Abstract
Background & objectives:
Plasmid has led to increase in resistant bacterial pathogens through the exchange of antimicrobial resistance (AMR) genetic determinants through horizontal gene transfer. Baseline data on the occurrence of plasmids carrying AMR genes are lacking in India. This study was aimed to identify the plasmids associated with AMR genetic determinants in ESKAPE pathogens.
Methods:
A total of 112 ESKAPE isolates including Escherichia coli (n=37), Klebsiella pneumoniae (n=48, including 7 pan-drug susceptible isolates), Acinetobacter baumannii (n=8), Pseudomonas aeruginosa (n=1) and Staphylococcus aureus (n=18) were analyzed in the study. Isolates were screened for antimicrobial susceptibility and whole genome sequencing of isolates was performed using Ion Torrent (PGM) sequencer. Downstream data analysis was done using PATRIC, ResFinder, PlasmidFinder and MLSTFinder databases. All 88 whole genome sequences (WGS) were deposited at GenBank.
Results:
Most of the study isolates showed resistant phenotypes. As analyzed from WGS, the isolates included both known and unknown sequence types. The plasmid analysis revealed the presence of single or multiple plasmids in the isolates. Plasmid types such as IncHI1B(pNDM-MAR), IncFII(pRSB107), IncFIB(Mar), IncFIB(pQil), IncFIA, IncFII(K), IncR, ColKP3 and ColpVC were present in K. pneumoniae. In E. coli, IncFIA, IncFII, IncFIB, Col(BS512), IncL1, IncX3 and IncH were present along with other types. S. aureus harboured seven different plasmid groups pMW2 (rep5), pSAS1 (rep7), pDLK1 (rep10), pUB110 (repUS12), Saa6159 (rep16), pKH12 (rep21) and pSA1308 (rep21). The overall incidence of IncF type plasmids was 56.5 per cent followed by Col type plasmids 18.3 per cent and IncX 5.3 per cent. Other plasmid types identified were <5 per cent.
Interpretation & conclusions:
Results from the study may serve as a baseline data for the occurrence of AMR genes and plasmids in India. Information on the association between phenotypic and genotypic expression of AMR was deciphered from the data. Further studies on the mechanism of antibiotic resistance dissemination are essential for enhancing clinical lifetime of antibiotics.
Keywords: Antimicrobial resistance, β-lactamase, col-horizontal gene transfer, IncF, plasmids
Nosocomial infections are life-threatening and are a significant cause of morbidity and mortality rates. ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter species) is a group of Gram-negative and Gram-positive bacterial pathogens causing severe nosocomial infections. Infections caused by these pathogens are difficult and challenge to treat due to the high rates of antimicrobial resistance (AMR) seen1.
Most of these pathogens carry AMR encoding genes on mobile genetic elements favouring the rapid dissemination of intra- and inter-species spread. The maintenance of AMR genes under the selection pressure determines the presence of plasmids in the host and has led to the evolution of plasmids over time2. Plasmids are typed based on the replicon/incompatibility types (Inc) in Gram-negative organisms3,4 and rep gene-based typing in Gram-positive organisms5. Some of the most common Inc plasmid types observed among Gram-negative bacterial pathogens are IncF, IncH, IncN, IncA/C, Incl, IncX, IncR, IncQ and Col plasmids6. Such plasmids are known to carry multiple AMR genes through accumulation by vertical and horizontal transfer, and this rapid dissemination phenomenon increases the multidrug-resistant (MDR), extremely drug resistant (XDR) and pan drug-resistant pathogen rates especially in ESKAPE pathogens.
AMR gene classes that are carried on plasmids include β-lactamases [extended-spectrum β-lactamases (ESBLs), AmpC and carbapenemases], aminoglycoside modifying enzymes, 16S rRNA methyltransferases (16S RMTases) and the recently reported mobile colistin resistance gene (mcr) are of concern. ESBLs (blaTEM, blaSHV, blaCTX-M), carbapenemase (blaSPM, blaIMP, blaVIM, blaNDM, blaKPC and blaOXA48) and plasmid-mediated quinolone resistance genes [qnrA, qnrB, qnrS, aac(6’)lb-cr and qepA] were most commonly reported worldwide in IncF type plasmids6. This includes the subtypes IncFIA, IncFIB and IncFII plasmids reported in Escherichia coli7,8. IncI type plasmids were reported predominantly in Europe with genes coding for resistance to aminoglycosides, tetracyclines and quinolones9. Another common plasmid among the Gram-negatives includes IncH type. These were regarded for their association with multidrug resistance as these carry ESBLs and other genes encoding for resistance to sulphonamides, aminoglycosides, tetracyclines and streptomycin, most commonly in Salmonella Typhi10. IncH plasmid includes its association with mcr-1 and mcr-3 plasmid-mediated colistin resistance genes in E. coli11,12. The most worrisome is the co-existence of multiple classes of AMR genes on the same plasmid compromising the use of two broad antimicrobials for therapy. This includes the co-existence of ESBLs and carbapenemases, carbapenemases and 16S RMTases, ESBLs and quinolone resistance13.
The information of plasmid profiles among ESKAPE pathogen is lacking, especially from India. Hence, we undertook this study to identify the common plasmid types among ESKAPE pathogens causing nosocomial infections in a tertiary care centre in south India using whole-genome sequencing. The objectives of this study were as follows: (i) identification of AMR genes associated with plasmids, (ii) identification and comparison of sequence types (STs) with AMR genes and plasmid types, and (iii) association of phenotypic expression and AMR genes profile.
Material & Methods
A total of 105 non-repetitive isolates including E. coli (n=30), K. pneumoniae (n=48, including 7 pan-drug susceptible isolates), Acinetobacter baumannii (n=8), Pseudomonas aeruginosa (n=1) and Staphylococcus aureus (n=18) from blood cultures were included in this study. The isolates were received from November 2015 to October 2017 at the department of Clinical Microbiology, Christian Medical College, Vellore, India. Identification of pathogens up to species level was done by using standard microbiological methods14.
Antimicrobial susceptibility testing (AST): Antimicrobial susceptibility testing was performed by Kirby-Bauer disk diffusion method as recommended by Clinical and Laboratory Standards Institute (CLSI)15, using cefotaxime (30 μg), ceftazidime (30 μg), cefepime (30 μg), piperacillin/tazobactam (100/10 μg), cefoperazone/sulbactam (75/30 μg), gentamicin (10 μg), amikacin (30 μg), netilmicin (30 μg), ciprofloxacin (5 μg), imipenem (10 μg), meropenem (10 μg), minocycline (30 μg) and tigecycline (15 μg) for E. coli and K. pneumoniae. In addition, aztreonam (30 μg), levofloxacin (5 μg) and tobramycin (10 μg) were tested for P. aeruginosa and A. baumannii. For S. aureus, the following antibiotics were tested: Cefoxitin (30 μg), gentamicin (10 μg), erythromycin (15 μg), clindamycin (2 μg), netilmicin (30 μg), trimethoprim-sulphamethoxazole (1.25/23.75 μg), rifampicin (5 μg) and linezolid (30 μg). Susceptibility to colistin and vancomycin was determined by using the broth microdilution method (BMD), according to the CLSI guidelines15. E. coli ATCC 25922, P. aeruginosa ATCC 27853 and S. aureus ATCC 29213 were used as the quality control strains.
Whole genome sequencing: Genomic DNA of the isolates was extracted with QIAamp DNA mini kit (Qiagen, Hilden, Germany). Whole genome sequencing was performed using Ion Torrent (PGM) sequencer with 400 bp read chemistry (Life Technologies, CA, USA) according to manufacturer's instructions. Assembly of the data was performed de novo using AssemblerSPAdes v5.0.0.0 embedded in Torrent suite server version 5.0.3 (Life Technologies). The sequence annotation was done using PATRIC, the bacterial bioinformatics database, and analysis resource16 and the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) (http://www.ncbi.nlm.nih.gov/genomes/static/Pipeline.html). ST was analyzed by MLST 2.0 tool (https://cge.cbs.dtu.dk//services/MLST/). Plasmid replicon identification was performed using PlasmidFinder 1.3 (https://cge.cbs.dtu.dk//services/PlasmidFinder/) and plasmid typing was done in pMLST 1.4 (https://cge.cbs.dtu.dk/services/pMLST/). Further downstream analysis was performed using Center for Genomic Epidemiology (CGE) server (http://www.cbs.dtu.dk/services), Rapid Annotation and Subsystem Technology (RAST) server v2.0 (http://rast.theseed.org/FIG/rast.cgi) and Pathosystems Resource Integration Center (PATRIC) v3.5.36 (https://www.patricbrc.org/). Resistance genes profile was analyzed using ResFinder 3.1 from the CGE server (https://cge.cbs.dtu.dk//services/ResFinder/). The genome and plasmid sequences were also screened for AMR genes in Antibiotic Resistance Genes Database (ARDB) and Comprehensive Antibiotic Resistance Database (CARD) through PATRIC. Plasmid groups were identified based on inc typing in Gram-negative pathogen and replicon (rep) typing in S. aureus. These whole genome sequences were deposited at GenBank.
Results
Among E. coli, two of the 30 isolates were identified as multidrug-resistant (MDR). Majority of the tested isolates were extreme drug resistant (XDR), which includes E. coli (n=30), K. pneumoniae (n=41), P. aeruginosa (n=1) and A. baumannii (n=8). In addition, seven K. pneumoniae were pan drug-susceptible, which were used as an internal control for plasmid analysis. Overall, nineteen K. pneumonaie and all A. baumannii isolates were resistant to colistin by BMD. All S. aureus (n=18) were identified as methicillin resistant S. aureus (MRSA), and all were susceptible to vancomycin and linezolid.
Genome analysis: Assembly of the raw reads of the isolates showed 70 average contigs (≥500 bp), with an average of 50X coverage. Each pathogen was identified with the unique STs, except in E. coli (n=3). ResFinder analysis showed different combination of AMR genes in each pathogen. All these isolates carrying AMR genes specifically coding for different classes of antibiotic were confirmed from ARDB and CARD. All the genome data were submitted in GenBank database.
Plasmid analysis: Plasmids in Gram-negative (E. coli and K. pneumoniae) pathogens and S. aureus were identified by using inc and rep, respectively. The analysis of sequenced genome for plasmid showed the presence of single or combination of plasmids. The distribution of plasmid was highly heterogeneous and ST-specific plasmids were not observed. E. coli and K. pneumoniae isolates had 133 and 113 plasmid types respectively, where both had only 15 STs. In S. aureus, seven plasmid types were seen with 10 STs.
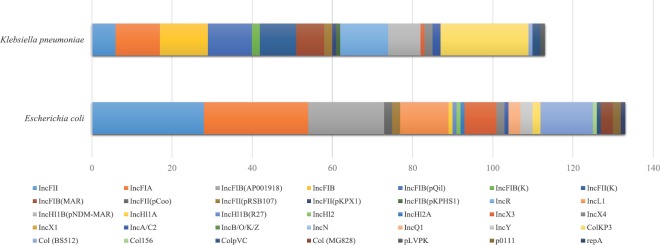
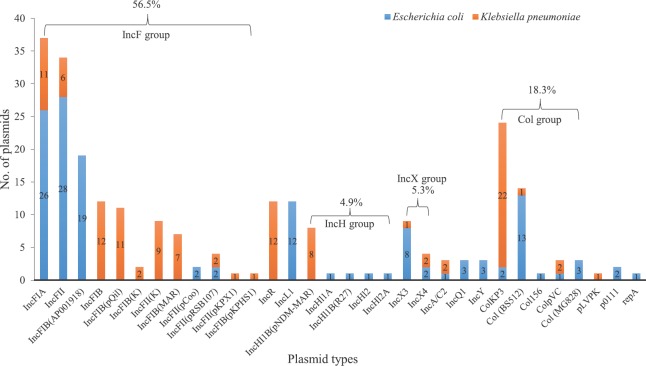
Majority of the E. coli isolates were found with IncFIA, IncFII, IncFIB, Col(BS512), IncL1, IncX3 and IncH plasmids (Fig. 1). While in K. pneumoniae, IncHI1B(pNDM-MAR), IncFII(pRSB107), IncFIB(Mar), IncFIB(pQil), IncFIA, IncFII(K), IncR, ColKP3, ColpVC were the most common plasmids. Overall, IncF (56.5%) was the predominant plasmid followed 18.3 per cent of Col and 5.3 per cent of IncX type plasmids. In addition, the frequency of other plasmid groups was found as <5 per cent among the tested isolates (Fig. 2). However, database for the screening of Inc plasmids in P. aeruginosa and A. baumannii using whole genome sequence was not available.
Fig. 1.
Species-wise distribution of plasmids among Escherichia coli and Klebsiella pneumoniae.
Fig. 2.
Distribution of plasmid replicon types identified among Escherichia coli and Klebsiella pneumoniae (n=246).
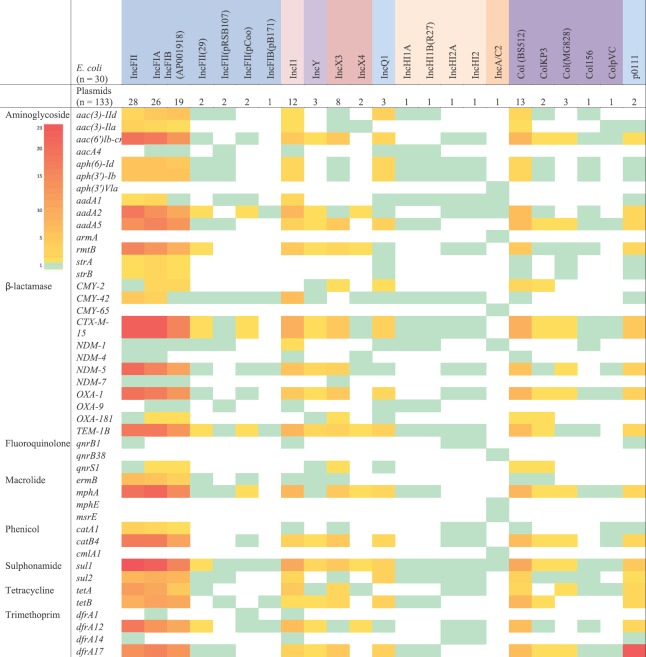
In most of the tested isolates, the presence of AMR genes correlated with phenotypic resistance pattern of tested antimicrobials. However, the level of agreement between the phenotypic and genotypic AMR positivity differed from species to species (Figs 3 and 4). The distribution and frequency of plasmids carrying different AMR genes in E. coli and K. pneumoniae are given in Figs. 5 and 6, respectively. All colistin resistant K. pneumoniae (n=19) isolates were negative for plasmid-mediated mcr gene coding for colistin resistance. Further, the genome of pan-drug susceptible K. pneumoniae (n=7) was found with IncHI1B(pNDM-MAR) plasmid.
Fig. 3.

Level of agreement between antimicrobial resistance genes and phenotypic resistance for Escherichia coli; genes present with no antimicrobial resistance correspond to non-expression of antimicrobial resistance genes. Full black circles represent total agreement between phenotype and genotype. Half-black circles represent lesser antimicrobial resistance genes than the phenotype. Empty circles represent even lesser antimicrobial resistance genes. Quarter black circle in aminoglycosides represents about 20 per cent non-expression of antimicrobial resistance genes, and quarter black circles in tetracyclines represent lesser genotype and phenotype compared to total numbers.
Fig. 4.
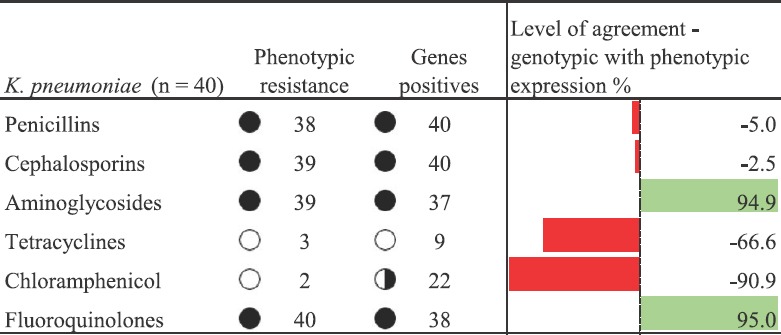
Level of agreement of antimicrobial resistance genes and phenotypic resistance for Klebsiella pneumoniae; genes present with no antimicrobial resistance correspond to non-expression of antimicrobial resistance genes. Full circles in penicillins, cephalosporins, aminoglycosides and fluoroquinolones represent total agreement between phenotype and genotype. Half circle in chloramphenicol represents less number of gene positives among K. pneumoniae. Empty circles in tetracyclines and chloramphenicol represent less antimicrobial resistance genes and phenotypic resistance in K. pneumoniae.
Fig. 5.
Heatmap showing association frequency of antimicrobial resistance genes and plasmids in Escherichia coli. IncFII, IncFIA and IncFIB (AP001918) had highest association with the antimicrobial resistance genes in comparison to other plasmid types. The grading numbers in color strip depicts the number of genes associated with the particular plasmid type.
Fig. 6.
Heatmap showing association frequency of antimicrobial resistance genes and plasmids in Klebsiella pneumoniae. ColKP3 followed by IncFII(K), IncHl1B and IncFIA had high association with antimicrobial resistance genes in comparison to other plasmid types. The grading numbers in color strip depicts the number of genes associated with the particular plasmid type.
In E. coli, β-lactamases encoding genes were predominant followed by aminoglycoside, trimethoprim, sulphonamide and macrolide resistance genes. Phenicol and tetracycline resistance genes were also seen. All these AMR genes were observed to carry on IncFII, IncFIA, IncFIB and Col(BS512) plasmids, while a similar observation was not seen with other plasmid types. Among β-lactamases, blaCTX-M-15 was common followed by blaNDM-5, blaTEM-1B and blaOXA-1 in IncF plasmids. sul1 gene for sulphonamide resistance was common than sul2 gene; tet A was frequently seen with IncFII plasmids, whereas tetB was noticed with IncFIB plasmids.
ColKP3 plasmid type was prevalent among K. pneumoniae and harboured almost all classes of AMR genes. This was followed by IncHI1B and IncF group plasmids [IncFII(K), IncFIA, IncFIB, IncFIB(pQil)] and IncR. Among β-lactamases, blaCTX-M-15 was most prevalent followed by blaOXA-232 and blaTEM-1B. Further, blaNDM-1 was common than blaNDM-5 and blaNDM-7. Rare genes such as blaLEN-12 (n=1), blaOKPB2 (n=1), blaTEM-124 (n=1), qepA (n=1) and qnrB66 (n=4) were also seen in this study among K. pneumoniae.
Of the tested S. aureus (n=18), rep gene was not found in five isolates. Among the 14 S. aureus isolates, a total of six rep families (rep5, rep7, rep10, rep16, repUS12, rep21) were assigned. Based on the combination of rep genes identified, seven different plasmid groups were identified in S. aureus. Each plasmid had a unique combination of rep gene sequences as follows: pMW2 (rep5), pSAS1 (rep7), pDLK1 (rep10), pUB110 (repUS12) and Saa6159 (rep16). The plasmids pKH12 and pSA1308 were found with same rep gene (rep21). This shows a recombination between pKH12 and pSA1308 plasmids.
Discussion
IncF plasmids were the commonest plasmids seen among E. coli and K. pneumoniae. IncF plasmids were reported from several countries including 49 per cent in Tanzania, 71 per cent in Germany and 45 per cent in Switzerland17. In Japan, the IncFIB (18.5%) was most commonly reported in E. coli followed by IncF (17.6%) and IncFIA (7.6%)18,19,20. In this study, majority of the isolates were identified with IncF type followed by Col group. Other types seen included IncX, IncH, IncR, IncL, IncA/C, IncY and IncQ plasmids.
The IncF plasmids were reported to carry varied and increased number of AMR genes than other plasmid types19. This includes clinically relevant ESBLs, such as blaCTX-M-15, plasmid-mediated AmpC genes (blaCMY and blaDHA), blaTEM-1 and blaOXA-1. In addition, IncF group plasmids were known to frequently harbour aminoglycoside and quinolone resistance genes qepA, armA, rmtB, aac(6’)-Ib-cr and qnr17.
In this study, IncFII, IncFIA and IncFIB type plasmids seen among E. coli harboured high number of blaCTX-M-15, blaNDM-5, blaTEM-1 and blaOXA-1 β-lactamase genes followed by aac(6′)-Ib-cr, aad and rmtB aminoglycoside resistance genes, aac(6′)-Ib-cr and qnr for quinolones resistance, tetA and tetB for tetracycline resistance, catB4 and catA1 for chloramphenicol resistance, and dfrA and sul genes for trimethoprim and sulphamethoxazole resistance. In K. pneumoniae, IncFII, IncFIA and IncFIB plasmids harboured blaCTX-M-15, blaNDM-5, blaNDM-1, blaTEM-1, blaOXA-232, blaOXA-181 and blaOXA-1 followed by aac(6′)-Ib-cr, aad, armA, rmtF, rmtB, qnrB1, qnrB66, tetA, tetB, tetD, catA1, catB3, dfrA and sul genes. The screening of blaNDM-5 may be necessary in Indian settings, as this variant is found higher than blaNDM-1 among E. coli. Among K. pneumoniae, blaNDM-5 and blaNDM-7 were seen in fewer isolates.
The common plasmid of IncHI1B type is pNDM-MAR. These are reported to carry the blaCTX-M-15, blaNDM and quinolone-resistant determinant qnrB120. In this study, E. coli harboured only one IncHI1B(R27) plasmid with blaNDM-1 and blaCTX-M-15. While in K. pneumoniae, eight isolates (including 7 pan-drug susceptible) (<5%) harboured IncHI1B(pNDM-MAR) plasmids with only one blaCTX-M-15. This highlights the risk of acquiring carbapenem-resistant gene (blaNDM) among pan-drug susceptible K. pneumoniae. Non-pNDM-MAR IncHI1B plasmids harboured blaCTX-M-15, blaNDM-1, qnrB1 and qnrB66. Plasmids reported to carry mcr-1 gene including IncFI and IncFIB were seen in this study. The presence of these plasmids in E. coli and K. pneumoniae may promote the acquisition of mcr gene and increase the threat of dissemination21.
Replicon groups IncA/C and Incl1 were frequently seen with Enterobacteriaceae and harboured multiple resistance genes including resistance for extended-spectrum cephalosporins (blaCMY) and carbapenems (blaNDM-1)17. In this study, Incl1 was identified nine per cent in E. coli with resistance genes to β-lactams, aminoglycosides, fluoroquinolones, tetracyclines, chloramphenicol and sulphonamides, while in K. pneumoniae none were seen. ColKP3 was reported as the most common plasmid type seen among K. pneumoniae and known to harbour AMR genes. In the present study, 19.5 per cent ColKP3 was found, and almost all had AMR genes for all classes of antibiotics.
Plasmids observed among Gram-positives are mostly common between S. aureus and Enterococcus spp. In S. aureus, the β-lactamase gene (blaZ) was encoded by pMW2 and pSAS1 plasmid, and both the plasmids belong to the pMW2 like plasmid group22. The present study showed pMW2 (rep5) plasmid carrying blaZ among MRSA. The plasmid pUB110 was reported with ant (4’)-1 which encodes for kanamycin and tobramycin resistance gene23. The pDLK1 plasmid had ermC gene encoding for erythromycin resistance24, and the qacC gene seen in pSA1308 plasmid encodes for multi-drug efflux pump encoding25. However, in the present study, pUB110 and pSA1308 were seen in a few MRSA isolates.
In this study, the plasmid types identified among the specified pathogens and AMR genes corroborated well with the available literature worldwide26. However, the plasmid replicon types identified among the study isolates were diverse than the STs. This might be due to the presence of multiple replicon types in the same organism harbouring plasmid27. The number of STs for each organism is higher than the number of plasmid types available for that organism. However, for tracking the AMR genes and to identify a plasmid outbreak in a hospital situation, plasmid typing is essential in a local setting.
Although most of the isolates harbour AMR genes, expression percentage differs widely. The non-expression of a few genes might be due to various factors such as mutations in the protein coding region, insertion elements in the gene promoter region, and within-gene recombination28. In this study among E. coli, >70 per cent association was observed between AMR genes and phenotypic expression, except for aminoglycosides, where 19 per cent were non-expressive.
In K. pneumoniae, among penicillins and cephalosporins, very few isolates possessed non-expressive genes (5 and 2.5%). Aminoglycosides and fluoroquinolones did not possess any non-expressive genes, whereas, tetracyclines and chloramphenicol had high non-expressive genes (66.6 and 90%). Similar case for tetracycline was observed by Gow et al28. Aminoglycosides and fluoroquinolones showed agreement (95%) between phenotypic expression and genotypic AMR findings. Plasmid-mediated genes for colistin resistance were not seen in any of the study isolates. Presence or absence of a genotype does not specify the isolate as resistance or susceptible. The mechanism behind the AMR is numerous and complex28. Hence, a phenotypic test is necessary from a diagnostic point of view for confirmation of the genotype. Studying the plasmid prevalence among Enterobacter, Enterococci, Pseudomonas and Acinetobacter would give a complete understanding of plasmid prevalence and dynamics of AMR dissemination among ESKAPE pathogens. This acts as a limitation to the present study.
Results from this pilot study may act as a baseline data for plasmid among E. coli and K. pneumoniae in India. Only isolated cases of plasmids have been reported previously by our group from India29,30,31. These findings substantiate that occurrence of a few common plasmid types among the hospital pathogens may lead to a considerable amount of horizontal gene exchange.
In conclusion, plasmids analysis revealed that among β-lactamases, blaCTX-M-15 was present both in E. coli and K. pneumoniae, followed by blaTEM-1B. In addition, blaNDM-5 and blaOXA-1 were also seen in E. coli, whereas blaNDM-1 and blaOXA-232 were common among K. pneumoniae. The study also showed the relation between phenotypic and genotypic expression of AMR for various classes. IncFII plasmid was observed in E. coli, while, ColKP3 followed by IncFII(K) was present in K. pneumoniae. Understanding on such vectors carrying the AMR genes could help in improving strategies on better control of AMR dissemination.
Footnotes
Financial support & sponsorship: The authors acknowledge the Indian Council of Medical Research, New Delhi, for providing the grant for this research (Ref. No: AMR/TF/55/13ECDII dated 23/10/2013) and Fluid Research Grant of Christian Medical College, Vellore (IRB min. no. 9616 dated 01.09.2015).
Conflicts of Interest: None.
References
- 1.Veeraraghavan B, Jesudason MR, Prakasah JAJ, Anandan S, Sahni RD, Pragasam AK, et al. Antimicrobial susceptibility profiles of Gram-negative bacteria causing infections collected across India during 2014-2016: Study for monitoring antimicrobial resistance trend report. Indian J Med Microbiol. 2018;36:32–6. doi: 10.4103/ijmm.IJMM_17_415. [DOI] [PubMed] [Google Scholar]
- 2.Boot M, Raadsen S, Savelkoul PH, Vandenbroucke-Grauls C. Rapid plasmid replicon typing by real time PCR melting curve analysis. BMC Microbiol. 2013;13:83. doi: 10.1186/1471-2180-13-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.García-Fernández A, Chiaretto G, Bertini A, Villa L, Fortini D, Ricci A, et al. Multilocus sequence typing of IncI1 plasmids carrying extended-spectrum beta-lactamases in Escherichia coli and Salmonella of human and animal origin. J Antimicrob Chemother. 2008;61:1229–33. doi: 10.1093/jac/dkn131. [DOI] [PubMed] [Google Scholar]
- 4.Hancock SJ, Phan MD, Peters KM, Forde BM, Chong TM, Yin WF, et al. Identification of IncA/C plasmid replication and maintenance genes and development of a plasmid multilocus sequence typing scheme. Antimicrob Agents Chemother. 2017;61 doi: 10.1128/AAC.01740-16. pii: e01740-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jensen LB, Garcia-Migura L, Valenzuela AJ, Løhr M, Hasman H, Aarestrup FM. A classification system for plasmids from enterococci and other Gram-positive bacteria. J Microbiol Methods. 2010;80:25–43. doi: 10.1016/j.mimet.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 6.Rozwandowicz M, Brouwer MSM, Fischer J, Wagenaar JA, Gonzalez-Zorn B, Guerra B, et al. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J Antimicrob Chemother. 2018;73:1121–37. doi: 10.1093/jac/dkx488. [DOI] [PubMed] [Google Scholar]
- 7.Kim J, Bae IK, Jeong SH, Chang CL, Lee CH, Lee K, et al. Characterization of IncF plasmids carrying the blaCTX-M-14 gene in clinical isolates of Escherichia coli from Korea. J Antimicrob Chemother. 2011;66:1263–8. doi: 10.1093/jac/dkr106. [DOI] [PubMed] [Google Scholar]
- 8.Nicolas-Chanoine MH, Bertrand X, Madec JY. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev. 2014;27:543–74. doi: 10.1128/CMR.00125-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fortini D, Fashae K, García-Fernández A, Villa L, Carattoli A. Plasmid-mediated quinolone resistance and β-lactamases in Escherichia coli from healthy animals from Nigeria. J Antimicrob Chemother. 2011;66:1269–72. doi: 10.1093/jac/dkr085. [DOI] [PubMed] [Google Scholar]
- 10.Dierikx C, van Essen-Zandbergen A, Veldman K, Smith H, Mevius D. Increased detection of extended spectrum beta-lactamase producing Salmonella enterica and Escherichia coli isolates from poultry. Vet Microbiol. 2010;145:273–8. doi: 10.1016/j.vetmic.2010.03.019. [DOI] [PubMed] [Google Scholar]
- 11.Falgenhauer L, Waezsada SE, Yao Y, Imirzalioglu C, Käsbohrer A, Roesler U, et al. Colistin resistance gene Mcr-1 in extended-spectrum β-lactamase-producing and carbapenemase-producing gram-negative bacteria in Germany. Lancet Infect Dis. 2016;16:282–3. doi: 10.1016/S1473-3099(16)00009-8. [DOI] [PubMed] [Google Scholar]
- 12.Yin W, Li H, Shen Y, Liu Z, Wang S, Shen Z, et al. Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. MBio. 2017;8 doi: 10.1128/mBio.00543-17. pii: e00543-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muthuirulandi Sethuvel DP, Anandan S, Devanga Ragupathi NK, Veeraraghavan B, Vinod O, Walia K, et al. Association of blaCTX-M-15 and qnr genes in multidrug-resistant Salmonella typhimurium and Shigella spp from India. J Infect Dev Ctries. 2015;9:1294–7. doi: 10.3855/jidc.6965. [DOI] [PubMed] [Google Scholar]
- 14.Koneman EW, Allen SD, Janda WM, Schreckenberger PC, Winn WC. 5th ed. Philadelphia: Lippincott-Raven publishers; 1997. Color atlas and textbook of diagnostic microbiology. [Google Scholar]
- 15.CLSI Document M100. 27th ed. Wayne, PA: CLSI; 2017. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. [Google Scholar]
- 16.Wattam AR, Abraham D, Dalay O, Disz TL, Driscoll T, Gabbard JL, et al. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 2014;42:D581–91. doi: 10.1093/nar/gkt1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lyimo B, Buza J, Subbiah M, Temba S, Kipasika H, Smith W, et al. IncF plasmids are commonly carried by antibiotic resistant Escherichia coli isolated from drinking water sources in Northern Tanzania. Int J Microbiol. 2016;2016:3103672. doi: 10.1155/2016/3103672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J, Stephan R, Karczmarczyk M, Yan Q, Hächler H, Fanning S. Molecular characterization of Bla ESBL-harboring conjugative plasmids identified in multi-drug resistant Escherichia coli isolated from food-producing animals and healthy humans. Front Microbiol. 2013;4:188. doi: 10.3389/fmicb.2013.00188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamamoto S, Nakano M, Kitagawa W, Tanaka M, Sone T, Hirai K, et al. Characterization of multi-antibiotic-resistant Escherichia coli isolated from beef cattle in Japan. Microbes Environ. 2014;29:136–44. doi: 10.1264/jsme2.ME13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Villa L, Poirel L, Nordmann P, Carta C, Carattoli A. Complete sequencing of an IncH plasmid carrying the blaNDM-1, blaCTX-M-15 and qnrB1 genes. J Antimicrob Chemother. 2012;67:1645–50. doi: 10.1093/jac/dks114. [DOI] [PubMed] [Google Scholar]
- 21.Poirel L, Jayol A, Nordmann P. Polymyxins: Antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev. 2017;30:557–96. doi: 10.1128/CMR.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kennedy AD, Porcella SF, Martens C, Whitney AR, Braughton KR, Chen L, et al. Complete nucleotide sequence analysis of plasmids in strains of Staphylococcus aureus clone USA300 reveals a high level of identity among isolates with closely related core genome sequences. J Clin Microbiol. 2010;48:4504–11. doi: 10.1128/JCM.01050-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buntaran L, Hatta M, Sultan AR, Dwiyanti R, Sabir M. Sccmec type II gene is common among clinical isolates of methicillin-resistant Staphylococcus aureus in Jakarta, Indonesia. BMC Res Notes. 2013;6:110. doi: 10.1186/1756-0500-6-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lozano C, García-Migura L, Aspiroz C, Zarazaga M, Torres C, Aarestrup FM. Expansion of a plasmid classification system for Gram-positive bacteria and determination of the diversity of plasmids in Staphylococcus aureus strains of human, animal, and food origins. Appl Environ Microbiol. 2012;78:5948–55. doi: 10.1128/AEM.00870-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wassenaar TM, Ussery DW, Ingmer H. The qacC gene has recently spread between rolling circle plasmids of Staphylococcus, indicative of a novel gene transfer mechanism. Front Microbiol. 2016;7:1528. doi: 10.3389/fmicb.2016.01528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58:3895–903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orlek A, Stoesser N, Anjum MF, Doumith M, Ellington MJ, Peto T, et al. Plasmid classification in an era of whole-genome sequencing: Application in studies of antibiotic resistance epidemiology. Front Microbiol. 2017;8:182. doi: 10.3389/fmicb.2017.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gow SP, Waldner CL, Harel J, Boerlin P. Associations between antimicrobial resistance genes in fecal generic Escherichia coli isolates from cow-calf herds in Western Canada. Appl Environ Microbiol. 2008;74:3658–66. doi: 10.1128/AEM.02505-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Devanga Ragupathi NK, Muthuirulandi Sethuvel DP, Gajendiran R, Daniel JL, Walia K, Veeraraghavan B, et al. First Indian report of IncX3 plasmid carrying blaNDM-7 in Escherichia coli from bloodstream infection: Potential for rapid dissemination. New Microbes New Infect. 2017;17:65–8. doi: 10.1016/j.nmni.2017.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pragasam AK, Shankar C, Veeraraghavan B, Biswas I, Nabarro LE, Inbanathan FY, et al. Molecular mechanisms of colistin resistance in Klebsiella pneumoniae causing bacteremia from India – A first report. Front Microbiol. 2016;7:2135. doi: 10.3389/fmicb.2016.02135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shankar C, Veeraraghavan B, Nabarro LEB, Ravi R, Ragupathi NKD, Rupali P, et al. Whole genome analysis of hypervirulent Klebsiella pneumoniae isolates from community and hospital acquired bloodstream infection. BMC Microbiol. 2018;18:6. doi: 10.1186/s12866-017-1148-6. [DOI] [PMC free article] [PubMed] [Google Scholar]