | 0 | 5 (100) | 0 |
| PET positive & CEA Normal* | 6 | 3 (50) | 2 (33) | 6 (100) |
| PET N or equivocal & CEA N | 10 | 1 (10) | 9 (90) | 4 (40) |
1 patient had no detectable mutation
Similarly, all 6 patients with normal CEA but positive FDG-PET findings were confirmed to have colorectal cancer metastases. Three of these patients were also ctDNA positive. Four of the patients with a normal CEA and normal or equivocal FDG-PET findings have relapsed; 1 had positive and 3 had negative ctDNA results. Of the remaining 6 patients, 5 were ctDNA negative and 1 had no mutation detected. Five remain alive in complete remission. One died within 30 days of enrolment (non-cancer related).
Two patients with a detectable ctDNA had not recurred at completion of the 2 year planned follow-up period. One of these patients had a ypT3N1 rectal adenocarcinoma resected in July 2012 and completed adjuvant chemotherapy in January 2013. Routine surveillance detected a change in a 7mm cystic liver lesion (normal CEA), which did not change further on subsequent imaging. The other patient had a ypT2N0 rectal adenocarcinoma resected in August 2012 after receiving neo-adjuvant oxaliplatin-based chemoradiation in a clinical trial. The patient completed adjuvant chemotherapy in January 2013 and was enrolled when multiple new non-specific pulmonary nodules (normal CEA) were detected on routine surveillance. These remained stable over the next two years and were considered most likely to represent granulomatous disease.
Discussion
At diagnosis, patients with colorectal cancer undergo a standard workup including CEA and CT scan, with these repeated at intervals during routine surveillance. The intent is the early detection of metastatic disease, where patients may benefit from curative intent surgery, where feasible, or may achieve a survival benefit from the early administration of palliative intent treatment.12, 13 There are however potential downsides associated with such investigations, which include anxiety for all patients awaiting results, which is further elevated when non-specific findings are present. Ultimately further investigation to immediately define the significance of these may not be possible or may not be definitive. Thus some patients will be left with indeterminate results where only follow-up over subsequent months will provide a definitive interpretation.
One of the many challenges clinicians face in the care of patients undergoing routine surveillance for early stage colorectal cancer is defining the optimal approach to equivocal results, which are not infrequent. In a series of patients with stage II or III colon cancer that ultimately remained disease free, Chao et al. found that 53% had at least one abnormal investigation over the period of routine surveillance that could have represented metastatic disease. This included 20% who had a transiently elevated CEA, 40% with non-specific CT findings and 13% who had both together or at separate times.4
Whilst there is a role for FDG-PET in recurrent colorectal cancer, particularly when considering loco-regional therapies including metastatectomy, studies evaluating FDG-PET in the specific context of indeterminate CEA or CT findings are limited.14 Despite this, FDG-PET is now arguably the standard further investigation in this context. Downsides of FDG-PET imaging are many, including patient discomfort due to the duration of the imaging and exposure to radiation. False negative results can occur, and are of particular relevance in colorectal cancer due to the incidence of mucinous tumors. False positive results are also common, particularly in certain scenarios. In a retrospective series of colorectal cancer patients with indeterminate pulmonary nodules who then proceeded to lung resection, 60% of patients with histologically benign lesions had undergone pre-operative FDG-PET. In all instances FDG-avid lesions were reported.15 Notably, small lung nodules was the most common indeterminate finding in the study reported by Chao et al.4 Furthermore, FDG-PET scans may also be reported as equivocal, as was the case in 9/30 (30%) of our cohort of patients, resulting in additional tests and potentially increased patient anxiety.16 Another concern with the widespread use of FDG-PET imaging is the associated substantial cost, several thousand dollars in some jurisdictions. While the cost of ctDNA analysis is also significant currently, it is projected to fall to several hundred dollars per sample in the near future.17 It is also a simple and non-invasive test, only requiring a blood draw.
Here, we have evaluated ctDNA as a diagnostic marker in patients with at least one routine investigation suggesting the possibility of metastatic disease. This is clearly a different question to the one asked in other studies of patient cohorts with early stage disease where serial ctDNA analysis was routinely performed alongside standard CEA and CT scans, such studies finding ctDNA could be detected many months before recurrence was otherwise evident. In the current study, as shown in Figure 3a, ctDNA analysis defined a patient subset at significantly higher risk of recurrence (ctDNA detectable) or lower risk of recurrence (ctDNA undetectable) than the overall population. Amongst the 15 patients with an equivocal or negative FDG-PET scan, the sole patient with detectable ctDNA had confirmed relapse, suggesting the value of ctDNA analysis might extend to this specific patient subset. It is worth noting that FDG-PET scanning was at the discretion of the treating clinician, so the value if all patients had undergone FDG-PET may be greater or lesser than what was seen in our series. We would expect the former as FDG-PET scans were selectively used and less likely to be performed where they were considered unlikely to be informative, such as for patients with lung nodules measuring a few millimetres in diameter where FDG-PET has limited sensitivity. Clinicians may also have chosen not to perform FDG-PET where the primary tumor was known to be mucinous.
As well as the specific clinical scenario, the value and interpretation of any additional test result needs to be considered in the context of the level of clinical suspicion. Of the 13 patients with a high clinical suspicion of metastatic disease, all except one had later confirmed relapse, with the remaining patient dying within 30 days of enrolment due to a cerebrovascular accident. This indicates that clinicians are quite accurate in predicting the ultimate outcome, however the median time to confirmed recurrence was 88 days, meaning for all these patients a considerable time period with an uncertain future. Earlier confirmation of metastatic disease via ctDNA detection may have led to earlier intervention but that could not be examined in our study. In the patient subset assessed as having a low or intermediate likelihood of having metastatic disease, the hazard ratio for recurrence associated with a detectable ctDNA was similar to the overall population and also reached statistical significance (Figure 3b).
Of particular interest are the 2 patients with a detectable ctDNA who have not developed recurrent disease. As outlined above, the clinical scenarios are patients with rectal adenocarcinoma who were enrolled for non-specific CT findings (change in a cystic liver lesion and new pulmonary lesions respectively), which have remained stable over time. Neither of these patients had an elevated CEA at enrolment or at any time during the follow-up period. Extremely slow growing metastatic disease remains a possibility in the patient with indeterminate lung nodules, with lung metastases from colorectal cancer sometimes having a very indolent behaviour. Notably the longest time to progression in our series was 617 days, not far short of 2 years. However, most likely the two cases are false positive results and we would suggest for future studies the utility of ctDNA analysis might be improved by incorporating repeat analysis where the initial ctDNA result was positive, particularly if the clinical suspicion is low. Analysis for multiple mutated DNA fragments rather than choosing just one, as was performed in our study, is another strategy that might improve the test accuracy.
This study indicates a potential role for ctDNA in a subset of patients where routine investigations reveal findings equivocal for recurrence and/or metastatic disease. This adds to the recent data indicating a potential role of serial ctDNA analysis in the surveillance of patients with colorectal cancer. In the context explored in our study, a negative or a positive ctDNA could be used to guide the intensity and frequency of further investigation. Ultimately, the role of ctDNA analysis in routine surveillance or in further investigating patients with indeterminate findings will be defined by further studies enrolling larger numbers of patients and using more precise protocols for further investigation. As ctDNA analysis becomes cheaper the use of this test will be more attractive from a cost-effectiveness point of view, particularly if results can inform the use of FDG-PET imaging or other further invasive investigations. A benefit to patients in terms of moderating anxiety levels should also not be underappreciated.
Limitations of this study include the modest sample size, the heterogenous nature of the patients enrolled and the absence of a defined protocol for further patient investigation and follow-up. A prescriptive protocol for further investigation was not put in place as there is no one size fits all strategy and necessarily patients underwent a range of appropriate further investigations at varying time intervals and informed by the clinical scenario, reflective of current real-world practice. However, the majority of cases did undergo FDG-PET scans, which we would note were widely accessible and funded for this indication at the time the study was conducted. Our definition of an elevated CEA of >5 μg/L did not account for smoking status, which was not recorded at baseline. However, this is unlikely to have impacted on results as of the 6 patients who were enrolled for an isolated elevated CEA, all except one were subsequently found to have abnormal radiological findings. The remaining patient had a normal CT prior to enrolment and a CEA >10 μg/L which would be considered elevated even in a current smoker.
To our knowledge this is the first study to prospectively evaluate the potential utility of ctDNA analysis in a specific subset of patients with a history of colorectal cancer. We have demonstrated that ctDNA analysis is a significant predictor of recurrence and of remaining cancer free. Further studies are required to further explore this potential and to define the optimal strategy for patient follow-up, which ultimately could be a combination of CEA, CT, FDG-PET and ctDNA, with the latter either as a routine test or informing the use of the other available modalities.
Brief description: “Novelty and Impact”.
Patients with resected early stage colorectal cancer undergoing surveillance often present with routine findings considered indeterminate for recurrence. We explored the role of ctDNA in a subgroup of these patients and demonstrated that ctDNA analysis was a significant predictor of recurrence and of remaining cancer-free. As a potentially effective, minimally invasive test, further studies to determine the optimal role of ctDNA in colorectal cancer surveillance as an adjunct or as a routine test are warranted.
Acknowledgements
We thank the Victorian Cancer Biobank for sample processing.
Funding: This study is supported by Australian National Health and Medical Research Council (GNT1060804), Victorian Cancer Agency (J.T.), the National Institute of Health (CA62924, GM07309, and P30-CA006973), Virginia and D.K. Ludwig Fund for Cancer Research, The John Templeton Foundation, The Conrad R. Hilton Foundation, The Sol Goldman Sequencing Facility at Johns Hopkins and the Marcus Foundation.
Abbreviations
- CAP
Chest, abdomen and pelvis
- CEA
Carcinoembryonic antigen
- CT
Computed tomography
- ctDNA
Circulating tumor deoxyribonucleic acid
- FDG-PET
18Fluorodeoxyglucose positron emission tomography
Footnotes
Author disclosures:
CT receives royalty payments from PapGene Inc. NP, KK and BV are founders of PapGene and Personal Genome Diagnostics. NP is a member of the Scientific Advisory Board of NeoPhore Ltd. KK is a member of the Scientific Advisory Boards of Eisai-Morphotek, Sysmex Inostics, NeoPhore Ltd and CAGE. BV is a member of the Scientific Advisory Boards of Eisai-Morphotek, Sysmex Inostics, Neophore Ltd, Exelixis Gp (Camden Partners) and CAGE. These companies, as well as other companies, have licensed technologies from Johns Hopkins University, on which NP, KK and BV are inventors. These licences and relationships are associated with equity or royalty payments to NP, KK and BV. For CT, NP, KK and BV the terms of these arrangements are being managed by Johns Hopkins University in accordance with its conflict of interest policies.
References
- 1.Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2016. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A, Jemal A. Colorectal cancer statistics, 2017. CA: A Cancer Journal for Clinicians 2017;67: 177–93. [DOI] [PubMed] [Google Scholar]
- 3.Cancer Council Australia Colorectal Cancer Guidelines Working Party. Clinical practice guidelines for the prevention, early detection and management of colorectal cancer. Sydney: Cancer Council Australia. In: Council NHaMR, ed., 2017. [Google Scholar]
- 4.Chao M, Gibbs P. Caution is required before recommending routine carcinoembryonic antigen and imaging follow-up for patients with early-stage colon cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2009;27: e279–80; author reply e81. [DOI] [PubMed] [Google Scholar]
- 5.Sobhani I, Tiret E, Lebtahi R, Aparicio T, Itti E, Montravers F, Vaylet C, Rougier P, Andre T, Gornet JM, Cherqui D, Delbaldo C, et al. Early detection of recurrence by 18FDG-PET in the follow-up of patients with colorectal cancer. British journal of cancer 2008;98: 875–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berger KL, Nicholson SA, Dehdashti F, Siegel BA. FDG PET Evaluation of Mucinous Neoplasms. American Journal of Roentgenology 2000;174: 1005–8. [DOI] [PubMed] [Google Scholar]
- 7.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, Antonarakis ES, Azad NS, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6: 224ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tie J, Kinde I, Wang Y, Wong HL, Roebert J, Christie M, Tacey M, Wong R, Singh M, Karapetis CS, Desai J, Tran B, et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Annals of oncology : official journal of the European Society for Medical Oncology 2015;26: 1715–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tie J, Wang Y, Tomasetti C, Li L, Springer S, Kinde I, Silliman N, Tacey M, Wong H-L, Christie M, Kosmider S, Skinner I, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Science Translational Medicine 2016;8: 346ra92–ra92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tie J, Cohen JD, Wang Y, Li L, Christie M, Simons K, Elsaleh H, Kosmider S, Wong R, Yip D, Lee M, Tran B, et al. Serial circulating tumour DNA analysis during multimodality treatment of locally advanced rectal cancer: a prospective biomarker study. Gut 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci U S A 2011;108: 9530–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shiono S, Okumura T, Boku N, Hishida T, Ohde Y, Sakao Y, Yoshiya K, Hyodo I, Mori K, Kondo H. Outcomes of segmentectomy and wedge resection for pulmonary metastases from colorectal cancer. European journal of cardio-thoracic surgery : official journal of the European Association for Cardio-thoracic Surgery 2017;51: 504–10. [DOI] [PubMed] [Google Scholar]
- 13.Dhir M, Sasson AR. Surgical Management of Liver Metastases From Colorectal Cancer. Journal of Oncology Practice 2016;12: 33–9. [DOI] [PubMed] [Google Scholar]
- 14.Sanli Y, Kuyumcu S, Ozkan ZG, Kilic L, Balik E, Turkmen C, Has D, Isik G, Asoglu O, Kapran Y, Adalet I. The utility of FDG-PET/CT as an effective tool for detecting recurrent colorectal cancer regardless of serum CEA levels. Annals of nuclear medicine 2012;26: 551–8. [DOI] [PubMed] [Google Scholar]
- 15.Jung E-J, Kim S-R, Ryu C-G, Paik JH, Yi JG, Hwang D-Y. Indeterminate pulmonary nodules in colorectal cancer. World Journal of Gastroenterology : WJG 2015;21: 2967–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fraum TJ, Fowler KJ, McConathy J, Dehdashti F. Indeterminate Findings on Oncologic PET/CT: What Difference Does PET/MRI Make? Nuclear Medicine and Molecular Imaging 2016;50: 292–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yi X, Ma J, Guan Y, Chen R, Yang L, Xia X. The feasibility of using mutation detection in ctDNA to assess tumor dynamics. International journal of cancer 2017;140: 2642–7. [DOI] [PMC free article] [PubMed] [Google Scholar]