Abstract
Glyphosate, the active compound of Roundup, is one of the most used pesticides in the world. Its residues are often detected in animal feed, but the impact on the animal gut microbiota and on pathogens of the intestine has not intensively been investigated. In this study, we analyzed the minimum inhibitory concentration (MIC) of glyphosate isopropylamine salt and a common glyphosate-containing herbicide formulation in 225 Salmonella enterica isolates by broth microdilution. A bacteriostatic effect of glyphosate on Salmonella growth was detected at the concentration range of 10 to 80 mg/mL for both the active ingredient and the ready-to-use formulation. Time/year of isolation, host species, and serovars revealed a statistically significant influence on MIC values. Recently collected Salmonella isolates had significantly higher MIC values for glyphosate and the glyphosate-containing product compared with isolates collected between 1981 and 1990. Isolates from pigs showed significantly higher MIC values compared with isolates from poultry, and isolates of the Salmonella serovar Typhimurium had significantly higher MIC values than Salmonella Enteritidis and Infantis isolates.
Keywords: glyphosate, enterobacteriaceae, Salmonella, minimum inhibitory concentration, MIC, historic isolates
Introduction
Glyphosate (N-(phosphonomethyl)glycine) is one of the most widely used herbicides in the world [1]. Its usage has increased significantly since the first genetically glyphosateresistant agricultural crops were introduced to the market in the 1990s [2]. In parallel to the increase of the usage of glyphosate-containing herbicides (GCH), the discussion about its toxicity on higher organisms and other impacts on the environment, e.g., on freshwater communities [3–5] or soil organisms [6, 7], came intensively into the focus of public interest.
The target structure of glyphosate is the penultimate step in the shikimate pathway and well described [8–10]. The herbicide binds to the complex of 5-enolpyruvylshikimate-3-phosphate (EPSP) synthase and its substrate and inhibits the production of aromatic amino acids. This metabolic pathway is present in plants, bacteria, some fungi, and unicellular parasites but not in vertebrates [11]. In bacteria, 3 variants of the targeted enzyme gene have been described so far, accounting for the distinct effect of glyphosate in microbiology. Two of the gene variants show a high similarity, while the third one, which has only recently been found, differs [12]. The resulting variations of the target enzyme provide a conceivable reason for diverging sensitivities to glyphosate, acting as an antimicrobial substance in bacteria.
Nowadays, genetically modified plants are commonly grown and highly polluted with glyphosate in many countries in the world, because the pesticide can be applied throughout the whole growth phase.
Residues of glyphosate were found in soy [13, 14] and maize [15], which both play an important role in animal nutrition. Hence, livestock-associated microbiota can get in contact with glyphosate through residues in animal feed as demonstrated by glyphosate detection in urine and feces of dairy cows [16].
If glyphosate and GCHs cause adverse effects on the environment remains unclear, especially regarding complex bacterial communities.
It was postulated by Krüger et al. [17] and Ackermann et al. [18] that a daily intake of glyphosate with animal feed can lead to dysbiosis with an increased number of pathogenic bacteria surviving in the intestine, depending on the sensitivity to glyphosate. Regarding a ruminal setting, these findings could not be confirmed [19]. Shehata et al. [20] determined differing minimum inhibitory concentrations (MIC) for glyphosate and a formulation of glyphosate for Enterobacteriaceae of farm animal origin. In the case of food microorganisms, results show differences in the susceptibility between glyphosate and glyphosate formulations. While glyphosate itself did not affect microbial growth, an inhibitory effect was determined when using the formulation [21].
In general, chances for survival and reproduction of pathogens in the intestine are higher in an imbalanced microbiological environment. Salmonella species are pathogens causing subclinical infections in pigs and leading to chronic carriage in poultry. Its shedding and spread from farm animals into the environment is of major concern in food hygiene, due to its ability to cause foodborne diseases. The World Health Organization ranks non-typhoidal salmonellosis as one of the four key global causes of diarrheal diseases [22]. Majowicz et al. [23] estimated the worldwide number of Salmonella infections to 93.8 million. Following poultry, pork is considered the second leading source of Salmonella infections in humans [24]. Non-species specific serovars pose zoonotic potential and occur in host species in different frequencies. The most important livestock-associated serovars are Salmonella Typhimurium, Salmonella Enteritidis and Salmonella Infantis.
For this study, we investigated MICs for glyphosate and a commonly used formulation of glyphosate to quantify potential inhibitory effects on different strains of the major foodborne zoonotic pathogen Salmonella enterica isolated from livestock animals.
The study aimed to i) determine the MIC for glyphosate and the glyphosate-containing formulation Roundup LB plus in Salmonella isolates originating from farm animals, ii) compare the trend of MIC development within the last three decades comprising the period before and after the immense utilization of glyphosate in agriculture, and iii) reveal the influence of the animal host and the serovar on the MIC of the Salmonella isolates.
Materials and Methods
Two hundred twenty-five Salmonella enterica isolates were selected (Table 1), belonging to the serovars Typhimurium, Enteritidis, or Infantis. They originated from fecal samples of pigs and poultry. One hundred twenty of the isolates were sampled between 2014 and 2016 (recent isolates). Inclusion criteria for this study consisted of widespread sampling locations representing strains from all over Germany and a variability in antibiotic susceptibility. Sixty isolates originated from pigs and chicken, respectively. Each serovar was represented by 40 isolates. One hundred five isolates were isolated from 1981 until 1990 (historic isolates). Thirty-six of these isolates were assigned to serovar Typhimurium, 37 to serovar Enteritidis, and 32 to serovar Infantis. Forty-eight isolates were isolated from pigs, and 57 from poultry.
Table 1.
Overview of isolates. Number and distribution of tested isolates for the different collection times, species, and serovars. Historic isolates have been collected between 1981 and 1990; recent isolates have been collected between 2014 and 2016. All isolates were provided by the German Federal Institute of Risk Assessment
| Serovar | Poultry | Pig | Total | ||
|---|---|---|---|---|---|
| Historic | Recent | Historic | Recent | ||
| S. Typhimurium | 19 | 10 | 17 | 30 | 76 |
| S. Enteritidis | 25 | 30 | 12 | 10 | 77 |
| S. Infantis | 14 | 20 | 18 | 20 | 72 |
| Total | 58 | 60 | 47 | 60 | 225 |
MICs were determined in 96-well plates by broth microdilution, described by Wiegand et al. [25]. The concentration of glyphosate was calculated based on the concentration of glyphosate isopropylamine salt in a 40% solution, purchased from Sigma-Aldrich (Sigma-Aldrich Chemie GmbH, Taufenkirchen, Germany) with a purity of 98% (GLY) or in the herbicide formulation Roundup LB Plus (German license number: 024142–00, Monsanto) (RU). Starting from a concentration of 80 mg/mL, a twofold dilution series was prepared down to a concentration of 1.25 mg/mL. Ready-to-use MIC plates were stored at –80 °C. Nutrient rich Mueller Hinton medium (MH) (Oxoid GmbH, Wesel, CM0405) was used.
Inoculum was prepared as an overnight culture in Mueller Hinton broth and aerobically incubated for 16 h at 37 °C, with shaking. Subsequently, optical density of the overnight cultures was measured, adjusted to a number of 1 × 106 colony forming units (cfu) per milliliter. An inoculum of 5 μL for each well and isolate was transferred into prepared 96-well plates (with conical bottom; Sarstedt GmbH, Nürnberg) resulting in a final number of 5 × 104 cfu per well. All samples were processed in technical triplicates. Every isolate was tested for GLY and RU. The 96-well plates were aerobically incubated over night at a temperature of 37 °C in a humidity chamber [26].
The MIC was defined as the lowest concentration without visible growth in at least 2 out of 3 technical replicates. It was determined visually by using an impinging light and a mirror (SensiTouch by Sensititre).
Statistical analysis was performed with IBM SPSS statistics Version 24. Prior to calculation, data were transformed to an ordinal scale (10 mg/mL = 1; 20 mg/mL = 2; 40 mg/mL = 3; 80 mg/mL = 4). Because data were not normally distributed, comparisons between groups were calculated using the Mann–Whitney U test for two groups or the Kruskal–Wallis test for more than two groups, respectively.
Multivariable analysis of variance model was used to investigate the influence of isolation time (historical or recent isolates), serovar (S. Typhimurium, S. Infantis, or S. Enteritidis) and host (poultry or pig) on MIC values. Further, proportional-odds ordinal regression models were run with the MIC values as dependent variable and isolation time, serovar, and hosts as independent variables.
All two-way-interactions between influence factors were included in the initial models and removed if not statistically significant. P-values below 0.05 were considered statistically significant. Model diagnostics included check for normality and homoscedasticity of residuals. Ordinal regression models were additionally tested for the assumption of proportional odds.
Ethics
All isolates were provided by German Federal Institute of Risk Assessment. Non of the samples from which the Salmonella enterica were isolated were collected directly from animals from the investigating institute. Therefore no ethical approval was necessary.
Results
MIC for glyphosate isopropylamine salt (GLY) and for glyphosate salt within the formulation Roundup LB plus (RU) was investigated for 225 isolates of Salmonella enterica from fecal samples from pigs and poultry. The MICs varied slightly between replicates and between RU and GLY for the particular isolate. Hence, the MIC data comprised a very narrow range and were not normally distributed. The results of the statistical analysis are summarized in Table 2.
Table 2.
Statistical comparisons. Statistical comparison between the different groups with univariable Mann–Whitney U1 rpt. Kruskal–Wallis2 test as non-parametric test for not normally distributed data and the multivariable models of analysis of variance and ordinal regression. (* = statistically significant). Groups with higher MICs in bold
| Glyphosate isopropylamine salt (GLY) | Roundup LB plus (RU) | |||||
|---|---|---|---|---|---|---|
| Mann–Whitney U1/Kruskal–Wallis2 univariable | Multivariable analysis of variance | Multivariable ordinal regression | Mann–Whitney U1/Kruskal–Wallis2 univariable | Multivariable analysis of variance | Multivariable ordinal regression | |
| Isolation time: recent vs. historical |
P = 0.0121* | P = 0.006* | P = 0.008* | P < 0.001* | P < 0.001* | P < 0.001* |
| Host: pig vs. poultry |
P = 0.0271* | P = 0.030* | P = 0.031* | P = 0.021* | P = 0.174 | P = 0.097 |
| Serovars | P < 0.0012* | P < 0.001* | - | P <0.0012* | P < 0.001* | - |
| Typhimurium vs Enteritidis | P = 0.1471 | P = 0.238 | P = 0.381 | P = 0.002* | P < 0.001* | P = 0.003* |
| Typhimurium vs Infantis | P <0.0011 | P < 0.001* | - | P < 0.001* | P < 0.001* | - |
| Enteritidis vs. Infantis | P <0.0011* | P = 0.001* | P < 0.001* | P = 0.030* | P = 0.030* | P = 0.006* |
GLY vs. RU
The distribution of MICs of all isolates investigated for GLY and RU is shown in Figure 1. The determined values for GLY ranged from 10 mg/mL (lowest MIC) to 80 mg/mL (highest MIC). The median and the mode for the whole dataset were 40 mg/mL. For RU, MICs had a range of 20 mg/mL to 80 mg/mL. The median and the mode were 40 mg/mL. The 95% cutoff for both GLY and RU was located at 80 mg/mL (Figure 1).
Figure 1.
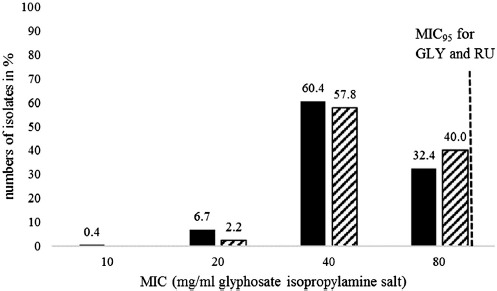
Distribution of the MIC values of the investigated Salmonella enterica isolates in percentage for glyphosate isopropylamine salt (GLY) (black) and Roundup LB plus (RU) (cross hatched). Dashed line marks 95% epidemiological cutoff for both GLY and RU
Comparison of Recent and Historic Isolates
The distribution of MICs expressed as percentage for the investigated Salmonella isolates separated according to historic and recent isolates to GLY and RU is presented in Figure 2. One hundred twenty of the investigated Salmonella enterica isolates where collected between 2014 and 2016. One hundred five isolates were collected between 1981 and 1990, prior to the intensive usage of glyphosate in agriculture. MIC values for the isolates collected before 1991 showed a distribution of 20 to 80 mg/mL for both GLY and RU. In general, equal ranges were determined for recent isolates regarding RU. For GLY, recent isolates showed a distribution of MIC ranging from 10 mg/mL to 80 mg/mL. The median was 40 mg/mL for both GLY and RU. In addition, the mode for both datasets was 40 mg/mL as well. The statistical analysis and the two statistical models used showed that the obtained differences of MICs between historical and recent isolates were highly significant (Table 2; Mann–Whitney U test P = 0.012; univariate analysis of variance P = 0.006; ordinal regression P = 0.008). Historical isolates had lower MIC against GLY compared with recent isolates (P = 0.006). For RU, the statistically significant influence of isolation time was also determined by univariable tests. Historical isolates had lower MIC compared with recent isolates (P < 0.001).
Figure 2.
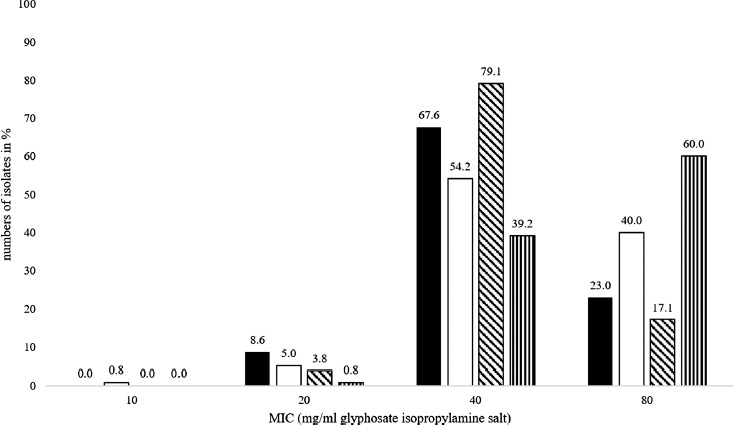
MIC values for the investigated Salmonella enterica isolates for glyphosate isopropylamine salt (GLY) and Roundup LB plus (RU) in comparison, differentiated between historic isolates collected before 1991 and recent isolates collected between 2014 and 2016. GLY before 1991 in black, GLY between 2014 and 2016 in white, RU before 1991 oblique crosshatched, and RU between 2014 and 2016 vertical crosshatched
Comparison of Host Species
A comparison of the MIC values of GLY and RU expressed as percentage and separated according to host species is shown in Figure 3. Regarding Salmonella isolates originating from pigs, a MIC of 80 mg/mL was more frequently determined than for those originating from poultry. This was the case for both GLY and RU. By contrast, the percentage of isolates from poultry showing low MICs of 20 mg/mL or 10 mg/mL for RU, respectively, was higher compared to that from pigs. The univariate analysis of variance confirmed that MICs of pig isolates were significantly higher than those obtained for poultry isolates (Table 2; P = 0.030). For RU, statistical significance could also be shown in the non-parametric tests (P = 0.021), whereas the analysis of variances and the ordinal regression model did not reveal a statistically significant influence of the host (P = 0.174; P = 0.097).
Figure 3.
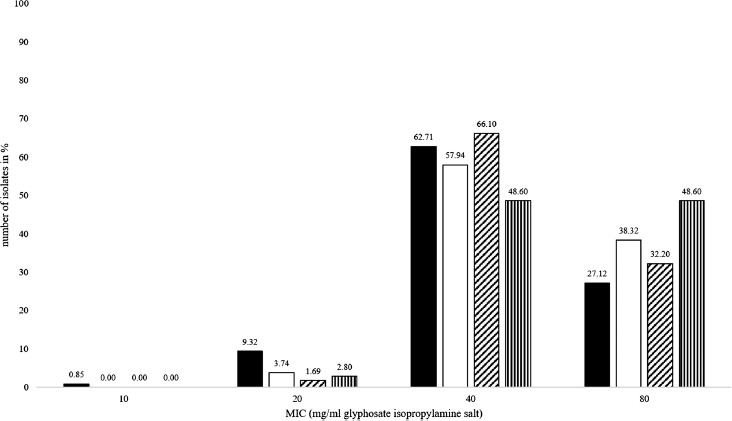
MIC values of Salmonella isolates from pig for glyphosate isopropylamine salt (white) and poultry for glyphosate isopropylamine salt (black) and for pig in Roundup LB plus (vertical crosshatched) and poultry in Roundup LB plus (oblique crosshatched)
Comparison of Salmonella Serovars
A comparison of the datasets obtained for the different serovars included in the study revealed that with the exception of the serovar Typhimurium, MICs of 40 mg/mL were most frequently determined. This accounts for both GLY and RU. For GLY, the obtained differences of MICs according to serovars were classified as significant by the Kruskal–Wallis test for more than 2 groups (P < 0.001).
In the statistical model for GLY, the comparison of all 3 serovars also revealed significant differences in MIC values (analysis of variance P < 0.001). Post-hoc pairwise comparison revealed a significance of the difference obtained for Typhimurium and Infantis (P < 0.001; higher values for Typhimurium than for Infantis), whereas that between the serovars Enteritidis and Typhimurium was rated as not significant by the modeling (P = 0.238). In the two calculated models, the serovar Typhimurium had highest MICs, followed by the serovar Enteritidis. The lowest MICs were obtained by the serovar Infantis. The differences between Enteritidis and Infantis and Typhimurium and Infantis were statistically significant, whereas between Typhimurium and Enteritidis, they were not (Table 2).
For RU, the Kruskal–Wallis test showed statistically significant differences between all 3 serovars. This correlated with the results of the analysis of variance and the ordinal regression model. MIC values of the serovar Enteritidis were significantly higher than those of the serovar Infantis (P = 0.030). Between the serovars Enteritidis and Typhimurium, a statistically significant difference (P < 0.001) was detected, where the serovar Typhimurium showed higher values than Enteritidis. This was also reflected by the ordinal regression (P = 0.003 Typhimurium vs. Enteritidis; P = 0.006 Enteritidis vs. Infantis). In summary, the comparison of the 3 serovars in RU revealed the highest MICs for Typhimurium compared to Enteritidis and Infantis, while the serovar Enteritidis showed higher values than Infantis.
Discussion
For the first time, we aimed to systematically investigate different Salmonella enterica isolates from food-producing animals for their susceptibility against the herbicide glyphosate (GLY) and a glyphosate-containing tallow amine free formulation, Roundup LB Plus (RU). Furthermore, we intended to analyze the potential impact of time of isolation, host species, and serovar on susceptibility to glyphosate.
MIC Values
In general, the range of MIC values determined for all isolates included in the study was rather narrow. Nonetheless, a statistical analysis of the generated study data revealed a significant impact of some parameters that were considered as potential factors beforehand and thus investigated.
In comparison with data published before, the mean MIC value of 40 mg/mL isopropylamine glyphosate in both the herbicide and the formulation as determined by our study has to be considered as rather high. Shehata et al. [20] determined MICs of 5.0 mg/mL glyphosate for 3 different Salmonella strains using the formulation “Roundup UltraMax”. Within the study conducted by Kurenbach et al. [27], MIC was investigated for one Salmonella Typhimurium isolate, resulting in a value of 6.19 mg/mL. The group used the formulation “Roundup Weed killer”. Only the work of Nielsen et al. [28] revealed comparable high MIC values, for example, a MIC of 80 mg/mL for the formulation “Glyfonova 450 plus” for two E. coli isolates. Unfortunately, they had no Salmonella enterica isolates in their dataset to compare with.
Different Formulations
The differences between reported MIC values and those determined by our study could be due to the usage of different glyphosate formulations. An impact of the formulation was considered by different authors [21, 29], as well as the usage of different media [28]. Further deviations were occurring because of a lack of standardized methods. For example, the formulation “Roundup UltraMax” used in some studies contained tallow amine as a surfactant, which is known to be more toxic than glyphosate [29, 30]. This remarkable difference underlines the issue of an insufficient comparability of results obtained for variable glyphosate formulations. Generally, little is known about the additional ingredients and their potential interactions with bacteria [31]. Glyphosate is known to reduce the amount of manganese, magnesium, and calcium in plants due to chelation [32]. This renders a possible interaction of glyphosate with the media used for MIC determination.
Different Media
Since bacteria could balance a lack of aromatic amino acids with an uptake of free amino acids from a nutrient-rich media to some extent, differences in media composition could also lead to different results. Also, the usage of different media and different methods in general is a possible factor, generating the big differences in MIC values [28]. Zucko et al. [33] investigated 488 prokaryotes sequences for the completeness of the genes for production of aromatic amino acids and came to the conclusion that host-associated bacteria may not process a complete shikimate pathway but instead gather aromatic amino acids from their host environment. While susceptibility testing in our study was conducted in Mueller Hinton broth, Kurenbach et al. [27] used LB media. Within the publication of Shehata et al. [20], it is not stated which media were used. The most comparable MICs, as mentioned before, were obtained by Nielsen et al. [28], who cultivated bacteria anaerobically and used Brain Heart Infusion broth and Reinforced Clostridial Medium. Overall, comparability of the results has to be considered to be limited if they were produced using different herbicide formulations and according to different protocols.
Time of Isolation
We compared MICs of Salmonella enterica isolates before and after the rise of the herbicide glyphosate in the 1990s [1]. Isolates collected before 1981 had significant lower MIC values for GLY and RU than those collected from 2014 to 2016. However, this does not necessarily imply that the decrease in sensitivity against both tested solutions is due to the vast increase of glyphosate usage leading to more resistant recent Salmonella enterica strains compared to historical strains.
Antimicrobials and other potential stressors like residues of pesticides [27] and biocides [34] are able to cause an increase in persistence and a decrease in susceptibility in microorganisms against different antibiotic agents [27]. The decrease in susceptibility does not necessarily require a change in the specifically targeted structures. It could be due to general mechanisms against stressors as well. The occurrence of co- and cross-resistance enables a decrease in susceptability against certain agents even when they are absent. Hence, not only an increase in residues of glyphosate itself within the environment could have led to a decrease of susceptibility against the herbicide, but also the increasing burden of residues of further active substances.
For example, MIC increase could be an increased tolerance due to unspecific modifications, like overexpression of multidrug-resistance efflux pumps [35]. This was recently shown for Enterobacteriaceae and biocides [36]. Bailey et al. [37] revealed an increase in tolerance of Salmonella Typhimurium after short time exposure to the biocide triclosan, which was due to an overexpression of efflux pumps.
Similar to glyphosate, the use of biocides increased in many environments within the last decades [38]. Especially in food [39] and farm surroundings, low biocides concentrations could be detected. It was stated that the presence of residues provides an environment, in which the selection of isolates with increased tolerance to different agents is potentially favored [40, 41]. As shown by Karatzas et al. [34], Salmonella species that survive at low doses of biocides are more likely to be resistant against antibiotics. The group concluded that a high MIC of biocides in Salmonella can lead to a selection for antibiotic-resistant isolates. This can for example happen due to co-selection, also in food-borne pathogens [42]. Furthermore, Parikh et al. [43] even detected a linked resistance between biocides and antibiotics. Whitehead et al. [44] showed also that high doses of biocides could lead to the selection of multidrug-resistant Salmonella enterica strains.
Another example specific for the livestock sector is the use of heavy metals like copper and zinc as growth promotors [45]. The heavy usage of these animal food components can lead to different modifications in bacteria, for example co-resistances [46] and enrichment of certain bacteria in the animal intestine due to other mechanisms [47].
Differences in MIC for Different Host Species
Our study revealed significantly higher MICs for glyphosate in pigs than in poultry. However, the differences are not highly significant. Moreover, host-dependent differences in MICs were not detected when using the formulation RU. An explanation for higher MIC values in pigs than in poultry could be a greater uptake of glyphosate residues with feed. Compared to poultry, daily feed rations for pigs are much higher, and they additionally have a much longer lifespan. A comparison of the results for RU as a complex formulation is always more complicated than with glyphosate as a pure substance due to the lack of knowledge of the additional ingredients in the formulations. In general, the results obtained for the different host species are very similar. A further investigation, comprising also species-specific Salmonella serovars (e.g. S. Pullorum for poultry or S. Derby for pigs), the analysis of a bigger sample set, or the comparison of the pig isolates with samples from only one poultry species (e.g. chicken) could potentially lead to more pronounced differences in results.
Differences in MIC for the Different Serovars of Salmonella enterica
Classification of Salmonella enterica according to serovars is a very important tool that relies on phenotypic properties only. For most serovars, the specific genetic background causing the phenotypic differences is not investigated. Nonetheless, the classification of the serovars is based on the differences in O and H antigens, which results in modified lipopolysaccharides (LPS) on the cell surface or flagella. This can lead to variations in polarization of cell surface [48] or other differences, which change the susceptibility for chemicals and other active substances [49]. In this study, the serovar Typhimurium provided higher MICs compared with the other two serovars Enteritidis and Infantis. However, based on the data obtained within the study presented here, this assumption cannot sufficiently be investigated. For proving the possible impact of differences of the cell surface, further investigations, like a comparison of MICs of closely related serovars, should be carried out. Also, whole genome sequencing would be required to properly address this issue.
Since the serovar Typhimurium is the predominant serovar in pigs [50] but not in poultry [51], differences in the serovars can be biased by this fact. This explanation would be in concordance with the assumption that glyphosate intake of pigs is higher than that of poultry, due to the longer lifespan and higher daily feed rations. Hence, it is conceivable that Enterobacteriaceae get in contact with glyphosate more frequently and at potentially higher doses, leading to a higher risk for the development of resistance. In addition, this matches our results of MICs being higher in pigs than in poultry. Underlining that our statistical models should exclude the mutual influences within the analyzed variables the dataset could still be too small to show this relationship.
However, the glyphosate residues detected in poultry and pig feed [20, 52] were considerably lower than the MIC.
Conclusion
To the best our knowledge, this is the first large scale study that systematically assessed glyphosate sensitivity in Salmonella enterica isolates of animal origin. We demonstrated significant differences between the MICs before and after the massive increase of glyphosate usage in agriculture. Further investigations are needed to show causality between the increase of glyphosate tolerance and the usage of glyphosate or other chemicals, antibiotics, or heavy metals in agriculture. Furthermore, resistance mechanisms behind the increased MIC should be clarified, and whether they are transferable between bacteria. In addition, investigations are needed to determine the impact of an increased resistance against glyphosate on the occurrence of pathogenic, zoonotic, and commensal bacteria, as well as on the composition of bacterial communities, especially for food-producing animals and for the environment.
Acknowledgements
We would like to thank the German Federal Institute for Risk Assessment for providing the Salmonella isolates, especially Dr. Istvan Szabo and the National Reference Laboratory for the Analysis and Testing Zoonoses (Salmonella). Many thanks to Nicole Roschanski for scientific advice and Maja Thieck and Diana Steinke for excellent technical support.
Footnotes
Funding Sources
This project was supported by funds of the Federal Ministry of Food and Agriculture (BMEL) based on a decision of the Parliament of the Federal Republic of Germany via the Federal Office for Agriculture and Food (BLE) (funding code 2815HS015–19). Furthermore, we acknowledge support by the German Research Foundation and the Open Access Publication Fund of the Freie Universität Berlin.
Authors’ Contribution
JP performed the experiments, collected, analyzed and interpreted the data, drafted the manuscript and figures with critical evaluation, and supported of all other authors. KB performed the experiments and collected the data. RM contributed to the statistical data analysis and wrote sections of the manuscript. OM and UR conceived and designed the study, as well as critically revised the manuscript. All authors approved the final version to be published.
Conflict of Interest
The authors declare that there is no conflict of interest.
References
- 1.Duke SO, Powles SB. Mini-review Glyphosate: a once-in-a-century herbicide. Pest Manag Sci. 2008;64:319–25. [DOI] [PubMed] [Google Scholar]
- 2.Duke SO. The history and current status of glyphosate. Pest Manag Sci. 2017:9–24. [DOI] [PubMed] [Google Scholar]
- 3.Vera MS, Di Fiori E, Lagomarsino L, Sinistro R, Escaray R, Iummato MM, et al. Direct and indirect effects of the glyphosate formulation Glifosato Atanor® on freshwater microbial communities. Ecotoxicology. 2012;21:1805–16. [DOI] [PubMed] [Google Scholar]
- 4.Pizarro H, Vera MS, Vinocur A, Pérez G, Ferraro M, Menéndez Helman RJ, et al. Glyphosate input modifies microbial community structure in clear and turbid freshwater systems. Environ Sci Pollut Res. 2015;23:5143–53. [DOI] [PubMed] [Google Scholar]
- 5.Pérez AGL, Torremorell A, Mugni H, Rodriguez P, Vera MS, Allende L, et al. Effects of the Herbicide Roundup on Freshwater Microbial Communities: A Mesocosm study. Ecol Appl. 2007;17:2310–22. [DOI] [PubMed] [Google Scholar]
- 6.Sánchez-de León Y, De Melo E, Soto G, Johnson-Maynarrd J, Lugo-Pérez J. Earthworm Populations, Microbial Biomass and Coffee Production in Different Experimental Agroforestry Management Systems in. Caribb J Sci. 2006;42:397–409. [Google Scholar]
- 7.García-Pérez JA, Alarcón-Gutiérrez E, Perroni Y, Barois I. Earthworm communities and soil properties in shaded coffee plantations with and without application of glyphosate. Appl Soil Ecol. 2014;83:230–7. [Google Scholar]
- 8.Steinrücken HC, Amrhein N. The herbicide glyphosate is a potent inhibitor of 5-enolpyruvylshikimic acid-3-phosphate synthase. Biochem Biophys Res Commun. 1980;94:1207–12. [DOI] [PubMed] [Google Scholar]
- 9.Schönbrunn E, Eschenburg S, Shuttleworth WA, Schloss J V, Amrhein N, Evans JN, et al. Interaction of the herbicide glyphosate with its target enzyme 5-enolpyruvylshikimate 3-phosphate synthase in atomic detail. Proc Natl Acad Sci U S A. 2001;98:1376–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amrhein N, Johänning D, Schab J, Schulz A. Biochemical basis for glyphosate-tolerance in a bacterium and a plant tissue culture. Fed Eur Biochem Soc. 1983;157. [Google Scholar]
- 11.Herrmann KM, Weaver LM. The Shikimate Pathway. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:473–503. [DOI] [PubMed] [Google Scholar]
- 12.Fei YY, Gai JY, Zhao TJ. Identification of regulated genes conferring resistance to high concentrations of glyphosate in a new strain of Enterobacter. FEMS Microbiol Lett. 2013;349:135–43. [DOI] [PubMed] [Google Scholar]
- 13.Lorenzatti E, Maitre M, Lenardón A, Lajmanovich R, Peltzer P, Anglada M. Pesticide residues in immature soybean in Argentina croplands. Fresen Environ Bull. 2004;13:675–8. [Google Scholar]
- 14.Bøhn T, Cuhra M, Traavik T, Sanden M, Fagan J, Primicerio R. Compositional differences in soybeans on the market: Glyphosate accumulates in Roundup Ready GM soybeans. Food Chem. 2014;153:207–15. [DOI] [PubMed] [Google Scholar]
- 15.Dill GM. Glyphosate-resistant crops: History, status and future. Pest Manag Sci. 2005;61:219–24. [DOI] [PubMed] [Google Scholar]
- 16.Krüger M, Schledorn P, Schrödl W, Hoppe H-W, Lutz W, Shehata AA. Detection of Glyphosate Residues in Animals and Humans. J Environ Anal Toxicol. 2014;04:1–5. [Google Scholar]
- 17.Krüger M, Shehata AA, Schrödl W, Rodloff A. Glyphosate suppresses the antagonistic effect of Enterococcus spp. on Clostridium botulinum. Anaerobe. 2013;20:74–8. [DOI] [PubMed] [Google Scholar]
- 18.Ackermann W, Coenen M, Schrödl W, Shehata AA, Krüger M. The influence of glyphosate on the microbiota and production of botulinum neurotoxin during ruminal fermentation. Curr Microbiol. 2014;70:374–82. [DOI] [PubMed] [Google Scholar]
- 19.Riede S, Toboldt A, Breves G, Metzner M, Köhler B, Bräunig J, et al. Investigations on the possible impact of a glyphosate-containing herbicide on ruminal metabolism and bacteria in vitro by means of the ‘Rumen Simulation Technique. J Appl Microbiol. 2016;121:644–56. [DOI] [PubMed] [Google Scholar]
- 20.Shehata AA, Schrödl W, Aldin AA, Hafez HM, Krüger M. The effect of glyphosate on potential pathogens and beneficial members of poultry microbiota in vitro. Curr Microbiol. 2013;66:350–8. [DOI] [PubMed] [Google Scholar]
- 21.Clair E, Linn L, Travert C, Amiel C, Séralini GE, Panoff JM. Effects of roundup® and glyphosate on three food microorganisms: Geotrichum candidum, Lactococcus lactis subsp. cremoris and lactobacillus delbrueckii subsp. bulgaricus. Curr Microbiol. 2012;64:486–91. [DOI] [PubMed] [Google Scholar]
- 22.(http://www.who.int/news-room/fact-sheets/detail/salmonella-(non-typhoidal)27.06.2018).
- 23.Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O’Brien SJ, et al. The Global Burden of Nontyphoidal Salmonella Gastroenteritis. Clin Infect Dis. 2010;50:882–9. [DOI] [PubMed] [Google Scholar]
- 24.Hauser E, Tietze E, Helmuth R, Junker E, Blank K, Prager R, et al. Pork contaminated with Salmonella enterica serovar 4,[5], 12:i:-, an emerging health risk for humans. Appl Environ Microbiol. 2010;76:4601–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiegand I, Hilpert K, Hancock REW. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc. 2008;3:163–75. [DOI] [PubMed] [Google Scholar]
- 26.Walzl A, Kramer N, Mazza G, Rosner M, Falkenhagen D, Hengstschläger M. A Simple and Cost Efficient Method to Avoid Unequal Evaporation in Cellular Screening Assays, Which Restores Cellular Metabolic Activity. Int J Appl Sci Technol. 2012;2:17–22. [Google Scholar]
- 27.Kurenbach B, Marjoshi D, Amábile-cuevas CF, Ferguson GC, Godsoe W, Gibson P, et al. Sublethal Exposure to Commercial Formulations of the Herbicides Changes in Antibiotic Susceptibility in Escherichia coli and Salmonella enterica serovar Typhimurium. MbioAsmOrg. 2015;6:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nielsen LN, Roager HM, Casas ME, Frandsen HL, Gosewinkel U, Bester K, et al. Glyphosate has limited short-term effects on commensal bacterial community composition in the gut environment due to sufficient aromatic amino acid levels. Environ Pollut. 2018;233:364–76. [DOI] [PubMed] [Google Scholar]
- 29.Mesnage R, Defarge N, Spiroux de Vendômois J, Séralini GE. Potential toxic effects of glyphosate and its commercial formulations below regulatory limits. Food Chem Toxicol. 2015;84:133–53. [DOI] [PubMed] [Google Scholar]
- 30.Powell HA, Kerbby NW, Rowell P. Natural tolerance of cyanobacteria to the herbicide glyphosate. New Phytol. 1991;119:421–6. [Google Scholar]
- 31.Benbrook CM. Why Regulators Lost Track and Control of Pesticide Risks: Lessons From the Case of Glyphosate-Based Herbicides and Genetically Engineered-Crop Technology. Curr Environ Heal Reports. 2018;1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cakmak I, Yazici A, Tutus Y, Ozturk L. Glyphosate reduced seed and leaf concentrations of calcium, manganese, magnesium, and iron in non-glyphosate resistant soybean. Eur J Agron. 2009;31:114–9. [Google Scholar]
- 33.Zucko J, Dunlap WC, Shick JM, Cullum J, Cercelet F, Amin B, et al. Global genome analysis of the shikimic acid pathway reveals greater gene loss in host-associated than in free-living bacteria. BMC Genomics. 2010;11:628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karatzas KAG, Randall LP, Webber M, Piddock LJV, Humphrey TJ, Woodward MJ, et al. Phenotypic and proteomic characterization of multiply antibiotic-resistant variants of Salmonella enterica serovar typhimurium selected following exposure to disinfectants. Appl Environ Microbiol. 2008;74:1508–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blanco P, Hernando-Amado S, Reales-Calderon J, Corona F, Lira F, Alcalde-Rico M, et al. Bacterial Multidrug Efflux Pumps: Much More Than Antibiotic Resistance Determinants. Microorganisms. 2016;4:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karatzas KAG, Webber MA, Jorgensen F, Woodward MJ, Piddock LJV, Humphrey TJ. Prolonged treatment of Salmonella enterica serovar Typhimurium with commercial disinfectants selects for multiple antibiotic resistance, increased efflux and reduced invasiveness. J Antimicrob Chemother. 2007;60:947–55. [DOI] [PubMed] [Google Scholar]
- 37.Bailey AM, Constantinidou C, Ivens A, Garvey MI, Webber MA, Coldham N, et al. Exposure of Escherichia coli and Salmonella enterica serovar Typhimurium to triclosan induces a species-specific response, including drug detoxification. J Antimicrob Chemother. 2009;64:973–85. [DOI] [PubMed] [Google Scholar]
- 38.Scenhir Scientific Committee on Emerging and Newly Identified Health Risks. Eur Commisson Brussles, Belgium. 2009:1–87. [Google Scholar]
- 39.Condell O, Iversen C, Cooney S, Power KA, Walsh C, Burgess C, et al. Efficacy of biocides used in the modern food industry to control Salmonella enterica, and links between biocide tolerance and resistance to clinically relevant antimicrobial compounds. Appl Environ Microbiol. 2012;78:3087–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abrantes MC, Fátima Lopes Mde, Kok J. Impact of manganese, copper and zinc ions on the transcriptome of the nosocomial pathogen Enterococcus faecalis V583. PLoS One. 2011;6:e26519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coelho JR, Carrico JA, Knight D, Martinez JL, Morrissey I, Oggioni MR, et al. The Use of Machine Learning Methodologies to Analyse Antibiotic and Biocide Susceptibility in Staphylococcus aureus. PLoS One. 2013;8:e5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wales A, Davies R. Co-Selection of Resistance to Antibiotics, Biocides and Heavy Metals, and Its Relevance to Foodborne Pathogens. Antibiotics. 2015;4:567–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parikh SL, Xiao G, Tonge PJ. Inhibition of InhA, the enoyl reductase from Mycobacterium tuberculosis, by triclosan and isoniazid. Biochemistry. 2000;39:7645–50. [DOI] [PubMed] [Google Scholar]
- 44.Whitehead RN, Overton TW, Kemp CL, Webber MA. Exposure of salmonella enterica serovar Typhimurium to high level biocide challenge can select multidrug resistant mutants in a single step. PLoS One. 2011;6:e22833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu W, Li L, Chen M, Zhou Z, Zhang W, Ping S, et al. Genome-wide transcriptional responses of Escherichia coli to glyphosate, a potent inhibitor of the shikimate pathway enzyme 5-enolpyruvylshikimate-3-phosphate synthase. Mol Biosyst. 2013;9:522–30. [DOI] [PubMed] [Google Scholar]
- 46.Yazdankhah S, Rudi K, Bernhoft A. Zinc and copper in animal feed - development of resistance and co-resistance to antimicrobial agents in bacteria of animal origin. Microb Ecol Heal Dis. 2014;1:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bednorz C, Oelgeschläger K, Kinnemann B, Hartmann S, Neumann K, Pieper R, et al. The broader context of antibiotic resistance: Zinc feed supplementation of piglets increases the proportion of multi-resistant Escherichia coli in vivo. Int J Med Microbiol. 2013;303:396–403. [DOI] [PubMed] [Google Scholar]
- 48.Hermansson M, Kjelleberg S, Korhonen TK, Stenström T-A. Hydrophobic and Electrostatic Characterization of Surface Structures of Bacteria and Its Relationship to Adhesion to an Air-Water Interface. Arch Microbiol. 1982;308–12.6217795 [Google Scholar]
- 49.Matamouros S, Miller S. S. Typhimurium strategies to resist killing by cationic antimicrobial peptides. Biochim Biophys Acta – Biomembr. 2015;1848:3021–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fedorka-Cray PJ, Gray JT, Wray C. Salmonella Infections in Pigs. Wray C, Wray A, editors. Salmonella in Domestic Animals; New York; 2000. p. 191–207. [Google Scholar]
- 51.Poppe C. Salmonella Infections in the Domestic Fowl. Wray C, Wray A, editors. Salmonella in Domestic Animals; New York; 2000. p. 107–32. [Google Scholar]
- 52.(EFSA) EFSA. Evaluation of the impact of glyphosate and its residues in feed on animal health. EFSA J. 2018. May 1;16:e05283. [DOI] [PMC free article] [PubMed] [Google Scholar]


