Abstract
Background & objectives:
Infection from fluoroquinolone-resistant extra-intestinal Escherichia coli is a global concern. In this study, isolation and characterization of fluoroquinolone-resistant extra-intestinal E. coli isolates obtained from hospital samples were undertaken to detect plasmid-mediated quinolone resistance (PMQR) genes.
Methods:
Forty three isolates of E. coli obtained from patients with extra-intestinal infections were subjected to antibiogram to detect fluoroquinolone resistance. The mechanism of fluoroquinolone resistance was determined by the detection of PMQR genes and mutations in quinolone resistance determining region (QRDR).
Results:
Of the 43 isolates, 36 were resistant to nalidixic acid (83.72%) and 28 to ciprofloxacin (65.11%). Eight E. coli isolates showed total resistance to both the antimicrobials without any minimum inhibitory concentration. The detection of PMQR genes with qnr primers showed the presence of qnrA in two, qnrB in six and qnrS in 21 isolates. The gene coding for quinolone efflux pump (qepA) was not detected in any of the isolates tested. The presence of some unexpressed PMQR genes in fluoroquinolone sensitive isolates was also observed.
Interpretation & conclusions:
The detection of silent PMQR genes as observed in the present study presents a risk of the transfer of the silent resistance genes to other microorganisms if present in conjugative plasmids, thus posing a therapeutic challenge to the physicians. Hence, frequent monitoring is to be done for all resistance determinants.
Keywords: Antibiotic resistance, plasmid-mediated quinolone resistances, quinolone resistance determining regions
The increasing trend of antibiotic resistance among bacterial pathogens is a cause of global concern1. Escherichia coli, a member of the family Enterobacteriaceae, is known to cause extra-intestinal infections frequently showing resistance to fluoroquinolones2. Although a normal flora in the intestinal tract of human and animals, pathogenic strains of E. coli cause intestinal infections such as gastroenteritis and extra-intestinal conditions such as urinary tract infection (UTI), meningitis, septicaemia, nosocomial pneumonia, osteomyelitis and wound infections. Virulence factors, such as adhesins and exotoxins, play an important role in the pathogenesis of this microorganism3. Although the major reservoir of extra-intestinal pathogenic E. coli (ExPEC) causing infection remains the alimentary tract, other sources, such as contaminated food, are also incriminated4. ExPEC is known to harbour specialized virulence factors to cause extra-intestinal disease particularly UTI infections5,6. Such isolates can pose a significant threat to public health since these can also harbour resistance determinants that can make a pathogen resistant to multiple antimicrobial classes including present generation cephalosporins and fluoroquinolones7.
Fluoroquinolones are an effective class of drugs used by clinicians for treating the infections of Gram-negative pathogens including UTI and hospital-acquired infections. However, the indiscriminate use of these antimicrobials has increased the prevalence of quinolone and fluoroquinolone resistance in bacterial pathogens usually mediated by point mutations in topoisomerase II (gyrA and gyrB) and topoisomerase IV (parC and parE) genes, as well as by the overexpression of efflux pumps8. In addition, plasmid-mediated quinolone resistance (PMQR) genes (qnrA, qnrB, qnrC, qnrS, qnrD, qnrE and qnrVC), associated with a modified aminoglycoside acetyltransferase gene [aac (6′)-1b- cr] and a specific quinolone efflux pump qepA and oqxAB have also been described in Enterobacteriaceae2,9,10. The PMQR genes in bacteria are known to display reduced susceptibility to fluoroquinolones; however, these may not present mutations in quinolone resistance determining region (QRDR)11.
Plasmid-mediated quinolone resistance mechanism is a cause of concern since PMQR genes are located on conjugative plasmids and have been shown to disseminate fluoroquinolone resistance in E. coli isolates. The pentapeptide protein complex encoded by qnr determinants are thought to bind to topoisomerase II preventing it from the action of fluoroquinolones, aac(6’)-lb-cr is known to modify fluoroquinolones with piperazinyl moiety, while plasmid-mediated qepA encodes an efflux pump of major facilitator family12.
Material & Methods
Bacterial strains & determination of antimicrobial susceptibility testing: Phenotypic identification of E. coli isolates (n=43) from clinical samples obtained from Madras Medical Mission, Department of Microbiology, Mogappair and Chennai, India, over the period of two years (2015-2017) was done and these isolates were subjected to antibiotic sensitivity testing. The isolates were analyzed for quinolone/fluoroquinolone susceptibility using disc diffusion assay according to the Clinical and Laboratory Standards Institute (CLSI) guidelines13. Briefly, the bacterial isolates with the density of 0.5 McFarland turbidity were swabbed onto the pre-poured and dried Mueller Hinton agar (HiMedia Laboratories Pvt. Ltd., Mumbai). The antibiotic discs of ciprofloxacin (CIP) (5 μg) and nalidixic acid (NA) (30 μg) (HiMedia) were placed on the bacterial lawn using a sterile applicator. After overnight incubation at 37°C, inhibition zone diameters were measured and interpreted as resistant, sensitive or intermediate sensitive as per CLSI guidelines13. E. coli (ATCC 25922) was used as a quality control strain.
Characterization of PMQR genes: All the 43 isolates were checked for genus specific gene and for the presence of PMQR genes using PCR14,15. PCR was performed in 30 μl reaction volumes containing 3 μl of 10 × buffer [100 mM Tris-HCl (pH 9), 1.5 mM MgCl2, 50 mM KCl and 1% gelatine], 100 μM of four deoxyribonucleotide triphosphates each (dATP, dGTP, dCTP and dTTP), 10 pmol of each forward and reverse primers and 1.0 U of Taq DNA polymerase with 2 μl of template DNA. All the isolates were tested for the presence of PMQR genes using primers listed in the Table I. The amplified PCR products were further purified using a QIAquick PCR purification Kit (Qiagen, Hilden, Germany).
Table I.
Oligonucleotide primers used for determining gyrase and topoisomerase IV target genes
| Primers | Sequence (5’- 3’) | Annealing temperature | References |
|---|---|---|---|
| Genus specific primer | |||
| uidA | F-AAAACGGCAAGAAAAAGCAG R-ACGCGTGGTTACAGTCTTGCG | 63ºC | 14 |
| QRDR mutation detection primers | |||
| gyrA | F-GAC CTT GCG AGA GAA ATT ACA C R-GAT GTT GGT TGC CAT ACC TAC G | 55°C | 16 |
| parC | F-CGG AAA ACG CCT ACT TAA ACT A R-GTG CCG TTA AGC AAA ATG T | 55°C | 16 |
| MAMA gyrA83 | R-TCG TGT CAT AGA CCG GGC | 55°C | 16 |
| MAMA gyrA87 | R-GCG CCA TGC GGA CGA TCG TTT C | 55°C | 16 |
| MAMA parC80 | R-ATC GCT TCA TAA CAG GCT CT | 55°C | 16 |
| MAMA parC84 | R- CCA TCA GGA CCA TCG CCT C | 55°C | 16 |
| gyrB | F-GTG AAA TGA CCC GCC GTA AA R-TGA TAA GCG TCG CCA CTT CC | 55°C | 17 |
| ParE | F-TCT GGC CGG ATG AAA CCT TC R-TTT CAG TGG CAT GAT CGC CT | 55°C | 17 |
| MAMA gyrB 447 | R-GAC GTT GAG GAT TTT ACC CTC | 55°C | 17 |
| MAMA parE 416 | R-GCG GAG TCA CCT TCC ACT AG | 55°C | 17 |
| PMQR detection primers | |||
| qnrA | F: ATTTCTCACGCCAGGATTTG R: GATCGGCAAAGGTTAGGTCA | 55°C | 15 |
| qnrB | F: GATCGTGAAAGCCAGAAAGG R: ACGATGCCTGGTAGTTGTCC | 55°C | 15 |
| qnrS | F: ACGACATTCGTCAACTGCAA R: TAAATTGGCACCCTGTAGGC | 55°C | 15 |
| qepA | F: CGTGTTGCTGGAGTTCTTC R: CTGCAGGTACTGCGTCATG | 59°C | 15 |
PMQR, plasmid-mediated quinolone resistance; QRDR, quinolone resistance determining region; MAMA, mismatch amplification mutation assays
Detection of mutation in QRDR using mismatch amplification mutation-MAMA-PCR: A MAMA assay was performed on the 43 isolates to detect the point mutations in the QRDR16. The known point mutations at amino acid position 83 and 87 of gyrA, 80 and 84 of parC, 447 of gyrB and 416 amino acid position of parE were targeted17. The primers used in the study are outlined in Table I.
PCR was performed in 30 μl reaction volumes containing 3 μl of 10X Taq buffer, 83 μM of four deoxyribonucleotide triphosphates, 30, 20 and 10 picomoles of forward, MAMA reverse and control reverse primers, respectively and 1 U of Taq DNA polymerase with 2 μl of DNA as a template. PCR amplification was carried out in a thermal cycler (Bio-Rad, USA) with the initial denaturation at 95°C for 5 min followed by 35 cycles of denaturation at 94°C, annealing at 55°C and extension at 72°C for 40 seconds respectively, and a final extension at 72°C for 10 min. PCR products were visualized on two per cent agarose gel stained with ethidium bromide (0.5 μg/ml) in 1× tris-acetate EDTA (TAE) buffer loaded with 10 μl of the reaction mixture and observed under UV light in a Gel Documentation system (Bio-Rad, USA).
DNA sequence analysis: The purified PCR products were sequenced in an automated ABI 3100 Genetic analyser (Applied Biosystems, USA) using fluorescent label dye terminators. The nucleotide sequences were analyzed using BLAST programmes, blastn and blastp (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The amino acids deduced from the DNA sequences were obtained through a web-based programme Expasy Translate tool (https://web.expasy.org/translate/) following which the novel sequences were submitted to the GenBank.
Results
Forty-three extra-intestinal isolates of E. coli obtained from biological samples from patients with UTIs (21), wound infections (6), neonatal meningitis (5), septicaemia (5), nosocomial pneumonia (4) and osteomyelitis (2) from all age groups confirmed by phenotypic tests were reconfirmed as E. coli by PCR-based genotypic test for uidA gene.
Antibiogram analysis: Of the 43 quinolone/fluoroquinolone-resistant isolates, 28 (65.11%) were found to be resistant to ciprofloxacin while 36 (83.72%) were resistant to nalidixic acid. Seven isolates were sensitive to both the antibiotics and three showed intermediate sensitivity to ciprofloxacin.
Characterization of PMQR genes: Results of plasmid-mediated quinolone resistance genes identified by PCR using PMQR primers revealed 29 of the 43 isolates (67.44%) harbouring PMQR genes. Among these, two (6.89%), six (20.68%) and 21 (72.41%) were positive for qnrA, qnrB and qnrS, respectively (Fig. 1A and B). Seven (16.27%) isolates which were sensitive to both the antibiotics by phenotypic test, possessed plasmid-mediated quinolone resistance genes, one of which was (qnrB) sequenced and submitted to the GenBank.
Fig. 1.
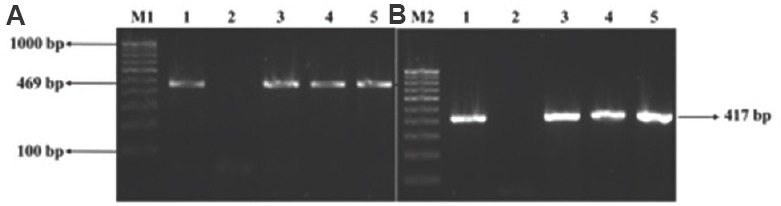
Agarose gel electrophoresis image of PMQR genes (A) M1: 100 bp DNA ladder; lane 1: qnrB PCR product; lane 2: negative control; lanes 3-5: isolates positive for qnrB. (B) M2: 100 bp DNA ladder; lane 1: qnrS PCR product; lane 2: negative control; lanes 3-5: isolates positive for qnrB. PMQR, plasmid-mediated quinolone resistance.
Detection of mutations in QRDR using MAMA-PCR: The MAMA PCR in the absence of mutations(s) in sensitive isolates generated two PCR products from the wild-type gene using universal forward/ reverse and MAMA reverse primers whereas, a single amplicon was produced in resistant isolates with QRDR mutation(s) due to the inhibition of PCR in the presence of two or more mismatches at the 3’end of the MAMA primer (Fig. 2A–D). Nine (20.9%) of the 43 isolates resistant to both the antimicrobial agents harboured mutation only at amino acid position 83 of gyrA while one of the isolates possessed mutation only at position 87 of the same gene. Thirteen isolates (30.23%) resistant to both the antibiotics presented mutations at gyrA 83, 87 and at parC 80 regions. Two (4.65%) of the isolates which were resistant to only nalidixic acid harboured mutations at both gyrA position 83 and 87. Four (9.30%) isolates showing resistance to both the antibiotics harboured mutation at amino acid position 83 of gyrA and 80 of parC while four (9.30%) isolates had mutation at position 83 of gyrA and 84 of parC. An isolate of E. coli resistant to both the antibiotics displayed mutation at all the four important positions of QRDR (gyrA 83, 87 and parC 80, 84) (Table II). An isolate resistant to nalidixic acid did not harbour any mutation in the QRDR. None of the isolates showed mutation in the gyrB and parE regions. The PCR products of gyrA and parC of one of the representative E. coli isolate were sequenced and analyzed. The partial sequences with possible QRDR mutations were submitted to the GenBank and were assigned GenBank accession numbers MF288967 for gyrA and MF2889868 for parC.
Fig. 2.
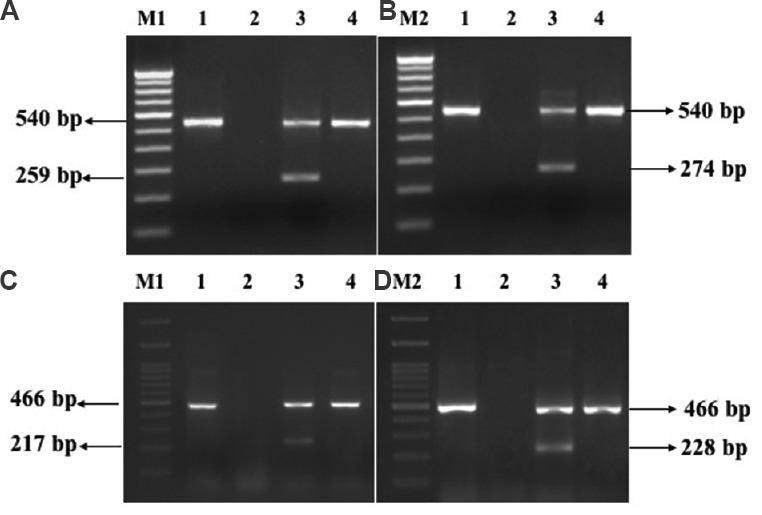
PCR products of duplex MAMA-PCR assays (A) MAMA gyrA 83. M1: 100 bp DNA ladder; lane 1: gyrA PCR; lane 2: negative control; lane 3: gyrA 83 without mutation; lane 4: gyrA 83 with mutation. (B) MAMA gyrA 87. M2:100 bp DNA ladder; lane 1: gyrA PCR; lane 2: negative control; lane 3: gyrA 87 without mutation; lane 4: gyrA 87 with mutation. (C) MAMA parC 80. M1:100 bp DNA ladder; lane 1: parC PCR; lane 2: negative control; lane 3: parC 80 without a mutation; lane 4: parC 80 with a mutation. (D) MAMA parC 84. M2:100 bp DNA ladder; lane 1: parC PCR; lane 2: negative control; lane 3: parC 84 without mutation; lane 4: parC 84 without mutation. MAMA-PCR, mismatch amplification mutation assays-polymerase chain reaction.
Table II.
Mutation status of Escherichia coli isolates at topoisomerase targets
| Total number of mutations | Number (%) of isolates | Alterations detected by MAMA-PCR | |||||
|---|---|---|---|---|---|---|---|
| gyrA 83 | gyrA 87 | parC 80 | parC 84 | gyrB 447 | parE 416 | ||
| 1 | 9 (20.93) | Mutation | None | None | None | None | None |
| 1 (2.32) | None | Mutation | None | None | None | None | |
| 2 | 4 (9.30) | Mutation | None | Mutation | None | None | None |
| 2 (4.65) | Mutation | Mutation | None | None | None | None | |
| 4 (9.30) | Mutation | None | None | Mutation | None | None | |
| 3 | 13 (30.23) | Mutation | Mutation | Mutation | None | None | None |
| 4 | 1 (2.32) | Mutation | Mutation | Mutation | Mutation | None | None |
MAMA, mismatch amplification mutation assays
Discussion
Fluoroquinolones are a major class of antimicrobial drugs used widely for the treatment of infections caused by Gram-negative bacterial pathogens. E. coli being one of the major causes of several extra-intestinal and hospital-acquired infections is recognized as a major problem to tackle as it shows resistance to most of the quinolones and fluoroquinolones. Mechanisms underlying fluoroquinolone resistance were earlier thought to be confined to vertical inheritance due to the spontaneous occurrence of point mutations in QRDR regions18. However, now it is reported to be spread horizontally using plasmid-mediated qnr genes and efflux pump genes19. Although, in most of the cases, fluoroquinolone resistance is attributed to mutations in the QRDR regions, a reasonable percentage of isolates in this study also harboured PMQR genes and is in agreement with the reports of Kao et al20. Among the PMQR positive isolates the highest percentage harboured qnrS (77.77%) followed by qnrB (22.22%) and qnrA (11.11%); however, qepA was not observed in any of the isolates. In contrast, a study from China21 showed presence of qnrS in isolates followed by qepA with the absence of qnrA and qnrB. Another study from China22 showed the presence of qnr, aac(6′)-Ib-cr, qepA and oqxAB in 2.7, 24.5, 11.9 and 6.3 per cent, respectively of fluoroquinolone-resistant E. coli isolates. In a study from Korea, Yang et al23 observed PMQR genes in 73.8 per cent of ciprofloxacin-resistant E. coli isolates. Although, in our study, the sample size was small, yet our results showed the presence of PMQR genes in 67.44 per cent of the E. coli isolates included. PMQRs, such as qepA and aac-(6′)-Ib, were found to be dominant in the aquatic environments24. This observation supported the notion that the aquatic environment might constitute the original source of PMQR genes25. Our study also demonstrated that extra-intestinal E. coli isolates might carry silent antibiotic resistance genes, since a few fluoroquinolone sensitive isolates (16.27%) of E. coli harboured PMQRs such as qnrB and qnrS. Perhaps this gene did not express in these isolates as phenotypic resistance. The reason for the silent nature of the qnrS and qnrB of the sensitive isolates needs further study. Silencing of antibiotic resistance genes may be a phenomenon that has not received much attention. Enne et al26 reported silencing of several plasmid-borne (pVE46) antibiotic resistance genes such as blaoxa-2, aadA1, sul1 and tetA in E. coli isolated from pig following oral inoculation of organic piglets. There was no deletion of genes or promoter regions. However, the silent resistance genes were expressed again when the plasmid carrying resistance genes transferred to a new host. This suggested that the silencing phenomenon was due to the chromosomal effects of the host. Later, it was also found that the silencing was reversed at a low frequency of 10−6-10−10 in the original host26. Deekshit et al27 suggested that the deletion of promoter region was the main reason for unexpressive nature of the chloramphenicol acetyltransferase (catA) gene in Salmonella Weltevreden. These reports show the future risk associated with the global emergence of plasmid-borne resistance pattern of clinical pathogens and these PMQRs can also contribute to the elevated levels of ciprofloxacin minimum inhibitory concentrations (MICs) in clinical isolates.
All the fluoroquinolone-resistant isolates harbouring PMQR genes also had QRDR mutations. A MAMA-PCR was used to detect mutations at four major QRDRs (gyrA 83, gyrA 87, parC 80 and parC 84). It has been used to detect point mutations in gyrA and parC regions in fluoroquinolone-resistant bacterial pathogens28,29,30. The point mutation S83L at gyrA 83 was the most commonly observed change31,32 followed by mutation at gyrA 87. In the present study, 76.44 per cent (33/43) of the isolates harboured point mutation at gyrA 83 and 39.53 per cent (17/43) at gyrA 87. Thus, it is important to analyze these regions to check the multiple mutation status of resistant pathogens.
Among the 43 isolates of E. coli, seven different patterns of QRDR mutations were observed. Although, in the present study, all the mutations at gyrA 87 were associated with mutation at gyrA 83, one nalidixic acid-resistant isolate (S5) harboured mutation only at gyrA position 87. Higher levels of quinolone/fluoroquinolone resistance with increased MICs in many bacterial pathogens are usually associated with double mutations in gyrA region33. Similarly, in our study, the isolates resistant to both the antibiotics harboured double mutation at gyrA and other regions. Some of the nalidixic acid-resistant isolates did not show any mutation in the QRDRs as reported by us earlier also34. Since PMQRs are mainly known to increase the MIC in resistant isolates, its exact relevance as a sole mechanism of fluoroquinolone resistance needs further study.
In conclusion, our study showed the occurrence of plasmid-mediated resistance as the second most encountered fluoroquinolone resistance mechanism among clinical isolates of E. coli. In addition, the detection of silent PMQR genes in sensitive isolates could pose a future risk of these isolates transforming into resistant forms upon antibiotic challenge. Hence, it is advisable to check for the presence of resistance determinants even in phenotypic sensitive isolates.
Footnotes
Financial support & sponsorship: Financial support received by the last author (IK) from Nitte University intramural research fund and Indian Council of Medical Research (AMR/37/2011-ECD-1) towards this study is acknowledged.
Conflicts of Interest: None.
References
- 1.Jones-Dias D, Manageiro V, Graça R, Sampaio DA, Albuquerque T, Themudo P, et al. QnrS1- and Aac(6’)-Ib-cr-producing Escherichia coli among isolates from animals of different sources: Susceptibility and genomic characterization. Front Microbiol. 2016;7:671. doi: 10.3389/fmicb.2016.00671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aoike N, Saga T, Sakata R, Yoshizumi A, Kimura S, Iwata M, et al. Molecular characterization of extraintestinal Escherichia coli isolates in Japan: Relationship between sequence types and mutation patterns of quinolone resistance-determining regions analyzed by pyrosequencing. J Clin Microbiol. 2013;51:1692–8. doi: 10.1128/JCM.03049-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vincent C, Boerlin P, Daignault D, Dozois CM, Dutil L, Galanakis C, et al. Food reservoir for Escherichia coli causing urinary tract infections. Emerg Infect Dis. 2010;16:88–95. doi: 10.3201/eid1601.091118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manges AR, Smith SP, Lau BJ, Nuval CJ, Eisenberg JN, Dietrich PS, et al. Retail meat consumption and the acquisition of antimicrobial resistant Escherichia coli causing urinary tract infections: A case-control study. Foodborne Pathog Dis. 2007;4:419–31. doi: 10.1089/fpd.2007.0026. [DOI] [PubMed] [Google Scholar]
- 5.Johnson L, Sabel A, Burman WJ, Everhart RM, Rome M, MacKenzie TD, et al. Emergence of fluoroquinolone resistance in outpatient urinary Escherichia coli isolates. Am J Med. 2008;121:876–84. doi: 10.1016/j.amjmed.2008.04.039. [DOI] [PubMed] [Google Scholar]
- 6.Guo S, Brouwers HJ, Cobbold RN, Platell JL, Chapman TA, Barrs VR, et al. Fluoroquinolone-resistant extraintestinal pathogenic Escherichia coli, including O25b-ST131, isolated from faeces of hospitalized dogs in an Australian veterinary referral centre. J Antimicrob Chemother. 2013;68:1025–31. doi: 10.1093/jac/dks515. [DOI] [PubMed] [Google Scholar]
- 7.Motayo BO, Ogiogwa IJ, Okerentugba PO, Innocent-Adiele HC, Nwanze JC, Onoh CC, et al. Antimicrobial resistance profile of extra-intestinal Escherichia coli infections in a South-Western Nigerian City. J Microbiol Res. 2012;2:141–4. [Google Scholar]
- 8.Hawkey PM. Mechanisms of quinolone action and microbial response. J Antimicrob Chemother. 2003;51(Suppl 1):29–35. doi: 10.1093/jac/dkg207. [DOI] [PubMed] [Google Scholar]
- 9.Albornoz E, Tijet N, De Belder D, Gomez S, Martino F, Corso A, et al. qnrE1, a member of a new family of plasmid-located quinolone resistance genes, originated from the chromosome of Enterobacter species. Antimicrob Agents Chemother. 2017;61 doi: 10.1128/AAC.02555-16. pii: e02555-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xia R, Guo X, Zhang Y, Xu H. QnrVC-like gene located in a novel complex class 1 integron harboring the ISCR1 element in an Aeromonas punctata strain from an aquatic environment in Shandong province, China. Antimicrob Agents Chemother. 2010;54:3471–4. doi: 10.1128/AAC.01668-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Veldman K, Cavaco LM, Mevius D, Battisti A, Franco A, Botteldoorn N, et al. International collaborative study on the occurrence of plasmid-mediated quinolone resistance in Salmonella enterica and Escherichia coli isolated from animals, humans, food and the environment in 13 European countries. J Antimicrob Chemother. 2011;66:1278–86. doi: 10.1093/jac/dkr084. [DOI] [PubMed] [Google Scholar]
- 12.Jacoby GA, Gacharna N, Black TA, Miller GH, Hooper DC. Temporal appearance of plasmid-mediated quinolone resistance genes. Antimicrob Agents Chemother. 2009;53:1665–6. doi: 10.1128/AAC.01447-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.CLSI Document M100. 27th ed. Wayne, PA: CLSI; 2017. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. [Google Scholar]
- 14.Bej AK, DiCesare JL, Haff L, Atlas RM. Detection of Escherichia coli and shigella spp. in water by using the polymerase chain reaction and gene probes for uid. Appl Environ Microbiol. 1991;57:1013–7. doi: 10.1128/aem.57.4.1013-1017.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robicsek A, Jacoby GA, Hooper DC. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect Dis. 2006;6:629–40. doi: 10.1016/S1473-3099(06)70599-0. [DOI] [PubMed] [Google Scholar]
- 16.Qiang YZ, Qin T, Fu W, Cheng WP, Li YS, Yi G, et al. Use of a rapid mismatch PCR method to detect gyrA and parC mutations in ciprofloxacin-resistant clinical isolates of Escherichia coli. J Antimicrob Chemother. 2002;49:549–52. doi: 10.1093/jac/49.3.549. [DOI] [PubMed] [Google Scholar]
- 17.Jazeela K, Chakraborty G, Shetty SS, Rohit A, Karunasagar I, Vijaya Kumar D, et al. Comparison of mismatch amplification mutation assay PCR and PCR-restriction fragment length polymorphism for detection of major mutations in gyrA and parC of Escherichia coli associated with fluoroquinolone resistance. Microb Drug Resist. 2019;25 doi: 10.1089/mdr.2017.0351. doi 10.1089/mdr.2017.0351. [DOI] [PubMed] [Google Scholar]
- 18.Weigel LM, Steward CD, Tenover FC. gyrA Mutations associated with fluoroquinolone resistance in eight species of Enterobacteriaceae. Antimicrob Agents Chemother. 1998;42:2661–7. doi: 10.1128/aac.42.10.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nordmann P, Poirel L. Emergence of plasmid-mediated resistance to quinolones in Enterobacteriaceae. J Antimicrob Chemother. 2005;56:463–9. doi: 10.1093/jac/dki245. [DOI] [PubMed] [Google Scholar]
- 20.Kao CY, Wu HM, Lin WH, Tseng CC, Yan JJ, Wang MC, et al. Plasmid-mediated quinolone resistance determinants in quinolone-resistant Escherichia coli isolated from patients with bacteremia in a university hospital in Taiwan, 2001-2015. Sci Rep. 2016;6:32281. doi: 10.1038/srep32281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia LN, Li L, Wu CM, Liu YQ, Tao XQ, Dai L, et al. A survey of plasmid-mediated fluoroquinolone resistance genes from Escherichia coli isolates and their dissemination in Shandong, China. Foodborne Pathog Dis. 2010;7:207–15. doi: 10.1089/fpd.2009.0378. [DOI] [PubMed] [Google Scholar]
- 22.Zhao L, Zhang J, Zheng B, Wei Z, Shen P, Li S, et al. Molecular epidemiology and genetic diversity of fluoroquinolone-resistant Escherichia coli isolates from patients with community-onset infections in 30 Chinese county hospitals. J Clin Microbiol. 2015;53:766–70. doi: 10.1128/JCM.02594-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang HY, Nam YS, Lee HJ. Prevalence of plasmid-mediated quinolone resistance genes among ciprofloxacin-nonsusceptible Escherichia coli and Klebsiella pneumoniae isolated from blood cultures in Korea. Can J Infect Dis Med Microbiol. 2014;25:163–9. doi: 10.1155/2014/329541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan L, Liu D, Wang XH, Wang Y, Zhang B, Wang M, et al. Bacterial plasmid-mediated quinolone resistance genes in aquatic environments in China. Sci Rep. 2017;7:40610. doi: 10.1038/srep40610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poirel L, Cattoir V, Nordmann P. Plasmid-mediated quinolone resistance; interactions between human, animal, and environmental ecologies. Front Microbiol. 2012;3:24. doi: 10.3389/fmicb.2012.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Enne VI, Delsol AA, Roe JM, Bennett PM. Evidence of antibiotic resistance gene silencing in Escherichia coli. Antimicrob Agents Chemother. 2006;50:3003–10. doi: 10.1128/AAC.00137-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deekshit VK, Kumar BK, Rai P, Srikumar S, Karunasagar I, Karunasagar I, et al. Detection of class 1 integrons in Salmonella weltevreden and silent antibiotic resistance genes in some seafood-associated nontyphoidal isolates of Salmonella in South-West coast of India. J Appl Microbiol. 2012;112:1113–22. doi: 10.1111/j.1365-2672.2012.05290.x. [DOI] [PubMed] [Google Scholar]
- 28.Al-Marzooq F, Mohd Yusof MY, Tay ST. Molecular analysis of ciprofloxacin resistance mechanisms in Malaysian ESBL-producing Klebsiella pneumoniae isolates and development of mismatch amplification mutation assays (MAMA) for rapid detection of gyrA and parC mutations. Biomed Res Int. 2014;2014:601630. doi: 10.1155/2014/601630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen TN, Hotzel H, El-Adawy H, Tran HT, Le MT, Tomaso H, et al. Genotyping and antibiotic resistance of thermophilic Campylobacter isolated from chicken and pig meat in Vietnam. Gut Pathog. 2016;8:19. doi: 10.1186/s13099-016-0100-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cui M, Wu C, Zhang P, Wu C. Development of multiplex-mismatch amplification mutation-PCR assay for simultaneous detection of Campylobacter jejuni and mutation in gyrA gene related to fluoroquinolone resistance. Foodborne Pathog Dis. 2016;13:642–5. doi: 10.1089/fpd.2016.2169. [DOI] [PubMed] [Google Scholar]
- 31.Onseedaeng S, Ratthawongjirakul P. Rapid detection of genomic mutations in gyrA and parC genes of Escherichia coli by multiplex allele specific polymerase chain reaction. J Clin Lab Anal. 2016;30:947–55. doi: 10.1002/jcla.21961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deekshit VK, Kumar BK, Rai P, Karunasagar I, Karunasagar I. Differential expression of virulence genes and role of gyrA mutations in quinolone resistant and susceptible strains of Salmonella Weltevreden and Newport isolated from seafood. J Appl Microbiol. 2015;119:970–80. doi: 10.1111/jam.12924. [DOI] [PubMed] [Google Scholar]
- 33.Jurado S, Orden JA, Horcajo P, De La Fuente R, Ruiz-Santa-Quiteria JA, Martínez-Pulgarín S, et al. Characterization of fluoroquinolone resistance in Escherichia coli strains from ruminants. J Vet Diagn Invest. 2008;20:342–5. doi: 10.1177/104063870802000314. [DOI] [PubMed] [Google Scholar]
- 34.Santhosh KS, Deekshit VK, Venugopal MN, Karunasagar I, Karunasagar I. Multiple antimicrobial resistance and novel point mutation in fluoroquinolone-resistant Escherichia coli isolates from Mangalore, India. Microb Drug Resist. 2017;23:994–1001. doi: 10.1089/mdr.2016.0142. [DOI] [PubMed] [Google Scholar]


