Abstract
The yeast “two-component” osmotic stress phosphorelay consists of the histidine kinase, Sln1p, the phosphorelay intermediate, Ypd1p and two response regulators, Ssk1p and Skn7p, whose activities are regulated by phosphorylation of a conserved aspartyl residue in the receiver domain. Dephospho-Ssk1p leads to activation of the hyper-osmotic response (HOG) pathway, whereas phospho-Skn7p presumably leads to activation of hypo-osmotic response genes. The multifunctional Skn7 protein is important in oxidative as well as osmotic stress; however, the Skn7p receiver domain aspartate that is the phosphoacceptor in the SLN1 pathway is dispensable for oxidative stress. Like many well-characterized bacterial response regulators, Skn7p is a transcription factor. In this report we investigate the role of Skn7p in osmotic response gene activation. Our studies reveal that the Skn7p HSF-like DNA binding domain interacts with a cis-acting element identified upstream of OCH1 that is distinct from the previously defined HSE-like Skn7p binding site. Our data support a model in which Skn7p receiver domain phosphorylation affects transcriptional activation rather than DNA binding to this class of DNA binding site.
INTRODUCTION
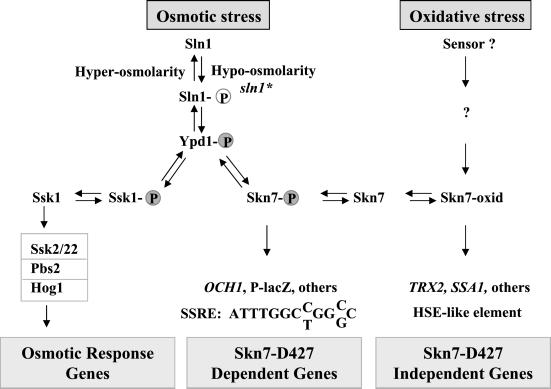
Although two-component signal transduction is a common mechanism for environmental sensing in eubacteria, its use in the yeast, Saccharomyces cerevisiae is restricted to the osmotic and oxidative stress pathways. The two-component molecules in S. cerevisiae work together in a phosophorelay pathway consisting of the sensor-kinase, Sln1p, the phosphorelay molecule, Ypd1p, and a pair of response regulators, Ssk1p and Skn7p, whose activities are modulated by phosphorylation of a conserved aspartyl residue within the receiver domain (Maeda et al., 1994; Posas et al., 1996; Ketela et al., 1998; Li et al., 1998).
One branch of the pathway (SLN1-YPD1-SSK1; Figure 1) is important for the response to hyper-osmotic stress. Under hyper-osmotic stress, Sln1p kinase activity is dampened, and the resultant accumulation of Ssk1p in the unphosphorylated form leads to activation of the HOG1 MAP kinase pathway whose targets include genes involved in the biosynthesis of glycerol, an important compatible osmolyte in yeast, as well as genes involved in other aspects of the osmotic response (Posas et al., 1996). The second branch of the pathway (SLN1-YPD1-SKN7; Figure 1) is activated in response to hypo-osmotic stress (Tao et al., 1999) and is known to be involved in cell wall integrity (Brown et al., 1993, 1994) and cell cycle (Morgan et al., 1995; Bouquin et al., 1999). Under these conditions, the Sln1p kinase is presumed to cause increased levels of phosphorylation of Ypd1p and then the response regulators, Ssk1p and Skn7p (Fassler et al., 1997). Accumulation of phospho-Skn7p leads to the activation of the Mcm1-dependent lacZ reporter gene known as P-lacZ (Li et al., 1998) as well as undefined native targets of the pathway that presumably play a role in adaptation of yeast cells to hypo-osmotic environments (Li et al., 1998).
Figure 1.
Different phospho-forms of Sln1 regulate two separate pathways. HOG1–dependent gene expression is activated upon accumulation of dephosphorylated Sln1 (Sln1) and SKN7-dependent gene expression is activated upon accumulation of phosphorylated Sln1 (Sln1-P) and aspartyl phosphorylation of Skn7p. In response to oxidative stress, Skn7p may undergo an uncharacterized modification or conformational change to generate Skn7-oxid. As described in the body of this article, the two forms of Skn7p, Skn7-P, and Skn7-oxid appear to bind different sequence elements. These are represented at the bottom of the figure as the SSRE (sln1 star [sln*] response element) and HSE (heat shock element).
In addition to its role in the SLN1–SKN7 osmotic response pathway, Skn7p plays a distinct role in the oxidative stress response pathway (Krems et al., 1996). Its role in the two signal transduction pathways appears to involve different activation mechanisms, because the oxidative stress pathway is independent of the phospho-accepting Asp-427 (D427), whereas the SLN1–SKN7 pathway is dependent on this residue (Morgan et al., 1997; Li et al., 1998). Work described in this article supports the conclusion depicted in Figure 1 that the DNA sequence element required for the SLN1-dependent role of Skn7p is distinct from the element involved in the SLN1-independent role of Skn7p in oxidative stress.
The majority of bacterial response regulators have a DNA binding domain in addition to a receiver domain. In response to aspartyl phosphorylation, these receivers undergo a change in conformation that alters their function and accounts for the novel pattern of gene expression that occurs in response to a specific stimulus (Hakenbeck and Stock, 1996). A small number of bacterial response regulators do not have associated transcription factor activity but are instead coupled to an enzymatic activity. For example, CheB, a response regulator in the chemotaxis pathway, has phosphodiesterase activity (Amsler and Matsumura, 1995). Alternatively, a response regulator may mediate protein–protein interaction. For example, phosphorylation of the CheY response regulator regulates its interaction with the motor protein complex (Macnab, 1995; Shukla et al., 1998). In the case of the yeast response regulator, Ssk1p, aspartyl phosphorylation controls the accessibility of its protein interaction interface with the downstream MEK kinases, Ssk2p and Ssk22p. Dephospho Ssk1p interacts with and stimulates Ssk2p and Ssk22p activity (Posas and Saito, 1998).
In contrast to Ssk1p, the yeast response regulator Skn7p has a DNA binding domain. This domain was initially recognized by its similarity to the DNA binding domain of heat shock factor, Hsf1p (Brown et al., 1993, 1994; Morgan et al., 1995). Thus, the Skn7p response regulator is a eukaryotic example of the transcription-factor–coupled response regulator common in eubacteria. Skn7p binds to the promoters of the oxidative stress response gene, TRX2, and to genes such as SSA1 containing heat shock elements (Morgan et al., 1997; Raitt et al., 2000). In both cases Skn7p mediates activation in response to oxidative stress. However, oxidative stress activation of SSA1 and TRX2 is independent of the Skn7p receiver domain aspartate D427 whose phosphorylation defines its activity as an Sln1p effector (Morgan et al., 1997; Raitt et al., 2000).
To understand the effect of D427 phosphorylation on the activity of Skn7p, it was first necessary to identify a DNA binding site from which activation would require D427. In this study we report the identification of such a site in the promoter of the OCH1 gene. OCH1 encodes an α-1,6 mannosyltransferase involved in N-linked glycoprotein maturation (Nakayama et al., 1992; Nakanishi-Shindo et al., 1993; Lehle et al., 1995), and OCH1 mutants display reduced cell wall integrity (Lee and Elion, 1999). The SLN1 response element in the OCH1 promoter maps to the 13-base pair (bp) repeated sequence: ATTTGGCC/TGGC/GCC, a sequence that is distinct from the previously defined binding site(s) in the SKN7-regulated promoters of oxidative and heat stress genes. The identification of OCH1, encoding an α-1,6 mannosyl transferase, as a gene whose expression is elevated in response to activation of the SLN1 pathway suggests that cell wall modifications may be one important aspect of the response of yeast cells to hypo-osmotic stress. We further find that D427 is not required for binding of Skn7p to the OCH1 promoter, although its activation is D-dependent, suggesting that D427 dependence of gene expression is specified at a step subsequent to DNA binding.
MATERIALS AND METHODS
Strains and Media and Yeast Techniques
Strains used in this work are summarized in Table 1. Media were prepared as described by Sherman et al. (1986). Synthetic complete medium (SC-aa) lacked the specified amino acids (e.g., SC-leucine). Yeast cultures were grown at 30°C. Hygromycin B was added to YPD plates to 50 or 70 μg/ml. Liquid YPD media for Ca2+ induction experiments was adjusted to pH 5.5 by addition of succinate to 0.5 M. Yeast transformation was performed by a modified LiOAc method (Ito et al., 1983).
Table 1.
Strains used in this study
| Strain name | Genotype |
|---|---|
| JF1434 | MATa his4-917 lys2-128δ leu2 trp1Δ1 ura3-52 |
| JF1435 | MATa sln1-21a his4-917 lys2-128δ leu2 trp1Δ1 ura3-52 |
| JF1565b | MATα his3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2 Δ 202 canR cyhR |
| JF1755 | MATa skn7Δ::TRP1 his4-917 lys2-128δ leu2 trp1Δ1 ura3-52 |
| JF1756 | MATa skn7Δ::TRP1 sln1-21 his4-917 lys2-128δ leu2 trp1Δ1 ura3-52 |
| JF1904b | MATα skn7Δ::TRP1 his3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2Δ202 canR cyhR |
| JF1910b | MATα sln1-22 his3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2 Δ 202 canR cyhR |
| JF2052b | MATα skn7Δ::TRP1 sln1-21 his3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2Δ202 canR cyhR |
| JF2054b | MATα sln1-21 his3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2Δ202 canR cyhR |
| JF2123b | MATa fps1Δ::LEU2 his3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2Δ202 |
| JF2202b | MATα crz1Δ::kanR skn7Δ::TRP1 his3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2Δ202 canR cyhR |
| JF2203b | MATα crz1Δ::kanR sln1-21 skn7Δ::TRP1 his3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2Δ202 canR cyhR |
| JF2206b | MATα swi5Δ::kanR skn7Δ::TRP1 his3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2Δ202 canR cyhR |
| JF2207b | MATα swi5Δ::kanR sln1-21 skn7Δ::TRP1 his3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2Δ202 canR cyhR |
sln1-21 is one of several activating alleles in SLN1 collectively referred to as sln1* alleles. The sln1* strains used in this work are strains containing the sln1-21 mutation consisting of a T550I change in Sln1p or the sln1-22 mutation consisting of a P1148S change.
Isogenic strains created by transformation of JF1565 or its CAN CYH parent.
Plasmids
Plasmids were constructed for this study or are part of the Fassler laboratory collection. Plasmid pSL232 is pRS315 (LEU2, CEN; Sikorski and Hieter, 1989) in which the SKN7 ORF plus 243 base pairs upstream and 535 base pairs downstream was introduced as a 3.5-kb SalI-HindIII fragment. Some flanking sequence (including the subcloning sites) of the Yep13 library vector carrying SKN7 is included. pCLM669 and pCLM700 are SKN7 plasmids that have been altered so that D427 encodes N (pCLM699) or E (pCLM700). Each SKN7 construct complemented the oxidative stress sensitivity phenotype of skn7Δ mutants.
Plasmid pSL1091 is a pRS315 derivative lacking a PstI site (pSL1090) with a 3.7-kb SalI-HindIII fragment containing SKN7 and a deletion of a 608-bp PstI-NsiI including the putative DBD. To construct pSL1108, the 608-bp PstI-NsiI fragment of pSL1090 was subcloned into Litmus 28 (NEB, Beverly, MA) to use as a template in site-directed PCR mutagenesis (QuikChange Site-Directed Mutagenesis Kit; Stratagene, La Jolla, CA) to generate S137A and R140A changes. The mutated fragment replaced the original PstI-NsiI fragment in pSL1090.
Plasmid pSEYC102 is a CEN URA3 plasmid with a 3.3-kb lacZ insert lacking the first five amino acids of the lacZ ORF (gift from S. Moye-Rowley, University of Iowa). Plasmid pSL1156, pSL1157, and pSL1158 were created by insertion of BamHI-BglII fragments containing different amounts of OCH1 upstream regulatory sequence. OCH1 fragments were generated by PCR amplification using forward primers including terminal BamHI and reverse primers including terminal BglII sites. After digestion, fragments were cloned into BamHI and BglII sites of Litmus 28, confirmed by sequencing and subcloned into BamHI cut pSEYC102 so as to retain the upstream BamHI site. pSL1156 contains OCH1 sequences −154 to +26; pSL1157 contains −526 to +26 and pSL1158 contains −355 to +26. pSL1249 was made by inserting the BamHI-BglII OCH1 −526 to −355 fragment into the BamHI site of pSL1156.
pSL1265, pSL1266, pSL1267, pSL1276, and pSL1277 are derivatives of pSL1156 in which the putative HSE and SCB sites were deleted (hse1Δ is a deletion of −251 to −243; hse2Δ is a deletion of −167 to −159 and scbΔ is a deletion of −296 to −290) and replaced with an NsiI restriction site by site-directed mutagenesis (QuikChange; Stratagene) of a Litmus-28 subclone carrying OCH1 −355 to −154. Double mutants were created by repeating the mutagenesis step. After sequencing, 200-bp BamHI-BglII fragments containing various mutated OCH1 promoter regions were cloned into the BamHI site of pSL1156.
pZL1320-pZL1323 are derivatives of pSEYC102 into which we cloned different OCH1 fragments generated by PCR amplification with 5′ primers including terminal EcoRI sites and 3′ primers including terminal BamHI sites. Constructs were confirmed by sequencing. pZL1320 contains OCH1 sequences −336 to +26; pZL1321 contains −314 to +26; pZL1322 contains −285 to +26; pZL1323 contains −265 to +26.
pZL1331 contains OCH1 sequences −314 to −261 and −154 to +26. It was constructed by insertion of a PCR fragment encompassing OCH1 −314 to −261 into the EcoRI/BamHI sites of pSL1156. pZL1332 contains −285 to −261 and −153 to +26 and was constructed by replacing the 2.5-kb BglII/BamHI vector fragment in pSL1156 with a PCR fragment amplified from pZL1322. pZL1333 was constructed in two steps. +605 to + 1354 of Litmus 28 was amplified with a 5′ primer containing terminal SmaI and BamHI sites followed by the sequence ATTTGGCCGGCC, a HindIII site and homology to the Litmus vector; and a 3′ primer containing a terminal BglII site, the complement of ATTTGGCCGGCC, a HindIII site and homology to the Litmus vector. After digestion, the PCR fragment was cloned into the SmaI and BamHI sites of pSL1156 to create pZL1335. Sequencing revealed a 1-bp deletion in the PCR product such that one ATTTGGCCGGCC repeat in the construct had three consecutive T's and the other had two. pZL1333 was generated by HindIII digestion of pZL1335 to remove the Litmus sequence. pZL1369 and pZL1370 constructs were generated in two steps. The first step was PCR amplification using reverse primers consisting of a 5′ tail including a BglII site and repeat A (−311 to −300) or repeat B (−287 to −275) followed by sequences upstream of OCH1 in pSL1156. A forward primer consisted of a BamHI site followed by sequences complementary to the pSL1156 vector. Fragments were digested with BamHI and BglII and cloned into the corresponding sites in pSL1156.
pZL1369m construction was conducted in several steps. The first step consisted of one round of PCR using linearized Litmus 28 as a template and a forward primer containing in order, eight random base pairs, an EcoRI site, the first eight positions of repeat A, two degenerate positions, positions 11 and 12 of repeat A, a HindIII site and homology to Litmus 28 starting at base pairs 606. After gel purification, the product was amplified using a forward primer complementary to the first 22 positions of the forward primer of the first step and a reverse primer complementary to Litmus 28 starting at base pairs 1350 and containing added HindIII and BamHI sites at the 5′ end. The pool of randomly mutagenized fragments was digested with EcoRI and BamHI, cloned into the OCH1 minimal reporter, pSL1156, and then digested with HindIII and self-ligated to remove Litmus 28 sequences.
The HIS4-based OCH1-lacZ plasmid, pZL1354, is a derivative of plasmid x-52 (Nagawa and Fink, 1985) consisting of HIS4 sequences −688 to −314, an XhoI site and HIS4 −144 to +34 fused to lacZ. OCH1 −314 to −261 was cloned into the XhoI site.
Plasmid pZL1357 was constructed by insertion of an EcoRI-BamHI fragment containing FKS2 sequences −785 to −690 generated by genomic PCR into the EcoRI-BamHI sites of pSL1156.
Electophoretic Mobility Shift Analysis
Preparation of yeast cell extracts, protein DNA binding reactions, and electrophoretic fractionation of complexes were performed essentially as described previously (Yu and Fassler, 1993). Binding was performed in 20 μl and included a constant amount (5 or 10 μg) protein extract and 1 μg poly(dI-dC) in EMSA buffer (25 mM Tris-HCl, pH 7.5, 50 mM NaCl, 2 mM EDTA, 7 mM MgCl2, 10% glycerol) and 0.5–1 ng (5–20 cps) DNA. Reactions were incubated at 37°C for 15 min. DNA probes were end-labeled with T4 polynucleotide kinase and [γ-32P]ATP. In supershift experiments, antibody was added at a 1:20 dilution. The Skn7 antibody was the generous gift of L. Johnston (National Institute for Medical Research, London) is highly specific and has been used to demonstrate the presence of Skn7p in other DNA complexes (Morgan et al., 1997; Raitt et al., 2000). (Gels were 4% polyacrylamide [29:1 acrylamide:bisacrylamide] in 0.5× TBE [89 mM Tris, 89 mM borate, 2.4 mM EDTA, pH 8.0]). Electrophoresis at 150–200 V for ∼2 h followed a 1 h 100-V preelectrophoresis step. Gels were dried on Whatman 3MM paper and subject to autoradiography and phosphoimage analysis.
β-Galactosidase Assays
Cultures for β-galactosidase assays were grown in SC media and harvested by centrifugation at 1–2 × 107 cells/ml. For analysis of calcium responsiveness, cultures were grown in SC media and subcultured to low pH YPD containing CaCl2. The calcineurin inhibitor FK520 was added to a concentration of 3 μg/ml. Extracts were prepared using glass bead lysis and cleared by centrifugation. Activities were calculated in Miller units (Miller, 1972) and are expressed as the average of four to six assays using at least three independent colonies or transformants.
Hygromycin B Assay
Cultures were grown to log phase in SC medium. Dilutions were made in the same medium. Five microliters of each dilution was spotted onto fresh plates and allowed to dry before incubation at 30°C for 90 h.
RNA Analysis
Total RNA was isolated by glass bead lysis from 200- to 500-ml SC cultures grown to 1–2 × 107 cells/ml. mRNA was prepared from ∼1 mg total RNA using the PolyA tract mRNA isolation system (Promega Biotech, Madison, WI). Radioactive probes were generated by random priming (Feinberg and Vogelstein, 1983, 1984) in the presence of [32P]dATP. RNA was separated on 1% GTG agarose (Seakem) gels containing 10% vol/vol formaldehyde and transferred by capillary blotting (5 h, 10× SSC) to 0.2-μm Nytran filters. After UV cross-linking, blots were hybridized at 65°C in PerfectHyb Plus solution (Sigma) for ≥5 h, and washed in 2× SSC, 0.1% SDS (2 times), and 0.5× SSC 0.1% SDS (two times). Quantitation was performed by PhosphorImager analysis using the Molecular Dynamics 445 SI PhosphorImager (Sunnyvale, CA) and ImageQuant software.
Calcium Treatment
Strains were cultured overnight in SC-Ura and then subcultured in the same medium for 3 h before being shifted to YPD (pH 5.5) containing 50 mM CaCl2. Where indicated, FK520 (3 μg/ml) was added at the time of calcium addition. Incubation in YPD was for 3 h. Cultures were harvested at midlog phase.
RESULTS
OCH1, A Mannosyltransferase Gene, Is a Target of the SLN1–SKN7 Pathway
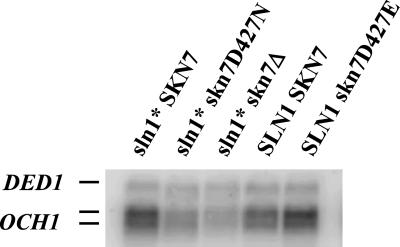
The SLN1–SKN7 pathway was previously defined with respect to its ability to activate SKN7-dependent reporter genes (Ketela et al., 1998; Li et al., 1998). Because these synthetic reporters have no counterpart in the yeast genome, we sought a natural yeast gene whose transcript levels would similarly reflect the activity of the pathway. We find that OCH1 transcript levels are also elevated in a strain bearing activated alleles of the SLN1 gene (sln1*). OCH1 levels were elevated 2.4-fold in sln1-21 strains (Figure 2) and 2.9-fold in sln1-22 strains (not shown) relative to isogenic SLN1 strains. The increase in OCH1 expression in the sln1* mutant depends on the presence of the receiver domain aspartate (D427) of Skn7p, as can be seen by comparing the sln1* skn7D427N strain to the sln1* SKN7 strain (lanes 1 and 2). The skn7D427E mutation is known to cause a constitutively active phenotype (Brown et al., 1994; Krems et al., 1996; Li et al., 1998). Consistent with this observation, OCH1 levels are elevated in this mutant even in the absence of an sln1* mutation (lanes 4 and 5). The SKN7D427 dependent phenotype of sln1* activation of OCH1 suggests that OCH1 is a natural target of the SLN1–SKN7 pathway, and that its expression should be elevated upon exposure of cells to low-osmotic strength environments.
Figure 2.
OCH1 transcript levels are dependent on SLN1 and SKN7. Representative Northern in which equivalent amounts of mRNA prepared from SLN1+ skn7Δ (JF1904) and sln1* skn7Δ (JF2052) strains carrying various SKN7 plasmids were subjected to electrophoresis. Plasmids were pSL232, SKN7+; pCLM699, skn7D427N; pCLM700, skn7D427E and pRS315, empty vector. The RNA blot was then hybridized with a radiolabeled OCH1 probe and a probe to the SLN1-independent DED1 gene for normalization. Because analysis of OCH1 hybridization patterns in an och1 null mutant (our unpublished results) suggested that both bands of the doublet are due to OCH1 hybridization, both bands were used in quantitation. Normalized expression was calculated by dividing OCH1 values by DED1 values in each lane after phosphoimage analysis. The average increase in OCH1 levels in the sln1-21 strain was 2.4-fold (n = 9). Shown is a single representative autoradiogram.
OCH1 Expression Is Activated by Mutations in the Major Glycerol Channel, Fps1
To verify that OCH1 expression is elevated upon hypo-osmotic stress, as expected for targets of the SLN1–SKN7 pathway (Tao et al., 1999), expression of the gene was measured in response to mutations that cause a perturbation of normal osmotic balance. The FPS1 gene encodes the major glycerol channel in yeast and is responsible for the release of excess intracellular glycerol into the medium. Loss-of-function fps1 mutations were shown previously to activate the SLN1–SKN7 pathway in a SLN1- and SKN7-dependent manner by causing an imbalance in the normal osmotic gradient (Tao et al., 1999). The imbalance caused by fps1 mutation mimics hypo-osmotic stress and was used here to examine the response of OCH1 to this type of stress by comparing the activity of an OCH1-lacZ reporter (−355 to +26) that responds to the SLN1 pathway in FPS1 and fps1Δ strains. Activity of the reporter was elevated twofold in the fps1 mutant. As expected from previous studies, the fps1 mutant showed reduced elevation in reporter activity when grown in the presence of 1 M sorbitol (Table 2). Thus, fps1 stimulation of OCH1 expression is due to osmotic imbalance. These results indicate that the stimulus previously defined (using the P-lacZ reporter) as activating the SLN1–SKN7 pathway (Tao et al., 1999) also activates OCH1 gene expression.
Table 2.
Osmotic imbalance activates the OCH1-lacZ reporter
| Relevant genotypea | β-Galactosidase activity OCH1-lacZb
|
|
|---|---|---|
| Normal medium | Sorbitol medium | |
| Wild type | 133.1 ± 13.6 | 101.5 ± 13.9 |
| sln1* | 448.9 ± 80.1 | 479.6 ± 68.0 |
| fps1Δ | 271.3 ± 26.4 | 154.7 ± 10.8 |
Strains JF1565 (wild type), JF2054 (sln1-21), and JF2123 (fps1Δ) each carrying pSL1158, a CEN-based lacZ reporter driven by OCH1 −355 to +26.
β-Galactosidase values, expressed as Miller units, are the average ± SD of six trials involving at least three individual colonies or transformants.
The DNA Binding Domain of Skn7 Is Required for Activation of SLN1–SKN7 Target Genes
To confirm the importance of the Skn7p DNA binding domain in Skn7p in SLN1–SKN7 signaling, we used the OCH1-lacZ reporter (−355 to +26). This reporter mimics the expression of the native gene both in its response to sln1* mutations and in the dependence of that response on Skn7D427 (Table 3). The presence of a domain within the Skn7 protein that resembles the DNA binding domain (DBD) of heat shock factor (Hsf1p) combined with the predominantly nuclear localization of the protein had previously led to the conclusion that Skn7p functions as a transcription factor. Evidence that Skn7p binds the promoters of the SKN7-regulated genes, TRX2 and SSA1, has been reported; however, the importance of the putative DNA binding domain of Skn7 has not been experimentally verified. Furthermore, SKN7-dependent activation from the reported binding sites within these genes is independent of the receiver domain D427 (Morgan et al., 1997; Raitt et al., 2000). We examined the relevance of the Skn7p DNA binding domain in the D427-dependent SLN1–SKN7 pathway by measuring the effect of DNA binding domain mutations on OCH1-lacZ expression. Both deletion of the Hsf1p-like DNA binding domain (Skn7-hsfΔ) and mutations at S137 and R140 known to be critical for Hsf1p binding and activation (Hubl et al., 1994) eliminated sln1* activation (Table 3). Western analysis confirmed that both mutant Skn7 proteins were present at levels indistinguishable from wild type (our unpublished results). Hence, we conclude that a functional DNA binding domain is required for sln1* activation of the OCH1-lacZ reporter.
Table 3.
OCH1-lacZ reporter activity depends on the conserved D427 of the Skn7p receiver and on the DNA binding domain
| Relevant SKN7 genotype | Relative β-galactosidase activity OCH1-lacZ
|
|
|---|---|---|
| SLN1a | sln1*a | |
| SKN7b | 100.0 (8.7) | 368 (38.3) |
| skn7-D427N | 24.4 (2.6) | 23.0 (6.1) |
| skn7-hsfΔ | 14.0 (3.3) | 15.3 (2.6) |
| skn7-S137A, R140A | 10.1 (1.9) | 14.2 (2.7) |
| skn7Δ | 8.5 (1.0) | 11.3 (1.0) |
SLN1 skn7Δ (JF1755) and sln1* skn7Δ (JF1756) strains carrying the OCH1-lacZ reporter (pSL1158) (−355 to +28) were transformed with each of five different plasmids and the β-galactosidase activity measured in cultures from four to six individual transformants from each strain. Values are expressed relative to the SKN7 SLN1 strain, which was set at 100. The actual average value in Miller units of the SKN7 SLN1 strain was 107.9. Values are given with the standard deviation of the mean in parentheses.
SKN7 plasmids were pSL232, SKN7+; pCLM699, skn7D427N; pSL1090, skn7 hsfΔ; and pSL1108, skn7 S137A R140A. The skn7Δ strain carries an empty vector.
Skn7 Protein Binds to Sequences Upstream of OCH1 in a D427-independent Manner
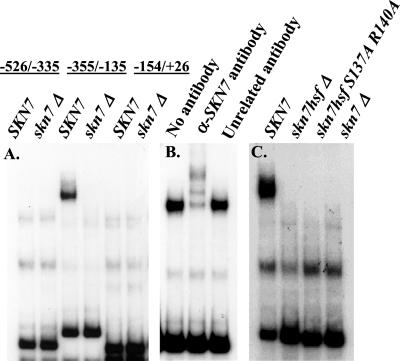
To localize the Skn7 binding site in OCH1, we used electrophoretic mobility shift analysis (EMSA) to look for Skn7p complex formation with fragments from the OCH1 promoter. The OCH1 promoter was initially divided into three fragments (−526 to −335; −355 to −135; and −154 to +26). Whole cell extracts were prepared from Δskn7 cells carrying a SKN7 plasmid or an empty vector and added to reactions containing each of the three labeled OCH1 fragments. A Skn7p-specific band was detected in reactions containing the −355 to −135 probe and not with either of the other two probes (Figure 3A) and was not detected in reactions containing skn7Δ extract.
Figure 3.
Skn7p forms a complex with the −355 to −135 region of OCH1. Electrophoretic mobility shift assays were carried out (A) with 5 μg extracts prepared from the skn7Δ strain (JF1755) carrying the SKN7 plasmid, pSL232 or the empty vector, pRS315. Radiolabeled OCH1 fragments were as indicated; (B) with 5 μg extracts SKN7+ extract as in A, plus no added antibody, polyclonal α-Skn7p or α-Gal11p (unrelated) antibody (1:20); (C) with 5 μg extract prepared from a skn7Δ strain (JF1755) carrying the SKN7 plasmids, pSL1108 (skn7- S137A, R140A), pSL1091 (skn7 hsf-Δ), pSL232 (SKN7) or the empty vector, pRS315. Radiolabeled −355 to −135 OCH1 fragment was used as probe in B and C.
To establish that Skn7 protein is present within this complex, polyclonal antisera directed against Skn7p (generous gift from L. Johnston, National Institute for Medical Research, London) was added to the reaction mixture at the same time as the protein extract (Figure 3B). A shift in the mobility of the complex was seen in reactions to which Skn7p antibody was added. In contrast, no shift in the mobility of the complex was seen in reactions to which a control antisera (anti-Gal11p) was added. As expected from the results of the reporter analysis, mutations in the HSF-like DBD of Skn7p eliminated binding (Figure 3C). The possibility that the loss of DNA binding was an indirect result of loss of Skn7p localization was ruled out by showing that DNA binding domain mutations do not alter the nuclear localization of GFP-Skn7 derivatives (our unpublished results).
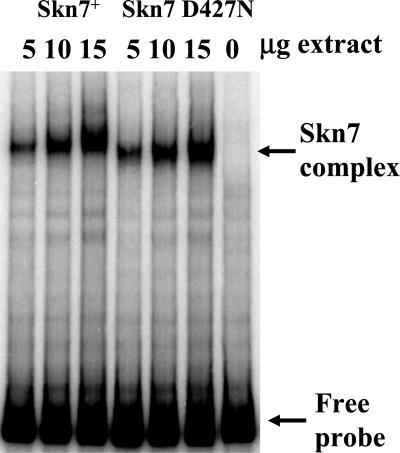
To examine whether D427 phosphorylation is required for binding to targets of the SLN1–SKN7 pathway, EMSA was conducted using extracts from the nonphosphorylatable skn7D427N strain. Extracts containing only D427N Skn7p showed no major changes in the amount of SKN7-dependent complex formation (Figure 4). Interestingly, however, the mobility of the Skn7D427Np complex appears to be slightly faster than that of the Skn7p complex, indicating a possible change in the composition of the complex. The basis for the apparent change in mobility is currently being investigated and is considered further in the discussion. These results suggest that changes in Skn7p conformation due to phosphorylation do not regulate binding but rather some other aspect of SKN7-dependent activation of gene expression.
Figure 4.
The role of aspartate 427 (D427) in Skn7 binding to the OCH1 promoter. Binding of Skn7D427 and the unphosphorylatable mutant derivative, Skn7D427N, from crude extracts to the −355 to −135 fragment of OCH1 was examined by electrophoretic mobility shift assays. Extracts were prepared from strain JF1755 carrying the SKN7 plasmid, pSL232, or the skn7D427N plasmid, pCLM699. Binding was assessed in the presence of increasing amounts (5, 10, and 15 μg) of each extract. The right-most lane contains no extract.
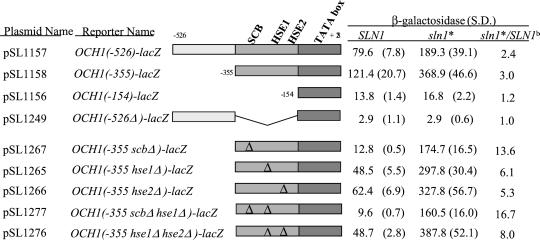
The sln1* Response Element in OCH1 Is Not an HSE
The sln1* response element (SSRE) was further mapped by measuring sln1* activation of OCH1-lacZ reporter derivatives containing various deletions of the OCH1 promoter. Results of this analysis (Figure 5, rows 1–4) localized the sln1* response to the −355 to −154 region of the OCH1 promoter, consistent with the results of EMSA. Skn7p was previously found to bind the HSE2 element from the SSA1 promoter (Raitt et al., 2000) and the OCH1 promoter has two HSE-like sites: one at −163 (TTCTTCGAA) and the other at −250 (GAAAAGTCC). To examine the role of these elements in OCH1 regulation, the sites were deleted singly and in combination (Figure 5). Although mutations of the HSEs reduced the level of basal expression of the OCH1 gene, the magnitude of sln1* activation was not reduced. Expression of OCH1 in the hse1Δ hse2Δ double mutant was eight times higher in a sln1* strain than a SLN1 wild-type strain. Hence, the putative HSEs are not responsible for sln1* activation of OCH1 expression.
Figure 5.
Sequences between −355 to −154 of OCH1 are required for sln1* activation in a minimal OCH1-lacZ reporter. Activation by subclones of the −526 to −154 OCH1 fragment and by mutant derivatives of the −355 to −154 OCH1 fragment was assessed in the context of the minimal (−154 to +26) OCH1-lacZ reporter. Each reporter was introduced into SLN1+ SKN7+ (JF1434) and sln1-21 (sln1*) SKN7+ (JF1435) strains and β-galactosidase activity (Miller units) measured in extracts prepared from four to six transformants. Similarities to known transcription factor binding sequences present within this fragment are listed. Delta symbols represent the positions of mutations (see MATERIALS AND METHODS) that were introduced to destroy the corresponding consensus binding sites. The ratio of activity in sln1* relative to SLN1+ strains reflects the extent to which the remaining sequences retain sln1* activation capabilities. S.D., SD.
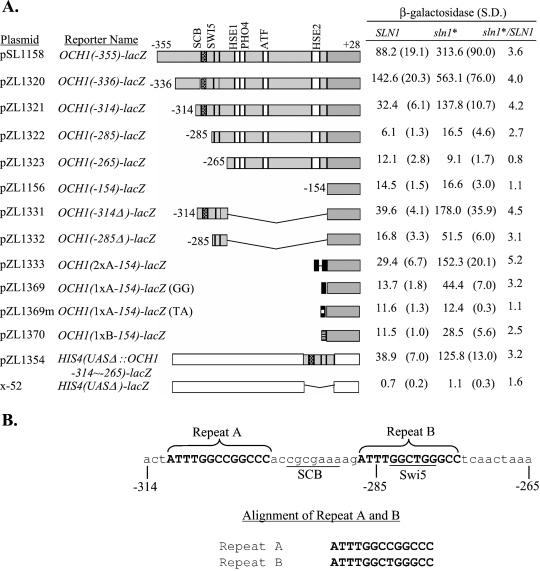
The sln1* Response Element in OCH1 Maps to a 13-bp Repeated Sequence Flanking an SCB-like Element
To localize the sln1* response element more precisely, a series of deletions was constructed. Partial sln1* activation was retained in all constructs containing the sequences between −285 and −265, and full activity was evident in constructs containing −314 to −265. Sequences on either side of this region were dispensable, but deletions within this region such as in the −265 reporter (Figure 6A) exhibited sln1* activation ratios of ≤1.1. To verify that the −314 to −265 region is sufficient for sln1* activation, the −314 to −265 fragment was subcloned into the promoter sequences of an HIS4-lacZ fusion gene from which the HIS4 UAS had been deleted. The new construct exhibited 3.2-fold induction by sln1* (Figure 6A), thus confirming that the sequences from −314 to −265 are sufficient for the sln1* activation effect.
Figure 6.
Localization of the sln1* response element (SSRE). (A) Measurements of the β-galactosidase activities (Miller units) of 5′ and internal deletions of the −355 to −154 OCH1 fragment in SLN1 (JF1434) and sln1* (JF1435) strains localized the SSRE to the region between −314 and −265. Plasmid pZL1354 contains the same OCH1 sequences (−314 to −261) as pZL1331, except inserted into a UAS-less HIS4-lacZ reporter. Plasmids pZL1333, pZL1369, and pZL1370 consist of two copies of repeat A, 1 copy of repeat A, and 1 copy of repeat B inserted, respectively (see part B of this figure). pZL1369m is a mutant derivative of pZL1369 in which positions 9 and 10 have been mutated from G to T and A, respectively. Values reported are the average of six measurements involving at least three colonies or transformants with the exception of pZL1369 whose activity is the average of three measurements. S.D., SD. (B) DNA sequence of the SSRE (−314 to −265 of OCH1) with putative SCB and Swi5p binding sites shown. A repeated motif is shown above the sequence alignment of repeat A and B is shown below the sequence.
Within the −314 to −265 region are two repeats (labeled A and B) separated by an SCB-like sequence (Figure 6B). The SCB-like sequence is a putative binding site for the Swi4p-Swi6p complex. An SCB-like site was present in the 23-bp fragment shown to mediate Skn7p binding to TRX2 (Morgan et al., 1997). Deletion of the SCB-like element in OCH1 reduced basal expression of the reporter, but did not reduce sln1* activation (Figure 5). Hence, the SCB-like element is not required for the sln1* response. To examine the role of the repeats in sln1* activation of OCH1, a reporter (pZL1333) was constructed in which two A repeats were substituted for sequences upstream of −153 (Figure 6A). The two repeats are separated by a 6-bp HindIII site that does not contain similarity to an SCB element. This construct showed 5.2-fold sln1* activation, compared with 4.5-fold activation seen using the −314 reporter (pZL1331). These results indicate that the repeat A sequence is sufficient for this response. Reporters with a single repeat also mediated sln1* activation, but only to 50% of the two-repeat reporters (Figure 6A, 1xA, pZL1369; 1xB, pZL1370)
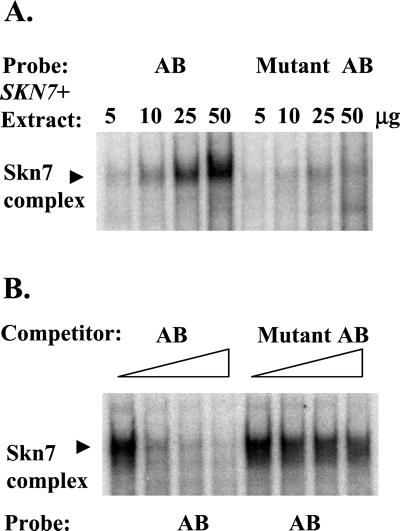
To assess the importance of the repeat sequence, a 2-bp substitution was introduced into the 1xA reporter (pZL1369) converting two conserved G residues at positions 9 and 10 to TA. β-Galactosidase analysis revealed that sln1* activation was abolished in the mutated reporter (Figure 6A, pZL1369m). The effect of these mutations was also examined using gel shift analysis. Two probes were synthesized, one containing OCH1 sequences from −316 to −271 and a second, mutant derivative, containing the same sequences but with positions 9 and 10 in each repeat (−277, −278, −302, −303 in the OCH1 promoter) mutated from GG to TA. Figure 7 shows the formation of a single complex using the wild-type probe. This complex is Skn7 dependent because it is absent in reactions performed with extracts prepared from skn7Δ strains (our unpublished results).
Figure 7.
Skn7 binding is affected by mutations in the A and B repeats. Electrophoretic mobility shift assays were carried out with extract prepared from a skn7Δ strain (JF1755) carrying the SKN7 plasmid, pSL232. Fifty-base-pair oligonucleotides consisting of OCH1 sequences −316 to −271 and a mutant derivative in which positions G9 and G10 of repeats A and B were mutated to T and A, respectively, were radiolabeled and used as probes or as competitors. (A) Increasing amounts (5, 10, 25, and 50 μg) of extract was incubated with wild-type (AB) and mutant (Mutant AB) oligonucleotide probes; (B) 5 μg of wild-type extract was incubated with the wild-type oligo (AB). Increasing amounts (0-, 2.5-, 5-, and 10-fold molar excess) of unlabeled AB oligo (AB) or unlabeled mutant AB oligo (mutant AB) were added as competitor to the binding reactions. The position of the Skn7 complex is indicated to the left. The lane to which no competitor was added was duplicated and is represented twice (lanes 1 and 5) in the figure.
Levels of Skn7p complex formation with the wild-type or mutant oligonucleotide probe were compared. Complex formation was much less efficient when the mutant oligonucleotide was used as probe (Figure 7A). Likewise the wild-type oligonucleotide could be shown to compete for binding to the wild-type probe, but the mutant oligonucleotide competed less efficiently (Figure 7B). This set of experiments indicate that Skn7p binding and D427-dependent activation are both mediated by the repeat sequences in the OCH1 promoter.
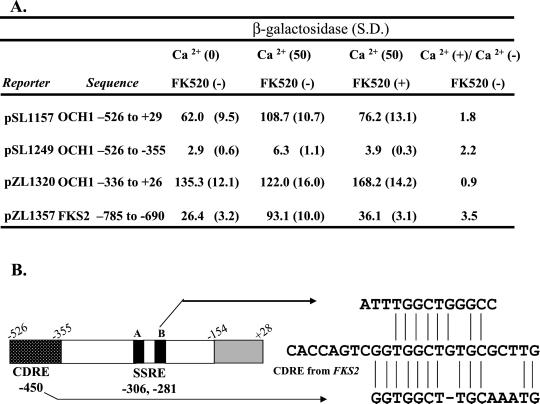
The sln1* Response Element Resembles a Calcineurin-dependent Response Element (CDRE) and a Swi5 Binding Site
The B repeat sequence of the SSRE in OCH1 contains a perfect match to the 4-bp putative core sequence (GGCT) as well as some flanking sequence similarity to the calcium-dependent response element (CDRE) to which the Crz1 transcription factor binds (Stathopoulos and Cyert, 1997; Kim Williams and Martha Cyert, personal communication). Because Crz1p has recently been shown to interact with Skn7p (Williams and Cyert, 2001), it was of interest to determine if the SSRE is a binding site for Crz1 transcription factor. To address this issue, we tested whether OCH1 reporters containing the SSRE sequence could mediate calcium-dependent transcriptional activation. A long OCH1-lacZ reporter (−526 to +26; pSL1157) was inducible by calcium treatment. However, a shorter (−336 to +26; pSL1320) OCH1 reporter that responds to sln1* was not induced by calcium treatment (Figure 8A). The effect of a crz1Δ mutation on OCH1-lacZ expression was also examined. The presence or absence of the CRZ1 gene did not influence OCH1-lacZ expression (our unpublished results). Results of this analysis indicate that the sln1* response element is distinct from the CDRE and that the calcium response element for OCH1 localizes to the −526 to −355 fragment. Inspection of the sequences in this fragment reveals a sequence with some similarity to the CDRE from FKS2 (Stathopoulos and Cyert, 1997; Figure 8B).
Figure 8.
The sln1* response element (SSRE) and the CDRE are separable. (A) The CDRE of OCH1 is located upstream of the sln1* response region. β-Galactosidase measurements are shown in Miller units for three different OCH1-lacZ reporters and one FKS2-lacZ reporter (pZL1357) in the SLN1+ strain (JF1434). Cultures were untreated, treated with 50 mM Ca2+ or treated with 50 mM Ca2+ plus 3 mM FK520 (Sigma Ascomycin), a calcineurin inhibitor. β-Galactosidase values are the average of six measurements on a minimum of three independent colonies. S.D., SD. (B) The relative positions of the CDRE and sln1* response elements within OCH1 promoter sequences are shown schematically. The numbers below the figure indicate the position of the central base of each recognition element. Sequence alignment of the sln1* response element (SSRE) and the calcium-dependent response element (CDRE) is shown to the right. The sequences of the sln1* response element from OCH1 and the CDRE-like sequence from OCH1 were aligned relative to the CDRE from FKS2. Vertical lines show identities. A short horizontal dash represents a gap introduced to optimize the alignment
Possible contributions to sln1* activation by Swi5p were also tested. The SSRE (repeat B) contains a consensus site (KGCTGR) for the binding of the Swi5p transcription factor. Isogenic swi5 and sln1* swi5 deletion strains were examined using both EMSA and OCH1-lacZ reporter assays. The absence of Swi5 had no effect on the mobility or the abundance of the Skn7p-dependent complex in gel shift assays and the activity of the OCH1-lacZ reporter was unaffected by swi5 deletion (our unpublished results). These results suggest that Swi5p is not an integral component of the Skn7p-dependent complex and has no role in SKN7-dependent activation of the OCH1 gene.
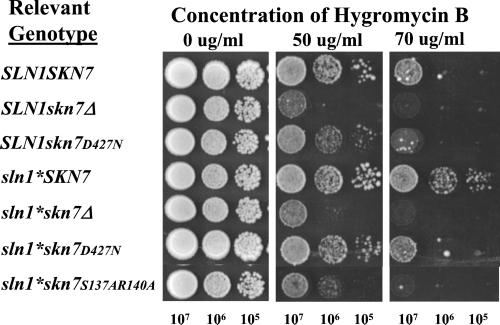
Hygromycin B Sensitivity of skn7 Mutants Suggests a Role for the SLN1–SKN7 Pathway in Cell Wall Integrity
Many mutants with defects in glycosylation exhibit sensitivity to the drug hygromycin B (Dean, 1995). Because OCH1 encodes a protein involved in mannosylation and och1 deletion mutants are themselves modestly hygromycin B sensitive (Lee and Elion, 1999), we tested whether mutations in the SLN1–SKN7 pathway might also exhibit hygromycin B sensitivity. The sln1* mutation caused a modest resistance to a dosage of drug that is highly toxic to wild-type cells, whereas skn7Δ strains were more sensitive than wild type. The skn7D427N mutant and the DNA binding domain mutant, skn7 S137A, R140A, both exhibited sensitivity intermediate to wild-type and skn7Δ strains (Figure 9). The hygromycin phenotype of mutations in components of the SLN1–SKN7 pathway supports the idea that OCH1, and other cell wall integrity genes are targets of the pathway and part of the cellular response to hypo-osmotic stress.
Figure 9.
The sensitivity of cells to hygromycin B is related to the activity of the SLN1-SKN7 pathway. Cells with designated genotypes were diluted in YPD medium as indicated, and 5 μl was spotted onto YPD plates with indicated concentrations of hygromycin B. All plates were incubated at 30°C for 90 h. All strains were grown and spotted simultaneously on the same batch of plates. Strains were SLN1skn7Δ (JF1904) or sln1-21 skn7Δ, (JF2052) carrying various SKN7 plasmids including: pRS315 (skn7Δ); pSL232 (SKN7); pCLM699 (skn7D427N); and pSL1108 (skn7-S137A, R140A).
DISCUSSION
The SLN1–SKN7 pathway has thus far been assigned no clear-cut function, and despite genetic links to the cell wall and cell cycle, few genes have been identified whose expression is dependent on the receiver domain aspartate, D427. OCH1 is one of the first reports of a gene whose expression responds to Skn7D427p and is therefore a candidate target for the SLN1–SKN7 pathway. Our demonstration that Skn7p binds to the OCH1 promoter strongly suggests that OCH1 expression is directly regulated by Skn7p in a d-dependent manner. In addition to providing insight into the function of the pathway, the identification of OCH1 as a target lends support to previous reports that the pathway is linked to cell wall integrity (Brown et al., 1993; Ketela et al., 1999). Based on our identification and characterization of the Skn7p binding site in the promoter of OCH1, it will now be possible not only to identify additional targets but also to elucidate the mechanism of d-dependent activation by Skn7p.
The OCH1 Gene Is a Target of the SLN1–SKN7 Pathway and Is Activated by Hypo-osmotic Stress Caused by Mutation of the Fps1p Glycerol Channel
The OCH1 gene is positively regulated by the activity of the SLN1–SKN7 pathway. OCH1 mRNA levels are increased in the presence of activating alleles of SLN1, and this increase is dependent on D427 of Skn7p. These effects are also seen using OCH1-lacZ reporter constructs. In the reporter assays one can easily recognize several different components of Skn7p-mediated regulation of OCH1. In addition to the 3.6-fold D427 dependent sln1* stimulation, it is apparent that basal expression of OCH1 also requires Skn7D427p. OCH1 levels decrease 4-fold in the SLN1 skn7D427N mutant relative to the SLN1 SKN7 strain (Table 3). In addition, we observe a D427 independent contribution to OCH1 expression because levels are reduced by another 3-fold in skn7 deletion or the skn7 DBD mutants (Table 3).
Microarray analysis shows that OCH1 expression is also modestly regulated by the cell cycle, peaking in G1; regulated both positively and negatively in diauxic shift experiments; and downregulated 5 h into sporulation (DeRisi et al., 1997; Chu et al., 1998; Spellman et al., 1998). Although many of the cis-acting regulatory sites identified by computational consensus element identification (Figures 5 and 6) may be nonfunctional, it is clear that the OCH1 promoter is complex. Despite this complexity, we have been able to dissect this promoter and define the sequences involved in regulation by the SLN1–SKN7 pathway.
Yeast cells normally retain a shallow osmotic gradient across the membrane with the intracellular environment being somewhat more concentrated than the extracellular environment. This gradient is important for growth and morphogenesis (Cosgrove, 1986; Dale and Sutcliffe, 1986; Martinez de Maranon et al., 1996). fps1 mutant cells exhibit a larger than normal gradient due to the intracellular accumulation of glycerol (Luyten et al., 1995; Tamas et al., 1999). The fps1 mutant therefore mimics hypotonic stress. This stimulus was previously shown to activate the SLN1–SKN7 pathway. The effect is specific to the SKN7 branch of the pathway because deletion of ssk1 did not reduce fps1 activation (Tao et al., 1999). The identification of OCH1 as a gene that can be activated by fps1Δ brings added detail to our understanding of the SLN1–SKN7 pathway and continues to suggest that the SLN1 “osmosensor” is more accurately a sensor of osmotic balance.
Skn7 Is a Multifunctional Transcription Factor
Several studies have contributed to the conclusion that Skn7p is a transcription factor. In addition to its nuclear localization (Brown et al., 1994; Raitt et al., 2000) and the presence of an HSF-like putative DNA binding domain, Brown et al. (1994) showed that Skn7-Gal4 DBD fusion proteins were capable of activating transcription of Gal4-dependent reporter genes. Krems et al. (1996) used similar constructs to show that transcriptional activation by Skn7p requires the receiver domain. In both studies activation by the Skn7-Gal4 DBD protein was at least partially dependent on D427. Morgan et al. (1997) were the first to show sequence-specific binding of native Skn7p to a 23-bp stretch in the upstream regulatory region of the oxidative stress inducible gene, TRX2. Recently, Raitt et al. (2000) showed binding of Skn7p to a heat shock element (HSE2) in the promoter of the heat shock gene, SSA1, as well as SKN7-dependent oxidative stress induction of a synthetic HSE-lacZ reporter. Based on similarity between the sequences involved in Skn7p binding to the upstream regulatory regions of TRX2 and SSA1, it has been suggested that the sites in the SSA1 and TRX2 genes are variations of a consensus Skn7p binding site, although its precise sequence has not yet been elucidated (Raitt et al., 2000).
Although the evidence is strong that Skn7p is a transcription factor in the oxidative stress pathway, it was not obvious that Skn7p would play the same function in SLN1–SKN7 signaling. The Sln1 histidine kinase is a plasma membrane localized protein (Ostrander and Gorman, 1999; C. Malone and R.J. Deschenes, unpublished results). Because Sln1p is part of a phosphorelay pathway consisting of Ypd1p and the response regulators Ssk1p and Skn7p, a pool of each of these proteins might be expected to be localized to the inner leaflet of the plasma membrane to be regulated by Sln1. We therefore entertained the possibility that Skn7p might have a cytoplasmic function with respect to SLN1–SKN7 target gene activation despite its predominantly nuclear localization, and binding activity with respect to certain oxidative stress and heat shock genes (Brown et al., 1994; Morgan et al., 1997; Raitt et al., 2000). For example, a small pool of cytoplasmically localized Skn7p might stimulate the activity of a second transcription factor upon phosphorylation of D427. In this case, the DNA binding domain of Skn7p might be dispensable for activation of OCH1 and other SLN1–SKN7 target genes. Our analysis of mutations in the Hsf1-like domain of Skn7p argues against this. These and numerous other mutations (S. Li and J. S. Fassler, unpublished data) in the Skn7p DNA binding domain that might have been expected to discriminate between nuclear and cytoplasmic roles of Skn7p showed equivalent defects with respect to oxidative and SKN7D427 dependent phenotypes. Likewise, although it is formally possible that Skn7 might associate with DNA by interacting with other DNA binding proteins, this seems unlikely based on the effects of mutations in the Skn7p DNA binding domain. Consistent with the conclusion that Skn7p plays the role of a transcription factor in the SLN1–SKN7 pathway, we found that both Skn7p binding and SKN7D427 dependent activation localize to the same region of the OCH1 promoter.
Skn7 Recognizes a Novel Site in OCH1
The Skn7p binding sites previously identified in the TRX2 and SSA1 genes bear a resemblance to HSEs, as might be expected based on the HSF-like DNA binding domain present in Skn7p. However, Skn7p binding and activation with respect to the TRX2 and SSA1 genes are independent of D427 and hence do not reflect the activity of the SLN1–SKN7 pathway. Several observations suggested that SLN1-dependent activation would not be mediated by the previously identified element. First, although there is both an SCB-like site including the sequence CGAAA thought to be important for Skn7p binding in TRX2 and two HSE type Skn7p binding sites (identified in SSA1) in the region of the OCH1 promoter to which we localized the sln1* response, several constructs in which these sites were not preserved continued to mediate sln1* activation and Skn7p binding. Likewise, in earlier studies, mutation of the CG in the CGAAA motif known to be required for efficient Skn7p binding in TRX2-lacZ reporters (Morgan et al., 1997) did not eliminate sln1* activation (G. Gingerich, S. Dean, and J. S. Fassler, unpublished data). In both OCH1-lacZ and TRX2-lacZ reporters, elimination of the CGAAA motif reduced “basal” reporter activation by a factor of 10 (see Figure 5) but did not compromise the induction normally seen in a sln1* mutant. Similarly, the HSE elements in the OCH1 promoter are important for basal OCH1 expression (Figure 5) but are not required for sln1* activation. Interestingly, the extent of sln1* activation is increased in reporters lacking SCB or HSE sites suggesting that SLN1 pathway activity may be influenced by additional physiological signals.
Identification of the d-dependent Skn7 binding site, which we have called the SSRE, was accomplished using both reporter analysis and electrophoretic mobility shift assays. The response element was localized to sequences between −335 and −265 of the OCH1 promoter. Each of the known consensus binding sequences present in this region including a putative SCB, SWI5 site and CRZ1 site was experimentally ruled out as responsible for sln1* activation. The relevance of the repeat sequence observed in this region to sln1* activation was demonstrated using various reporters. Reporters consisting of one copy and two copies of repeat A in front of the minimal and otherwise inactive OCH1 promoter were responsive, as were reporters consisting of repeat B. β-Galactosidase measurements indicate that repeat A and B are nearly equally responsive and that their combined activities in the OCH1 promoter are additive. Furthermore, mutation of two conserved G's within repeat A eliminated sln1* activation. Likewise, gel shift analysis using a 50-bp fragment containing the two repeats plus the intervening SCB site showed Skn7p binding, and binding was substantially diminished using the probe containing the GG substitution found to eliminate activation in our reporter analysis. Binding was not affected, however, when the SCB site was deleted (our unpublished results). In summary, we have defined a 13-bp sequence that binds Skn7p and that is distinct from previously defined Skn7p binding sites.
The Role of Skn7 D427-dependent Phosphorylation
Among bacterial response regulators, phosphorylation of the receiver domain modulates the function of the associated output domain. For example, phosphorylation of OmpR, a response regulator involved in osmotic stress sensing in Escherichia coli, affects its DNA binding activity (Aiba et al., 1989). Similarly, receiver domain inhibition of the DNA binding domain is alleviated upon phosphorylation of the E. coli NarL response regulator, which mediates changes in gene expression in response to availability of nitrate and nitrite (Baikalov et al., 1996, 1998). NtrC, a response regulator involved in nitrogen regulation, undergoes a requisite aspartyl phosphorylation-dependent oligomerization before binding DNA (Wyman et al., 1997). Phosphorylation of the Bacillus SpoOA response regulator, in contrast, allows conversion of a transcriptionally incompetent polymerase-promoter competent complex to one that is transcriptionally competent (Spiegelman et al., 1995). What aspect of Skn7p function is changed by aspartyl phosphorylation to allow transcription at the OCH1 and other SKN7-dependent promoters is a matter of intense interest. The failure to observe any effect of the D427N mutation on Skn7p binding to TRX2 (Morgan et al., 1997) or OCH1 indicates that the binding step is unlikely to be regulated by phosphorylation. Subcellular localization of the Skn7 protein to the nucleus was also ruled out because a GFP-tagged Skn7p show no differences in Skn7D427Np and Skn7p wild-type localization to the nucleus (J. Lu, S. Li, and J. Fassler, unpublished results; Brown et al., 1994; Raitt et al., 2000). Other possible points of regulation include Skn7p interaction with auxiliary proteins as hinted at by the slight change in mobility of the Skn7D427Np complex and/or with the basal transcriptional machinery.
Skn7 Binding Does Not Involve the Crz1 Transcription Factor
Binding sites for the Crz1 transcription factor have been identified in several promoters. Based on an alignment of these elements, the core binding sequence may be GGCT (K. Williams and M. Cyert, personal communication). An exact match to the core and similarity to flanking sequences is found in Repeat B of the SSRE. The resemblance between the Crz1p binding site and the newly determined sln1* response element, led us to test the idea that Crz1p might be a Skn7p partner. We conclude that it is unlikely that Crz1p is a Skn7p partner at the OCH1 promoter based on the following observations: (1) Reporter genes with minimal sln1* response elements failed to exhibit calcium-induced activation (Figure 8); (2) There were no changes in the mobility of the Skn7p-OCH1 (−355 to −135) complex in crz1Δ extracts; and (3) There was no effect of the crz1Δ mutation on OCH1-lacZ reporter gene expression (S. Dean and J. Fassler, unpublished data).
Skn7-dependent Activation of Distinct Genes in Response to Stress
Our data suggest that Skn7p binds to at least two types of cis-acting elements and that its binding is independent of D427. Hence, in the steady state, the Skn7p pool may be distributed among the promoters of both oxidative and osmotic response genes. Although more detailed analysis of the DNA-protein interaction would be required to rule out the possibility that the Skn7p DBD might be capable of distinct interactions with different sequences, as a working model, we assume that Skn7p recognition of distinct sites is mediated in part by interacting proteins. Based on the small mobility shift in Skn7p vs. Skn7D427Np complexes, we further speculate that the phosphorylation of D427 in SSRE-based complexes leads to association of additional auxiliary factors required not for binding but rather for activation. Further characterization of SSRE- and HSE-based complexes in the presence of Skn7p and Skn7D427Np is in progress to test the various premises of this model.
The Physiological Role of the SLN1–SKN7 Pathway
The identification of hypotonic stress as a stimulus for the SLN1–SKN7 pathway suggests that the targets of this pathway will be required for adaptation to this stress. The observation that skn7 deletion mutants are not lethal may indicate that hypotonic stress is not life-threatening as long as the wall is strong. Alternatively, pathways such as the PKC MAP kinase cascade may provide some important redundancy as is suggested by the synthetic inviability of a skn7 pkc1 double mutant (Brown et al., 1994). The STE vegetative growth pathway also appears to play a partially redundant role in cell wall integrity (Lee and Elion, 1999).
The OCH1 gene encodes an α-1,6-mannosyltransferase, which is involved in initiation of mannose outer chain elongation of N-linked oligosaccharides (Nakayama et al., 1992; Nakanishi-Shindo et al., 1993; Lehle et al., 1995). Mannan is an important structural component of the cell wall, and och1 null mutants display reduced cell wall integrity and are hypersensitive to calcofluor white, hygromycin B, and SDS (Lee and Elion, 1999). In addition, och1 mutants have defects in cell division. The frequency of multibudded, multinucleate, and anucleate cells were increased in och1 cells (Mondesert et al., 1997; Lee and Elion, 1999). Furthermore both the STE vegetative growth (SVG) pathway, which plays a role parallel to that of the PKC pathway in maintaining cell wall integrity, and the pheromone response pathway are activated in och1 mutants (Lee and Elion, 1999; Cullen et al., 2000). Hence the elevated expression of OCH1 in response to hypotonic stress potentially has widespread consequences for the cell.
skn7 mutants are hypersensitive to hygromycin B, a phenotype associated with glycosylation mutants. The D427 dependence of the hygromycin hypersensitive phenotype in skn7 mutants and the hyperresistance phenotype of sln1* mutants suggests that one or more targets of the SLN1–SKN7 pathway are required for normal glycosylation. Preliminary experiments in which the expression of OCH1 in high-copy failed to rescue the hygromycin hypersensitivity of a skn7 deletion mutant point to the existence of additional glycosylation genes as targets of this pathway. Genome-wide expression experiments are in progress that will test this hypothesis.
ACKNOWLEDGMENTS
The authors thank Scott Moye-Rowley, Bob Malone, Cheryl Malone, and Mei-Yeh Jade Lu for critical reading of the manuscript; members of the Deschenes and Fassler laboratory for lively debate; and Lee Johnston for providing the Skn7 antibody. This work was supported by grant GM56719 from the National Institutes of Health.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.01-09-0434. Article and publication date are at www.molbiolcell.org/cgi/10.1091/mbc.01–09-0434.
REFERENCES
- Aiba H, Nakasai F, Mizushima S, Mizuno T. Evidence for the physiological importance of the phosphotransfer between the two regulatory components, EnvZ and OmpR, in osmoregulation in Escherichia coli. J Biol Chem. 1989;264:14090–14094. [PubMed] [Google Scholar]
- Amsler CD, Matsumura P. In: Chemotactic signal transduction in Escherichia coli and Salmonella typhimurium. In: Two-Component Signal Transduction. Hoch J A, Silhavy T J, editors. Washington, DC: ASM; 1995. pp. 89–103. [Google Scholar]
- Baikalov I, Schroder I, Kaczor-Grzeskowiak M, Cascio D, Gunsalus RP, Dickerson RE. NarL dimerization? Suggestive evidence from a new crystal form. Biochemistry. 1998;37:3665–3676. doi: 10.1021/bi972365a. [DOI] [PubMed] [Google Scholar]
- Baikalov I, Schroder I, Kaczor-Grzeskowiak M, Grzeskowiak K, Gunsalus RP, Dickerson RE. Structure of the Escherichia coli response regulator NarL. Biochemistry. 1996;35:11053–11061. doi: 10.1021/bi960919o. [DOI] [PubMed] [Google Scholar]
- Bouquin N, Johnson AL, Morgan BA, Johnston LH. Association of the cell cycle transcription factor Mbp1 with the Skn7 response regulator in budding yeast. Mol Biol Cell. 1999;10:3389–3400. doi: 10.1091/mbc.10.10.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JL, Bussey H, Stewart RC. Yeast Skn7p functions in a eukaryotic two-component regulatory pathway. EMBO J. 1994;13:5186–5194. doi: 10.1002/j.1460-2075.1994.tb06849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JL, North S, Bussey H. SKN7, a yeast multicopy suppressor of a mutation affecting cell wall β-glucan assembly, encodes a product with domains homologous to prokaryotic two component regulators and to heat shock transcription factors. J Bacteriol. 1993;175:6908–6915. doi: 10.1128/jb.175.21.6908-6915.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I. The transcriptional program of sporulation in budding yeast [published erratum appears in Science 1998 Nov 20;282(5393):1421] Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- Cosgrove D. Biophysical control of plant cell growth. Annu Rev Plant Physiol. 1986;37:377–405. doi: 10.1146/annurev.pp.37.060186.002113. [DOI] [PubMed] [Google Scholar]
- Cullen PJ, Schultz J, Horecka J, Stevenson BJ, Jigami Y, Sprague GF., Jr Defects in protein glycosylation cause SHO1-dependent activation of a STE12 signaling pathway in yeast. Genetics. 2000;155:1005–1018. doi: 10.1093/genetics/155.3.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale JE, Sutcliffe JF. Water relations of plant cells. Plant Physiology. A Treatise. Vol. IX: Water and Solutes in Plants. New York: Academic Press; 1986. pp. 1–48. [Google Scholar]
- Dean N. Yeast glycosylation mutants are sensitive to aminoglycosides. Proc Natl Acad Sci USA. 1995;92:1287–1291. doi: 10.1073/pnas.92.5.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–6. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Fassler JS, Gray WM, Malone CL, Tao W, Lin H, Deschenes RJ. Activated alleles of yeast SLN1 increase Mcm1-dependent reporter gene expression and diminish signaling through the Hog1 osmosensing pathway. J Biol Chem. 1997;272:13365–13371. doi: 10.1074/jbc.272.20.13365. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B. A technique for radiolabelling DNA restriction fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B. A technique for radiolabelling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1984;137:266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Hakenbeck R, Stock JB. Analysis of two-component signal transduction systems involved in transcriptional regulation. Methods Enzymol. 1996;273:281–300. doi: 10.1016/s0076-6879(96)73026-4. [DOI] [PubMed] [Google Scholar]
- Hubl ST, Owens JC, Nelson HC. Mutational analysis of the DNA-binding domain of yeast heat shock transcription factor. Nat Struct Biol. 1994;1:615–620. doi: 10.1038/nsb0994-615. [DOI] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketela T, Brown JL, Stewart RC, Bussey H. Yeast Skn7p activity is modulated by the Sln1p-Ypd1p osmosensor and contributes to regulation of the HOG pathway. Mol Gen Genet. 1998;259:372–378. doi: 10.1007/s004380050824. [DOI] [PubMed] [Google Scholar]
- Ketela T, Green R, Bussey H. Saccharomyces cerevisiae mid2p is a potential cell wall stress sensor and upstream activator of the PKC1-MPK1 cell integrity pathway. J Bacteriol. 1999;181:3330–3340. doi: 10.1128/jb.181.11.3330-3340.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krems B, Charizanis C, Entian K-D. The response regulator-like protein Pos9/Skn7 of Saccharomyces cerevisiae is involved in oxidative stress resistance. Curr Genet. 1996;29:327–334. doi: 10.1007/BF02208613. [DOI] [PubMed] [Google Scholar]
- Lee BN, Elion EA. The MAPKKK Ste11 regulates vegetative growth through a kinase cascade of shared signaling components. Proc Natl Acad Sci USA. 1999;96:12679–12684. doi: 10.1073/pnas.96.22.12679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehle L, Eiden A, Lehnert K, Haselbeck A, Kopetzki E. Glycoprotein biosynthesis in Saccharomyces cerevisiae: ngd29, an N-glycosylation mutant allelic to och1 having a defect in the initiation of outer chain formation. FEBS Lett. 1995;370:41–45. doi: 10.1016/0014-5793(95)00789-c. [DOI] [PubMed] [Google Scholar]
- Li S, Ault A, Malone CL, Raitt D, Dean S, Johnston LH, Deschenes RJ, Fassler JS. The yeast histidine protein kinase, Sln1p, mediates phosphotransfer to two response regulators, Ssk1p and Skn7p. EMBO J. 1998;17:6952–6962. doi: 10.1093/emboj/17.23.6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luyten K, Albertyn J, Skibbe WF, Prior BA, Ramos J, Thevelein JM, Hohmann S. Fps1, a yeast member of the MIP family of channel proteins, is a facilitator for glycerol uptake and efflux and is inactive under osmotic stress. EMBO J. 1995;14:1360–1371. doi: 10.1002/j.1460-2075.1995.tb07122.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnab R. Flagellar switch. In: Hoch JA, Silhavy TJ, editors. Two-Component Signal Transduction. Washington DC: ASM; 1995. pp. 181–199. [Google Scholar]
- Maeda T, Wurgler-Murphy SM, Saito H. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature. 1994;369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- Martinez de Maranon M, Marechal PA, Gervais P. Passive response of Saccharomyces cerevisiae to osmotic shifts: cell volume variations depending on the physiological state. Biochem Biophys Res Commun. 1996;227:519–523. doi: 10.1006/bbrc.1996.1539. [DOI] [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- Mondesert G, Clarke DJ, Reed SI. Identification of genes controlling growth polarity in the budding yeast Saccharomyces cerevisiae: a possible role of N-glycosylation and involvement of the exocyst complex. Genetics. 1997;147:421–434. doi: 10.1093/genetics/147.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan BA, Banks GR, Toone WM, Raitt D, Kuge S, Johnston LH. The Skn7 response regulator controls gene expression in the oxidative stress response of the budding yeast Saccharomyces cerevisiae. EMBO J. 1997;16:1035–1044. doi: 10.1093/emboj/16.5.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan BA, Bouquin N, Merrill GF, Johnston LH. A yeast transcription factor bypassing the requirement for SBF and =DSC1/MBF in budding yeast has homology to bacterial signal transduction proteins. EMBO J. 1995;14:5679–5689. doi: 10.1002/j.1460-2075.1995.tb00255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagawa F, Fink GR. The relationship between the “TATA” sequence and transcription initiation sites at the HIS4 gene of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1985;82:8557–8561. doi: 10.1073/pnas.82.24.8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi-Shindo Y, Nakayama KI, Tanaka A, Toda Y, Jigami Y. Structure of the N-linked oligosaccharides that show the complete loss of alpha-1,6-polymannose outer chain from och1, och1 mnn1, and och1 mnn1 alg3 mutants of Saccharomyces cerevisiae. J Biol Chem. 1993;268:26338–26345. [PubMed] [Google Scholar]
- Nakayama K, Nagasu T, Shimma Y, Kuromitsu J, Jigami Y. OCH1 encodes a novel membrane bound mannosyltransferase: outer chain elongation of asparagine-linked oligosaccharides. EMBO J. 1992;11:2511–2519. doi: 10.1002/j.1460-2075.1992.tb05316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrander DB, Gorman JA. The extracellular domain of the Saccharomyces cerevisiae Sln1p membrane osmolarity sensor is necessary for kinase activity. J Bacteriol. 1999;181:2527–2534. doi: 10.1128/jb.181.8.2527-2534.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posas F, Saito H. Activation of the yeast SSK2 MAP kinase kinase kinase by the SSK1 two-component response regulator. EMBO J. 1998;17:1385–1394. doi: 10.1093/emboj/17.5.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posas F, Wurgler-Murphy SM, Maeda T, Witten EA, Thai TC, Saito H. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell. 1996;86:865–875. doi: 10.1016/s0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- Raitt DC, Johnson AL, Erkine AM, Makino K, Morgan B, Gross DS, Johnston LH. The skn7 response regulator of Saccharomyces cerevisiae interacts with hsf1 in vivo, and is required for the induction of heat shock genes by oxidative stress. Mol Biol Cell. 2000;11:2335–2347. doi: 10.1091/mbc.11.7.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks JB. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1986. pp. 163–167. [Google Scholar]
- Shukla D, Zhu XY, Matsumura P. Flagellar motor-switch binding face of CheY and the biochemical basis of suppression by CheY mutants that compensate for motor-switch defects in Escherichia coli. J Biol Chem. 1998;273:23993–23999. doi: 10.1074/jbc.273.37.23993. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegelman GB, Bird TH, Voon V. Transcription regulation by the Bacillus subtilis response regulator Spo0A. In: Hoch JA, Silhavy TJ, editors. Two-Component Transduction. Washington, DC: ASM; 1995. pp. 159–179. [Google Scholar]
- Stathopoulos AM, Cyert MS. Calcineurin acts through the CRZ1/TCN1-encoded transcription factor to regulate gene expression in yeast. Genes Dev. 1997;11:3432–3444. doi: 10.1101/gad.11.24.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamas MJ, Luyten K, Sutherland FC, Hernandez A, Albertyn J, Valadi H, Li H, Prior BA, Kilian SG, Ramos J, Gustafsson L, Thevelein JM, Hohmann S. Fps1p controls the accumulation and release of the compatible solute glycerol in yeast osmoregulation. Mol Microbiol. 1999;31:1087–1104. doi: 10.1046/j.1365-2958.1999.01248.x. [DOI] [PubMed] [Google Scholar]
- Tao W, Deschenes RJ, Fassler JS. Intracellular glycerol levels modulate the activity of Sln1p, a Saccharomyces cerevisiae two-component regulator. J Biol Chem. 1999;274:360–367. doi: 10.1074/jbc.274.1.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams KE, Cyert MS. The eukaryotic response regulator Skn7p regulates calcineurin signaling through stabilization of Crz1p. EMBO J. 2001;20:3473–3483. doi: 10.1093/emboj/20.13.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman C, Rombel I, North AK, Bustamante C, Kustu S. Unusual oligomerization required for activity of NtrC, a bacterial enhancer-binding protein. Science. 1997;275:1658–1661. doi: 10.1126/science.275.5306.1658. [DOI] [PubMed] [Google Scholar]
- Yu G, Fassler JS. SPT13/GAL11 of Saccharomyces cerevisiae negatively regulates activity of the MCM1 transcription factor in Ty1 elements. Mol Cell Biol. 1993;13:63–71. doi: 10.1128/mcb.13.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]