Abstract
The Indian Council of Medical Research, in 2013, initiated the Antimicrobial Resistance Surveillance & Research Network (AMRSN) to enable compilation of data on six pathogenic groups on antimicrobial resistance from the country. The overarching aim of this network was to understand the extent and pattern of antimicrobial resistance (AMR) and use this evidence to guide strategies to control the spread of AMR. This article describes the conception and implementation of this AMR surveillance network for India. Also described are the challenges, limitations and benefits of this approach. Data from the Network have shown increasing resistance in Gram-negative bacteria in the hospitals that are part of this network. Combined resistance to third-generation cephalosporins and fluoroquinolones and increasing carbapenem resistance are worrisome, as it has an important bearing on the patients’ outcome and thus needs to be addressed urgently. Data generated through this Network have been used to develop treatment guidelines, which will be supportive in harmonizing treatment practices across the tertiary level healthcare institutions in the country. While, the major benefit of having a surveillance system is the collection of real-time accurate data on AMR including the mechanisms of resistance, representativeness to community, sustaining the current effort and expanding the current activities to next levels of healthcare settings are the major challenges. The data emanating from the network besides providing evidence, expose several gaps and lacunae in the ecosystem and highlight opportunities for action by multiple stakeholders.
Keywords: AMRSN, antimicrobial resistance, carbapenems, colistin, data management, Gram-negative, Indian Council of Medical Research, surveillance network
Introduction
Antimicrobial resistance (AMR) poses both health and economic burden for patients and healthcare systems, globally. India has a large burden of infectious diseases and is one among the largest consumers of antibiotics in the world1. The efficacy of several antibiotics is threatened by the emergence of resistant microorganisms. Multiple interlinked factors including high burden of disease, poor public health infrastructure, lack of appropriate diagnostic support, poor infection control practices and the tendency of clinicians to continue empirical treatment practices, have amplified the crisis of AMR in India2. Unregulated over-the-counter availability of antibiotics and non-compliance to the recommended treatment duration have been recognized as key drivers for the emergence of resistance in India3. The resistant bacterial strains emerging out of selection pressure spread either through the hospital-acquired infections or from the community. Non-availability of nationwide data on estimates of the extent of drug resistance, greatly limits the concerted response against AMR in India. Most of the AMR data available in the past have been from individual hospitals and from small networks, which did not represent the national picture.
To address this missing link, the Indian Council of Medical Research (ICMR), New Delhi, initiated the Antimicrobial Resistance Surveillance & Research Network (AMRSN) in 2013 to collate nationally representative data. Understanding of the molecular mechanisms of bacterial resistance, how bacteria evolve, acquire and transmit antibiotic resistance is vital for forecasting and addressing the problem4. Thus, the current ICMR effort also includes understanding the molecular mechanisms of resistance as part of the objectives of the Network.
The main goals of ICMR-AMRSN are to (i) establish network of hospitals to monitor trends in the antimicrobial susceptibility profile of clinically important bacteria and fungi limited to human health; (ii) include comprehensive molecular studies for identifying the clonality of drug-resistant pathogens and their transmission dynamics to enable a better understanding of AMR in the Indian context and develop suitable interventions; (iii) disseminate information on AMR in pathogenic organisms to stakeholders to promote interventions that reduce AMR; and (iv) create data management system for data collection and analysis.
In this report, the experience with the setting up and implementation of a hospital/laboratory-based surveillance system at major referral tertiary care hospitals of India is shared and also as to how the data being generated are/will be used to guide interventions.
ICMR-AMRSN
Constitution of an Advisory Group and finalization of plan
An Advisory Group with experts from clinical microbiology, infectious diseases, epidemiology, statistics, and having experience specifically in surveillance in the above contexts, was constituted in 2011 for developing a comprehensive plan on AMR surveillance. The experts brainstormed and identified the following six pathogens as focus areas for ICMR-AMRSN: (i) Enterobacteriaceae causing sepsis, (ii) Gram-negative non-fermenters, (iii) Enteric fever pathogens, (iv) Diarrhoeagenic bacterial organisms, (v) Gram-positives: staphylococci and Enterococci, and (vi) Fungal pathogens (excluded in the WHO priority pathogens)-yeasts (Candida and Cryptococcus spp.) and mycelial fungi (Aspergillus spp. and Zygomycetes spp.).
The pathogenic groups identified by the Advisory Group also aligned well with the WHO priority pathogen list released in 20175.
Selection of study sites and target patient populations
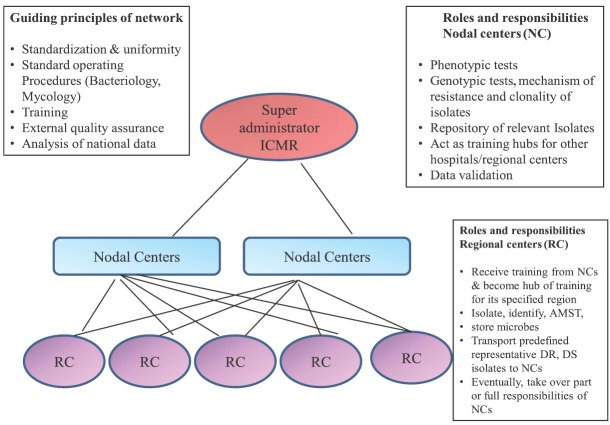
Six nodal centres (NCs) for each pathogenic group were identified in four tertiary care hospitals6 (Fig. 1 and Box 1). The principal investigators were identified for each NC and their roles and responsibilities were defined. The NC has been assigned the responsibility for antimicrobial susceptibility testing (AST), carrying out in-depth studies on resistance mechanisms and genetic marker analysis, provide training and also act as repository for isolates relevant to their pathogenic group. The surveillance network is managed by the coordinating centre at ICMR Headquarters, New Delhi, along with their NC6 (Fig. 1).
Fig. 1.
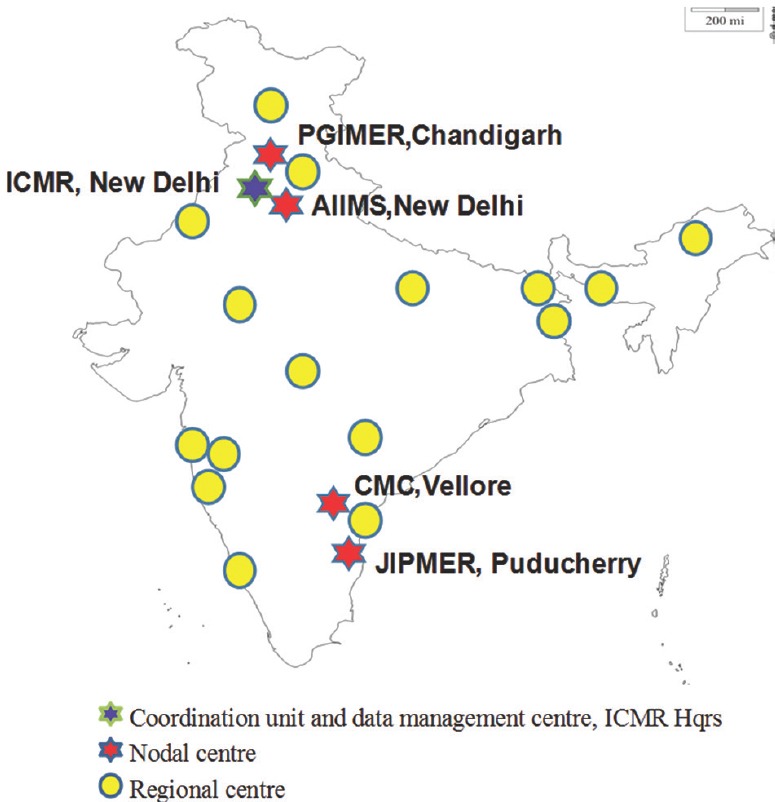
Nodal and regional centres for Antimicrobial Resistance Surveillance & Research Network System. AIIMS, All India Institute of Medical Sciences; CMC, Christian Medical College; ICMR, Indian Council of Medical Research; JIPMER, Jawaharlal Institute of Postgraduate Medical Education & Research; PGIMER, Postgraduate Institute of Medical Education and Research. Source: http://iamrsn.icmr.org.in/index.php/amrsn/amrsn-network.
Box 1.
Time line of expansion of AMRSN network
| Phase 1: Establishment of six Nodal Centres in 2013-2014 |
|---|
| AIIMS, New Delhi: Enteric pathogens |
| CMC, Vellore: GNNF, Diarrhoeal pathogens |
| JIPMER, Puducherry: Gram-positives |
| PGIMER, Chandigarh: Enterobacteriaceae, Fungal pathogens |
| Phase 2: Inclusion of six Regional Centres in 2017 |
| AFMC, Pune |
| Apollo, Chennai |
| Hinduja Hospital, Mumbai |
| MGIMS, Sevagaram |
| Sir Ganga Ram Hospital, New Delhi |
| Tata Medical Center, Kolkata |
| Phase 3: Inclusion of 10 Regional Centres in 2018 |
| AIIMS, Bhopal |
| AIIMS, Jodhpur |
| Assam Medical College, Dibrugarh |
| IPGMER, Kolkata |
| KGMU, Lucknow |
| Kasturba Medical College, Manipal |
| Nizam Hospital, Hyderabad |
| Regional Institute of Medical Sciences, Imphal |
| LTMGH, Sion, Mumbai |
| SKIMS, Srinagar |
GNNF, Gram-negative non-fermenters; JIPMER, Jawaharlal Institute of Postgraduate Medical Education and Research; RCs, regional centres; CMC, Christian Medical College; PGIMER, Post-graduate Institute of Medical Education and Research; IPGMER, Institute of Postgraduate Medical Education and Research; LTMGH, Lokmanya Tilak Municipal General Hospital and Medical College SKIMS, Sher-i-Kashmir Institute of Medical Sciences; AFMC, Armed Forces Medical College; AIIMS, All India Institute of Medical Sciences; MGIMS, Mahatma Gandhi Institute of Medical Sciences; KGMU, King George’s Medical University
Sixteen regional laboratories (regional centres, RCs) were included from tertiary care hospitals to provide data and fixed number of isolates for each pathogenic group6. The participating hospital sites were chosen based on the population attending the hospital, number of faculty, number of samples received and processed at the microbiology laboratory and to ensure that a pan Indian geographic representation of isolates was attained. The regional laboratories provide data for all the pathogenic groups, carry out AST and transfer fixed number of isolates to NCs periodically.
One of the main objectives of establishing this Network was to bring about harmonization and uniformity in the AMST procedures being followed for bacteriology and mycology. This was accomplished by formulating standard operating procedures (SOPs) on bacteriology and mycology, based on the Clinical Laboratory Standards Institute (CLSI) guidelines7,8. The SOPs are revised periodically to include the changes proposed by CLSI and are used for trainings of all the participating hospitals.
Sampling sites, sample inclusion and exclusion criteria and organisms
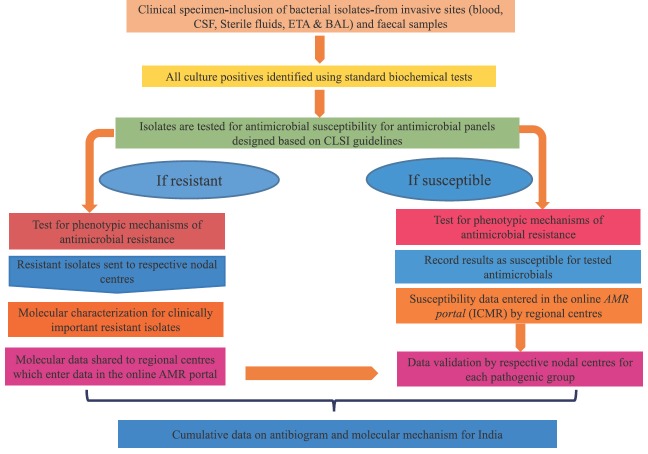
Schematic view of workflow for AMRSN system is depicted in Fig. 2A. All hospitals included in network are tertiary care hospitals having established clinical microbiology laboratories. Both the NCs and RCs follow SOPs formulated by the ICMR to collect resistance data9,10. While the RCs carry out only AMST, the NCs focus on the identified resistant organisms and carry out detailed molecular studies on the respective group of pathogens (Fig. 2B).
Fig. 2A.
Flow diagram depicting the study algorithm for AMRSN system. AMR, antimicrobial resistance; BAL, bronchoalveolar lavage; CLSI, Clinical & Laboratory Standards Institute; CSF, cerebrospinal fluid; ETA, endotracheal aspirate; ICMR, Indian Council of Medical Research.
Fig. 2B.
The structure of Indian Council of Medical Research Antimicrobial Resistance Surveillance & Research network, guiding principles and roles and responsibilities of nodal center and regional centers. AMST, Antimicrobial Susceptibility Testing; NC, Nodal Center; RC, Regional Center; DR, drug resistant; DS, drug sensitive.
Antimicrobial susceptibility testing
All the network laboratories perform routine microbiological investigations, using standard biochemical identification (up to species level) and carry out antimicrobial susceptibility/resistance pattern on all clinical isolates received using SOPs. The Network captures quantitative data, i.e. minimum inhibitory concentration (MICs) or zone diameters in disc diffusion tests which are more significant than the qualitative data (interpretations as susceptible, intermediate or resistant), that indicate only broad trends for many drug-organism combinations. Phenotypic assays for the detection of mechanisms of resistance are performed for isolates at each centre11. Each NC and RC determines the antibiogram of the isolates against panel (available antimicrobials of choice) with breakpoints recommended by ICMR SOPs (Table I). All data are validated by the NCs for each pathogenic group. All laboratories conduct internal quality control and routinely participate in External Quality Assurance Systems (EQAS - bacterial identification and AST) conducted by NCs assigned by ICMR. The laboratories in surveillance network are part of the National EQAS conducted by the Indian Association of Medical Microbiologists collaborating centres (Sir Ganga Ram Hospital, New Delhi for northern India and CMC, Vellore for southern India).
Table I.
Target bacteria and antibiotics: Pathogen-drug combinations for antimicrobial susceptibility testing (AST)
| Pathogen | Antibacterial agents used for AST |
|---|---|
| Enterobacteriaceae | Amikacin, cefepime, cefotaxime, cefoperazone-sulbactam, piperacillin-tazobactam, ceftazidime, chloramphenicol, ciprofloxacin, doxycycline, ertapenem, imipenem, meropenem, nitrofurantoin, norfloxacin, colistin/polymyxin B, trimethoprim-sulphamethoxazole |
| Salmonella Typhi and Paratyphi | Ampicillin, trimethoprim-sulphamethoxazole, ciprofloxacin, ofloxacin, chloramphenicol, ceftriaxone, cefixime |
| Pseudomonas aeruginosa | Ceftazidime, cefepime, ciprofloxacin, levofloxacin, gentamicin, tobramycin, amikacin, polymyxin B, netilmicin, piperacillin-tazobactam, aztreonam, imipenem, meropenem |
| Acinetobacter baumannii | Ceftazidime, cefepime, piperacillin/tazobactam, cefoperazone-sulbactam, levofloxacin, amikacin, tetracycline, netilmicin, imipenem, meropenem |
| Shigella spp., Diarrhoeagenic Escherichia coli, Salmonella spp., Vibrio spp. | Ampicillin, tetracycline, trimethoprim-sulphamethoxazole, nalidixic acid, norfloxacin, ciprofloxacin, cefixime, azithromycin, clindamycin |
| Staphylococcus spp. | Cefoxitin, ciprofloxacin, clindamycin, co-trimoxazole, daptomycin, erythromycin, linezolid, mupirocin high level, penicillin, tetracycline, tigecycline, vancomycin, teicoplanin |
| Enterococcus spp. | Ampicillin, daptomycin, gentamicin, nitrofurantoin, ciprofloxacin, teicoplanin, vancomycin, linezolid |
Molecular characterization of antimicrobial resistance
Molecular mechanism of resistance gives insights on the sources of AMR genes, mobile DNA elements and mutations responsible for resistance and clones prevalent in India. One of the objectives of AMRSN is to create baseline data for AMR and molecular epidemiology in India. Molecular characterization of the resistance mechanisms is performed by corresponding NCs for pathogens. Sixty resistant isolates per species, per year, are shared by RCs for molecular characterization with NCs. Each NC tests the isolates received from RCs and other NCs for AMR genes. Molecular data are shared with the respective RCs and entered in online AMR portal6.
Data management and analysis in the laboratory surveillance system
An important component of integrated AMR surveillance is the informatics solution/suite for collection, storage and analysis of surveillance data, which can enforce both quality AMST in laboratories and provide analytics to support the development of national policies on antimicrobial usage. Data need to be communicated as rapidly as possible to a diverse range of stakeholders, including those who submitted the basic data12. Considering the features of an ideal AMR surveillance tool and the limitations of existing tools, a centralized online IT-enabled computerized data entry portal i-AMRSS, was specifically designed which is a user-friendly web-based tool to facilitate integrated collection, management, analysis and reporting of surveillance findings transparent and accessible (http://bmi.icmr.org.in/amr/). The i-AMRSS requires only web browser and is ideally suited for implementation in countrywide AMR surveillance. The collected data are analyzed based on patient's age, sex, locality, unit/ward, site of sample collection, antibiotics consumed and antibiotic susceptibility pattern.
Data collected from national network of nodal centres can be used to monitor the extent of resistance, variations in resistance rates, detecting the emergence of new resistance traits and can be utilized in future to measure the impact of any interventions. Periodic discussions and training sessions for the investigators and data entry operators are conducted by the ICMR on data collection, data entry and analysis, as the system is dynamic and continuously evolving.
Highlights of antimicrobial resistance surveillance data
A snapshot of some key findings emanating from ICMR-AMRSN, which is currently targeting major clinically important bacterial and fungal pathogens, is presented in this section. Pathogens isolated from the samples during 2015-2018 are included. Isolates tested resistant or intermediate against an antibiotic were classified as non-susceptible. In contrast to western data, ICMR data showed clear dominance of Gram-negative pathogens, with Gram-negatives responsible for 90 per cent of respiratory, 86 per cent of urine, 73 per cent of CSF, 76 per cent of sterile body fluids and 55 per cent of blood isolates in severe hospital-acquired infections. This will have a bearing in empiric choice of antibiotics (ICMR-AMRSN 2016-2018).
Enterobacteriaceae
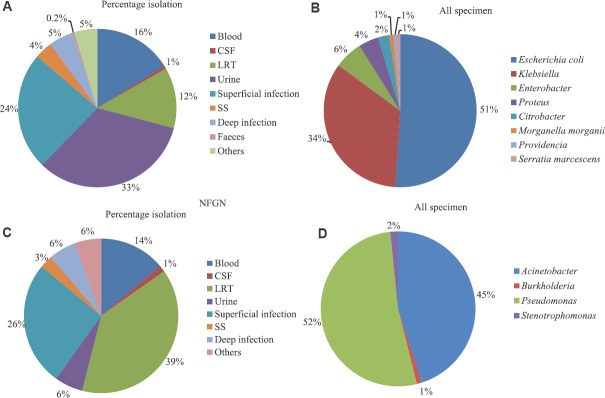
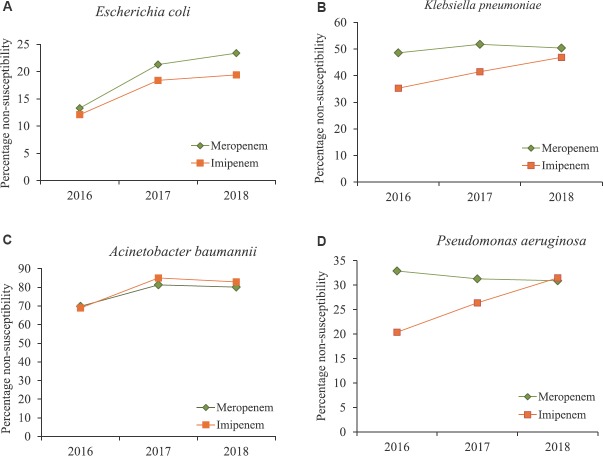
Among Enterobacteriaceae, Escherichia coli (51%) was the most commonly isolated pathogen, followed by Klebsiella (34%), Enterobacter species (6%) and Proteus species (4%) (Fig. 3A,B). Higher rates of resistance to cephalosporins and aminoglycosides were observed among Enterobacteriaceae spp. Non-susceptibility to cephalosporins was found to be 73-77 per cent in K. pneumoniae and 76-80 per cent in E. coli. Both K. pneumoniae and E. coli also showed non-susceptibility rates of >65 per cent to fluoroquinolones (FQs) (Table II). Resistance was lower on the addition of β-lactamase inhibitor (against cefoperazone/sulbactam). Significant resistance to carbapenems was also seen. K. pneumoniae showed higher non-susceptibility to meropenem (49.3%) than imipenem (42.7%) (Table II, Fig.4A, B). In general, Klebsiella spp. followed by Enterobacter spp. were found to be more resistant as compared to E. coli.
Fig. 3.
Isolation rates of non-fermenting Gram-negative bacteria and Enterobacteriaceae from different clinical specimens (ICMR- AMRSN 2016-2018). (A) Isolation rates of Enterobacteriaceae from different specimens. (B) Isolation rates of different Enterobacteriaceae. (C) Isolation rates of non-fermenting Gram-negative bacteria from different specimens. (D) Isolation rates of different non-fermenting Gram-negative bacteria. Note: (1) Blood includes: Blood-central catheter, blood-peripheral and peripheral catheter-blood. (2) LRT (lower respiratory tract) includes: Bronchoalveolar lavage, sputum, lung aspirate, endotracheal aspirate and lobectomy tissue (lung tissue). (3) Superficial infection includes: Skin and soft tissue, pus/exudate, wound swab, superficial biopsy and superficial tissue. (4) Deep infection includes: Abscess aspirate, pus aspirate, deep biopsy and deep tissue. (5) Sterile sites (SS) includes: Fluid from sterile spaces, abdominal fluid, intracostal tube fluid, pancreatic drain fluid, pericardial fluid, peritoneal fluid and pleural fluid.
Table II.
Non-susceptible percentages* of Gram-negative bacilli isolated from all specimen (except faeces)
| Antimicrobial agent | E. coli | K. pneumoniae | A. baumannii | P. aeruginosa |
|---|---|---|---|---|
| Amikacin | 18.6 | 48.5 | 77.8 | 31.2 |
| Gentamicin | - | - | - | 39.3 |
| Ciprofloxacin/levofloxacin† | 76.3 | 65.8 | 73.2 | 41.4 |
| Cefotaxime/cefepime† | 80 | 77.2 | 87.1 | 37.8 |
| Ceftazadime | 76.6 | 73.1 | 87 | 34.7 |
| Imipenem | 22.5 | 42.7 | 82.2 | 29.6 |
| Meropenem | 26.9 | 49.3 | 78.3 | 31.3 |
| Piperacillin-tazobactam | 38.6 | 57.5 | 83 | 30.1 |
| Colistin | 1.1 | 8.8 | - | 3.2 |
*Non-susceptibility data include both I and R data. †Ciprofloxacin and cefotaxime tested in E. coli and K. pneumoniae. Levofloxacin and cefepime tested in A. baumannii and P. aeruginosa. K. pneumoniae, Klebsiella pneumoniae; A. baumannii, Acinetobacter baumannii; P. aeruginosa, Pseudomonas aeruginosa; I, intermediate; R, resistant; E. coli, Escherichia coli Source: ICMR-AMRSN data 2016-2018
Fig. 4.
Carbapenem non-susceptibility pattern among Gram-negative bacteria isolated from blood cultures (2016-2018).
Source: ICMR-AMRSN data 2016-2018.
Molecular characterization of β-lactamase in EB-Gram-negative bacteria (GNB): The percentages of extended-spectrum β-lactamases (ESBL)-producing E. coli and K. pneumoniae reported to ICMR network remained high and predominant in 2016. ESBL production conferred by the presence of OXA-1, TEM and AmpC, was often seen in combination with other acquired resistance mechanisms, conferring resistance to other important treatment alternatives such as FQs and aminoglycosides. TEM (54%) was the most prevalent followed by OXA-1 (22%) and SHV (16%). K. pneumoniae with carbapenemase, NDM, OXA-48-like or VIM production has been reported from all the centres13. Overall, NDM was the most prevalent (27%) carbapenemase followed by VIM (19%), IMP (15%) and KPC (15%) and the prevalence of AmpC β-lactamases was below 10 per cent.
Non-fermenting Gram-negative bacteria (NF-GNB)
ICMR network data reported an increasing prevalence of non-fermenting Gram-negative bacteria (NF-GNB) causing hospital-acquired infections14. Pseudomonas species was the most commonly isolated pathogen (52%) followed by Acinetobacter species (45%) (Fig. 3C, D). Treatment of multidrug-resistant (MDR) (cephalosporin, carbapenems, aminoglycosides and quinolones) and pan-drug resistance in NF-GNB is a real challenge to the treating intensivist. Pseudomonas species were resistant to several classes of antimicrobials tested, although lesser than Acinetobacter spp. Acinetobacter baumannii was the most resistant organism and showed >70 per cent non-susceptibility to most of the antibiotics tested except colistin (Table II). Among carbapenems, non-susceptibility to imipenem was higher (82.2%) than meropenem (78.3%) (Fig. 4C, D). Non-susceptibility rates were higher in the invasive specimens such as lower respiratory tract, blood and CSF compared to other specimens tested against different classes of antibiotics. Higher non-susceptibility rates were observed for cephalosporins such as cefepime and ceftazidime (87%) across all the specimens followed by piperacillin-tazobactam (83%), amikacin (77.8%) and levofloxacin (73.2%) in A. baumannii (Table II). About ten centres also reported the isolation of Burkholderia cepacia and Strenotrophomonas maltophila.
Non-susceptibility to anti-pseudomonal agents did not show any increase over the years and was seen to be 30-40 per cent. Piperacillin/tazobactam and carbapenems showed promising results with 70 per cent susceptibility. P. aeruginosa showed 41.4 per cent non-susceptibility to ciprofloxacin, 39.3 per cent to gentamycin and 34-38 per cent to cephalosporins (Table II). Isolates from ICU demonstrated higher resistance rates compared to isolates from non-ICU settings. Among the carbapenem-resistant isolates, piperacillin/tazobactam and colistin showed better activity than other agents.
Molecular characterization in P. aeruginosa and A. baumannii: Molecular characterization of drug resistance genes in P. aeruginosa provided β-lactamases profile across the four centres in India15. Co-occurrence of ESBLs and carbapenemase was seen around 15-25 per cent in these isolates. Although, combination agents are used for treatment, co-occurrence of ESBLs and carbapenemases results in poor clinical outcome, if β-lactams are used. In A. baumannii, PER (20-50%) was the predominant ESBL followed by TEM (10-38%) contributing to β-lactam resistance. Among the carbapenemases, a class D oxacillinases, OXA-23-like gene (97-100%) was the predominant followed by NDM (15-30%) (ICMR-AMRSN 2016-2018). Currently, 99 per cent of A. baumannii are susceptible to colistin (ICMR-AMRSN 2016-2018), and is considered to be the promising treatment option.
Antimicrobial susceptibility in Salmonella (typhoidal and non-typhoidal)
In 2016, majority of enteric fever was caused by S. Typhi (62%), followed by S. Paratyphi A (28%). MDR was seen in around 10 per cent in S. Typhi and 19 per cent in S. Paratyphi A. Across all the centres, susceptibility to ciprofloxacin (Fig. 5) and nalidixic acid was 15 and 1 per cent, respectively. In this scenario, ceftriaxone became the first line of treatment, but recently, there have been sporadic reports of clinical failure to ceftriaxone in S. Typhi and ESBL-producing non-typhoidal Salmonella. Although all isolates were susceptible to third-generation cephalosporins (Fig. 5), a creeping MIC was observed over the years (Fig. 6). In 2016, ceftriaxone-resistant S. Typhi strain was reported from AIIMS (received from RC from the western part of India) which was confirmed by next-generation sequencing (NGS) at CMC, Vellore16. Spread of such resistance would further limit available therapeutic options with only azithromycin as a treatment option. Data from ICMR AMRS Network showed that in S. Typhi multidrug resistance to older antibiotics ampicillin, chloramphenicol and trimethoprim-sulphamethoxazole (TMP-SMX) was showing a downwards trend over a period 2013-2018 and resistance to FQs was increasing (Fig. 5A). Based on the data, ciprofloxacin should not be used as the first-line drug. The use of third-generation cephalosporins should be restricted to complicated or ciprofloxacin-resistant S. enterica serovar Typhi.
Fig. 5.
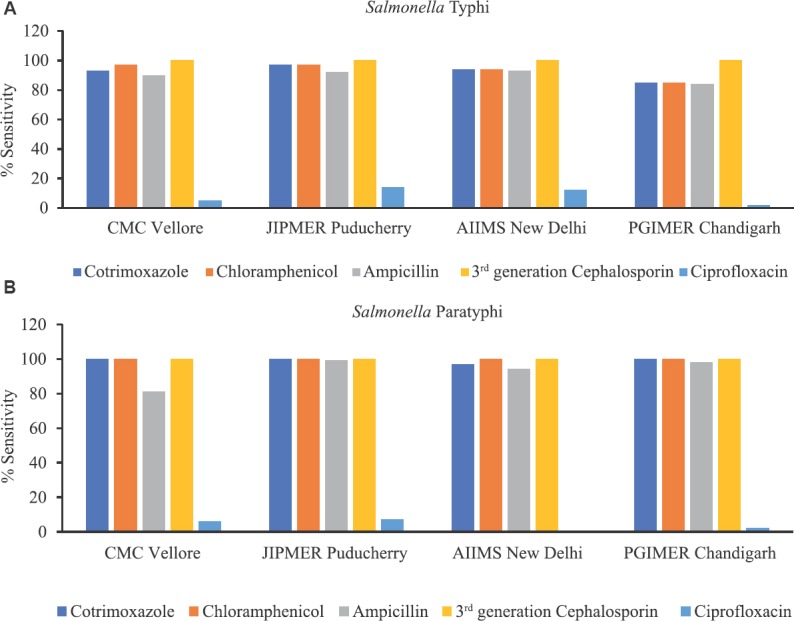
(A) Percentage susceptibility in Salmonella Typhi isolated from four nodal centers in Indian Council of Medical Research-Antimicrobial Resistance Surveillance & Research network AMRSN from 2013 to 2018 (N=650). (B) Percentage susceptibility in Salmonella Paratyphi A isolated from four nodal centers in ICMR-AMRS Network from 2013 to 2018 (N=227).
Fig. 6.
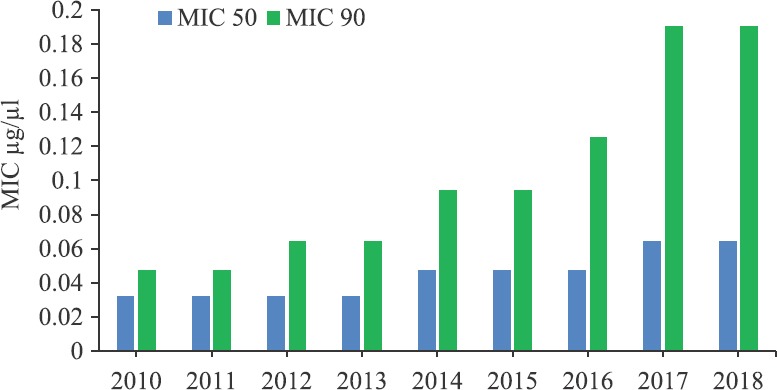
Creeping minimum inhibitory concentrations (MIC) for ceftriaxone in Salmonella Typhi and Salmonella Paratyphi A isolated from four nodal centers in ICMR-AMRSN from 2013 to 2018 (N=877).
Molecular mechanism of resistance in Salmonella serovars: Salmonella isolates were screened for mutations in quinolone resistance determining regions, and tested for plasmid-mediated quinolone resistance genes (qnrB and qnrS). All non-susceptible isolates of Salmonella (~80%) had mutations in gyrA and parC genes. The common mutations were Ser83→Phe/Tyr and Asp87 → Asn/Gly/Tyr. Mutations in efflux pump encoding genes and qnr genes were not detected in any of the tested isolates. All S. Typhi isolates were clustered into two sequence types – ST1 and ST2. All S. Paratyphi A were clustered in ST85 and ST129, based on the mutation in sucA gene. These results were in concordance with the previously published reports from CMC, Vellore17,18. Bloodstream infections (BSIs) due to Salmonella species other than typhoidal Salmonella were reported from Chandigarh and Puducherry19.
Diarrhoeal pathogens
The susceptibility pattern for enteropathogenic E. coli and Vibrio cholera did not show any change in AMR trend over the years. Shigella spp. was the second common diarrhoeal pathogen (37%), next to Aeromonas spp. (46.9%). Shigella flexneri was the predominant serotype (62%) isolated followed by Shigella sonnei (35%). S. flexneri showed increased non-susceptibility to ampicillin (57.4%) and norfloxacin (48.5%). Percentage non-susceptibility to nalidixic acid (96.8%) and TMP-SMX (82.5%) was found to be high. Susceptibility to ampicillin was lesser in S. flexneri (40%) than S. sonnei (70%) (ICMR-AMRSN 2016-2018).
Further, the transmission of AMR genes carrying plasmids between the clinical pathogens was investigated. The plasmid-containing blaCTX-M-15 and qnrS1 genes was successfully transferred from MDR S. flexneri to a susceptible Salmonella spp. The sequencing revealed the association of qnr and blaCTX-M genes and the possibility of rapid dissemination of these genes in Shigella spp. increases the threat for spread of cephalosporin resistance among Enterobacteriaceae20 (Table III).
Table III.
Occurrence of antimicrobial resistance (AMR) genes among Shigella spp.
| Profile, AMR genes | January 2014 to December 2016, number of positives (%), (n=103) |
|---|---|
| ESBLs | |
| blaOXA | 48 (47) |
| blaTEM | 18 (17) |
| blaCTX-M-1 | 6 (6) |
| AmpC | |
| ampC | 10 (10) |
| Sulphonamides | |
| dhfr1a | 95 (92) |
| sul II | 73 (71) |
| FQs | |
| qnr A | 0 (0) |
| qnr B | 1 (1) |
| qnr S | 14 (14) |
ESBLs, extended-spectrum β-lactamases; FQs, fluoroquinolones
Source: Christian Medical College, Vellore (2014-2016)
Trends in Gram-positive pathogens
Staphylococci: Among Staphylococci, marginal reduction in the proportion of methicillin-resistant Staphylococcus aureus (MRSA) was observed during 2016-2017 (33.78%) from 2015 (42.6%). Mupirocin resistance and inducible clindamycin resistance in MRSA have remained relatively constant over the past three years. Among the MRSA, 4.6 per cent were non-susceptible to mupirocin. About 93 per cent of Staphylococcus aureus isolated from all specimens were non-susceptible to penicillin and 79 per cent were non-susceptible to ciprofloxacin (Table IV).
Table IV.
Non-susceptible percentages* of Gram-positive cocci isolated from all specimen
| Antimicrobial agent | S. aureus | MSSA | MRSA | CoNS | E. faecalis | E. faecium |
|---|---|---|---|---|---|---|
| Cefoxitin | 35.3 | 0 | 99.9 | 68.9 | - | - |
| Ciprofloxacin | 78.8 | 72.7 | 89.9 | 58.3 | 87.6 | 94.6 |
| Clindamycin | 23.6 | 14 | 40.3 | 43.6 | - | - |
| Erythromycin | 53.2 | 41.2 | 74.6 | 74.6 | - | - |
| Linezolid | 0.8 | 0.3 | 1.6 | 2.2 | 1.7 | 4.8 |
| Mupirocin/gentamicin (high level)† | 2.2 | 1.2 | 4.6 | - | 47.7 | 72.2 |
| Penicillin/ampicillin† | 92.8 | 89.3 | 98.9 | 85.4 | 30.2 | 79.6 |
| Teicoplanin | 1.4 | 0.6 | 2.6 | 3.4 | 4.2 | 20.8 |
| Tetracycline | 10.9 | 7.3 | 17.5 | 18.5 | - | - |
| Tigecycline | 0.6 | 0.2 | 1.2 | 2.1 | - | - |
| Trimethoprim-sulphamethoxazole | 35.5 | 31.2 | 43.4 | 53.2 | - | - |
| Vancomycin | 1.5 | 0.5 | 3.8 | 2.3 | 3.9 | 22.1 |
*Non-susceptibility data include both I and R data, †Mupirocin and penicillin tested for S. aureus, MSSA, MRSA, CoNS. Gentamicin and ampicillin tested for E. faecalis and E. faecium. S. aureus, Staphylococcus aureus; MSSA, methicillin-susceptible Staphylococcus aureus; MRSA, methicillin-resistant Staphylococcus aureus; CoNS, coagulase-negative Staphylococcus aureus; E. faecalis, Enterococcus faecalis; E. faecium, Enterococcus faecium; I, intermediate; R, resistant
Source: ICMR-AMRSN data 2016-2018
Although full blown vancomycin resistance has not been reported in S. aureus or coagulase-negative S. aureus (CoNS) from our Network, occasional isolates of vancomycin intermediate S. aureus (VISA) and vancomycin intermediate coagulase-negative S. aureus (VICoNS) have been observed. More worrying was that heterogeneous vancomycin intermediate S. aureus (hVISA) rates detected by population analysis profile-area under the curve (AUC-profile) increased from 9.6 per cent (2015) to 14.4 per cent (2016) among which, the highest rates were observed in ICU isolates (2016), suggesting a cumulative effect of overuse of vancomycin in the hospital setting. The other worrisome finding was the steadily increasing non-susceptibility to linezolid in S. aureus (0.8%) and CoNS (2.2%) (Table IV).
Staphylococcus haemolyticus and S. epidermidis were the more common species of CoNS, with S. haemolyticus showing higher levels of drug resistance (MRSH 86.6%, MRSE 61%) isolated mostly from blood and pus. S. hominis, S. lugdunensis and S. saprophyticus were other species of CoNS identified in lesser numbers. Overall, increasing levels of resistance were observed commonly among CoNS, particularly S. haemolyticus and S. hominis [e.g. methicillin resistance among CoNS was 66% (2015-2017)]. The non-susceptibility rates of S. haemolyticus were 93 per cent to penicillin, 87 per cent to cefoxitin and 81 per cent to ciprofloxacin (ICMR AMRSN data 2016-2018).
Enterococci
Vancomycin-resistant Enterococci (VRE) is an emerging threat in hospital settings21 that have been observed in 11.9 per cent (34 of 286) of isolates. Higher non-susceptibility to vancomycin was observed in Enterococcus faecium (22%) than E. faecalis (3.9%) (Table IV). The main source of VRE in hospitals is cross-contamination among patients through healthcare workers and admission of already gut colonized patients in the ICU21. However, VRE genes can be transmitted to other Enterococci species through plasmids, while the spread of MRSA is primarily chromosomally mediated and thus, there is no interspecies transfer. The prevention of VRE colonization in the gut is an important method to control VRE infections in hospitalized patients21.
Although vancomycin non-susceptibility has been gradually increasing over the last decade, there has been a concomitant shift in the relative ratio of the two species from 80:20 to 20:80, thereby practically raising the overall glycopeptide resistance in Enterococci from <15 per cent to near 30 per cent over the same period. E. faecium isolates showed higher non-susceptibility to ampicillin (79.6%) and gentamicin (72.2%).
Molecular mechanism of AMR in Gram-positive cocci: Resistance to methicillin was confirmed by the presence of mecA gene in 512 randomly selected S. aureus isolates from 2015 to 2017. The isolates also included those received at JIPMER from the RCs under the EQAS programme. However, in 16 per cent of the isolates, which were identified phenotypically as MRSA [by both cefoxitin disc diffusion (CDD) and MRSA latex agglutination test (LAT)], mecA gene was negative by PCR. On the other hand, there were 18 isolates which were identified as methicillin-susceptible S. aureus by CDD but were positive by both mecA gene and MRSA LAT. Although there are non-mecA gene mediated mechanisms of MRSA, these are very rarely encountered in clinical practice. There could be some minor variations in the mecA gene near the region of primer annealing leading to negative PCR results though functionally the gene may be unaffected. Such an occurrence can be confirmed using multiple primers targeting different regions of the mecA gene22. The erythromycin- and clindamycin-resistant S. aureus isolates carried erm genes, ermC being the most common. The MRSA isolates from AIIMS and PGIMER were positive exclusively for ermC genes. However, the numbers of isolates with phenotypic resistance to macrolides submitted for EQAS were too small to draw any conclusions. Mupirocin resistance in S. aureus was coded by the mupA gene in a majority of the isolates. Nine of the ten linezolid-resistant isolates carried the cfr gene23. Almost all the VRE carried the vanA gene (Table V).
Table V.
Distribution of resistance genes among the phenotypic-resistant Gram-positive cocci
| Organism | Phenotypic resistance | Resistance genes detected |
|---|---|---|
| Staphylococcus aureus | Cefoxitin (512) | mecA (496) |
| Erythromycin (210) | erm genes (erm A-48, erm B-25, erm C-90) | |
| Clindamycin (66) | (erm A-19, erm C-38) | |
| Mupirocin (47) | mupA (38) | |
| Cefoxitin (278) | mecA (265) | |
| Linezolid (10) | Cfr (9) | |
| Enterococcus | Vancomycin (214) | vanA (212) |
Values in parenthesis indicate number of positives
Source: Jawaharlal Institute of Postgraduate Medical Education & Research, Puducherry
MLST was performed on MRSA isolates from JIPMER revealed 19 different sequence types and eight major clonal complexes (CCS). ST772 (also called the Bengal Bay clone) belonging to CC1, was the most common CA-MRSA clone observed in our isolates similar to reports from India and elsewhere in the world24.
Fungal infections
From the national perspective, Candida tropicalis (39.6%) was the major yeast isolated followed by C. albicans (32.8%), C. glabrata (10%), C. parapsilosis (7.2%), C. utilis (4.7%), C. krusei (2.6%), and C. auris (1.1%). C. abicans isolates showed non-susceptibility to caspofungin (9.2%), fluconazole (7.4%), followed by voriconazole (3.6%), micafungin (2%) and anidulafungin (1.7%). For C. tropicalis, the highest non-susceptibility was seen against caspofungin (9.1%), followed by fluconazole (6.4%), anidulafungin (2.4%), micafungin (2.3%) and voriconazole (1.6%). Higher percentage of C. glabrata (25.3%) and C. parapsilosis (15.8%) showed non-susceptibility to flucanozole (ICMR-AMRSN data 2016-2018).
Molecular characterization of AMR in pathogenic yeast: A total of 38 per cent isolates of C. tropicalis showed amino acid substitution (Y132F and S154F) in ERG11 gene due to A395T and C461T mutation. Amino acid substitutions were present at the active drug binding site indicating interference in binding. Other isolates showed an azole-resistant phenotype in the absence of any amino acid substitution. A total of 229 clinical Aspergillus fumigatus isolates were screened for the presence of transcriptional enhancer and mutation TR34/L98H. The presence of TR34 mutation in upstream promoter region of cyp51A was seen in only one isolate. In clinical A. flavus isolates, cyp51A overexpression was seen and could be one of the mechanisms responsible for azole resistance. The efficacy of voriconazole against susceptible and resistant A. flavus in murine model showed that susceptible isolates required a higher drug exposure than resistant strains for the same treatment effect25. In addition, high MIC to terbinafine in Trichophyton mentagrophytes and Trichophyton rubrum isolates with F397L substitution in the squalene epoxidase gene was identified (ICMR-AMRSN data 2016-2018).
Discussion
ICMR-AMRSN was designed with an aim to provide essential information about AMR in critical pathogens and over time it has developed into an efficient and flexible platform for detecting and characterizing emerging resistance threats in humans. This network was instrumental in bringing about harmonization and uniformity in the AST procedures followed for bacteriology and mycology through formulation of SOPs for AST and development of a web-based data management system for collection and analysis of AMR data. This nation-wide surveillance study documented the high and ever-increasing prevalence of Gram-negative bacteria with increasing resistance to last resort drugs such as carbapenems and colistin. Non-susceptibility to the broad-spectrum antibiotics including third-generation cephalosporins and FQs was 75-80 per cent in E. coli, 65-77 per cent in K. pneumoniae, 73-87 per cent in A. baumannii and around 40 per cent in P. aeruginosa. The data (2016-2018) from the network also showed that majority of the Gram-negative isolates were MDR (ICMR-AMRSN data 2016-2018). The proportion of carbapenem resistance was high in Gram-negative bacteria during the surveillance. Higher non-susceptibility to meropenem was observed in A. baumannii (69.8, 81.3, 80.1%) followed by K. pneumonia (48.6, 51.8, 50.4%), P. aeruginosa (32.9, 31.3, 30.9%) and E. coli (13, 21 and 23%) in 2016, 2017 and 2018, respectively (Fig. 4 A–D). Infections due to pathogens-producing ESBL and carbapenemases will be challenging and difficult to treat.
Resistance to carbapenems is conferred predominantly by metallo-β-lactamases (VIM and NDM) and carbapenem hydrolyzing class D β-lactamases (OXA48-like). Enterobacteriaceae isolates showing resistance to β-lactams were found to be carrying blaNDM and blaOXA48 across the centres. All of the sequenced NDM and OXA-48-like were identified as NDM-1 and OXA-181 variants26,27.
Colistin is a last resort antibiotic used for treating severe Gram-negative infections. In this study, 13 per cent of K. pneumoniae isolates were non-susceptible to carbapenems and colistin (ICMR-AMRSN data 2016-2018). The resistance situation for K. pneumoniae therefore remains problematic as seen in light of ICMR surveillance data from 2015 to 2018 (ICMR-AMRSN data 2016-2018).
While nosocomially acquired Acinetobacter spp., is susceptible only to polymyxins, carbapenems do not work on Stenotrophomonas and colistin does not work on Burkholderia. On the contrary, cotrimoxazole, minocycline and levofloxacin, not favoured by majority, shows high susceptibility to the latter two pathogens. Since each species of NF-GNB has its own signature susceptibility profile, our results emphasize the need to identify Burkholderia and Stenotrophomonas correctly, which are usually misidentified as Pseudomonas by most laboratories, so that treatment with appropriate antibiotics can be initiated.
The typhoid data from the ICMR network shows increasing MIC to ceftriaxone and increased susceptibility to co-trimoxazole, ampicillin and chloramphenicol suggesting that bacterial resistance is reversible. In the present global scenario of changing AMR pattern, continuous surveillance and molecular characterization of Salmonella spp. is important to map the geographical areas, which need immediate alterations in treatment strategies with regard to the travel and deciding vaccine strategies. The recent outbreak of ceftriaxone-resistant Salmonella Typhi in Hyderabad, Pakistan, was identified through AMR surveillance, which led to the implementation of appropriate control measures to contain the outbreak28. ICMR-NC on typhoid at AIIMS, New Delhi, reported four cephalosporin-resistant Salmonella enterica serovar Typhi isolates (429038, 430040, 7830 and 458426) isolated from blood. Whole-genome sequencing revealed AMR genes blaSHV-12 and qnrB7 for 429038, 430040 and 458426, whereas 7830 had the blaCMY-2 gene, thus confirming the mechanism of cephalosporin resistance in these isolates. Some isolates were found to harbour the IncX3 plasmid17.
Among the Gram-positive pathogens, the network data point towards increased susceptibility to gentamicin and co-trimoxazole in S. aureus suggesting that these two antibiotics can be good alternatives for treating community acquired S. aureus infections. The data from ICMR network document that CoNS are far more resistant than S. aureus and are causing more clinical infections, particularly central line-associated blood stream infection and catheter-related BSI. This is in line with earlier reports, which highlight CoNS as agents of BSI29.
Several studies from India have highlighted the occurrence of candidaemia as a nosocomial BSI associated with central venous catheters30,31. C. tropicalis was most commonly isolated yeast and non-susceptibility in C. tropicalis and C. albicans against fluconazole and caspofungin was a major concern. Fungal pathogens are, therefore, included as priority pathogen in ICMR Network.
Robust AMR surveillance systems have been found useful to detect significant changes and shifts in susceptibility to antibiotics in a timely manner to prevent and reduce further development of resistance32. Many such programmes such as National Healthcare Safety Network, Japan Nosocomial infections Surveillance, SENTRY Antimicrobial Surveillance Program and European Antimicrobial Resistance Surveillance System have been found useful by countries to capture changes in trends of resistance and antimicrobial usage and guide policies to reduce resistance by instituting a variety of prevention measures32. The results of the ICMR-AMRSN provide valuable data, which can form baseline for targeted antibiotic policy (speciality-wise, syndrome-wise and category-wise) and implementation of antibiotic stewardship in tertiary care medical facilities in our country. It is well documented from India that prescribers do not de-escalate to narrow antimicrobials despite culture data supporting the same33. Since most doctors start with high dose, de-escalation can impact cost, antimicrobial consumption, patient outcomes and resistance rates. Hence, guidance de-escalation should be emphasized under the antimicrobial stewardship programme (AMSP).
Another observation is high incidence of resistant infections in ICUs, followed by wards and out patient department (OPD)34. High drug resistant infections in ICUs of both Government and private hospitals underscores need for better infection control and AMSP practices to reduce antibiotic selection pressure. Realizing that the gap of poor infection control practices cannot be bridged with antimicrobial use, ICMR network of hospitals is also part of Centers for Disease Control and Prevention (CDC) funded Global Health Security Agenda (GHSA) project, which aims to reduce infection rates in hospitals35.
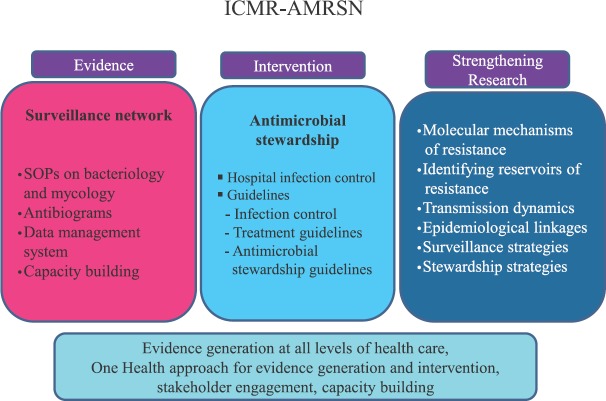
AMR surveillance data serve as an important tool that influence clinical decision-making and choice of antibiotics for empirical treatment32. The data generated by ICMR's AMR Network were utilized to frame treatment guidelines for antimicrobial use in 201736. An AMSP has also been initiated in the ICMR Network hospitals to encourage hospitals to develop hospital specific antibiograms and treatment guidelines and establish of the structure and process of AMSP with close collaboration of microbiologists, clinicians and hospital administrators. It is envisaged that having the local evidence-based treatment guidelines will be helpful in rationalizing antibiotic prescriptions in hospitals. ICMR has published Hospital Infection Control Guidelines37, and Antimicrobial Stewardship Guidelines38 to enable hospitals to create a comprehensive AMS programme (Fig. 7). The next phase ICMR's AMR Network will focus on using next-generation sequencing (NGS) for in-depth studies on dynamics of transmission and creating a “One Health” platform for integrated surveillance. Understanding the reservoirs and the relative contribution of different reservoirs to the burden of AMR (Fig. 7) requires cross-sectoral collaboration between the veterinary and food production sectors. ICMR is working closely with Indian Council of Agricultural Research (ICAR) to better understand the inter-relationship between antimicrobial use and AMR across India. The emergence of resistance to colistin, is a major concern39. Colistin use in livestock sector is being addressed through engagement with ICAR as colistin is used in animal feed among poultry as growth promoter and for therapeutic purposes in veterinary sector.
Fig. 7.
The activities of Indian Council of Medical Research Antimicrobial Resistance Surveillance & Research Network.
The journey to development of this surveillance network has been fraught with many challenges (Box 2). Despite having some of the best Indian hospitals in the network and SOPs, it was challenging to streamline the quality of data collection. Data entry operators provided to each laboratory to facilitate data entry have been useful in streamlining data entries. Thus, dedicated human resource will be one of the major challenges to sustenance of this network beyond completion of project duration and would require ownership at hospital level to provide resources for this activity. Countries like Japan and European countries that have well-functioning AMR networks collect AMR through laboratory information systems (LIS) and antimicrobial usage data through hospital information systems (HIS)40. Unfortunately, even best government hospitals in India do not have HIS/LIS. The absence of information systems impacts the data collection which is usually incomplete and makes it challenging to correlate AMR rates with antimicrobial consumption rates and clinical outcomes. It has been reported that increase in resistance to a particular class of antibiotic is directly related to its consumption in hospital and it has been documented that restricting antimicrobials, specially, carbapenems, even for a short duration, serves as an effective strategy for addressing the problem of carbapemen resistance41. Until HIS/LIS is introduced in all hospitals, there is a need to find a way to sustain antimicrobial consumption data collection mechanism. The next essential component for sustaining quality data collection is periodic trainings, which is part of ICMR efforts and the latest example is that of training on colistin resistance assays. In light of a joint recommendation by the CLSI and EUCAST released in 2016 recommending standard broth microdilution (BMD) method for MIC testing of colistin, all ICMR network hospitals were trained on BMD42.
Box II.
Challenges of sustaining surveillance network
| Only 20 hospitals-Not representative of entire country |
| Prior antibiotic exposure data difficult to capture |
| Expansion, coordination and improvement of the diverse elements of surveillance |
| Continued training support for their staff in order to undertake quality antimicrobial susceptibility testing (AST) and surveillance |
| Reflection of tertiary care settings and not general community hospitals |
| Funding to sustain the quality data and molecular data over a long period |
Many improvements have been made to ICMR-AMRSN over the time since it was started in 2013, and the next phase will focus on understanding the transmission dynamics, importance of mobile genetic elements, identifying reservoirs of drug resistant pathogens etc. Besides, there will be efforts to create convergence of the surveillance, infection control and stewardship activities so that these activities work like loops feeding each other required data and culminating into meaningful outcome and evidence which is used by hospitals to handle AMR crisis. As of now the ICMR effort is limited to tertiary care hospitals, but the need to capture AMR data at primary, secondary levels and also community at large is well recognized and will be addressed through future programmes. The ICMR network has also initiated activities towards ‘One Health’ approach by engaging the relevant stakeholders. Having evidence from veterinary and environmental sectors will provide a holistic picture of AMR in India, enhance our understanding on the epidemiological linkages and identify opportunities for intervention to control AMR in the country.
Conclusion
ICMR-AMR Surveillance Network fulfils an important gap by providing evidence for trends of AMR in tertiary care hospitals in India. There is a need to create tools that help us draw meaningful outcomes from the data being collected. The challenge would be to achieve the scale which is truly representative of the country and to percolate to next levels of healthcare system, both in government and private hospitals while maintaining the quality and the rigor. Equally important would be to sustain efforts initiated towards ‘One Health’ initiative and utilize the latest scientific advancements to understand the importance of each of the components of ecosystem contributing towards the AMR problem and devise suitable interventions.
Footnotes
Financial support & sponsorship: None.
Conflicts of Interest: None.
References
- 1.Laxminarayan R, Chaudhury RR. Antibiotic resistance in India: Drivers and opportunities for action. PLoS Med. 2016;13:e1001974. doi: 10.1371/journal.pmed.1001974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ganguly NK, Arora NK, Chandy SJ, Fairoze MN, Gill JP, Gupta U, et al. Rationalizing antibiotic use to limit antibiotic resistance in India. Indian J Med Res. 2011;134:281–94. [PMC free article] [PubMed] [Google Scholar]
- 3.Walia K. Emerging problem of antimicrobial resistance in developing countries: Intertwining socioeconomic issues. Reg Health Forum. 2003;7:1–10. [Google Scholar]
- 4.Landers TF, Cohen B, Wittum TE, Larson EL. A review of antibiotic use in food animals: Perspective, policy, and potential. Public Health Rep. 2012;127:4–22. doi: 10.1177/003335491212700103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO/EMP/IAU/2017.12. Geneva: WHO; 2017. World Health Organization. Prioritization of pathogens to guide discovery, research and development of new antibiotics for drug-resistant bacterial infections, including tuberculosis. [Google Scholar]
- 6.Indian Council of Medical Research: Antimicrobial Resistance Surveillance & Research Initiative. [accessed on January 31, 2018]. Available from: http://iamrsn.icmr.org.in/index.php/amrsn/amrsn-network .
- 7.CLSI document M07-A10. 10th ed. Wayne, PA: CLSI; 2015. Clinical and Laboratory Standards Institute. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard. [Google Scholar]
- 8.CLSI supplement M100. 27th ed. Wayne, PA: CLSI; 2017. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. [Google Scholar]
- 9.Indian Council of Medical Research. Standard operating procedures: Bacteriology. Antimicrobial Resistance Surveillance and Research Network. New Delhi: ICMR; 2015. [accessed on January 31, 2018]. Available from: https://www.icmr.nic.in/sites/default/files/guidelines/Standard_Operating_Procedures.pdf . [Google Scholar]
- 10.Indian Council of Medical Research. Standard operating procedures; Mycology laboratories: Antimicrobial Resistance Surveillance and Research Network. New Delhi: ICMR; 2015. [accessed on January 31, 2018]. Available from: http://icmr.nic.in/sites/default/files/guidelines/Standard_Operating_Procedures_Mycology.pdf . [Google Scholar]
- 11.Pragasam AK, Raghanivedha M, Anandan S, Veeraraghavan B. Molecular characterization of imipenem-resistant, meropenem-susceptible Pseudomonas aeruginosa with blaVIM-2 phenotype: Potential for dissemination. Jpn J Infect Dis. 2016;69:159–60. doi: 10.7883/yoken.JJID.2015.273. [DOI] [PubMed] [Google Scholar]
- 12.Karp BE, Tate H, Plumblee JR, Dessai U, Whichard JM, Thacker EL, et al. National antimicrobial resistance monitoring system: Two decades of advancing public health through integrated surveillance of antimicrobial resistance. Foodborne Pathog Di s. 2017;14:545–57. doi: 10.1089/fpd.2017.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Veeraraghavan B, Shankar C, Karunasree S, Kumari S, Ravi R, Ralph R. Carbapenem resistant Klebsiella pneumoniae isolated from bloodstream infection: Indian experience. Pathog Glob Health. 2017;111:240–6. doi: 10.1080/20477724.2017.1340128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vijayakumar S, Gopi R, Gunasekaran P, Bharathy M, Walia K, Anandan S, et al. Molecular characterization of invasive carbapenem-resistant Acinetobacter baumannii from a tertiary care hospital in South India. Infect Dis Ther. 2016;5:379–87. doi: 10.1007/s40121-016-0125-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pragasam AK, Vijayakumar S, Bakthavatchalam YD, Kapil A, Das BK, Ray P, et al. Molecular characterisation of antimicrobial resistance in Pseudomonas aeruginosa and Acinetobacter baumannii during 2014 and 2015 collected across India. Indian J Med Microbiol. 2016;34:433–41. doi: 10.4103/0255-0857.195376. [DOI] [PubMed] [Google Scholar]
- 16.Das S, Samajpati S, Roy I, Sankar S, Gaind R, Deb M, et al. Molecular subtyping of Salmonella enterica serovar Typhi by pulsed-field gel electrophoresis and multiple-locus variable-number tandem-repeat analysis in India: Their association with antimicrobial resistance profiles. Jpn J Infect Dis. 2017;70:536–43. doi: 10.7883/yoken.JJID.2016.478. [DOI] [PubMed] [Google Scholar]
- 17.Rodrigues C, Kapil A, Sharma A, Devanga Ragupathi NK, Inbanathan FY, Veeraraghavan B, et al. Whole-genome shotgun sequencing of cephalosporin-resistant Salmonella enterica serovar typhi. Genome Announc. 2017;5 doi: 10.1128/genomeA.01639-16. pii: e01639-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharma P, Dahiya S, Balaji V, Kanga A, Panda P, Das R, et al. Typhoidal salmonellae: Use of multi-locus sequence typing to determine population structure. PLoS One. 2016;11:e0162530. doi: 10.1371/journal.pone.0162530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dahiya S, Sharma P, Kumari B, Pandey S, Malik R, Manral N, et al. Characterisation of antimicrobial resistance in Salmonellae during 2014-2015 from four centres across India: An ICMR antimicrobial resistance surveillance network report. Indian J Med Microbiol. 2017;35:61–8. doi: 10.4103/ijmm.IJMM_16_382. [DOI] [PubMed] [Google Scholar]
- 20.Muthuirulandi Sethuvel DP, Anandan S, Devanga Ragupathi NK, Veeraraghavan B, Vinod O, Walia K. Association of blaCTX-M-15 and qnr genes in multidrug-resistant Salmonella typhimurium and Shigella spp from India. J Infect Dev Ctries. 2015;9:1294–7. doi: 10.3855/jidc.6965. [DOI] [PubMed] [Google Scholar]
- 21.Amberpet R, Sistla S, Parija SC, Thabah MM. Screening for intestinal colonization with vancomycin resistant enterococci and associated risk factors among patients admitted to an adult intensive care unit of a large teaching hospital. J Clin Diagn Res. 2016;10:DC06–9. doi: 10.7860/JCDR/2016/20562.8418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rajkumar S, Sistla S, Manoharan M, Sugumar M, Nagasundaram N, Parija SC, et al. Prevalence and genetic mechanisms of antimicrobial resistance in Staphylococcus species: A multicentre report of the Indian council of medical research antimicrobial resistance surveillance network. Indian J Med Microbiol. 2017;35:53–60. doi: 10.4103/ijmm.IJMM_16_427. [DOI] [PubMed] [Google Scholar]
- 23.Brijwal M, Dhawan B, Rawre J, Sebastian S, Kapil A. Clonal dissemination of linezolid-resistant Staphylococcus haemolyticus harbouring a G2576T mutation and the cfr gene in an Indian hospital. J Med Microbiol. 2016;65:698–700. doi: 10.1099/jmm.0.000279. [DOI] [PubMed] [Google Scholar]
- 24.Nadig S, Namburi P, Raghunath D, Arakere G. Genotyping of methicillin-resistant Staphylococcus aureus isolates from Indian hospitals. Curr Sci. 2006;91:1364. [Google Scholar]
- 25.Rudramurthy SM, Seyedmousavi S, Dhaliwal M, Chakrabarti A, Meis JF, Moutonb JW. Pharmacodynamics of voriconazole against wild-type and azole-resistant Aspergillus flavus isolates in a nonneutropenic murine model of disseminated Aspergillosis. Antimicrob Agents Chemother. 2017;61:e01491–16. doi: 10.1128/AAC.01491-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma A, Bakthavatchalam YD, Gopi R, Anandan S, Verghese VP, Veeraraghavan B. Mechanisms of carbapenem resistance in K. pneumonia and E. coli from bloodstream infections in India. J Infect Dis Ther. 2016;4:293. [Google Scholar]
- 27.Mairi A, Pantel A, Sotto A, Lavigne JP, Touati A. OXA-48-like carbapenemases producing Enterobacteriaceae in different niches. Eur J Clin Microbiol Infect Dis. 2018;37:587–604. doi: 10.1007/s10096-017-3112-7. [DOI] [PubMed] [Google Scholar]
- 28.Yousafzai MT, Qamar FN, Shakoor S, Saleem K, Lohana H, Karim S, et al. Ceftriaxone-resistant Salmonella typhi outbreak in Hyderabad city of Sindh, Pakistan: High time for the introduction of typhoid conjugate vaccine. Clin Infect Dis. 2019;68:S16–21. doi: 10.1093/cid/ciy877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wattal C, Raveendran R, Goel N, Oberoi JK, Rao BK. Ecology of blood stream infection and antibiotic resistance in intensive care unit at a tertiary care hospital in North India. Braz J Infect Dis. 2014;18:245–51. doi: 10.1016/j.bjid.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parameswaran R, Sherchan JB, Varma DM, Mukhopadhyay C, Vidyasagar S. Intravascular catheter-related infections in an Indian tertiary care hospital. J Infect Dev Ctries. 2011;5:452–8. doi: 10.3855/jidc.1261. [DOI] [PubMed] [Google Scholar]
- 31.Patil HV, Patil VC, Ramteerthkar MN, Kulkarni RD. Central venous catheter-related bloodstream infections in the intensive care unit. Indian J Crit Care Med. 2011;15:213–23. doi: 10.4103/0972-5229.92074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Masterton R. The importance and future of antimicrobial surveillance studies. Clin Infect Dis. 2008;47(Suppl 1):S21–31. doi: 10.1086/590063. [DOI] [PubMed] [Google Scholar]
- 33.Singh S, Menon VP, Mohamed ZU, Kumar VA, Nampoothiri V, Sudhir S, et al. Implementation and impact of an antimicrobial stewardship program at a tertiary care center in South India. Open Forum Infect Dis. 2019;6:ofy290. doi: 10.1093/ofid/ofy290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Indian Council of Medical Research. Annual Report. AMR surveillance network data. January 2017- December 2017. New Delhi: Indian Council of Medical Research; 2017. [Google Scholar]
- 35.Swaminathan S, Prasad J, Dhariwal AC, Guleria R, Misra MC, Malhotra R, et al. Strengthening infection prevention and control and systematic surveillance of healthcare associated infections in India. BMJ. 2017;358:j3768. doi: 10.1136/bmj.j3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Indian Council of Medical Research. Treatment Guidelines for Antimicrobial Use in Common Syndromes. New Delhi: ICMR; 2017. [accessed on January 31, 2018]. Available from: http://iamrsn.icmr.org.in/images/pdf/STG270217.pdf . [Google Scholar]
- 37.Indian Council of Medical Research. Hospital Infection Control Guidelines. New Delhi, India: ICMR; 2016. [accessed on January 31, 2018]. Available from: http://iamrsn.icmr.org.in/images/pdf/Hospital%20Infection%20control%20guidelines.pdf . [Google Scholar]
- 38.Indian Council of Medical Research. Antimicrobial Stewardship Program Guidelines. New Delhi: ICMR; 2018. [accessed on January 31, 2018]. Available from: http://iamrsn.icmr.org.in/images/pdf/AMSP_Guidelines_final.pdf . [Google Scholar]
- 39.Kaur A, Gandra S, Gupta P, Mehta Y, Laxminarayan R, Sengupta S. Clinical outcome of dual colistin- and carbapenem-resistant Klebsiella pneumoniae bloodstream infections: A single-center retrospective study of 75 cases in India. Am J Infect Control. 2017;45:1289–91. doi: 10.1016/j.ajic.2017.06.028. [DOI] [PubMed] [Google Scholar]
- 40.Nyasulu P, Paszko C, Mbelle N. A Narrative Review of the Laboratory Information System and Its Role in Antimicrobial Resistance Surveillance in South Africa. Adv Microbiol. 2014;4:692–6. [Google Scholar]
- 41.Abdallah M, Badawi M, Amirah MF, Rasheed A, Mady AF, Alodat M. Impact of carbapenem restriction on the antimicrobial susceptibility pattern of Pseudomonas aeruginosa isolates in the ICU. J Antimicrob Chemother. 2017;72:3187–90. doi: 10.1093/jac/dkx273. [DOI] [PubMed] [Google Scholar]
- 42.European Committee on Antimicrobial Susceptibility Testing. Recommendations for MIC Determination of Colistin (polymyxin E): As recommended by the joint CLSI-EUCAST Polymyxin Breakpoints Working Group. 2016. Mar 22, [accessed on January 31, 2018]. Available from: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/General_documents/Recommendations_for_MIC_determination_of_colistin_March_2016.pdf .