Abstract
A G‐rich sequence found in the regulatory region of the RANKL gene, which is associated with homeostasis of bone metabolism, folds into a two‐quartet basket‐type G‐quadruplex stabilized by A⋅G⋅A and G⋅G⋅G base‐triads. Perusal of local structural features together with G/A‐to‐T modifications uncovered the critical role of A5 for the formation of a distinct antiparallel two‐quartet topology and not the three‐quartet topology that would be expected based on the sequence with four GGG‐tracts alone. The structural changes induced by the A5‐to‐T5 modification include a switch in orientation and relative positions of G‐strands that together with anti to syn reorientation of G12 provide insights into the complexity of the interactions that influence the folding of G‐rich DNA. Understanding the impact of loop residues on the stability and formation of G‐quadruplexes advances our knowledge and ability to predict structures adopted by G‐rich sequences, which are involved in regulatory mechanisms in the cell, and may also facilitate drug design.
Keywords: DNA, folding, G-quadruplexes, NMR spectroscopy, structure
G‐rich DNA sequences have been shown to fold into four‐stranded G‐quadruplex structures in vitro and in cells, which points to their biological significance.1 Sequences able to form G‐quadruplexes are predominantly localized in human telomeres and regulatory regions of many (onco)genes, in which they can modulate telomere maintenance, gene expression, DNA replication, and other essential cellular processes.2 The structures of G‐quadruplexes, apart from various environmental conditions, such as cation concentration, pH, temperature, and molecular crowding, highly depend on the particular G‐rich sequence.3 The most characteristic structural element of G‐quadruplexes is the G‐quartet, a planar alignment of four Hoogsteen‐type hydrogen‐bonded guanines. G‐quartets demonstrate stacking of guanine residues in several geometries that affect topological properties, cation binding affinities, and stability of the overall structure, and at the same time, pertain to the orientation of the four G‐rich strands constituting the G‐quadruplex. However, the great structural polymorphism of G‐quadruplexes predominantly arises from the numerous combinations of regions that connect the G‐quartet‐forming guanines and comprise different types of loops, which can vary in length and sequence.1, 4 Minor modifications in loop sequences can result in dramatic changes of structure,5 which is not surprising owing to the frequent involvement of loop residues in interactions such as base‐pairing and/or stacking.6 Approximately a decade ago, the first sequences consisting of four tracts of three consecutive guanines were shown to form two‐quartet G‐quadruplexes.6b,6c The overall folding of these structures is defined by the interactions of extra G‐quartet residues rather than by maximizing the number of G‐quartets.
A G‐rich sequence has been located in the regulatory region of the RANKL gene, whose excessive activity leads to unbalanced bone remodeling and can influence the incidence of osteoporosis.7 The wild‐type sequence, d(G 4AG 3AGCG 3AGAG 3), consists of three G‐tracts with three successive guanine residues and one G‐tract with four guanine residues, which presumably causes the structural polymorphism that prohibits structural studies of individual G‐quadruplexes (Supporting Information, Figure S1). Simple G4‐to‐T4 modification results in a G‐rich sequence with four GGG‐tracts, d(G 3TAG 3AGCG 3AGAG 3), hereafter designated as Ran4, which folds into a single G‐quadruplex in the presence of K+ ions. A high‐resolution structure of Ran4 reveals the formation of a two‐quartet G‐quadruplex with numerous base‐pairing and stacking interactions involving loop residues on both sides of the G‐quartet core. Structural changes induced by the modifications of the extra G‐quartet residues with thymine residues revealed that the two‐quartet antiparallel topology of Ran4 critically depends on the interactions of a single‐loop adenine. Modulation of the specific loop interactions involving the adenine residue leads to exciting structural changes including a two‐to‐three‐quartet structural switch.
A 1D 1H NMR spectrum of Ran4 recorded at 5 °C in the presence of 70 mm KCl and 15 mm potassium phosphate buffer (pH 7.0) shows six resolved signals between δ=11.5 and 12.0 ppm corresponding to eight imino proton resonances consistent with the formation of a single two‐quartet G‐quadruplex (Figure 1 A). Additional resonances at δ=10.7, 10.9, 11.3 and 12.9 ppm imply the involvement of respective residues in hydrogen bonds thus defining extra G‐quartet structural elements. The latter four signals broaden to the baseline upon increase of sample's temperature presumably owing to accelerated exchange of imino protons with the solvent, while the signals between δ=11.5 and 12.0 ppm remain narrow even at 45 °C, which is in agreement with a more robust structure of the G‐quartet core (Figure S2). G‐quartets consist of residues G1⋅G13⋅G19⋅G7 and G2⋅G6⋅G18⋅G14 with clockwise and anti‐clockwise hydrogen bond directionalities, respectively (Figure 1; for unambiguous assignment of imino protons see Figure S3). The antiparallel G‐strands of Ran4 form wide, narrow, and two medium grooves (Figures 1 B) and are linked by one diagonal and two lateral loops (Figure 2 A). The first lateral loop progresses anti‐clockwise, while the progression of the second is clockwise, forming a (−ld+l) basket‐type topology.4 Intense intraresidual NOE cross‐peaks in the anomeric‐aromatic region of the NOESY spectrum and downfield chemical shift of C8 carbon atoms at δC≈140 ppm indicate syn glycosidic conformations for G1, G6, G13, and G18.
Figure 1.
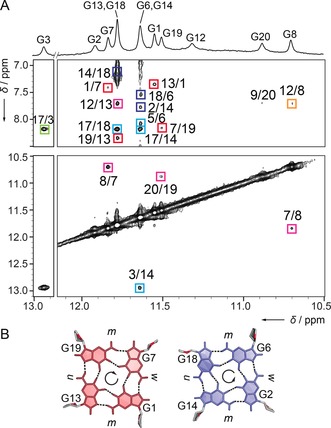
A) The imino region of 1D 1H NMR spectrum of Ran4 at 5 °C (top) and imino–aromatic and imino–imino regions of 2D NOESY spectrum (mixing time of 300 ms). Cross‐peaks labeled with green, blue, red, and orange rectangles correspond to the A5⋅G3⋅A17 base‐triad, G2⋅G6⋅G18⋅G14 quartet, G1⋅G13⋅G19⋅G7 quartet, and G20⋅G8⋅G12 base‐triad, respectively. Cross‐peaks labeled with cyan and magenta rectangles correspond to NOE connections between base‐triads and G‐quartets. B) Clockwise and anti‐clockwise hydrogen bond directionalities in G1⋅G13⋅G19⋅G7 (left) and G2⋅G6⋅G18⋅G14 (right) quartets, respectively. Labels m, w, and n stand for medium, wide, and narrow grooves, respectively.
Figure 2.
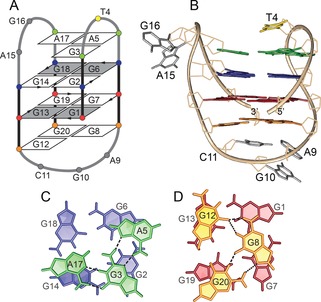
A) Schematic of a two‐quartet antiparallel basket‐type topology adopted by Ran4. Residues in syn and anti glycosidic conformations are represented with grey and white rectangles, respectively. G1⋅G13⋅G19⋅G7 and G2⋅G6⋅G18⋅G14 quartets are highlighted in red and blue, respectively. A5⋅G3⋅A17 and G20⋅G8⋅G12 base‐triads are presented in green and orange, respectively. T4 is shown in yellow, while other loop residues are coloured grey. B) The lowest‐energy structure of Ran4 (PDB id 6GZN). Stacking interactions between C) A5⋅G3⋅A17 base‐triad (green) and G2⋅G6⋅G18⋅G14 quartet (blue) and between D) G20⋅G8⋅G12 base‐triad (orange) and G1⋅G13⋅G19⋅G7 quartet (red). Hydrogen bonds connecting base‐triads are depicted with black dashed lines.
Several imino‐aromatic NOE contacts between the loop and G‐quartet‐forming residues corroborate formation of A5⋅G3⋅A17 and G20⋅G8⋅G12 base‐triads stacked on the G‐quartet core (Figures 1 A and 2; for a detailed presentation of NOE contacts and base‐pairing arrangements see Figure S4). Furthermore, formation of the A5⋅G3⋅A17 base‐triad is supported by the chemical shift of G3 H1 at δ=12.9 ppm and the NOE contact between G3 H1 and A17 H2. On the other hand, the NOE contact between G12 H8 and G8 H1 corroborates the relative orientations of these two residues within the G20⋅G8⋅G12 base‐triad. In addition to the residues constituting base‐triads, loop residues T4, A9, G10, and C11 are well defined and stacked either above A5⋅G3⋅A17 or under G20⋅G8⋅G12 base‐triads. On the other hand, residues A15 and G16 are exposed to the solvent and represent the most dynamic part of the molecule (Figure 2 B; for NMR restraints and structure statistics see Table S1; for an ensemble of 10 lowest energy structures see Figure S5). A two‐quartet G‐quadruplex core with two additionally stacked layers of base‐triads is in accord with the melting temperature of Ran4, which is higher by approximately 10 °C than that of a two‐quartet G‐quadruplex only, such as TBA8 (Table 1, Figure S6).
Table 1.
Ran4 and modified sequences with their melting temperatures.
| modification | sequence[a] | T m [°C][b] |
|---|---|---|
|
Ran4 |
1 5 10 15 20
GGGTAGGGAGCGGG AGAGGG |
59.5 |
| Ran4G3T | GG TTA GG GAGCG GG AGA GG G | 54.4 |
| Ran4G12T | GG GTA GG GAGCT GG AGA GG G | 48.0 |
| Ran4G3TG12T | GG TTA GG GAGCT GG AGA GG G | 50.0 |
| Ran4A17T | GG GTA GG GAGCG GG AGT GG G | 58.5 |
| Ran4G8T | GG GTA GG TAGCG GG AGA GG G | 50.5 |
| Ran4G20T | GG GTA GG GAGCG GG AGA GG T | 50.7 |
| Ran4A5T | GG GTT GG GAGCG GG AGA GG G | 60.2 |
[a] Guanine residues in bold correspond to G‐quartet‐forming residues in Ran4, while residues in italic participate in base‐triads. Modifications are underlined. [b] Melting temperatures were determined in 70 mm KCl, 10 mm potassium phosphate buffer, pH 7.0 at 295 nm.
Intrigued by the structural details of Ran4, we individually modified adenine and guanine residues from A5⋅G3⋅A17 and G20⋅G8⋅G12 base‐triads to thymine residues to gain insights into the specific roles these residues play in a formation of a two‐quartet G‐quadruplex (Table 1). The results of an attempt to obtain more quantitative insights into the interactions of base‐triads with G‐quartets using a simplified two‐state approximation demonstrated a high level of enthalpy–entropy compensation that does not follow G/A‐to‐T modifications in a straightforward manner and/or suggests intermediates in the folding process (see below).9 However, perusal of 1H NMR and CD spectra (Figure 3 and the Supporting Information, Figures S7–S9) enabled us to categorize the effects of modifications of Ran4 G‐quadruplex into three groups.
Figure 3.
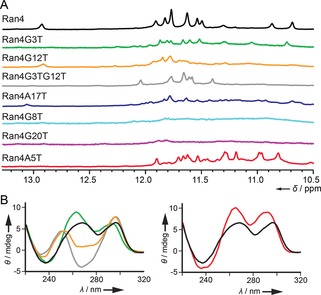
A) Imino regions of 1D 1H NMR spectra of Ran4 and its modified sequences. B) Comparison of CD spectra of Ran4 and modified sequences from the first (left) and third group of modifications (right). NMR and CD spectra of the same oligonucleotide are shown with matching colour.
The first group includes individual modifications of G3 and G12 that disrupt the respective base‐triad, while keeping the two‐quartet G‐quadruplex core intact. Inspired by these observations, we also synthetized an oligonucleotide with double G3‐to‐T3 and G12‐to‐T12 modifications that, interestingly, still formed a stable two‐quartet G‐quadruplex despite disruption of both base‐triads. The second group of modifications comprises G/A‐to‐T modifications of G8, A17, and G20, which were found to preclude the formation of Ran4 G‐quadruplex. The third group includes an A5‐to‐T5 modification, which results in a three‐quartet (3+1) hybrid G‐quadruplex substantiated by 12 signals in the imino region of the 1H NMR spectrum (Figure 3 A) and several imino–imino, imino–aromatic and aromatic–aromatic NOE connectivities (for a more in‐depth discussion of the topology determination, see Figures S10–S13).
Comparison of the Ran4 and Ran4A5T topologies discloses significant structural changes that arise upon A5‐to‐T5 modification and lead from a two‐quartet basket‐type G‐quadruplex to a three‐quartet (3+1) hybrid structure (Figure 4). Antiparallel G‐strands I and II connected with a lateral loop in Ran4 are in Ran4A5T oriented parallel to each other and are linked by a propeller‐type loop. Furthermore, the diagonal middle loop that connects G‐strands II and III in Ran4 adopts a lateral‐type topology in Ran4A5T. Changes in the orientation of the first and second loop upon A5‐to‐T5 modification are governed by the interactions correlating loop residues that bridge G‐strands I and II (Figure S14). Formation of the first lateral loop in Ran4 G‐quadruplex depends on the interactions of loop adenine with residues in A5⋅G3⋅A17 base‐triad. Although the A5⋅G3 base‐pair formally extends one side of the G‐quadruplex, guanine residues from G‐strand I of Ran4 can, owing to the involvement of G3 in the interaction with A5, participate in the formation of only two G‐quartets. Upon A5‐to‐T5 modification, no base‐pair interactions are observed between G3 and neighboring loop residues, allowing G3 to engage in a G‐quartet. Consequently, the shorter linker between G‐strands I and II in Ran4A5T composed of residues T4 and T5 leads to an energetically preferable propeller loop. As a result, G‐strands I and II in Ran4A5T change their orientations and relative positions with respect to the G‐strands III and IV. While G‐strand II in Ran4 is closer to G‐strand IV leading to a diagonal middle loop, it is adjacent to G‐strand III in Ran4A5T resulting in a lateral middle loop. Moreover, G3 becomes part of the G3⋅G8⋅G12⋅G20 quartet, which together with G1⋅G18⋅G14⋅G6 and G2⋅G7⋅G13⋅G19 quartets define the three‐layered G‐quartet core of Ran4A5T. The loop composed of residues A15–G16–A17 that connects G‐strands III and IV adopts a lateral‐type orientation and spans a narrow groove in both G‐quadruplexes.
Figure 4.
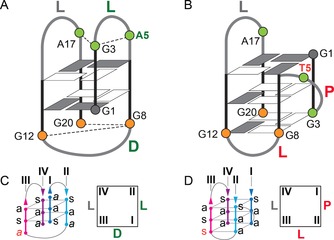
Comparison of A) Ran4 and B) Ran4A5T G‐quadruplex topologies and schematic of orientations and syn/anti patterns along G‐strands and birds‐eye view of C) Ran4 and D) Ran4A5T. Anti and syn guanine residues are presented in (A) and (B) as white and grey rectangles, respectively. Residues that form A5⋅G3⋅A17 base‐triad in Ran4 are coloured green, while residues from G20⋅G8⋅G12 base‐triad in Ran4 are shown in orange. Loop types are marked with L, D, and P for lateral, diagonal, and propeller loop, respectively. G‐strands in (C) and (D) are marked with I, II, III, and IV. Syn and anti glycosidic conformations are marked with s and a, respectively. Glycosidic conformations of guanine residues that are involved in base‐triads are in italic.
G‐quartets in Ran4 G‐quadruplex are characterized by syn:syn:anti:anti and anti:syn:syn:anti arrangements of glycosidic conformations. On the other hand, Ran4A5T exhibits a syn:syn:anti:syn and two anti:anti:syn:anti arrangements of glycosidic conformations within its G‐quartets (Figure 4). Interestingly, despite clear differences in the glycosidic bond orientations within G‐quartets, Ran4 and Ran4A5T share a very similar syn/anti distribution of guanine conformations along the G‐strands (Figure 4 C,D). Both structures exhibit syn—anti–anti glycosidic conformations along G‐stands I, II, and IV. The only reorganization is observed along G‐strand III, in which a conformational flip of G12 results in a change of anti–syn–anti glycosidic conformations in Ran4 to syn–syn–anti in Ran4A5T. Perusal of high‐resolution structure of Ran4 suggests that G12 could reorient easily to a syn glycosidic conformation without inducing major structural perturbations. Hypothetically, a simple anti–syn reorientation of G12 would bring its imino and amino protons in close proximity to G20 N7 and the carbonyl group. Such positioning of G12 and G20 is indeed observed in the Ran4A5T G‐quadruplex, in which they form a N1‐carbonyl, N7‐amino base‐pair as part of a G3⋅G8⋅G12⋅G20 quartet.
Details of the two topologies that, in fact, exhibit similar thermal stabilities (Table 1) are thought provoking. As G‐quadruplexes result from a kinetically driven assembly, it is exciting to consider the roles of structural elements in the stabilization of individual folds in relation to the crucial interactions that may be at the cross‐roads of assembly of four G‐strands into a well‐defined structure. Folding of the intramolecular G‐quadruplexes leading to antiparallel and (3+1) hybrid topologies may progress through various intermediates such as hairpins and G‐triplexes.10 Structural differences between G‐quadruplexes adopted by Ran4 and Ran4A5T inspired us to propose possible folding pathways that start from similar hairpins comprising G‐strands III and IV and are followed by addition of G‐strand II to form a G‐triplex (Figure S14). Involvement of A5 in the interactions with G3 finally defines the relative positions of G‐strands I and II in Ran4 and Ran4A5T G‐quadruplexes. Additionally, a nearly identical distribution of syn/anti guanine residues along G‐strands in Ran4 and Ran4A5T suggests that their folding routes diverge at the point where glycosidic conformations are already defined.10b,10e Similar modification‐induced changes in folding pathways that start from a common G‐triplex intermediate were observed for a G‐quadruplex with C2′‐fluorinated G‐quartet‐forming guanine.11
In‐depth knowledge of the effect of extra G‐quartet interactions on the G‐quadruplex folding is of special importance since our results, together with the recent literature data,12 imply that loop adenine residues may act as a universal structural switch that narrows a broad range of putative folds into an antiparallel conformation. Sequence‐dependent folding of constructs with AAA, TAA, TTA, and TTT loops revealed that adenine residues in the loops favor topologies with two pairs of antiparallel G‐strands.12a Substitutions of adenine residues involved in the TTA loops of G‐quadruplexes adopted by telomeric repeats of mammals with abasic sites lead to an increase in the number of G‐strands in parallel orientation.12b,12c Interestingly, helicases such as DHX36 exhibit conformation‐specific binding with preference for G‐quadruplexes with parallel G‐strands.12a, 13 The results of the present study suggest that adenine‐driven topological changes may considerably alter the molecular basis of G‐quadruplex recognition by helicases.13
Our structural study, to the best of our knowledge, is the first to suggest that expression of the RANKL gene, which is considerably upregulated in osteoporosis,7 may be regulated by putative folding of its G‐rich region into G‐quadruplexes. Moreover, structural polymorphism of the wild type sequence indicates that the formation of different G‐quadruplexes in the regulatory region of RANKL gene might act as a switch that can influence binding of various protein partners and consequently affect the incidence of bone‐related diseases.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary
Acknowledgements
The authors acknowledge the financial support from Slovenian Research Agency (research core funding No. P1‐242, J1‐6733 and J3‐7245) and the CERIC‐ERIC Consortium for access to experimental facilities.
M. Lenarčič Živković, J. Rozman, J. Plavec, Angew. Chem. Int. Ed. 2018, 57, 15395.
References
- 1.
- 1a. Phan A. T., Kuryavyi V., Luu K. N., Patel D. J. in Quadruplex Nucleic Acids (Eds.: S. Neidle, S. Balasubramanian), RSC Publishing, Cambridge, 2006, pp. 81–99; [Google Scholar]
- 1b. Burge S., Parkinson G. N., Hazel P., Todd A. K., Neidle S., Nucleic Acids Res. 2006, 34, 5402; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1c. Biffi G., Tannahill D., McCafferty J., Balasubramanian S., Nat. Chem. 2013, 5, 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.
- 2a. Todd A. K., Johnston M., Neidle S., Nucleic Acids Res. 2005, 33, 2901; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2b. Huppert J. L., Balasubramanian S., Nucleic Acids Res. 2005, 33, 2908; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2c. Huppert J. L., Balasubramanian S., Nucleic Acids Res. 2007, 35, 406; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2d. Chambers V. S., Marsico G., Boutell J. M., Di Antonio M., Smith G. P., Balasubramanian S., Nat. Biotechnol. 2015, 33, 877. [DOI] [PubMed] [Google Scholar]
- 3.
- 3a. Do N. Q., Phan A. T., Chem. Eur. J. 2012, 18, 14752; [DOI] [PubMed] [Google Scholar]
- 3b. Nakano S., Miyoshi D., Sugimoto N., Chem. Rev. 2014, 114, 2733; [DOI] [PubMed] [Google Scholar]
- 3c. Galer P., Wang B., Šket P., Plavec J., Angew. Chem. Int. Ed. 2016, 55, 1993; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2016, 128, 2033; [Google Scholar]
- 3d. Dickerhoff J., Weisz K., Angew. Chem. Int. Ed. 2015, 54, 5588; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2015, 127, 5680; [Google Scholar]
- 3e. Brčić J., Plavec J., Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1237; [DOI] [PubMed] [Google Scholar]
- 3f. Tateishi-Karimata H., Kawauchi K., Sugimoto N., J. Am. Chem. Soc. 2018, 140, 642. [DOI] [PubMed] [Google Scholar]
- 4.
- 4a. Webba da Silva M., Chem. Eur. J. 2007, 13, 9738; [DOI] [PubMed] [Google Scholar]
- 4b. Karsisiotis A. I., O'Kane C., Webba da Silva M., Methods 2013, 64, 28. [DOI] [PubMed] [Google Scholar]
- 5.
- 5a. Črnugelj M., Šket P., Plavec J., J. Am. Chem. Soc. 2003, 125, 7866; [DOI] [PubMed] [Google Scholar]
- 5b. Dai J., Carver M., Punchihewa C., Jones R. A., Yang D., Nucleic Acids Res. 2007, 35, 4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.
- 6a. Dai J., Punchihewa C., Ambrus A., Chen D., Jones R. A., Yang D., Nucleic Acids Res. 2007, 35, 2440; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6b. Lim K. W., Amrane S., Bouaziz S., Xu W., Mu Y., Patel D. J., Luu K. N., Phan A. T., J. Am. Chem. Soc. 2009, 131, 4301; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6c. Hu L., Lim K. W., Bouaziz S., Phan A. T., J. Am. Chem. Soc. 2009, 131, 16824; [DOI] [PubMed] [Google Scholar]
- 6d. Ghimire C., Park S., Iida K., Yangyuoru P., Otomo H., Yu Z., Nagasawa K., Sugiyama H., Mao H., J. Am. Chem. Soc. 2014, 136, 15537. [DOI] [PubMed] [Google Scholar]
- 7.
- 7a. Boyce B. F., Xing L., Curr. Osteoporos. Rep. 2007, 5, 98; [DOI] [PubMed] [Google Scholar]
- 7b. Wada T., Nakashima T., Hiroshi N., Penninger J. M., Trends Mol. Med. 2006, 12, 17; [DOI] [PubMed] [Google Scholar]
- 7c. Walsh M. C., Choi Y., Front. Immunol. 2014, 5, 511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Macaya R. F., Schultze P., Smith F. W., Roe J. A., Feigon J., Proc. Natl. Acad. Sci. USA 1993, 90, 3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.
- 9a. Dailey M. M., Miller M. C., Bates P. J., Lane A. N., Trent J. O., Nucleic Acids Res. 2010, 38, 4877; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9b. Mergny J.-L., Lacroix L., Oligonucleotides 2003, 13, 515. [DOI] [PubMed] [Google Scholar]
- 10.
- 10a. Mashimo T., Yagi H., Sannohe Y., Rajendran A., Sugiyama H., J. Am. Chem. Soc. 2010, 132, 14910; [DOI] [PubMed] [Google Scholar]
- 10b. Stadlbauer P., Trantirek L., T. E. Cheatham III , Koča J., Šponer J., Biochimie 2014, 105, 22; [DOI] [PubMed] [Google Scholar]
- 10c. Gray R. D., Trent J. O., Chaires J. B., J. Mol. Biol. 2014, 426, 1629; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10d. Rajendran A., Endo M., Hidaka K., Sugiyama H., Angew. Chem. Int. Ed. 2014, 53, 4107; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2014, 126, 4191; [Google Scholar]
- 10e. Bian Y., Tan C., Wang J., Sheng Y., Zhang J., Wang W., PLoS Comput. Biol. 2014, 10, e1003562; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10f. Stadlbauer P., Kührova P., Banaš P., Koča J., Bussi G., Trantirek L., Otyepka M., Šponer J., Nucleic Acids Res. 2015, 43, 9626; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10g. Marchand A., Gabelica V., Nucleic Acids Res. 2016, 44, 10999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dickerhoff J., Weisz K., ChemBioChem 2018, 19, 927. [DOI] [PubMed] [Google Scholar]
- 12.
- 12a. Tippana R., Xiao W., Myong S., Nucleic Acids Res. 2014, 42, 8106; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12b. Babinský M., Fiala R., Kejnovská I., Bendářová K., Marek R., Sagi J., Sklenář V., Vorlíčková M., Nucleic Acids Res. 2014, 42, 14031; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12c. Kejnovská I., Bednářová K., Renčiuk D., Dvořáková Z., Školáková P., Trantírek L., Fiala R., Vorlíčková M., Sagi J., Nucleic Acids Res. 2017, 45, 4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.
- 13a. Chen M. C., Murat P., Abecassis K., Ferré-D'Amaré A. R., Balasubramanian S., Nucleic Acids Res. 2015, 43, 2223; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13b. Tippana R., Hwang H., Opresko P. L., Bohr V. A., Myong S., Proc. Natl. Acad. Sci. USA 2016, 113, 8448; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13c. Mendoza O., Bourdoncle A., Boulé J.-B., R. M. Brosh, Jr. , Mergny J.-L., Nucleic Acids Res. 2016, 44, 1989; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13d. Chen M. C., Tippana R., Demeshkina N. A., Murat P., Balasubramanian S., Myong S., Ferré-D'Amaré A. R., Nature 2018, 558, 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary


