Abstract
Background
Calcium-activated chloride channel A4 (CLCA4) is known as a tumor suppressor which contributes to the progression of a number of types of malignant tumors. However, little is known about the functional roles of CLCA4 in colorectal cancer (CRC).
Material/Methods
In this study, the expression patterns and dysregulation of mRNAs in CRC tissues were profiled by analyzing GSE21510 datasets from Gene Expression Omnibus database which contains 104 primary hepatocellular carcinoma tissues and 24 normal liver tissues, and by performing Kaplan-Meier analysis of TCGA data. Additionally, immunohistochemistry and quantitative real-time polymerase chain reaction (qRT-PCR) were performed using clinical tissues collected at our institute. In order to explore the functional role of CLCA4, gain-of-function cell models were constructed in SW620 and LoVo cells. Wound healing assay and Transwell assay were carried out to access the cell migration and invasion ability.
Results
It was found that CLCA4 was an independent predictor for overall survival and lymph node metastasis. Additionally, immunohistochemistry and qRT-PCR results of the clinical tissues collected as part of our study further confirmed this correlation. In vitro experiments demonstrated that over-expression of CLCA4 could inhibit cell migration and invasion by suppressing epithelial-mesenchymal transition (EMT) via PI3K/ATK signaling and change the expression patterns of EMT markers in CLCA4-gain-of-function cell models.
Conclusions
CLCA4 inhibits migration and invasion by suppressing EMT via PI3K/ATK signaling and predicts favorable prognosis of CRC which may help to distinguish potential risk of lymph node metastasis in CRC.
MeSH Keywords: Cell Migration Inhibition, Colorectal Neoplasms, Epithelial-Mesenchymal Transition, Neoplasm Invasiveness
Background
Colorectal cancer (CRC) is considered as the third most common type of cancer worldwide and represent the second leading cause of cancer-related mortality globally [1,2]. Accumulating evidence suggests that molecular characteristics determine the response to treatment and prognosis in CRC [3]. The dysregulation of oncogenes or tumor suppressor genes in tumor cells results in alterations in cell growth, apoptosis, migration, invasion, and so on [4,5]. Therefore, identifying novel biomarkers and exploring key pathogenic genes could provide CRC patients with the most timely and reasonable diagnosis and treatment [6].
Lymph node metastasis is the most important prognostic factor in CRC [1]. Accurate preoperative prediction of lymph node status in CRC is important to find appropriate therapeutic decisions, such as the utilization of neoadjuvant or adjuvant chemotherapy for patients with lymph node metastasis, or the implementation of a more conservative approach to keep bowel resection for patients without lymph node metastasis [7,8]. However, the strategies adopted now in clinical practice usually have limited sensitivity and specificity in predicting lymph node metastasis [9,10]. Increasing evidence has shown that dysregulated mRNAs is a potential tumor markers to predict disease progression and metastasis. Our research group is trying to identify novel genes regulating CRC metastasis, including lymph node metastasis.
Calcium-activated chloride channel (CLCA) regulators are proteins which are characterized as a symmetrical multiple cysteine motif in amino terminal tail [11]. It has been shown that the expression of CLCA proteins was deregulated in a variety of cancers [12,13]. CLCA4 is a member of the CLCA family which has similar primary structure with CLCA1 and CLCA2 [14]. Downregulation of CLCA4 expression was observed in breast cancer, bladder cancer, and hepatocellular carcinoma and thought to facilitate tumor cells growth and metastasis by the regulation of epithelial-mesenchymal transition (EMT) [14–16]. When we tried to explore novel mRNAs differentially expressed between CRC cancer tissues and noncancerous tissues with potential research value based on GEO dataset GSE21510, we found that CLCA4 was one of the most differentially expressed mRNAs between tumor and normal groups. However, there are few studies on CLCA4 in the field of CRC besides a research mentioned that CLCA4 was decreased significantly in CRC patients [17], but the functional roles and underlining molecular mechanism of CLCA4 has not been well clarified yet.
In this study, we explore the expression of CLCA4 in CRC tissues and cells, explored the association of CLCA4 expression with clinicopathological features in CRC patients. By constructing CLCA4 over-expression cell models, it was found that the inactivation of the PI3K-AKT signaling and EMT mediated the exertion of biological function of CLCA4 in CRC cells.
Material and Methods
Cell culture and gene over-expression
SW620 and LoVo cell lines (both with relative higher metastatic potential) were obtained from Yearthbio Technology Co., Ltd. (Changsha, Hunan, China) and were cultured using DMEM/F12 medium supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Waltham, USA) under the condition of 5% CO2 at 37°C. CLCA4 over-expression plasmid (HG24887-UT) was purchased from Sino Biological (Beijing, China). CLCA4 over-expression lentivirus were constructed by Genomeditech (Shanghai, China).
RNA isolation and quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNAs were extracted from the cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and then were reverse transcribed using the Reverse Transcription Kit (Bio-Rad, Hercules, CA, USA). In order to detect the relative mRNA levels of CLCA4 and other related genes, quantitative real-time polymerase chain reaction (qRT-PCR) was performed using SYBR Green Supermix (Bio-Rad) in the Real-Time PCR Detection System (Bio-Rad, CFX96) based on the instructions of the manufacturer. All detections were implemented in 3 independent experiments. The relative quantification comparative Ct method was used to quantify the relative mRNA levels of target genes. β-actin was used as internal reference. The primer sequences were CLCA4-F: 5′-CCCTTCAGCTCGAAAGTAA-3′, CLCA4-R: 5′-AGATTGTATGCCCAAGTGCC-3′.
Western blotting
For the sake of detecting the protein expression of CLCA4 epithelial-mesenchymal transition markers and key elements of PI3K/AKT signaling, western blot was carried out. Cells were lysed and centrifuged. The supernatant was collected and denatured. Proteins were separated SDS-PAGE and blotted onto PVDF membrane. The membrane was then treated with TBST containing 50 g/L skimmed milk at room temperature for 4 hours, followed by incubation with the primary antibodies: anti-CLCA4 anti-E-cadherin (1: 1000, Proteintech), anti-N-cadherin (1: 1000, Proteintech), anti-Vimentin (1: 2000, Proteintech), anti-α-SMA (1: 500, Proteintech), anti-Snail (1: 200, Proteintech), anti-PI3K (1: 2000, Abcam), anti-p-PI3K (1: 1000, Abcam), anti-pan AKT (1: 500, Abcam), anti-p-AKT (1: 5000, Abcam), and anti-GAPDH (1: 1000, Proteintech) respectively, at 37°C for 1 hour. Membranes were rinsed and incubated for 1 hour with the correspondent peroxidase-conjugated secondary antibodies. Chemiluminescent detection was performed with the ECL kit (Pierce Chemical, Rockford, IL, USA). The amount of the protein of interest was normalized to the densitometric units of GAPDH.
Immunohistochemistry
A total of 64 cases of CRC paraffin tissue samples confirmed by histological examination were collected from the Department of General Surgery of Changde First People’s Hospital from January 2017 to December 2017. None of the patients received radiotherapy or chemotherapy. Normal tissues were taken from the mucosa more than 5 cm from the edge of the tumor. Normal colorectal mucosa was diagnosed by proportion. The expression of CLCA4 in tumor tissue and mucosa tissue were detected using immunohistochemistry with ElivisionTM plus Polyer HRP (Mouse/Rabbit) IHC Kit (MXB, KIT-9901) according to manufacturer’s instructions against anti-CLCA4 antibody (Abcam, ab197347). After DAB (3′-diaminobenzidine) coloration, the 5 slices of visual field were captured by Nikon light microscopy (200×). Two pathologists assessed and scored the staining results without knowing the clinical data of the samples. The positive staining of CLCA4 was mainly localized in the cytoplasmic brown-yellow granules of CRC tissues, and the expression level was determined according to the immunohistochemical staining intensity standard. The scoring criteria was as following: scored as 0 (negative), 1 (weak), 2 (medium), or 3 (strong). Extent of staining was scored as 0 (<5%), 1 (5–25%), 2 (26–50%), 3 (51–75%) and 4 (>75%) according to the percentages of the positive staining areas in relation to the whole carcinoma area. The immunoreactivity score (IRS) was generate by the percentage score multiplied the staining intensity score. High expression of CLCA4 was defined as detectable immunoreactions with IRS >4 [18].
Cell wound healing and Transwell assays
Wound healing assay was performed to estimate the migration ability of different CRC cell models. Cells were seeded and grown to 90% confluence in 6-well culture plates. The wounds on the monolayer which were created using 10 μL tips were washed with PBS 3 times. The gap width was measured at the presupposed time points by microscopy.
The invasiveness of CRC cells was assessed using Transwell assay. Total of 2×105 cells which were suspended in 100 μL medium with 0.1% serum were inoculated in the upper chambers of Transwell culture systems pre-coated with Matrigel (1: 8 dilution; BD Biosciences, San Jose, CA, USA), and medium with 10% FBS was added to the lower chambers. The invasive cells were fixed and stained with 0.5% crystal violet after incubation at 37°C for 24 hours, then the number was counted, and pictures were taken using a phase contrast microscope.
Statistical analysis
Three independent tests were set up for all experiments. Statistical analysis was performed and presented with GraphPad Prism 7.0 software. Differences between 2 independent groups were evaluated by Student’s t-tests. Differences for multiple comparisons were evaluated using one-way ANOVA. Overall survival (OS) was calculated using the Kaplan-Meier method; P<0.05 was considered significant differences. Data are presented as mean ± standard deviation.
Results
CLCA4 was decreased in CRC tissues compared with normal tissues in TCGA
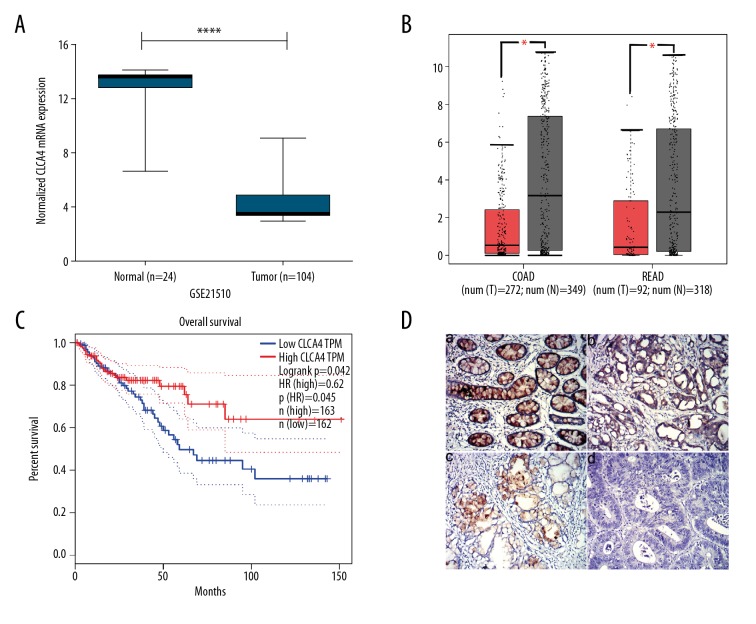
For the sake of tapping novel mRNAs deregulated between CRC cancer tissues and normal or adjacent tissues with potential research value, we explored GSE21510 dataset containing mRNA expression profiles of 104 CRC tissues and 24 noncancerous tissues. It was found that the expression of CLCA4 as remarkedly downregulated in tumor tissues compared with normal tissues and there was a highly significant difference in the mean expression between the 2 groups (Figure 1A). Meanwhile, the expression levels of CLCA4 in tumor tissues were significantly lower than that in normal tissues in both COAD and READ datasets when analyzed using GEPIA online software (http://gepia.cancer-pku.cn) (Figure 1B). Besides, the expression of CLCA4 was negatively corelated with the overall survival of CRC patients (Figure 1C).
Figure 1.
CLCA4 expression in CRC tissues and its correlation with overall survival of CRC patients. (A) Relative mRNA levels of CLCA4 in GEO dataset GSE21510, noncancerous tissues (normal), CRC cancer tissues (tumor). Mann-Whitney was used for statistical analysis. **** P<0.0001 versus normal. (B) The expression level of CLCA4 in tumor tissues was significantly lower than that in normal tissues in both COAD and READ datasets analyzed using GEPIA online software (http://gepia.cancer-pku.cn). (C) Expression of CLCA4 was negatively correlated with poor overall survival of CRC patients (analyzed using GEPIA, http://gepia.cancer-pku.cn). (D) The expression levels of CLCA4 were detected using immunohistochemistry in the clinical tissues collected ourselves. A scoring criteria was used for semi-quantification of immunohistochemistry. High expression of CLCA4 was defined as detectable immunoreactions with IRS >4. Representative pictures of different scores were displayed, a was adjacent tissue; b, c and d were moderately, weakly, and negatively expressed colorectal cancer tissues respectively. The magnification was 200×. IRS – immunoreactivity score.
To study the clinical significance of CLCA4 in CRC, the expression of CLCA4 was further detected using immunohistochemistry in the clinical tissues collected in our institute which included 64 cases of CRC paraffin tissue samples confirmed by histological examination from January 2017 to December 2017. Consistent with the GEO result, the CLCA4 expression in CRC tissues was obviously downregulated compared to that in the adjacent or noncancerous tissues (Figure 1D).
Correlation of CLCA4 expression and clinicopathological characteristics
Based on the IRS, the CRC specimens were divided into 2 groups according to relative expression levels of CLCA4. By incorporating the CLCA4 expression data with the clinicopathological features of the corresponding patients, single-factor statistical analysis was performed to explore the correlation of CLCA4 expression level with various pathological factors in CRC, including age, gender, tumor size, clinical stage, lymph node status and pathological differentiation status. It was found that CLCA4 expression was negatively correlated with lymphatic metastasis (P=0.037) but had no significance with age, gender, tumor size, TNM stage, or differentiation status (Table 1).
Table 1.
Clinicopathological association of CLCA4 expression in colorectal cancer patients.
| Clinicopathological features | No. cases | CLCA4 expression | P value | |
|---|---|---|---|---|
| High | Low | |||
| Age (years) | 0.874 | |||
| >60 | 29 | 13 | 16 | |
| ≤60 | 35 | 15 | 20 | |
| Gender | 0.728 | |||
| Male | 35 | 16 | 19 | |
| Female | 29 | 12 | 17 | |
| Tumor size | 0.506 | |||
| >5 cm | 29 | 14 | 15 | |
| ≤5 cm | 35 | 14 | 21 | |
| TNM stage | 0.050 | |||
| I–II | 34 | 11 | 23 | |
| III–IV | 30 | 17 | 13 | |
| Lymph node status | 0.037 | |||
| Absent | 34 | 19 | 15 | |
| Present | 30 | 9 | 21 | |
| Pathological differentiation | 0.594 | |||
| Well | 15 | 7 | 8 | |
| Moderately | 24 | 12 | 12 | |
| Poorly | 25 | 9 | 16 | |
CLCA4 was decreased in CRC cells compared to cells
In order to explore the functional role of CLCA4 in CRC, the endogenous expression of CLCA4 were detected in 4 CRC cells and colorectal epithelial cell HCoEpiC using western blot. The results showed that CLCA4 was downregulated in 4 CRC cell lines compared with HCoEpiC. Especially, the expression level of CLCA4 is even lower in LoVo and SW620, which have higher metastatic potential (Figure 2A). Then, LoVo and SW620 cells were chosen as parental cell lines to construct gain-of-function cell models for CLCA4. CLCA4 was augmented by infecting LoVo and SW620 with CLCA4 over-expression lentivirus. Augment of CLCA4 expression in the functional model cell lines was confirmed by western blot when compared to a negative control (NC) group in both SW620 and LoVo cells (Figure 2B).
Figure 2.
Gain-of-function cell models of CLCA4 were established in SW620 and LoVo cells. (A) Protein expression levels were detected in 4 CRC cell lines and colorectal epithelial cells HCoEpiC using western blot. (B) LoVo and SW620 were infected with CLCA4 over-expression lentivirus (CLCA4) and negative control lentivirus (NC) respectively. Then the protein expression of CLCA4 was analyzed using western blot to confirm that CLCA4 was over-expressed successfully in cell models. For relative quantification, GAPDH was used as the internal reference for western blot.
Enhanced expression of CLCA4 reduced migration and invasion of CRC cells
The functional role of CLCA4 was investigated in its gain-of-function cell models. Because the CLCA4 was negative correlation with lymphatic metastasis, we mainly paid attention to its regulation of metastasis and invasion, which had been implicated from clinical data. SW620 and LoVo cells were infected with CLCA4 over-expression lentivirus. Firstly, some changes had taken place, there was a decrease in mesenchymal morphology and an increase in epithelial cell morphology which suggested CLCA4 might participate in mesenchymal epithelial transformation (Figure 3A). Besides, CLCA4 over-expression significantly reduced the invasion ability of both SW620 and LoVo cells compared with NC group which was confirmed by Transwell assay (Figure 3B, 3D). On the other hand, CLCA4 over-expression obviously inhibited the migration capacity of both SW620 and LoVo cells compared with the NC group which was shown though the cell scratch test (Figure 3C, 3E). In order to further confirm if CLCA4 expression was correlated with the expression of EMT markers, western blot was performed to detect the alteration of related markers. It was found that CLCA4 over-expression could enhance the expression of epithelial cell marker E-cadherin but decrease the mesenchymal markers N-cadherin, Vimentin, Snail and α-SMA exactly (Figure 3F). Meanwhile, the western blot results showed that CLCA4 over-expression could inhibit the activation of PI3K-AKT signaling to some extent (Figure 3G).
Figure 3.
CLCA4 over-expression reduces abilities of migration and invasion of CRC cells. (A) CLCA4 over-expression resulted in the cell morphology change to some extent. There was a decrease in mesenchymal morphology and an increase in epithelial cell morphology (the magnification was 200×). (B) Transwell assay was used to assess the invasion ability of negative control lentivirus infected CRC cells (NC) and CLCA4 over-expressed CRC cells (CLCA4), meanwhile, the invasive cells were counted and compared (D). The magnification was 200×. * P<0.05 versus negative control lentivirus infected cell group (NC). (C) The wound healing assay was performed to detect the migration ability of CRC cells in negative control lentivirus infected cell group (NC) and CLCA4 over-expressed (CLCA4) CRC cells and the wound closure was measured (E). The magnification was 200×. * P<0.05 and ** P<0.01 versus negative control lentivirus infected cell group (NC). (F) Epithelial-mesenchymal transition marker proteins were detect using western blot in CLCA4 over-expressed cells and NC cells. For relative quantification, GAPDH was used as the internal reference. (G) Key components of PI3K-AKT pathway were detect using western blot in CLCA4 over-expressed cells and NC cells. For relative quantification, GAPDH was used as the internal reference.
PI3K-AKT signaling activation could abolish the inhibition of CLCA4 on the migration and invasion of CRC cells
PI3K-AKT signaling was partially inactivated after CLCA4 over-expression in CRC cells which suggests that PI3K-AKT is actually one of the important signal transduction pathways mediating CLCA4 to play biological roles. To further illustrate this point, insulin growth factor-1 (IGF-1) was used to activate PI3K-AKT signaling in CLCA4 over-expressed CRC cells and then Transwell and scratch assays were performed again to assess the invasion and migration capacity of CLCA4 gain-of-function model cells [19,20]. The results demonstrated that the activation of PI3K-AKT signaling by IGF-1 treatment could significantly restore the inhibition of CLCA4 on the invasion (Figure 4A, 4B) and migration (Figure 4C, 4D) of CRC cells. These results verified that CLCA4 exerts its metastasis inhibition function partially through PI3K-AKT signaling pathway.
Figure 4.
PI3K-AKT signaling activation abolishes the inhibition of CLCA4 on the migration and invasion of CRC cells. (A) Transwell assay was used to assess the invasion ability of CLCA4 over-expressed cells with (CLCA4 + IGF-1) or without IGF-1 treatment (CLCA4). The magnification was 200×. Meanwhile, the invasive cells were counted and compared (B). * P<0.05 versus CLCA4 over-expressed cells without IGF-1 treatment. (CLCA4). (C) The wound healing assay was performed to detect the migration ability of CLCA4 over-expressed cells with or without IGF-1 treatment. The magnification was 200× and (D) the wound closure was measured. *, P<0.05 versus CLCA4 over-expressed cells without IGF-1 treatment. (CLCA4).
Discussion
In this research, clinical analysis demonstrated that CLCA4 decrease was correlated with lymph node metastasis of CRC patients. Over-expression of CLCA4 could reduce the invasive and migration abilities of SW620 and LoVo cells through EMT inhibition. Meanwhile, these effects were exerted partially via inactivation the PI3K/AKT signaling. These results provide new insights into the functional role and molecular mechanism of CLCA4 deregulation in the progression of CRC.
CLCA proteins have been found not only to regulate inflammatory responses but also take part in carcinogenesis [15]. CLCA1 was reported to participate in FAK activation, metastatic growth of melanoma cells, and could be used as a predictor for tumor recurrence and poor survival in colorectal cancer [13,21]. Oxidative stress could induce CLCA2 expression in a p53-dependent manner to inhibit proliferation of breast cancer [22]. These findings implied that CLCA family proteins are involved in tumorigenesis and tumor progression.
As for CLCA4, it has been reported that loss of CLCA4 promotes epithelial-to-mesenchymal transition in breast cancer cells which suggested the regulation of CLCA4 on EMT for the first time [16]. Later, it was demonstrated that CLCA4 inhibits bladder cancer cell proliferation, migration, and invasion by suppressing the PI3K/AKT pathway [15]. Recently, researchers found that CLCA4 inhibits cell proliferation and invasion of hepatocellular carcinoma by suppressing EMT via PI3K/AKT signaling [14]. Nevertheless, the expression and functional role of CLCA4 in colorectal cancer are not well-established yet.
In this study, based on GEO dataset, it was found that the expression of CLCA4 significantly down-regulated in tumor tissues compared with normal tissues and the expression level of CLCA4 was negatively corelated with the overall survival of CRC patients. It is worth mentioning that the immunohistochemistry (IHC) results of CRC tissues collected at our institute showed that CLCA4 expression was negatively correlated with lymphatic metastasis (P=0.037) but there was no significant correlation with age, gender, tumor size, TNM stage, or differentiation status. Lymph node metastasis is the most important prognostic factor in CRC without hematogenous metastasis or peritoneal seeding [1]. In addition, CLCA4 has been found to regulate metastasis and invasion of bladder cancer and hepatocellular carcinoma cells [14,15]. Therefore, we focused on the regulation of CLCA4 on metastasis and invasion of CRC cells. We constructed CLCA4 over-expression cell models in SW620 and LoVo cell lines with relatively higher invasiveness and confirmed that CLCA4 was a negative regulator for metastasis and invasive ability of CRC cells. However, our experimental results were limited to the motility of cells which cannot truly reflect the regulation of CLCA4 on lymph node metastasis. It is regrettable that due to the limitation of funds and time, in vivo experiments could not be carried out, and can only be explored in our follow-up studies. In vivo, CLCA4 over-expressed CRC cells will be injected into nude mice using caudal vein injection. Then in vivo imaging can be used to observe the metastasis status at the preset time point.
EMT is one of the important molecular mechanisms of cell metastasis and literature has reported on the regulation of EMT by CLCA4 in other cancer types [16,17]. It was also verified that the regulation of CLCA4 on CRC cell metastasis was also mediated through the expression alterations of EMT markers in CLCA4 over-expression cell models. In addition, in order to explore the signal transduction pathway of CLCA4, we searched for the relative signal pathways which were reported to be involved in the regulation of cell migration and invasion of CLCA family proteins and then paid attention to FAK-ERK and PI3K-AKT signal pathways [14,15,21]. It was found that the PI3K-AKT signaling pathway was partially inactivated after over-expression of CLCA4. When CLCA4 over-expressed cells were treated with IGF-1 (activator of PI3K-AKT signaling), the migration and invasion capacity inhibited by CLCA4 was significantly restored. However, the key factors of FAK-ERK signaling did not show obvious alteration. These results demonstrated that CLCA4 plays inhibitory roles in invasion and migration through suppressing epithelial-mesenchymal transition via PI3K/AKT signaling of colorectal cancer.
Conclusions
In general, the present study showed that CLCA4 might inhibit CRC cell invasion and migration through suppressing EMT via the PI3K/AKT pathway. If this conclusion is further confirmed by in vivo research, CLCA4 might be a potential biomarker for predicting lymph node status in CRC patients for appropriate therapeutic decisions.
Footnotes
Source of support: Departmental sources
Conflict of interest
None.
References
- 1.Qu A, Yang Y, Zhang X, et al. Development of a preoperative prediction nomogram for lymph node metastasis in colorectal cancer based on a novel serum miRNA signature and CT scans. EBio Medicine. 2018;37:125–33. doi: 10.1016/j.ebiom.2018.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 3.Sridharan M, Hubbard JM, Grothey A. Colorectal cancer: How emerging molecular understanding affects treatment decisions. Oncology (Williston Park) 2014;28(2):110–18. [PubMed] [Google Scholar]
- 4.Liu W, Li H, Hong SH, et al. Olfactomedin 4 deletion induces colon adenocarcinoma in Apc(Min/+) mice. Oncogene. 2016;35(40):5237–47. doi: 10.1038/onc.2016.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ordonez-Moran P, Dafflon C, Imajo M, et al. HOXA5 counteracts stem cell traits by inhibiting WNT signaling in colorectal cancer. Cancer Cell. 2015;28(6):815–29. doi: 10.1016/j.ccell.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 6.Jiang W, Yang W, Yuan L, Liu F. Upregulation of AKIP1 contributes to metastasis and progression and predicts poor prognosis of patients with colorectal cancer. Onco Targets Ther. 2018;11:6795–801. doi: 10.2147/OTT.S151952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benson AB, 3rd, Venook AP, Cederquist L, et al. Colon cancer, Version 1.2017, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2017;15(3):370–98. doi: 10.6004/jnccn.2017.0036. [DOI] [PubMed] [Google Scholar]
- 8.van de Velde CJ, Aristei C, Boelens PG, et al. EURECCA colorectal: Multidisciplinary mission statement on better care for patients with colon and rectal cancer in Europe. Eur J Cancer. 2013;49(13):2784–90. doi: 10.1016/j.ejca.2013.04.032. [DOI] [PubMed] [Google Scholar]
- 9.Nakayama G, Tanaka C, Kodera Y. Current options for the diagnosis, staging and therapeutic management of colorectal cancer. Gastrointest Tumors. 2013;1(1):25–32. doi: 10.1159/000354995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shin SS, Jeong YY, Min JJ, et al. Preoperative staging of colorectal cancer: CT vs. integrated FDG PET/CT. Abdom Imaging. 2008;33(3):270–77. doi: 10.1007/s00261-007-9262-9. [DOI] [PubMed] [Google Scholar]
- 11.Xiao X, Tang YS, Mackins JY, et al. Isolation and characterization of a folate receptor mRNA-binding trans-factor from human placenta. Evidence favoring identity with heterogeneous nuclear ribonucleoprotein E1. J Biol Chem. 2001;276(44):41510–17. doi: 10.1074/jbc.M106824200. [DOI] [PubMed] [Google Scholar]
- 12.Walia V, Yu Y, Cao D, et al. Loss of breast epithelial marker hCLCA2 promotes epithelial-to-mesenchymal transition and indicates higher risk of metastasis. Oncogene. 2012;31(17):2237–46. doi: 10.1038/onc.2011.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang B, Cao L, Liu J, et al. Low expression of chloride channel accessory 1 predicts a poor prognosis in colorectal cancer. Cancer. 2015;121(10):1570–80. doi: 10.1002/cncr.29235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Z, Chen M, Xie LK, et al. CLCA4 inhibits cell proliferation and invasion of hepatocellular carcinoma by suppressing epithelial-mesenchymal transition via PI3K/AKT signaling. Aging (Albany NY) 2018;10(10):2570–84. doi: 10.18632/aging.101571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hou T, Zhou L, Wang L, et al. CLCA4 inhibits bladder cancer cell proliferation, migration, and invasion by suppressing the PI3K/AKT pathway. Oncotarget. 2017;8(54):93001–13. doi: 10.18632/oncotarget.21724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu Y, Walia V, Elble RC. Loss of CLCA4 promotes epithelial-to-mesenchymal transition in breast cancer cells. PLoS One. 2013;8(12):e83943. doi: 10.1371/journal.pone.0083943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang B, Cao L, Liu B, et al. The transition from proliferation to differentiation in colorectal cancer is regulated by the calcium activated chloride channel A1. PLoS One. 2013;8(4):e60861. doi: 10.1371/journal.pone.0060861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu X, Li YQ, Li QG, et al. Osteoglycin (OGN) reverses epithelial to mesenchymal transition and invasiveness in colorectal cancer via EGFR/Akt pathway. J Exp Clin Cancer Res. 2018;37(1):41. doi: 10.1186/s13046-018-0718-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao D, Tang T, Zhu J, et al. CXCL12 has therapeutic value in facial nerve injury and promotes Schwann cells autophagy and migration via PI3K-AKT-mTOR signal pathway. Int J Biol Macromol. 2019;124:460–68. doi: 10.1016/j.ijbiomac.2018.10.212. [DOI] [PubMed] [Google Scholar]
- 20.Wang J, Xie L, Wang S, et al. Azithromycin promotes alternatively activated macrophage phenotype in systematic lupus erythematosus via PI3K/Akt signaling pathway. Cell Death Dis. 2018;9(11):1080. doi: 10.1038/s41419-018-1097-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abdel-Ghany M, Cheng HC, Elble RC, Pauli BU. Focal adhesion kinase activated by beta(4) integrin ligation to mCLCA1 mediates early metastatic growth. J Biol Chem. 2002;277(37):34391–400. doi: 10.1074/jbc.M205307200. [DOI] [PubMed] [Google Scholar]
- 22.Walia V, Ding M, Kumar S, et al. hCLCA2 is a p53-inducible inhibitor of breast cancer cell proliferation. Cancer Res. 2009;69(16):6624–32. doi: 10.1158/0008-5472.CAN-08-4101. [DOI] [PMC free article] [PubMed] [Google Scholar]